Abstract
An invertebrate intestinal mucin gene, AgMuc1, was isolated from the malaria vector mosquito Anopheles gambiae. The predicted 122-residue protein consists of a central core of seven repeating TTTTVAP motifs flanked by hydrophobic N- and C-terminal domains. This structure is similar to that of mucins that coat the protozoan parasite Trypanosoma cruzi. Northern blot analysis indicated that the gene is expressed exclusively in the midgut of adult mosquitoes. A length polymorphism and in situ hybridization were used to genetically and cytogenetically map AgMuc1 to division 7A of the right arm of the second chromosome. The subcellular localization of the encoded protein in tissue culture cells was examined by using a baculovirus vector to express AgMuc1 protein tagged with the green fluorescent protein (GFP). The results indicated that this protein is found at the cell surface and that both hydrophobic domains are required for cell surface targeting. We propose that AgMuc1 is an abundant mucin-like protein that lines the surface of the midgut microvilli, potentially protecting the intestinal epithelium from the proteinase-rich environment of the gut lumen. An intriguing possibility is that, as an abundant surface protein, AgMuc1 may also interact with the malaria parasite during its invasion of the mosquito midgut.
Anopheles gambiae is the principal vector for transmission of human malaria, a disease that kills in excess of 2 million people worldwide (mostly children) every year. To be transmitted from one host to another, malaria parasites have to complete a complex life cycle in vector mosquitoes, starting in the midgut lumen, crossing through the midgut epithelial barrier, and finally invading the salivary glands, from where they can be inoculated into the next host during blood feeding.
The insect midgut is composed of a single layer of epithelial cells, which are lined at their basal side by a continuous extracellular layer, the basal lamina. On the apical side, the epithelial cell membranes are folded into numerous actin-filled microvilli. Microvilli greatly increase the surface area and play an important role in absorption of nutrients (1). The microvilli are exposed to the harsh environment of the gut lumen, and they are subjected to damage caused by food particle abrasion, digestive hydrolases, and attack by pathogens and parasites. Two extracellular structures have been proposed to provide protection to the microvilli: the peritrophic matrix and the glycocalyx (2–4). The peritrophic matrix is an extracellular sac, composed of chitin, proteins, and proteoglycans (2, 3), which completely surrounds the ingested food and is secreted by the gut epithelial cells. All the recently cloned peritrophic matrix proteins from Lucilia cuprina (5, 6), Trichoplusia ni (7), and the mosquito Anopheles gambiae (8) have at least two chitin-binding domains that are presumed to function in the cross-linking of the chitin fibrils. However, some of these proteins also have mucin-like domains (6, 7), suggesting that the insect peritrophic matrix resembles the vertebrate intestinal mucus, a structure largely composed of mucins. In addition to providing protection, the peritrophic matrix may also facilitate digestion by compartmentalization of digestive enzymes (9). Another protective structure is the glycocalyx (glyco = sweet/sugar, calyx = shell), which is an integral part of the microvillar membrane and appears as an electron-dense fuzzy coat on the outside of the microvillar surface (4, 10). The glycocalyx, including that of mosquitoes, is rich in carbohydrates, as it is recognized by a variety of lectins (11, 12). However, no experimental data are currently available on the molecular composition of the insect midgut glycocalyx. This is a subject of potential importance, because components of the glycocalyx may serve as receptors or attachment sites for invasion of parasites such as malaria.
Plasmodium development begins in the mosquito gut by formation of gametes, fertilization, meiosis, and differentiation into an ookinete. About 24 h later, the ookinete crosses the midgut epithelium from the luminal to the hemocoel side. Although the recognition of the gut epithelial cell surface by ookinetes is a crucial step in the life cycle of Plasmodium, little molecular information is available about this process. There is evidence to suggest that carbohydrate components of the gut epithelial cell surface are involved in interactions with the parasite (13, 14). However, no candidate glycosylated proteins from the adult Anopheles midgut have been identified.
Even in Anopheles species that can serve as vectors, not all strains support the development of Plasmodium with the same efficiency. Selected refractory mosquito strains exist in which the invading ookinetes are killed in the midgut epithelial cells either by lysis or, later, by melanotic encapsulation (15, 16). Other refractory mechanisms may act prior to or during midgut invasion by the ookinetes. The mechanisms controlling the demonstrated refractory traits remain unknown. Genetic mapping has shown that the melanotic encapsulation phenotype is controlled by three quantitative trait loci (17), whereas the lytic refractoriness is believed to be controlled by a different locus (15). The genetic elements controlling infection intensities at pre-encapsulation stages appear to be genetically unlinked to the encapsulation quantitative trait loci (QTL) and remain to be defined (17).
In this report, we describe the cDNA cloning of a mucin-like protein, named AgMuc1 for Anopheles gambiae mucin 1. Our results suggest that this putative mucin is an abundant surface-associated protein component of the midgut. Interestingly, a length polymorphism within the mucin domain was detected between strains that are susceptible and refractory to the malaria parasite. By use of this polymorphism, the AgMuc1 gene was mapped to a chromosomal region near but not at Pen1, the main locus controlling the melanotic encapsulation phenotype.
MATERIALS AND METHODS
cDNA Cloning and Sequencing.
One AgMuc1 cDNA clone, pm2, was obtained by screening an A. gambiae adult midgut Lambda ZAP cDNA library (18) with an antiserum against peritrophic matrix proteins; it was sequenced in both directions by using a dye-termination procedure. An identical AgMuc1 cDNA clone was obtained independently by screening a Lambda ZAP cDNA library from adult female abdomens (19) with a 32P-labeled dCTP trinucleotide repeat probe encoding a polythreonine stretch.
Northern Analysis.
Total RNA samples were isolated from A. gambiae adult midguts, adult carcasses (whole body minus midgut), pupae, larval guts, and larval carcasses. About 5 μg of total RNA from each sample was fractionated by electrophoresis on denaturing 1% agarose gels and transferred onto a nylon membrane. The full-length cDNA insert was labeled with [α-32P]dCTP by random labeling (20) and hybridized to the RNA blots at high stringency.
Isolation of the Muc1b Allele from the 4A r/r (Pink Eye) Strain.
A 318-bp PCR fragment was amplified by PCR from 4A r/r strain cDNA by using the AgMuc1-specific primers: MUCPA, 5′-AAAGTTGTTGTGGGACTAGTG-3′ and MUCPB, 5′-CGACATTGCCACGTATGCTCC-3′ with 26 cycles of 1 min at 94°C, 1 min at 56°C, and 1 min at 72°C. The fragment was cloned in a TOPO TA cloning vector (Invitrogen) prior to sequencing.
Genetic Mapping.
Genetic mapping of AgMuc1 was kindly performed by Rui Wang, using the same primers as for the cloning of the polymorphic allele, on the DNA of 55 individuals of families previously used in mapping the QTL for refractoriness of A. gambiae to Plasmodium cynomolgi B (17).
Cytogenetic Mapping.
The AgMuc1 cDNA clone was mapped by in situ hybridization to Suakoko strain Anopheles gambiae polytene chromosomes as previously described (21).
Constructs for Baculovirus-Based Protein Expression.
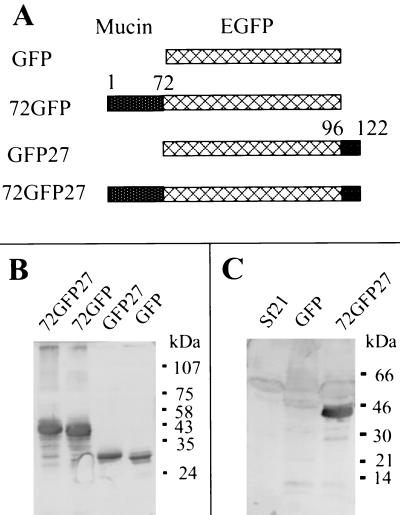
Four baculovirus transfer plasmid constructs were made (see Fig. 4A). A control green fluorescent protein (GFP) expression vector (pBacPak8-GFP) was constructed by excising the GFP-encoding insert from plasmid EGFP-C3 (CLONTECH) with NheI and PstI and cloning into the XbaI (XbaI and NheI fragments have compatible ends) and PstI sites of the pBacPak8 plasmid (CLONTECH). To obtain the pBacPak8–72GFP fusion construct, the cDNA fragment encoding the N-terminal 72 amino acid residues of AgMuc1 was amplified by PCR using cDNA clone pm2 as the template. The two PCR primers had additional sequences at their 5′ ends to provide restriction sites as follows. Forward primer: 5′-AGCCGCTAGCAACATGTTGAAAGTTGTT; reverse primer: 5′-AGCTACCGGTCCAGGTGCCACTGTG; the restriction sites for NheI and AgeI are underlined. This PCR product was digested with NheI and AgeI and was inserted into the EGFP-C3 plasmid digested with the same restriction enzymes. The 72GFP cDNA fragment was then excised with NheI and PstI and inserted into the XbaI and PstI sites of pBacPak8 plasmid. To construct pBacPak8-GFP27 and pBacPak8–72EGFP27, the cDNA fragment encoding the C-terminal 27 amino acids was amplified by PCR using a AgMuc1-specific primer (forward primer: 5′ GCTCAAGCTTTCAAGTGCCCCACAGG, HindIII site underlined) and a T3 primer. After digestion with HindIII and PstI (from the multiple cloning site of pBluescript), the PCR product was inserted into the HindIII and PstI sites of pBacPak8-GFP and pBacPak8–72GFP, respectively.
Figure 4.
Expression and characterization of mucin-GFP fusion proteins encoded by recombinant baculoviruses. (A) Diagrams showing recombinant protein structures. Lattice-patterned rectangles, GFP reporter protein; stippled rectangles, AgMuc1 sequences (see Fig. 1A). (B) Western blotting analysis of baculovirus-expressed GFP and GFP-mucin fusion proteins with an anti-GFP antibody. Total proteins from cells infected with different recombinant baculoviruses were loaded on each lane. A polyclonal antibody against GFP was used to probe the Western blots. The structure of the recombinant proteins indicated at the top of each lane is given in A. (C) Western blotting analysis of the baculovirus-expressed mucin-GFP protein with antiserum against total midgut microvilli. The Sf21 lane contains control cell lysate. Migration of marker proteins is indicated on the right side of B and C.
Cell Culture and Generation of Recombinant Viruses.
Sf21 (Spodoptera frugiperda) cells were grown at 27°C in Grace’s insect medium plus 10% fetal bovine serum. Recombinant baculoviruses were generated by homologous recombination by cotransfection of plasmid constructs and viral DNA digested with Bsu36 I (from the CLONTECH pBacPAK Baculovirus Expression System kit). The procedures recommended by the supplier were followed. Cells infected with recombinant viruses were easily identified with a UV dissection microscope to visualize GFP. Recombinant viruses were collected from the cell culture medium and used to infect Sf21 cells.
Preparation of Antibodies Against Microvilli.
Microvilli were prepared from midguts dissected from 5- to 10-day-old A. gambiae adult females according to English et al. (22). About 500 midguts were suspended in 2 ml of ice-cold 50 mM sucrose/0.1 mM PMSF/2 mM Tris⋅HCl, pH 7.4, and homogenized on ice with 15 strokes of a Dounce homogenizer. Then CaCl2 was added to a final concentration of 10 mM. After 15 min of shaking, the sample was centrifuged at 4,300 × g for 10 min at 4°C. The supernatant was collected and centrifuged at 26,000 × g for 10 min. The microvillar pellet was resuspended with PBS and used to immunize rabbits.
Analysis of Baculovirus-Expressed GFP Fusion Proteins.
Sf21 cells infected with the various recombinant viruses were collected 60 h after infection. Total proteins from the infected cells were first fractionated by electrophoresis on SDS/12% polyacrylamide gels and then blotted onto a nitrocellulose membrane. The fusion proteins were detected by Western blot analysis using polyclonal antibody against GFP (CLONTECH) or rabbit antiserum against microvilli.
Confocal Microscopy.
The subcellular localization of GFP or of GFP-fusion proteins was detected by the fluorescence of the GFP moiety by using a Bio-Rad MRC-600 laser scanning confocal imaging system with a 488-nm excitation wavelength.
Detection of 72GFP27 Protein on the Cell Surface.
Sf21 cells infected with 72GFP27 or 72GFP recombinant baculoviruses were collected by centrifugation at 2,000 × g for 2 min. The cells were then resuspended and incubated with basic insect medium containing rabbit anti-GFP antibody (CLONTECH, 1:500 dilution) and 1% BSA for 1 h at room temperature. The cells were then washed three times with basic insect cell medium and incubated for 30 min with basic insect medium containing 1% BSA and rhodamine-conjugated goat anti-rabbit antibody (Pierce; 0.02 mg/ml). The cells were washed three times and examined with a confocal fluorescence microscope. Exactly the same parameters were used to collect the fluorescence images of cells expressing 72GFP27 and 72GFP. Images from the green (inherent GFP fluorescence anywhere in the cell) and red (surface labeling by antibody) channels were merged.
RESULTS
Cloning of a Mucin cDNA.
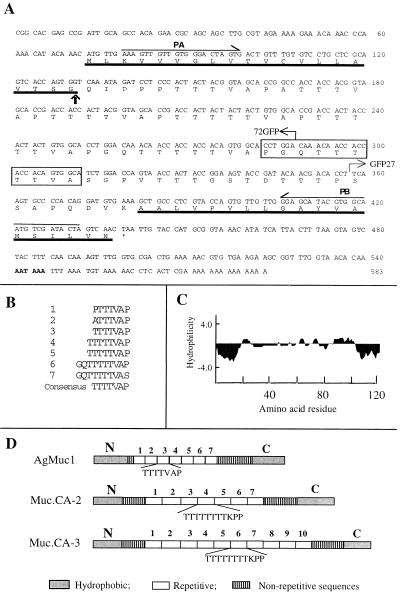
cDNA clones were obtained independently in our two laboratories by unrelated screens of A. gambiae midgut or abdominal Lambda ZAP cDNA expression libraries. One of the clones contains a 572-bp insert that includes an ORF encoding a protein of 122 amino acids (Fig. 1A). The cDNA has a putative polyadenylation signal 18 bp from the beginning of the poly(A) tail (AATAAA, boldface in Fig. 1A). The deduced protein has two hydrophobic fragments, one at the N terminus and the other at the C terminus flanking a central core of seven repeating TTTTVAP motifs, six of which are near perfect repeats (Fig. 1 B–D). The amino acid composition of the predicted protein is heavily biased, as it has 39 threonines (accounting for 44% of the residues in the central core), 11 prolines (13%), 9 alanines (11%), 9 valines (11%), 5 glycines (6%), 4 serines (5%), 4 glutamines (5%), 3 aspartic acids (4%), 1 isoleucine (1%), and 1 lysine (1%). The other 10 amino acids are absent. Repeating threonine- and proline-rich motifs are the hallmark of the extensively characterized vertebrate mucins (23–27). Hence, this protein was named AgMuc1, for Anopheles gambiae mucin.
Figure 1.
Nucleotide and predicted amino acid sequences of AgMuc1. (A) The N- and C-terminal hydrophobic protein domains are underlined. The putative cleavage site of the signal peptide is indicated by a vertical arrow and the borders of GFP fusion constructs by bent arrows. The putative polyadenylation signal sequence is in boldface. The polymorphic region missing in the Muc1b allele is boxed and the PCR primers used for genetic mapping (PA, PB) are indicated above the sequence. (B) The seven repeated motifs of the central protein core are aligned. The consensus repeat sequence is given at the bottom. (C) A Kyte–Doolittle hydropathy plot was generated with an average hydrophilicity window of 7 residues. (D) Diagrammatic comparison of the predicted amino acid sequence of the A. gambiae AgMuc1 mucin with that of Trypanosoma cruzi mucins, MUC.CA-2 and MUC.CA-3 (31, 32). AgMuc1 mucin contains TTTTVAP repeats, whereas the MUC.CA-2 and MUC.CA-3 mucins contain TTTTTTTTKPP motifs. All three mucins contain nonrepeated sequences between the repeat array and the N- and C-terminal hydrophobic sequences.
According to the rules of von Heijne (28), the N-terminal hydrophobic domain is likely to be a signal peptide with the cleavage site at the carboxyl side of Gly-20 (Fig. 1A). The C-terminal hydrophobic domain may serve to anchor the protein to the cell membrane. This sequence has the features of glycosyl-phosphatidylinositol (GPI) anchor signals: a region of 15–20 hydrophobic residues at the extreme C terminus preceded by a few hydrophilic residues (29), as in the case of GPI-anchored Trypanosoma cruzi mucins (30–32).
Although sequence similarity to known proteins was not detected by blast searches, the amino acid composition and sequence pattern of AgMuc1 show significant similarity to a mucin family isolated from the protozoan parasite T. cruzi (Fig. 1C). In contrast, vertebrate mucins are often much longer, have more numerous repetitive units, and lack an extreme C-terminal hydrophobic sequence. For instance, the human MUC2 has about 100 PTTTPITTTTTVTPTPTPTGTQT repeats, the number varying among different alleles (33). Both vertebrate and T. cruzi mucins are highly glycosylated by O-linkage to threonine or serine residues of the protein backbone (30, 34), suggesting that this may also be true for AgMuc1.
AgMuc1 Is Exclusively Expressed in Adult Midguts.
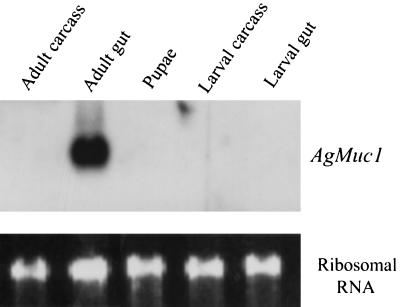
Developmental- and tissue-specific expression of the AgMuc1 gene was investigated by Northern blot analysis (Fig. 2). A single band of ≈0.8 kb was detected in the RNA isolated from adult midguts, but not in the RNA from adult carcasses, whole pupae, larval carcasses, or larval guts, suggesting that AgMuc1 is expressed only in adult midguts. The size of the mRNA detected on Northern blots agrees with the length of the cloned cDNA, assuming that the poly(A) tail contains ≈100 adenosine residues. The signal detected on Northern blots is very strong, suggesting that the mRNA is abundant in the adult gut.
Figure 2.
Developmental and tissue specificity of AgMuc1 expression. (Upper) Autoradiogram of a Northern blot of RNAs (≈5 μg per lane) isolated from the indicated tissues. The blot was hybridized overnight with a 32P-labeled AgMuc1 cDNA probe, washed, and exposed to film for 4 h. (Lower) Staining of ribosomal RNA with ethidium bromide to indicate the amount of RNA analyzed in each lane.
Polymorphic AgMuc1 Alleles and Mapping.
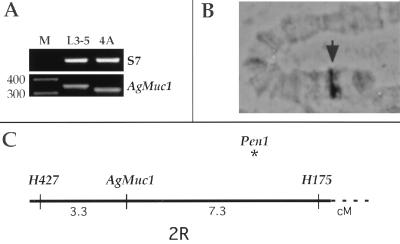
When reverse transcription–PCR primers flanking the repeat domain (Fig. 1A) were used a shorter AgMuc1 allele was detected in the malaria-susceptible A. gambiae 4A r/r strain (Fig. 3A). This allele, named AgMuc1b, lacks 30 bp that encode one repeat (PGQTTTTVA). All other A. gambiae laboratory strains tested (G3, Suakoko, and L3–5) were found to have the AgMuc1 allele. The L3–5 strain is refractory to malaria parasites. The invading ookinetes and early oocysts are encapsulated soon after crossing the midgut epithelium (16). The length polymorphism was used to map AgMuc1 genetically. The female F1 progeny of a cross between the susceptible 4A r/r and the refractory L3–5 strains were backcrossed with 4A r/r males, and segregation of AgMuc1 was followed in the backcross progeny. Recombination distances of AgMuc1 to previously mapped microsatellite markers (17) placed this gene in the general vicinity of the major refractory QTL, Pen1, on the right arm of the second chromosome (Fig. 3C). Cytogenetic mapping of the AgMuc1 cDNA clone to A. gambiae polytene chromosomes localized this gene to division 7A on the right arm of the second chromosome (Fig. 3B). This location correlates with the genetic and cytogenetic location of the adjacent microsatellite markers (data not shown) but is clearly separated from that of Pen1, which is most likely at 8C (F. Collins, personal communication).
Figure 3.
Polymorphism and genetic mapping of AgMuc1. (A) Ethidium bromide-stained agarose gel with the PCR-amplified AgMuc1 segment containing the length polymorphism from the refractory L3-5 strain and susceptible 4A r/r strain. Lane M, length markers. (B) The cytogenetic location of AgMuc1 in division 7A on the second chromosome is indicated by an arrowhead. (C) Genetic distances of AgMuc1 from adjacent microsatellite markers are shown in centimorgans (cM). The gene is within the same region as the Plasmodium refractory QTL Pen1, but the latter locus is distinct from AgMuc1 and is located closer to H175.
Expression of GFP-Mucin Fusion Proteins in Insect Cells.
GFP-AgMuc1 fusion proteins (Fig. 4A) were expressed in insect cells from a baculovirus vector. The recombinant proteins were detected by Western blot analysis using an anti-GFP antibody (Fig. 4B). The relative mobilities of the different recombinant proteins were those expected from their calculated molecular mass, indicating that the fusion proteins are faithfully expressed. However, the apparent molecular masses of the fusion proteins containing repeat sequences (72GFP and 72GFP27; Fig. 4A) were higher than the calculated molecular masses (34 and 36 kDa). This discrepancy may be due to glycosylation of the fusion proteins at the core mucin domain. However, the proteins may not be fully glycosylated (in humans carbohydrates make up 50% of the mass), either because of strong and rapid protein expression in the baculovirus system or because the Sf21 cells, which are ovarian in origin, do not have all the necessary enzymes.
An antiserum against midgut microvilli was used to probe control Sf21 cell lysates or lysates expressing recombinant proteins (Fig. 4C). A single band (of the same mobility as that detected by GFP antibody) was detected in cells expressing 72GFP27, but not in uninfected Sf21 cells or in cells expressing GFP. No bands were detected on similar blots with preimmune serum (data not shown). These results indicate that the antiserum is specific for the mucin sequences in the fusion protein and suggest that the mucin is a component of midgut microvilli.
Both Hydrophobic Domains Are Required to Target AgMuc1 to the Cell Surface.
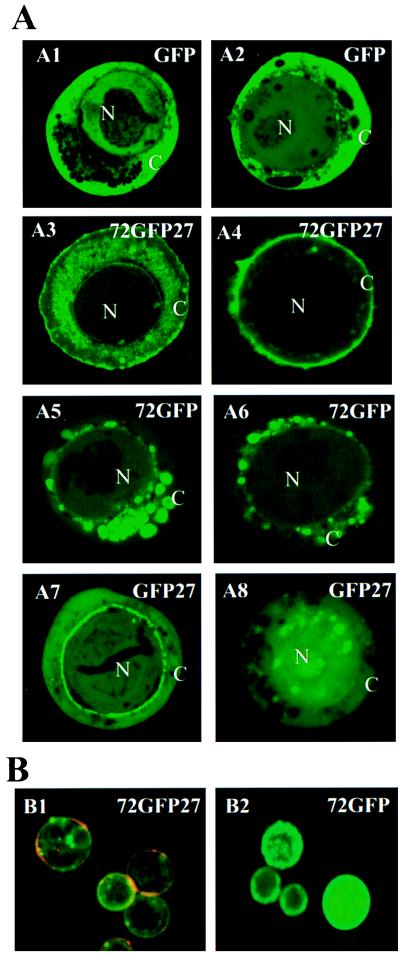
The products of all four fusion constructs shown in Fig. 4A could be detected and localized easily by fluorescence microscopy when expressed in Sf21 cells (Fig. 5A). This observation indicates that GFP fluorescence is not lost, even when this reporter protein is placed in the middle of the mucin protein (72GFP27). For all constructs, the fluorescence was strongly detectable at both 30 h and 60 h after infection, and it revealed final spatial localization by the latter time (A2, A4, A6, and A8 in Fig. 5A). In the control experiment, GFP was distributed throughout the cytoplasm and the nucleus (A1 and A2), except for some dark areas in the cytoplasm, which may correspond to secretory vesicles and/or lysosomes. In general, less GFP was detected in the nucleolus. Nuclear localization is consistent with the idea that GFP is a nonsecretory soluble protein that is able to diffuse into the nucleus because of its small size (≈28 kDa). The distribution of recombinant 72GFP27 protein, consisting of GFP fused on either side to the AgMuc1 N- and C-termini, differed dramatically from that of GFP alone. During the early stages of infection, the fusion protein was observed both at the cell surface and in the cytoplasm (A3). At later stages of infection, the protein was detected predominantly at the cell surface (A4). Therefore, the combination of AgMuc1 N- and C-terminal sequences is sufficient to direct the fusion protein to the cell surface. Fusion protein 72GFP accumulated predominantly in cytoplasmic vesicle-like structures and on the nuclear periphery (A5 and A6). The vesicle-like structures may be secretory vesicles, while fluorescence at the nuclear periphery may represent the condensed endocytoplasmic membrane as previously observed in baculovirus-infected cells (35). This pattern of distribution was expected because the 72 residues fused to GFP include the putative signal peptide. The GFP27 fusion protein showed a different distribution. At early times the protein appeared more or less uniformly distributed in the cytoplasm and in the nucleus. In many (but not all) cells the protein was concentrated at the periphery of the nuclear envelope (A7), suggesting that the fusion protein may be targeted to the endocytoplasmic membrane and/or the nuclear envelope. At later times, the protein tended to be more concentrated in the nucleus than in the cytoplasm (A8; compare with A2), indicating that the C-terminal sequences may contain a cryptic nuclear localization sequence. Hong et al. (35) found that a short N-terminal sequence of the occlusion-derived virus (ODV) envelope protein, composed predominantly of hydrophobic amino acids, is sufficient to direct proteins to cytoplasmic membranes, nuclear envelope, and nucleus when expressed in Sf9 cells from baculoviruses. It is possible that the C-terminal hydrophobic sequence of AgMuc1 is playing a similar role.
Figure 5.
Localization of baculovirus-expressed GFP fusion proteins in Sf21 cells. (A) Sf21 cells were infected with recombinant baculoviruses that express the GFP, 72GFP27, GFP27, and 72GFP recombinant constructs (Fig. 4A) under the control of the viral polyhedrin promoter. Representative images recorded at 30 h after infection (A1, A3, A5, and A7) and at 60 h after infection (A2, A4, A6, and A8) are shown. The green is due to fluorescence of GFP or of GFP fusion proteins. N, Nucleus; C, cytoplasm. (×1000.) (B) Immunological localization of mucin-GFP fusion proteins. Living Sf21 cells expressing fusion proteins 72GFP27 or 72GFP (as shown in A4 and A6, respectively) were incubated with a rabbit anti-GFP antibody followed by incubation with a rhodamine-conjugated anti-rabbit IgG secondary antibody, thus detecting GFP-containing proteins only if exposed to the surface (red channel). The distribution of GFP, independent of its location in the cell, was detected by its inherent fluorescence (green channel). The images shown in B1 and B2 were collected while using identical settings for the green and red channels, and images from the red and green channels were then merged. (×250.)
To further corroborate the association of the baculovirus-expressed mucin with the cell surface, rabbit anti-GFP antibody and rhodamine-conjugated goat anti-rabbit IgG were used to detect the GFP fusion protein on the surface of living cells infected with recombinant AgMuc1-GFP baculoviruses (red channel). The inherent fluorescence of GFP, anywhere in the cell, was also recorded (green channel). Fig. 5B shows the fluorescent images. A red or orange ring indicating the presence of externalized GFP-containing protein was observed around almost every cell expressing 72GFP27 (Fig. 5, B1). In contrast, with the same staining and detection conditions, no rhodamine signal was detected in any cells expressing 72GFP (Fig. 5, B2). These results indicate that the 72GFP27 protein is accessible to antibodies on the surface of living cells, whereas the 72GFP protein is not. In conclusion, both the N-terminal and the C-terminal AgMuc1 amino acid sequences are required to direct the protein to the plasma membrane. It appears that the N-terminal sequence targets the protein to the secretory vesicle pathway (Fig. 5, A5 and A6) and that sequences contained in the 27-amino acid C-terminal hydrophobic stretch subsequently target the protein to the cell surface (Fig. 5, A4 and B1).
DISCUSSION
We have isolated and molecularly characterized a cDNA encoding an invertebrate intestinal mucin whose properties suggest potential roles in midgut physiology. The high abundance of AgMuc1 mRNA and the probable membrane association of the protein raises the possibility of its involvement in mosquito–parasite interactions.
AgMuc1 is likely to be displayed on the surface of the microvilli of midgut epithelial cells. This possibility is strongly suggested by two of our experiments. First, the GFP-mucin fusion protein (72GFP27) is localized on the surface of Sf21 cells (Fig. 5); second, an antiserum raised against microvilli recognizes the recombinant mucin fusion protein (Fig. 4). Furthermore, certain sequence features distinguish AgMuc1 from vertebrate intestinal mucins and from insect peritrophic matrix mucins. Two vertebrate intestinal mucins, MUC1 and MUC2, lack putative GPI-anchor and hydrophobic C-terminal sequences, and have a significantly larger number of threonine-rich repeat units (25, 26). The A. gambiae Ag-Aper1 peritrophic matrix protein also lacks GPI-anchor sequences and contains cysteine-rich carbohydrate-binding domains which may bind to the chitinous fibers of the peritrophic matrix (8, 36). In contrast, AgMuc1 contains a putative GPI-anchor sequence, has hydrophobic domains at both termini which are separated from the central repeat region by nonrepetitive sequences, and lacks carbohydrate-binding domains. It shares these features with the mucins of the parasite T. cruzi, potentially defining a new family of membrane-associated mucins.
AgMuc1 is presumed to be highly glycosylated and may constitute an important component of the carbohydrate-rich layer previously observed by electron microscopy at the microvillar surface (10, 11). Similar membrane-associated intestinal mucins may also exist in vertebrates, but the membrane-associated mucins that have been characterized to date in vertebrates are nonintestinal and have different functions. For instance, human membrane-associated mucin MUC1 may play a role in tumor progression, in metastasis (37–39), and in cell–cell or cell–matrix interactions (40, 41).
Mucins play important roles in cell–cell interactions in multicellular organisms (reviewed in ref. 42) and have been implicated in the interactions between unicellular parasites with their hosts. For instance, the galactose-specific adherence protein from Entamoeba histolytica binds to human colonic mucins and epithelial cells (43). The mucin-type glycoproteins of T. cruzi have also been suggested to be involved in the interaction with and/or invasion of mammalian host cells (44, 45). Accumulating evidence suggests that the carbohydrate on the mosquito microvillar surface might serve as a receptor for the initial association between the malaria parasites and the gut epithelial cells. Ramasamy et al. (13, 14) found that chitotriose and antibodies against midgut glycoproteins inhibited malaria parasite development. Moreover, Shahabuddin and co-workers (46) found in an in vitro assay that P. gallinaceum ookinetes could not bind to Aedes aegypti midguts after chemical modification of epithelial cell surface carbohydrates. As a major component of the microvillar glycocalyx, AgMuc1 might prove to be involved in parasite–midgut interactions prior to invasion.
The existence of a length polymorphism within the AgMuc1 mucin domain enabled the genetic localization of this gene to a chromosomal region neighboring the determinants for refractoriness to the malaria parasite P. cynomolgi B. This finding is useful, as it provides a genomic landmark within this interesting chromosomal region. However, the cytogenetic localization of AgMuc1 does not coincide with the estimated cytological localization of Pen1 (F. Collins, personal communication) and is thus unlikely to directly affect the Pen1-determined encapsulation refractory trait. This is not surprising because the Pen1 QTL was shown to affect encapsulation capacity, but not other traits relating to Plasmodium infection (17). However, the lack of identity with Pen1 does not exclude the possibility that AgMuc1 might be involved in early stages of parasite–mosquito interactions. Polymorphic AgMuc1 proteins might exhibit altered binding affinity or specificity to the ookinete, affecting its capacity to interact with the midgut epithelial cell surface prior to invasion, and thus might lead to differences in the vectorial capacity in strains expressing different alleles. Interaction assays of ookinetes with polymorphic AgMuc1 proteins and genetic experiments monitoring potential cosegregation of limited infection intensity phenotypes and AgMuc1 alleles could test the potential role of this intestinal mucin in mosquito–parasite interactions. More broadly, it is relevant that the insect gut is an important target for pest control (47–49). Characterization of insect intestinal mucins could lead to the development of new strategies for disease and pest control.
Acknowledgments
We are grateful to Karen M. Gustashaw for assistance with confocal microscopy, Rui Wang for genetic mapping and, Claudia Blass for cytogenetic mapping of AgMuc1. This work was supported by grants from the John D. and Catherine T. MacArthur Foundation and from the National Institute of Arthritis and Infectious Diseases. G.D. was supported by a European Union Training and Mobility of Researchers (TMR) fellowship.
ABBREVIATIONS
- QTL
quantitative trait loci
- GFP
green fluorescent protein
- GPI
glycosyl-phosphatidylinositol
Footnotes
References
- 1.Billingsley P F, Lehane M J. In: Biology of the Insect Midgut. Lehane M J, Billingsley P F, editors. London: Chapman & Hall; 1996. pp. 3–25. [Google Scholar]
- 2.Jacobs-Lorena M, Oo M M. In: The Biology of Disease Vectors. Beaty B J, Marquardt W C, editors. Niwot: Univ. Press of Colorado; 1996. pp. 318–332. [Google Scholar]
- 3.Tellam R L. In: Biology of the Insect Midgut. Lehane M J, Billingsley P F, editors. London: Chapman & Hall; 1996. pp. 86–114. [Google Scholar]
- 4.Del Bene G, Dallai R, Marchini D. Int J Morphol Embryol. 1991;20:15–24. [Google Scholar]
- 5.Elvin C M, Vuocolo T, Pearson R D, East I J, Riding G A, Eisemann C H, Tellam R L. J Biol Chem. 1996;271:8925–8935. doi: 10.1074/jbc.271.15.8925. [DOI] [PubMed] [Google Scholar]
- 6.Casu R, Eisemann C, Pearson R, Riding J, East I, Donaldson A, Cadogan L, Tellam R. Proc Natl Acad Sci USA. 1997;94:8939–8944. doi: 10.1073/pnas.94.17.8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang P, Granados R R. J Biol Chem. 1997;272:16663–16669. doi: 10.1074/jbc.272.26.16663. [DOI] [PubMed] [Google Scholar]
- 8.Shen Z, Jacobs-Lorena M. J Biol Chem. 1998;273:17665–17670. doi: 10.1074/jbc.273.28.17665. [DOI] [PubMed] [Google Scholar]
- 9.Terra W R, Ferreira B P, Dillon R J. In: Biology of the Insect Midgut. Lehane M J, Billingsley P F, editors. London: Chapman & Hall; 1996. pp. 153–194. [Google Scholar]
- 10.Kitajima E W. Cytobiologie. 1995;11:299–303. [Google Scholar]
- 11.Rudin W, Hecker H. Parasitol Res. 1989;75:268–279. doi: 10.1007/BF00931811. [DOI] [PubMed] [Google Scholar]
- 12.Lane N J, Dallai R, Ashhurst D E. In: Biology of the Insect Midgut. Lehane M J, Billingsley P F, editors. London: Chapman & Hall; 1996. pp. 115–150. [Google Scholar]
- 13.Ramasamy R, Wanniarachchi I C, Srikrishnaraj K A, Ramasamy M S. Biochim Biophys Acta. 1997;1361:114–122. doi: 10.1016/s0925-4439(97)00020-3. [DOI] [PubMed] [Google Scholar]
- 14.Ramasamy M S, Kulasekera R, Wanniarachchi I C, Srikrishnaraj K A, Ramasamy R. Med Vet Entomol. 1997;11:290–296. doi: 10.1111/j.1365-2915.1997.tb00409.x. [DOI] [PubMed] [Google Scholar]
- 15.Vernick K D, Fujioka H, Seeley D C, Tandler B, Aikawa M, Miller L H. Exp Parasitol. 1995;80:583–595. doi: 10.1006/expr.1995.1074. [DOI] [PubMed] [Google Scholar]
- 16.Collins F H, Sakai R K, Vernick K D, Paskewitz S, Seeley D C, Miller L H, Collins W E, Campbell C C, Gwadz R W. Science. 1986;234:607–610. doi: 10.1126/science.3532325. [DOI] [PubMed] [Google Scholar]
- 17.Zheng L, Cornel A J, Wang R, Erfle H, Voss H, Ansorge W, Kafatos F C. Science. 1997;276:425–428. doi: 10.1126/science.276.5311.425. [DOI] [PubMed] [Google Scholar]
- 18.Lemos F J, Cornel A J, Jacobs-Lorena M. Insect Biochem Mol Biol. 1996;26:651–658. doi: 10.1016/s0965-1748(96)00014-8. [DOI] [PubMed] [Google Scholar]
- 19.Dimopoulos G, Seeley D, Wolf A, Kafatos F C. EMBO J. 1998;17:6115–6123. doi: 10.1093/emboj/17.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feinberg A P, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 21.Kumar V, Collins G H. Insect Mol Biol. 1996;3:41–47. doi: 10.1111/j.1365-2583.1994.tb00149.x. [DOI] [PubMed] [Google Scholar]
- 22.English L H, Readdy T L, Bastin A E. Insect Biochem. 1991;21:177–184. [Google Scholar]
- 23.Gum J R. Biochem Soc Trans. 1995;23:795–799. doi: 10.1042/bst0230795. [DOI] [PubMed] [Google Scholar]
- 24.Gendler S R, Spicer A P. Annu Rev Physiol. 1995;57:607–634. doi: 10.1146/annurev.ph.57.030195.003135. [DOI] [PubMed] [Google Scholar]
- 25.Gum J R, Jr, Hicks J W, Toribara N W, Siddiki B, Kim Y S. J Biol Chem. 1994;269:2440–2446. [PubMed] [Google Scholar]
- 26.Gum J R, Jr, Ho J J L, Pratt W S, Hicks J W, Hill A S, Vinall L E, Roberton A M, Swallow D M, Kim Y S. J Biol Chem. 1997;272:26678–26686. doi: 10.1074/jbc.272.42.26678. [DOI] [PubMed] [Google Scholar]
- 27.Spicer A P, Parry G, Patton S, Gendler S J. J Biol Chem. 1991;266:15099–15109. [PubMed] [Google Scholar]
- 28.von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Englund P T. Annu Rev Biochem. 1993;62:121–138. doi: 10.1146/annurev.bi.62.070193.001005. [DOI] [PubMed] [Google Scholar]
- 30.Schenkman S, Ferguson M A, Heise N, de Almeida M L, Mortara R A, Yoshida N. Mol Biochem Parasitol. 1993;59:293–303. doi: 10.1016/0166-6851(93)90227-o. [DOI] [PubMed] [Google Scholar]
- 31.Reyes M B, Pollevick G D, Frasch A C. Gene. 1994;140:139–140. doi: 10.1016/0378-1119(94)90745-5. [DOI] [PubMed] [Google Scholar]
- 32.Di Noia J M, Sanchez D O, Frasch A C. J Biol Chem. 1995;270:24146–24149. doi: 10.1074/jbc.270.41.24146. [DOI] [PubMed] [Google Scholar]
- 33.Gum J R, Jr, Hicks J W, Lagace R E, Byrd J C, Toribara N W, Siddiki B, Fearney F J, Lamport D T A, Kim Y S. J Biol Chem. 1991;266:22733–22738. [PubMed] [Google Scholar]
- 34.Di Noia J M, Pollevick G D, Xavier M T, Previato J O, Mendoca-Previato L, Sanchez D O, Frasch A C. J Biol Chem. 1996;271:32078–32083. doi: 10.1074/jbc.271.50.32078. [DOI] [PubMed] [Google Scholar]
- 35.Hong T, Summers M D, Braunagel S C. Proc Natl Acad Sci USA. 1997;94:4050–4055. doi: 10.1073/pnas.94.8.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen Z, Jacobs-Lorena M. J Mol Evol. 1999;48:341–347. doi: 10.1007/pl00006478. [DOI] [PubMed] [Google Scholar]
- 37.Burdick M D, Harris A, Reid C J. J Biol Chem. 1997;272:24198–24202. doi: 10.1074/jbc.272.39.24198. [DOI] [PubMed] [Google Scholar]
- 38.Cao Y, Blohm D, Ghadimi B M, Stosiek P, Xing P X, Karsten U. J Histochem Cytochem. 1997;45:1547–1557. doi: 10.1177/002215549704501111. [DOI] [PubMed] [Google Scholar]
- 39.Makiguchi Y, Hinoda Y, Imai K. J Cancer Res. 1996;87:505–511. doi: 10.1111/j.1349-7006.1996.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hilkens J, Vos H L, Wesseling J, Boer M, Storm J, van der Valk S, Calafat J, Patriarca C. Cancer Lett. 1995;90:27–33. doi: 10.1016/0304-3835(94)03674-8. [DOI] [PubMed] [Google Scholar]
- 41.Wesseling J, van der Valk S W, Vos H L, Sonnenberg A, Hilkens J. J Cell Biol. 1995;129:255–265. doi: 10.1083/jcb.129.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Klinken B J, Dekker J, Buller H A, Einerhand A W. Am J Physiol. 1995;269:913–627. doi: 10.1152/ajpgi.1995.269.5.G613. [DOI] [PubMed] [Google Scholar]
- 43.Ravdin J I, Shain D C, Kelsall B L. Vaccine. 1993;11:214–246. doi: 10.1016/0264-410x(93)90024-r. [DOI] [PubMed] [Google Scholar]
- 44.Serrano A A, Schenkman S, Yoshida N, Mehlert A, Richardson J M, Ferguson M A. J Biol Chem. 1995;270:27244–27253. doi: 10.1074/jbc.270.45.27244. [DOI] [PubMed] [Google Scholar]
- 45.Burleigh B A, Andrews N W. Annu Rev Microbiol. 1995;49:175–200. doi: 10.1146/annurev.mi.49.100195.001135. [DOI] [PubMed] [Google Scholar]
- 46.Zieler J, Nawrocki J, Shahabuddin M. J Exp Biol. 1999;202:485–495. doi: 10.1242/jeb.202.5.485. [DOI] [PubMed] [Google Scholar]
- 47.Vadlamudi R K, Weber E, Ji I, Ji T H, Bulla L A., Jr J Biol Chem. 1995;270:5490–5494. doi: 10.1074/jbc.270.10.5490. [DOI] [PubMed] [Google Scholar]
- 48.Francis B R, Bulla L A., Jr Insect Biochem Mol Biol. 1997;27:541–550. doi: 10.1016/s0965-1748(97)00029-5. [DOI] [PubMed] [Google Scholar]
- 49.Knight P J, Knowles B H, Ellar D J. J Biol Chem. 1995;270:17765–17770. doi: 10.1074/jbc.270.30.17765. [DOI] [PubMed] [Google Scholar]