Abstract
Bloom’s syndrome (BS) is a rare autosomal recessive disorder of humans characterized by severe pre- and postnatal growth deficiency, immunodeficiency, genomic instability, and a predisposition to a wide variety of neoplasms. The genomic instability is evidenced in BS somatic cells as a high incidence of gaps and breaks, chromatid exchanges, chromosome rearrangements, and locus-specific mutations. BS arises from a mutation in BLM, a gene encoding a protein with homology to the RecQ helicase family. Men with BS are sterile; women have reduced fertility and a shortened reproductive span. The current immunocytological study on mouse spermatocytes shows that the BLM protein is first evident as discrete foci along the synaptonemal complexes (SCs) of homologously synapsed autosomal bivalents in late zygonema of meiotic prophase. BLM foci progressively dissociate from the synapsed autosomal axes during early pachynema and are no longer seen in mid-pachynema. BLM colocalizes with the single-stranded DNA binding replication protein A, which has been shown to be involved in meiotic synapsis. However, there is a temporal delay in the appearance of BLM protein along the SCs relative to replication protein A, suggesting that BLM is required for a late step in processing of a subset of genomic DNA involved in establishment of interhomologue interactions in early meiotic prophase. In late pachynema and into diplonema, BLM is more dispersed in the nucleoplasm, especially over the chromatin most intimately associated with the SCs, suggesting a possible involvement of BLM in resolution of interlocks in preparation for homologous chromosome disjunction during anaphase I.
Keywords: meiosis, recombination, synapsis
Bloom’s syndrome (BS) is a rare autosomal recessive disorder characterized by growth deficiency, immunodeficiency, sun-sensitive facial erythema, genomic instability, and a greatly increased predisposition to a wide variety of cancers common in the general population (1). Somatic cells from BS individuals are hypermutable and have an excessive number of locus-specific mutations as well as a high frequency of microscopically visible chromatid gaps, breaks, and rearrangements, including quadriradials (the result of somatic crossing over). In BS cells, standard treatment with BrdUrd followed by staining with Hoechst 33258, which differentially labels sister chromatids, reveals an abnormally high incidence of sister chromatid exchanges as compared with normal cells (1).
The Bloom’s protein, BLM (2), belongs to a subfamily of DExH box-containing DNA and RNA helicases that includes the Escherichia coli protein RecQ (3), the Saccharomyces cerevisiae Sgs1p (4–6), the Schizosaccharomyces pombe rqh1p (7) or rad12p (8), and the human proteins RECQL (9, 10) and WRN, the protein mutated in Werner’s syndrome (11). Members of the RecQ family of helicases have been implicated in both DNA replication and recombination, as well as in chromosome segregation (for review, see refs. 12 and 13). Genomic instability associated with BS might therefore arise from a defect in one or more of these processes.
The range of cellular phenotypes associated with this class of RecQ helicases is not confined to mitotic cells. The meiotic phenotype of sgs1 mutants includes hyperrecombination, missegregation, and poor sporulation (4, 5). Men with BS (homozygotes or compound heterozygotes) produce no spermatozoa and consequently are sterile; women, although sometimes fertile, cease ovulation at unusually young ages (1). These defects clearly suggest involvement of BLM in one or more critical events in germ-cell production. Male carriers (heterozygotes) of BLM mutations are developmentally normal, but the spermatozoa of at least some of these men exhibit a higher than normal frequency of multiple chromosome breaks and rearrangements (14).
Replication protein A (RPA) is a heterotrimeric single-stranded (ss)DNA-binding protein that participates in DNA replication, repair, and recombination (for review, see ref. 15). It is present on meiotic prophase chromosomes and plays a role in both homologous synapsis and recombination (16). To investigate the possible role, or roles, for BLM in mammalian meiosis and to compare its localization pattern to that of RPA, we have used an antibody raised against a portion of the human BLM protein to examine BLM distribution during meiotic prophase in mouse spermatocytes. BLM localization is delayed relative to that of RPA, but its presence on synapsed bivalents in late zygotene and early pachytene nuclei suggests that it is involved in meiotic synapsis. Later in pachynema, BLM exhibits a more dispersed nucleoplasmic localization pattern over the chromatin, suggesting that it may play an additional role in the late stages of meiotic prophase.
MATERIALS AND METHODS
Mice and Preparation of Spermatocytes.
Over 200 nuclei were imaged from surface spreads (17) from a total of five C57BL/6 inbred mice. To increase the frequency of zygotene and early pachytene nuclei in our preparations, we used prepubertal mice (17–20 days old) in which the first meiotic wave of spermatocyte development had just begun (18).
Antigen and Antiserum Preparation.
The preparation of the BLM antigen and antiserum has been described (19). To visualize the axial/lateral elements of the synaptonemal complex, a goat polyclonal antibody against SCP3, a component of the axial/lateral element (20), was made and affinity-purified by using an ImmunoPure IgG purification kit (Pierce).
Antibody Detection.
Antibody incubation and detection procedures were as described (21). Fresh surface-spread spermatocyte preparations were incubated with primary antibodies against hsBLM (made in rabbit) and SCP3 (made in goat) followed by detection with anti-rabbit-FITC and anti-goat-rhodamine antibodies, respectively (Jackson ImmunoResearch). RPA (anti-rabbit) and BLM (anti-rabbit) colocalization was carried out by using sequential detection with a Fab-fragment secondary antibody (22). The BLM antibody was applied and detected first with Fab anti-rabbit IgG FITC (Jackson ImmunoResearch) followed by RPA detection with anti-rabbit IgG rhodamine. The Fab-fragment FITC secondary antibody binds to and blocks the antigen sites of the BLM antibody. Because the Fab fragment is nonantigenic, a second antibody (against RPA), also raised in rabbit, can be detected in a subsequent round of incubation with the RPA antibody followed by detection with a rhodamine-labeled anti-rabbit secondary antibody. Reverse staining was also done. All preparations were counterstained with 4′,6-diamino-2-phenylindole (DAPI, Sigma), and mounted in a diazabicyclo[2.2.2]octane (DABCO, Sigma) antifade solution.
Cytological Specificity.
A Western blot probed with the BLM antibody reveals a major and minor band on extracts from normal individuals (19). Although the major band is not present in cell extracts from BS individuals, the minor band persists (19). However, the antibody produces a focal pattern in somatic nuclei of cultured fibroblasts from normal individuals that is absent in similarly treated nuclei from BS individuals (19). Both the focal and nucleoplasmic staining in mouse spermatocytes was blocked by preincubation of BLM antibody with the BLM protein fragment, whereas the SCP3 signal remained positive on these cells (data not shown), confirming the specificity of both the focal and nucleoplasmic staining reactions for BLM.
Image Analysis.
All preparations were examined on a Nikon Eclipse E800 fluorescence microscope equipped with narrow band-pass filters. Each fluorochrome image was captured separately as an eight-bit source image by using a computer-assisted cooled charge-coupled device camera (Photometrics CH 350). The separate BLM/SCP3 or RPA/SCP3 images were 24-bit pseudocolored and merged in genejoin custom software developed by Tim Rand (23). Adobe photoshop was used to produce the merged BLM/RPA images. Although DAPI images provide information about the stage of meiotic prophase, they obscure many details of the relationships between the protein components, and so were not used in the final merged images shown here.
RESULTS
Background.
Meiotic prophase is divided into five stages: leptonema, zygonema, pachynema, diplonema, and diakinesis. During leptonema, the chromatin begins to condense, and proteinaceous axial elements begin to form between sister chromatids. As axial elements of homologous chromosomes align and come into contact during zygonema, a central element and transverse filaments form between homologues, completing the structure called the synaptonemal complex (SC). Homologous autosomes are fully synapsed throughout pachynema, the period during which reciprocal recombination (crossing over) occurs. During diplonema, the central element of the SC disassembles and homologous chromosomes begin to repel one another, but remain held together at chiasmata (crossing-over sites). Prophase concludes with diakinesis, during which further chromatin condensation occurs. Additional meiosis-specific structures include two types of meiotic nodules, termed early and late meiotic nodules, or recombination nodules, that are involved in meiotic synapsis and reciprocal recombination, respectively (see refs. 16, 24, and 25). Protein components of these structures have now been identified, and antibodies against them are available. SCP3, a component of the axial/lateral elements, allows visualization of these structures from the time they form in leptonema until they disassemble in diplonema (20). RPA becomes a component of early meiotic nodules once homologous chromosomes have synapsed and is also a component of late (recombination) nodules in the early stages of their development (16).
Leptonema.
Neither BLM nor RPA is detectable in leptotene nuclei as the chromatin begins to condense and axial elements begin to form.
Zygonema.
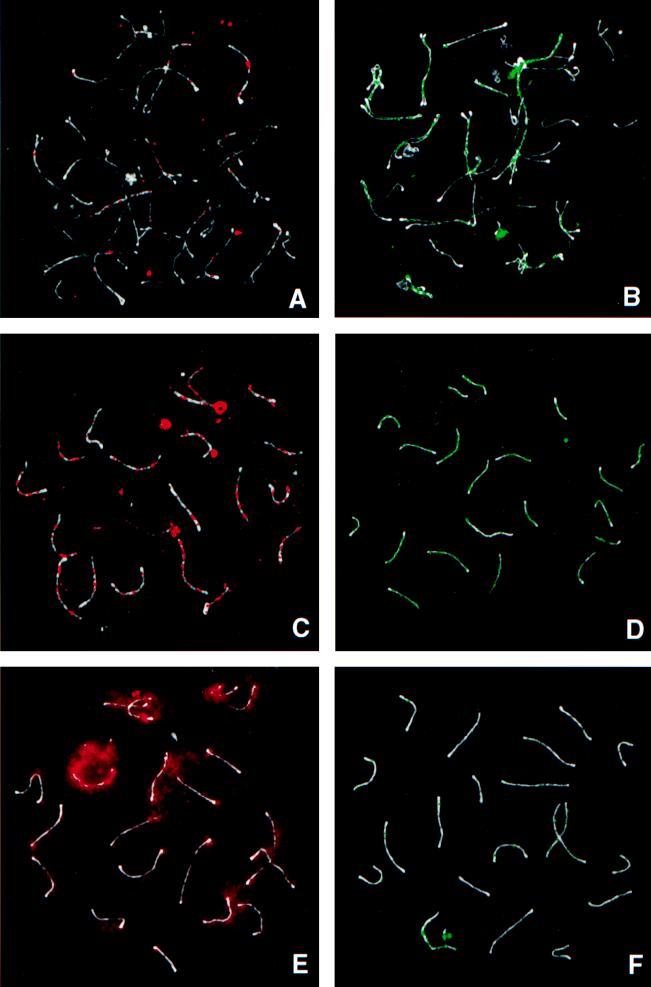
RPA is not present on asynapsed axial elements (16, 26). However, as soon as any portion of the bivalent has synapsed to form even a short stretch of SC, RPA foci are immediately apparent at multiple sites along its length (Fig. 1B) (16, 26). Because RPA binds to ssDNA, it is logical to assume that these sites are regions where a subset of DNA is present in a single-stranded form and available for direct interactions between homologous chromosomes. During zygonema, BLM also becomes associated with sites along the SCs. In contrast to RPA, BLM foci are not observable in early zygotene nuclei, even on those portions of the autosomal bivalents that have already synapsed. BLM foci only become evident in mid-late zygotene spermatocytes, and then only on a few of the synapsed autosomal axes (Fig. 1A). Both the number of BLM foci and the number of bivalents with foci vary greatly from nucleus to nucleus and reflect the degree of asynchrony within and between individual nuclei. This variation in pattern of BLM distribution on some, but not all, of the synapsed axes in late zygotene nuclei was observed repeatedly in our sample of over 50 zygotene spermatocytes.
Figure 1.
Comparison of the distribution of BLM (Left, red) and RPA (Right, green) at different times during meiotic prophase in mouse spermatocytes. The axial elements are stained with SCP3 (white). (A and B) Zygonema. Only some synapsed SCs in zygonema have BLM foci (A), whereas all synapsed SCs have RPA foci (B). (C and D) Early pachynema. SCs have multiple BLM foci (C) and RPA foci (D) along their length. (E and F) Mid-pachynema. Most BLM foci have disappeared from the SCs, and a more diffuse chromatin reaction is beginning to increase throughout the nucleoplasm. The build-up is most evident over the sex body (intense red area at 11 o’clock) and around the SCs (E). At a similar stage, the majority of RPA foci have disappeared (F).
Early Pachynema.
Moses (27) divided pachynema in mouse spermatocytes into five substages, based on distinct cytological criteria. Early pachynema, described here, corresponds to substages 1 and 2; mid- to late pachynema, described below, corresponds to substages 3–5. As described (16, 26), numerous RPA foci are present along the lengths of each autosomal SC bivalent in early pachytene spermatocytes (Fig. 1D). In the transition between early and mid-pachynema, RPA foci gradually begin to decrease in number (Fig. 1F) and disappear during mid-pachynema.
BLM foci also are present at multiple sites along the length of each autosomal bivalent in early pachynema. They too reach their maximum number and intensity on the autosomal axes during this substage (Fig. 1C). During the transition from early to mid-pachynema, BLM foci begin to disappear.
Mid-Late Pachynema.
As the BLM foci disappear during the mid-pachytene interval, a more diffuse nuclear localization pattern becomes increasingly prominent throughout the nucleus (delineated by the DAPI images; data not shown). However, it is present in a higher concentration around the SCs and in an even higher concentration throughout the sex body, the chromosomal domain of the X and Y chromosomes (Fig. 1E). The intensity of this diffuse localization pattern continues to build in the nucleoplasm throughout pachynema and persists into diplonema (data not shown).
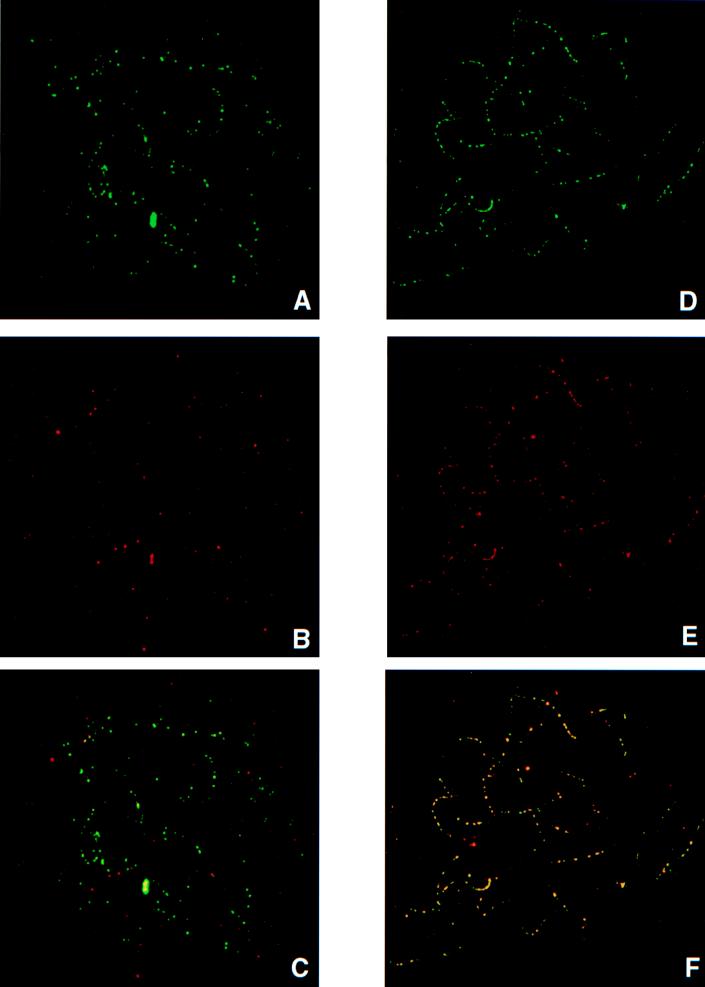
Colocalization.
As mentioned above, RPA becomes a component of early meiotic nodules as soon as homologous autosomes synapse (16), but BLM does not appear until later. To determine whether BLM is also a component of these structures, double-labeling experiments were carried out. When BLM foci begin to appear in late zygonema (Fig. 2 A–C), they colocalize with RPA. However, the reciprocal is not true; many linear arrays of RPA foci lack BLM foci, consistent with the delayed appearance of BLM relative to RPA described above. However, by early pachynema, BLM and RPA exhibit near perfect colocalization (Fig. 2 D–F).
Figure 2.
Colocalization of RPA (green) and BLM (red) on prophase spermatocytes. Colocalization produces a yellow signal. (A–C) Mid-zygotene nucleus with RPA (A), BLM (B), and RPA/BLM (yellow). Note that most of the foci in C are green, indicating that BLM is not yet present. (D–F) Early pachytene nucleus with RPA (D), BLM (E), and RPA/BLM (F). Note that most foci are yellow, indicating near perfect colocalization at this stage.
DISCUSSION
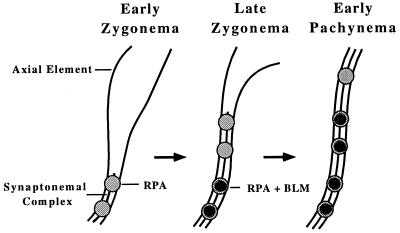
Meiotic chromatin is organized into loops, with only the chromatin at the base of these loops in contact with the axial/lateral element regions of the SCs (28). Some of the basal loop chromatin associated with the SCs is single-stranded, as evidenced by the binding of RPA (16, 26). RPA also is a component of early meiotic nodules (16), and it is at these sites that molecular interactions between homologues are thought to occur. The temporal localization pattern of BLM relative to RPA is illustrated in Fig. 3. BLM is a ssDNA helicase, a class of molecule that recognizes a single-stranded gap in a duplex, binds there, and then translocates into the duplex, creating more ssDNA with which RPA can bind. Therefore, the colocalization and coactivity of a ssDNA helicase and a ssDNA-binding protein could be predicted and supports the supposition that these foci mark sites of ssDNA synaptic-related meiotic activity. Although the exact function of BLM and the nature of the interactions remain unclear, some tentative conclusions can be deduced from the present observations.
Figure 3.
Diagram showing the change in RPA and BLM localization on a bivalent as it progresses from early to late zygonema and into early pachynema.
Technical Limitations.
First, it is important to understand that with current antibody technology, detection of a single molecule of a protein is impossible. Instead, 60 fluors have been calculated to be the minimum necessary for detection by fluorescence microscopy and charge-coupled device imaging techniques (S. G. Ballard, personal communication). A protein such as RPA polymerizes on ssDNA, and it is the presence of these multiple molecules that provides a sufficiently high concentration for detection. In contrast, models of helicase activity suggest that a single helicase molecule may be sufficient to unwind the DNA. Because it is unlikely that the BLM antibody used here had more than three or four binding sites for a secondary antibody which, in turn, had two to three attached fluors, it is reasonable to conclude that multiple BLM molecules must be present at each focus to be visualized.
Recombination.
RecQ helicase of E. coli suppresses illegitimate recombination (29) and can initiate and disrupt DNA recombination by unwinding several types of joint molecules (13). Loss of these RecQ ssDNA helicases appears to result in an increased frequency of recombination events, suggesting that these enzymes may prevent single-stranded regions of one chromatid or homologues from invading a neighboring DNA molecule and initiating recombination. As discussed above, if BLM were associated with a recombination intermediate in mammalian spermatocytes, the single molecule sufficient for its activity would be undetectable in our preparations. At no time during pachynema does the distribution pattern of BLM foci in pachytene spermatocytes correspond to the distribution pattern of MLH1, a protein implicated in meiotic reciprocal recombination (30, 31). Meiotic recombination is thought to be initiated by double-strand breaks. Although we cannot rule out the possibility of multiple breaks at each BLM focus, the severe genomic instability that would result from such multiple lesions per focus seems to preclude such an alternative. Moreover, if helicases such as BLM and Sgs1 were directly involved in recombination, one would predict that mutations in these genes would result in reduced recombination. Instead, cells that are homozygously defective for sgs1, rqh1, or BLM exhibit a hyperrecombination phenotype (1, 7, 12).
Replication.
In Lilium, delayed replication of a subset of genomic DNA, termed zygotene DNA (zygDNA), appears to play a critical role in synapsis (32) because blockage of this synthesis blocks synapsis (33). DNA synthesis during early meiotic prophase also has been observed in mammalian spermatocytes (34, 35). DNA replication in eukaryotes does not proceed one replicon at a time. Instead, the chromatin is organized into replicon clusters with multiple replicons within a cluster firing more or less synchronously (36). A model of organization of zygDNA into replicon clusters has been postulated to occur in mammalian meiotic nuclei (37). Under this model, the zygDNA would unwind and RPA would bind to the single-stranded sequences, which would then be used in a check for homology and establishment of interhomologue interactions before their replication. Multiple BLM molecules associated with the multiple ssDNA strands within these zygDNA replicon clusters would provide a sufficient concentration of molecules for fluorescence detection.
As discussed, RPA foci appear on SCs as soon as homologues synapse, whereas BLM foci appear later. Watt et al. (5) have suggested that Sgs1 of S. cerevisiae may release topological constraints on chromatin as replication forks converge rather than during the initial stages of unwinding for initiation of replication. If the BLM protein is playing a similar role in mammalian meiotic prophase nuclei, under the replication–cluster model discussed above, the delayed appearance of the antibody relative to RPA is consistent with its involvement in the terminal stages of zygDNA replication. Such a role would also be consistent with the observed defects in cultured BS cells that have been linked to replication errors (38). The microscopically visible sister chromatid exchanges and quadriradial homologous chromosome interchanges are also best explained as errors arising during S phase or G2 (1).
Initial expression of Blm mRNA in mouse testis occurs between days 12 and 14 after birth (39), as the first wave of spermatocytes reaches early pachynema (18) consistent with the peak of BLM foci we observe.
Recombination Reconsidered.
E. coli RecQ is a component of the RecF recombination pathway that participates both in recombination involving ssDNA gaps and in repair of broken replication forks (see ref. 13). It therefore seems likely that this BLM ortholog may link processes involved in DNA replication with those involved in repair and recombination. If multiple BLM protein molecules were first involved in zygDNA replication, and only one of these replicating sequences at any particular replication cluster were subsequently involved in reciprocal recombination, the single molecule of BLM involved in subsequent recombination would lie below the limits of the fluorescence detection methods discussed above. Thus, a role for BLM in crossover cannot be excluded, but as discussed above, is unlikely to be its sole function.
As discussed above, Sgs1p has been found to interact with three different yeast topoisomerases. Unmentioned and untested in these studies was Spo11p, the meiosis-specific protein that not only belongs to a unique family of topoisomerases, but initiates programmed double-stranded breaks in meiotic prophase of yeast (40). If Sgs1p does interact with Spo11p, the intriguing possibility exists that BLM might interact with the as-yet-unidentified mammalian SPO11 ortholog. Chester et al. (41) recently described an alternate Blm transcript in mouse testis. It is tempting to speculate that this meiosis-specific transcript of BLM might have evolved to better interact with mammalian SPO11.
Late Prophase Activity.
The dispersed nucleoplasmic distribution of BLM, especially its concentration along the SCs, may point to a second role for BLM in mammalian meiotic prophase. In addition to the interaction between Sgs1p and Top3p in S. cerevisiae (4), an interaction has also been demonstrated between Sgs1p and Top2p (5). Top2 is a type II topoisomerase that initiates double-strand breaks to disentangle intertwined DNA (42, 43) and to separate sister chromatids to insure proper segregation during mitosis as well as meiosis (reviewed in ref. 44). Mammalian TOP2 has been reported to accumulate on the SCs in mid-to late-pachytene nuclei (45), the same time during which we observed the increase in concentration of BLM in the nucleoplasm. The higher concentration of BLM around the chromatin near the SCs observed here suggests that these two proteins might be interacting. A likely function of BLM and TOP2 in late meiotic prophase as the primary spermatocyte nuclei make final preparations for the first meiotic division would be to resolve interlocks between sister chromatids. Although such a role would appear to be unrelated to either DNA replication or recombination, it is consistent with the helicase–topoisomerase interactions discussed above.
Cell Cycle Regulation.
ATM, the protein encoded by the gene mutated in ataxia telangiectasia, another human genetic disorder characterized by genomic instability (46), also has been shown to colocalize with RPA on synapsed axes of mammalian spermatocytes (16, 26). ATM, RPA, and BLM therefore all appear to be transient components of the same large protein complex on newly synapsed SCs. ATM has been implicated in detection of DNA damage and in cell cycle control (for review, see refs. 46 and 47). Although ataxia telangiectasia cells are defective at multiple cell cycle checkpoints, the most characteristic of these checkpoint defects is the inability to halt DNA synthesis after exposure to radiation damage (47–49). Despite the fact that BLM has not been implicated in cell cycle control, it is worth noting that the rqh1 gene of S. pombe, an ortholog of BLM, appears to couple physical completion of DNA replication with regulatory release from the S-phase checkpoint (7, 8). ATM, BLM, and RPA have all been implicated in both replication and repair activities of somatic cells. Further deciphering their individual and interactive roles on synapsed meiotic bivalents remains a challenge.
Acknowledgments
We thank James Ingles for the RPA antibody, Christa Heyting for the Scp3 cDNA, and Joanne Sweasy for expressing the SCP3 protein from which the SCP3 antibody was made. We thank Adelle Hack for her editorial suggestions and Lisa Webb for her computer expertise. We also appreciate the comments and suggestions of the anonymous reviewer. The work was supported by National Institutes of Health Grants GM47797 and GM55300 and a grant from the Ataxia Telangiectasia Medical Foundation to T.A. and funding from the New York Blood Center and National Institutes of Health Grant CA 50897 to J.G.
ABBREVIATIONS
- RPA
replication protein A
- BS
Bloom’s syndrome
- SC
synaptonemal complex
- ssDNA
single-stranded DNA
- zygDNA
zygotene DNA
References
- 1.German J, Ellis N A. In: The Genetic Basis of Human Cancer. Vogelstein B, Kenzler K W, editors. New York: McGraw–Hill; 1998. pp. 301–315. [Google Scholar]
- 2.Ellis N A, Groden J, Ye T Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 3.Nakayama K, Irono N, Nakayama H. Mol Gen Genet. 1985;200:266–271. doi: 10.1007/BF00425434. [DOI] [PubMed] [Google Scholar]
- 4.Gangloff S, McDonald J P, Bendixen C, Arthur L, Rothstein R. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watt P M, Louis E J, Borts R H, Hickson I D. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 6.Lu J, Mullen J R, Brill S J, Kleff S, Romeo A M, Sternglanz R. Nature (London) 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 7.Stewart E, Chapman C R, Al-Khodiary F, Carr A M, Enoch T. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey S, Han C S, Ramer S A, Klassen J C, Jacobson A, Eisenberger A, Hopkins K M, Lieberman H B, Freyer G G. Mol Cell Biol. 1998;18:2721–2728. doi: 10.1128/mcb.18.5.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puranam K L, Blackshear P J. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- 10.Seki M, Miyazawa H, Tada S, Yanagisawa J, Yamaoka T, Hoshino S-i, Ozawa K, Eki T, Nogami M, Okumura K, et al. Nucleic Acids Res. 1994;22:4566–4573. doi: 10.1093/nar/22.22.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu C E, Oshima J, Fu Y H, Wijsman E, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouaia S, et al. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 12.Watt P M, Hickson I D. Current Biol. 1996;6:265–267. doi: 10.1016/s0960-9822(02)00474-8. [DOI] [PubMed] [Google Scholar]
- 13.Harmon F G, Kowalczykowski S C. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin R H, Rademaker A, German J. Am J Hum Genet. 1994;55:1242–1246. [PMC free article] [PubMed] [Google Scholar]
- 15.Wold M S. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 16.Plug A W, Peters A H F M, van Breuklen B, Keegan K S, Hoekstra M, de Boer P, Ashley T. J Cell Sci. 1998;111:413–423. doi: 10.1242/jcs.111.4.413. [DOI] [PubMed] [Google Scholar]
- 17.Peters A H F M, Plug A W, van Vugt M J, de Boer P. Chromosome Res. 1997;5:66–71. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- 18.Goetz P, Chandley A C, Speed R M. J Cell Sci. 1984;65:249–263. doi: 10.1242/jcs.65.1.249. [DOI] [PubMed] [Google Scholar]
- 19.Neff N F, Ellis N A, Ye T Z, Noonan J, Huang K, Sanz M, Proytcheva M. Mol Biol Cell. 1999;10:665–676. doi: 10.1091/mbc.10.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lammers J H M, Offenberg H H, van Aalderen M, Vink A C G, Dietrich A J J, Heyting C. Mol Cell Biol. 1994;14:1137–1146. doi: 10.1128/mcb.14.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashley T, Plug A W, Xu J, Solari A J, Reddy G, Golub E I, Ward D C. Chromosoma. 1995;104:19–28. doi: 10.1007/BF00352222. [DOI] [PubMed] [Google Scholar]
- 22.Wessel G M, McClay D R. J Histochem Cytochem. 1986;34:703–706. doi: 10.1177/34.6.3084626. [DOI] [PubMed] [Google Scholar]
- 23.Ried T, Baldini A, Rand T, Ward D C. Proc Natl Acad Sci USA. 1992;89:1388–1392. doi: 10.1073/pnas.89.4.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stack S, Sherman J, Anderson L, Herickhoff L. In: Chromosomes Today. Summer A, Chandley A, editors. Vol. 11. London: Chapman & Hall; 1993. pp. 301–311. [Google Scholar]
- 25.Carpenter A T C. Cold Spring Harbor Symp Quant Biol. 1984;49:23–29. doi: 10.1101/sqb.1984.049.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Plug A W, Peters A H F M, Xu Y, Keegan K S, Hoekstra M F, Baltimore D, deBoer P, Ashley T. Nat Genet. 1997;17:457–461. doi: 10.1038/ng1297-457. [DOI] [PubMed] [Google Scholar]
- 27.Moses M J. In: Animal Models in Human Reproduction. Serio M, Martini L, editors. New York: Raven; 1980. pp. 169–190. [Google Scholar]
- 28.Weith A, Traut W. Chromosoma. 1980;78:275–291. [Google Scholar]
- 29.Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker S M, Plug A W, Prolla T A, Bronner C E, Harris A C, Yao X, Christie D-M, Monell C, Arnheim N, Bradley A, Ashley T, Liskay R M. Nature Genetics. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 31.Anderson L K, Reeves A, Webb L M, Ashley T. Genetics. 1999;151:1569–1579. doi: 10.1093/genetics/151.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hotta Y, Ito M, Stern H. Proc Natl Acad Sci USA. 1966;56:1184–1191. doi: 10.1073/pnas.56.4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth T F, Ito M. J Cell Biol. 1967;35:247–255. doi: 10.1083/jcb.35.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lima-de-Faria A, German J, Ghartnekan M, McGovern J, Anderson L. Hereditas. 1968;60:249–261. doi: 10.1111/j.1601-5223.1968.tb02205.x. [DOI] [PubMed] [Google Scholar]
- 35.Moses M J. Symp Soc Exp Biol. 1984;38:245–270. [PubMed] [Google Scholar]
- 36.Hand R. Cell. 1978;15:317–325. doi: 10.1016/0092-8674(78)90001-6. [DOI] [PubMed] [Google Scholar]
- 37.Plug A W, Xu J, Reedy G, Golub E I, Ashley T. Proc Natl Acad Sci USA. 1996;93:5920–5924. doi: 10.1073/pnas.93.12.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hand R, German J. Proc Natl Acad Sci USA. 1975;72:758–762. doi: 10.1073/pnas.72.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seki T, Seki M, Katada T, Enomoto T. Biochim Biophys Acta. 1998;1396:127–131. doi: 10.1016/s0167-4781(97)00192-9. [DOI] [PubMed] [Google Scholar]
- 40.Keeney S, Giroux C, Kleckner N. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 41.Chester N, Kuo F, Kozak C, O’Harra C D, Leder P. Genes Dev. 1998;12:3382–3393. doi: 10.1101/gad.12.21.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang T S F. Annu Rev Biochem. 1991;60:513–552. doi: 10.1146/annurev.bi.60.070191.002501. [DOI] [PubMed] [Google Scholar]
- 43.Holm C, Goto T, Wang J C, Botstein D. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 44.Watt P, Hickson I D. Biochem J. 1994;303:681–695. doi: 10.1042/bj3030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moens P B, Earnshaw W C. Chromosoma. 1989;98:317–322. doi: 10.1007/BF00292383. [DOI] [PubMed] [Google Scholar]
- 46.Shiloh Y. Eur J Hum Genet. 1995;3:116–138. doi: 10.1159/000472285. [DOI] [PubMed] [Google Scholar]
- 47.Meyn M S. Cancer Res. 1995;55:5991–6001. [PubMed] [Google Scholar]
- 48.Painter R B. In: Ataxia-telangiectasia. Gatti R A, Painter R B, editors. Heidelberg: Springer; 1993. pp. 257–268. [Google Scholar]
- 49.Beamish H, Williams R, Chen P, Lavin M F. J Biol Chem. 1996;271:20486–20493. doi: 10.1074/jbc.271.34.20486. [DOI] [PubMed] [Google Scholar]