Abstract
In this study, we compared the influence of two peptides on the selection of CD8αα and CD8αβ intraepithelial lymphocytes (IELs) of the intestine, which develop by a unique and partially thymus-independent process. Mice were used in which all T cells carried one transgenic T cell antigen receptor (TCR) (F5), and in which only well defined transgenic peptides were presented by H-2Db. The first peptide, for which the F5 TCR has a high affinity, derives from the influenza virus nucleoprotein (NP68). The second peptide, NP34, is an antagonistic variant of NP68 and is recognized by the F5 TCR with low affinity. To avoid presentation of endogenous peptides or production of T cells carrying alternative TCRs, F5 TCR transgenic mice were generated that were deficient for Tap-1 and Rag-1. In these mice, no CD3+CD8+ cells were found in lymph nodes, spleen, or intestine. Introduction of transgenes encoding either NP34 or NP68 along with an endoplasmic reticulum signal sequence enabled Tap-1-independent expression of each peptide in these mice. Positive selection of F5TCR+CD8+ thymocytes was not rescued by these transgenic peptides. However, the high-affinity NP68 peptide induced maturation of CD8αα IEL, whereas the low-affinity NP34 peptide stimulated development of both CD8αβ and CD8αα IEL, but in smaller numbers. When both peptides were present, CD8αβ T cells failed to develop and the number of CD8αα IELs was lower than in mice carrying the NP68 transgene alone. These data demonstrate that single ligands with a high or low affinity for TCR are capable of inducing or inhibiting the maturation of alternative subsets of IELs.
The thymus is the major source of mature T lymphocytes, and it is within this organ that the repertoire of antigenic specificities displayed by T cells is determined through positive and negative clonal selection. During these events, thymocytes encounter ligands made up of peptides associated with molecules of the major histocompatibility complex (MHC). In cases in which the interactions of the T cell antigen receptor (TCR) with these ligands are of high avidity, developing thymocytes undergo clonal deletion (negative selection), whereas in cases in which avidity is lower, thymocytes either fail to develop or undergo further maturation (positive selection).
It has long been suspected that other sites within the body may also act as microenvironments for T cell maturation and selection (1–4). The largest of such putative sites is the intestine, which harbors a phenotypically diverse array of predominantly CD8+ T lymphocytes (1–7). Most of the evidence supporting extrathymic T cell development has, until now, been provided principally by studies showing the presence of intestinal T cells in thymectomized radiation chimeras and in nude mice (1–7). More recently, T cell progenitors have themselves been identified in intestinal tissue within small cellular clusters termed cryptopatches, located within the lamina propria (LP) of the murine small intestine (8). Subsequently, it has been shown that CD4−CD8−CD3−c-Kit+Lin− lymphoid type cryptopatch cells could reconstitute the peripheral and intestinal T cell compartments upon adoptive transfer to immunodeficient mice (9).
Like their peripheral counterparts, the majority of intestinal αβTCR+ T cells recognize MHC class I molecules complexed with peptides. Thus, as with thymic T cell development, the emergence of αβTCR+ T cells within the intestine is considerably reduced in mice lacking either class I MHC-associated β2-microglobulin (β2m) or the peptide transporter protein (Tap-1), which are both required for assembly of peptide/MHC class I complexes (10–12). In support of a thymus-independent model of T cell maturation, it has also been demonstrated that CD8+ T cell development is chiefly dependent on extrathymic, rather than intrathymic, MHC class I expression (13). Notably, however, the criteria by which certain subsets of CD8+ intraepithelial lymphocytes (IELs) are selected appears distinct from those imposed in the thymus. In particular, it has been shown that CD8αα IELs that lack expression of the CD8β chain frequently express “forbidden” TCR Vβ chains specific for superantigens (14). Similarly, studies in TCR-transgenic mice have suggested that class I-restricted CD8αα IELs are developmentally dependent on peptide/MHC ligands that induce T cell deletion in the thymus (15–18). Thus, the encounter between a TCR clonotype with a high affinity self-ligand may lead to negative selection within the thymus but positive selection within the intestine.
To address the role of specific peptides in extrathymic T cell development we used genetically engineered mice that were Tap-1 deficient and coexpressed transgenes encoding α and β chains of the clonotypic F5 TCR along with Tap-independent forms of two peptides derived from the influenza virus nucleoprotein (NP). The first peptide, NP68, is recognized by the F5 TCR with high affinity and can induce profound deletion of thymocytes and expansion of peripheral CTL when injected into F5 TCR+ animals (19, 20). In contrast, the second peptide, NP34, is unable to mediate these effects and in fact shows weak antagonistic properties toward NP68-specific T cell responses. Furthermore, the NP34 peptide can inhibit in vivo positive selection (21) as well as negative selection of F5 TCR+ thymocytes in vitro (22). Here we show that both NP68 and NP34 can induce T cell development within the intestinal mucosa, but significant quantitative and qualitative differences exist between the T cell subsets selected by each peptide.
MATERIALS AND METHODS
Construction of the NP68 Transgene and Generation of Transgenic Mice.
To generate mice that expressed the NP68 peptide independently of Tap-1, a transgene was constructed that encoded the adenovirus E3/19K signal sequence fused to DNA encoding the NP68 peptide, under the control of the H-2K promoter. The construct was based on pHSE3′ (a kind gift of H. Pircher, Institute for Medical Microbiology, Freiburg, Germany). Two primers were synthesized encoding the 5′ and 3′ ends of the minigene (sense, ATG CAG GAT CCC CAC CAT GAG GTA CAT GAT TTT AGG CTT GCT GGC CCT TGC GGC AGT CTG CAG C; antisense, AGT TCT CAA GGA TTC TGA TCA CAT TGC GTC CAT TTC GTT GCT TGC TGC CGC GCT GCA GAC TGC CGC AAG GGC) that contained a 21-nucleotide overlap, each thus serving as the other’s template for PCR. PCR was performed using the Pfu DNA polymerase (Stratagene). The product was digested with BamHI, gel-purified, and ligated into the cloning site of pHSE3′. An additional alanine was introduced in the minigene between the E3/19K signal sequence and the NP68 peptide to avoid possible production of a nonfunctional octameric peptide caused by imprecise cleavage (21, 23, 24).
The minigene was excised by XhoI and KpnI digestion, gel-purified, and microinjected into fertilized F5+Tap-1−/− Rag-1+/− eggs obtained from Tap-1−/− females mated with F5+Tap-1−/−Rag-1−/− males. After overnight incubation, the eggs that matured into the two-cell stage were transferred to day 0.5 post coitum pseudopregnant BDF1 females. Three weeks after birth, genomic DNA was prepared from tail tissue, and the presence of the transgene was analyzed by Southern blotting using the 0.9-kb SalI/EcoRI fragment from the vector used for generating the transgenic mice.
Mice and PCR Typing.
Tap-1−/− (10), Rag-1−/− (25), and NP34 transgenic (NP34tg) (22) mice were produced in the laboratory of S.T. F5 TCR+ mice (19) were kindly provided by D. Kioussis (Mill Hill, U.K.). They were bred and maintained in a pathogen-free environment at the Massachusetts Institute of Technology. The presence of Tap-1, Rag-1, F5 TCR, and NP34tg was analyzed by PCR as previously described (21). The primers used for testing the presence of the NP68tg were 5′, GGA CGC TGG ATA TAA AGT CC, and 3′, AGT TCT CAA GAA TTC TGA TCA CAT T. Mice were analyzed between 8 and 12 weeks.
Lymphocyte Preparation.
IELs and LP lymphocytes (LPLs) were isolated as previously described (26). Briefly, the entire small intestine was removed from mice and flushed with ice-cold Mg2+- and Ca2+-free PBS (Sigma) + 2% FCS (Intergen, Purchase, NY). The intestine was then cut longitudinally and further washed before being cut into 0.5-cm sections, which were shaken in PBS/2% FCS for ½ hr each. Supernatant containing liberated cells was decanted and retained on ice, and the process was repeated. Cells harvested from each extraction were combined, pelleted, and resuspended in 5 ml of 44% (vol/vol) Percoll (Pharmacia). Five milliliters of 67% Percoll was underlaid in each fraction and centrifuged at 700 × g for 15 min. Lymphocytes accumulating at the 44%/67% interface were harvested and washed again in PBS/FCS and counted, using trypan blue exclusion to assess viable cells. LPLs were harvested from the remaining tissue by chopping it finely and incubating the tissue fragments for 1–2 hr under agitation with 0.02% collagenase B (Boehringer Mannheim) and 0.02% trypsin inhibitor (Sigma) in FCS-free RPMI medium 1640 (GIBCO/BRL) supplemented with 1 mM l-glutamine, 1 mM dithiothreitol, 1 mM penicillin/streptomycin (Sigma), and 25 mM Hepes (Sigma). Cells were washed free of collagenase and counted.
Antibodies and Cell Surface Staining.
Cells were stained with the following antibodies, all of which were purchased from PharMingen: biotin- or phycoerythrin (PE)- labeled anti-CD8α (clone 53–6.7), FITC-labeled anti-CD8β (clone Ly-3.2), biotin-labeled anti-CD3 (clone 145–2C11), biotin- or PE-labeled anti-αEβ7 (clone M293), FITC-labeled anti-Thy1.2 (clone 53–2.1), and PE-labeled anti-B220 (clone RA3–6B2). All biotinylated antibodies were used in combination with streptavidin red-670 (GIBCO/BRL). IELs and LPLs were incubated with purified anti-CD32/CD16 (clone 2.4G2) in 100 μl of PBS/FCS for 10 min on ice prior to staining, to prevent nonspecific binding of antibodies. Subsequently, 5 × 105 to 1 × 106 cells were incubated with relevant antibodies in 100 μl of PBS/FCS on ice for 15–30 min, when they were washed twice. Antibody-stained cells were analyzed by flow cytometry using a FACScan (Becton Dickinson) using CellQuest software (Becton Dickinson).
Immunohistochemistry and in Situ Cell Counting.
Immunohistochemical staining and cell counting were performed essentially as previously described (27). Sections were taken from the middle region of the intestine. After washing with PBS they were frozen in OCT freezing compound (Ames, Elkhart, IN) and stored at −80°C. Five-micrometer sections were cut and stained as previously described, using a purified anti-CD8α antibody (clone 53–6.7; PharMingen) followed by sequential incubations with biotinylated rabbit anti-rat Ig (Vector Laboratories) and avidin-biotinylated peroxidase reagent (Dako). Positively stained cells were counted, and the number of stained cells in the LP and the epithelial compartments were determined as a percentage of the total nucleated cells per villus in individual mice. Standard errors for each animal were computed from variation between counted fields.
RESULTS
Generation of NP34/NP68tg Mice on an F5+Rag-1−/−Tap-1−/− Background.
We obtained four lines of mice carrying the NP68 transgene, numbered 28 (5 copies), 91 (10 copies), 98 (3 copies), and 193 (3 copies). Mice carrying the NP34 transgene have been described earlier (21). NP68tg mouse line 193 was used for the experiments described in this study. In these F5+/−Tap-1−/− NP68tg mice, expression of the transgene could not be detected by Northern blot analysis but was detected by reverse transcription–PCR in all organs, including intestine (data not shown). The NP68 (or NP34) transgene was not expressed at levels sufficient to mediate stabilization of MHC class I molecules above background as analyzed by flow cytometry. Tap-1-independent presentation of NP68 was therefore confirmed by proliferation assays using F5 TCR+ lymph node cells as responder cells against whole Tap-1−/− NP68tg splenocyte stimulator cells. All F5 TCR+ mice used in this study were maintained on a Rag-1−/− background and hence had the following characteristics: (i) They expressed the TCR-α and -β chains of the F5 TCR that specifically recognizes the NP68 peptide presented by H-2Db. (ii) They were deficient for Rag-1 and therefore generated T cells expressing only the transgenic TCR chains. (iii) They were deficient for Tap-1 and therefore did not express MHC class I associated with endogenous peptides. (iv) They expressed NP34, NP68, or both peptides associated with H-2Db independently of Tap-1.
Transgenic Expression of Single Peptides in Tap-1−/− Mice Induces the Development of Intestinal F5 TCR+CD8+ T Cells.
As expected from previous studies, the Tap-1 gene mutation led to an almost complete loss of positive selection of F5 TCR+ thymocytes (10). This was reflected by the absence of single-positive (SP) CD8+ T cells in the thymus as well as in the spleens (Fig. 1A) and lymph nodes (21) as compared with their abundance in Tap-1+/+ mice. In some older Tap-1−/− animals small numbers of CD4+CD8+ (double-positive, DP) splenocytes were found, although the origin of these cells is not clear. Small numbers of (SP) CD8+ cells were detected in the epithelium of the small intestine of Tap-1−/− animals. In clear contrast to the situation in Tap-1+/+ mice, however, most of these cells lacked surface F5 TCR expression (Fig. 1B) and were thus reminiscent of immature TCR−CD8+ IELs previously observed in RAG-deficient mice (28, 29).
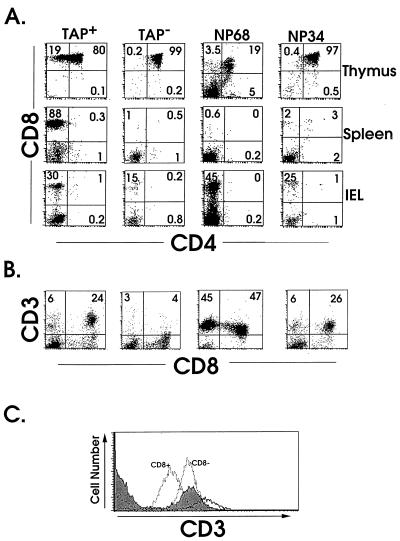
Figure 1.
Expression of NP transgenes restores intestinal CD8+ F5 TCR+ IEL, but not peripheral T cell, development in Tap-1-deficient mice. Thymocytes, splenocytes, and IELs were compared for CD8, CD4 and CD3 expression by using FACS analysis. (A) Two-color log plots are shown of CD4 and CD8 expression after gating on thymocytes, splenocytes, and IELs from Tap-1+/+, Tap-1−/−, and NPtg mice, designated by forward-scatter and side-scatter profiles. (B) Expression of CD3 versus CD8α on IELs from each of the lines of mice. (C) Histograms of log CD3 expression on IELs from F5 TCR+ mice on a Tap-1+/+ (filled), NP34tg (solid), and NP68tg background (dotted line). The CD8+ and CD8− cells from NP68tg mice are shown separately after analysis.
The lack of CD8+ T cells in the F5+Tap-1−/− animals facilitated the analyses of the effects of the transgenic peptides, which in the case of the NP68 transgene were measured by a conspicuous population of CD3+ IELs. Most of these cells were CD8+, but some lacked, or expressed intermediate levels of, CD8α (CD8αint) (Fig. 1B). The appearance of CD8+ IEL within the intestine contrasted strongly with the deletion of CD8+ thymocytes in the same animals, which had very small thymuses because of the loss of DP thymocytes. This loss was matched by an absence of SP CD8+ thymocytes and peripheral CD8+ T cells in NP68tg mice (Fig. 1A). A proportion of TCRloCD4−CD8− (DN) spleen cells were present in these animals, which is consistent with observations in other systems in which negative selection of transgenic TCR+ cells occurs (30). F5 TCR+ IELs were also detected in mice expressing the NP34 transgene, although they were present at a lower frequency compared with those in NP68tg mice and contained a much smaller percentage of DN IELs (Fig. 1B). The lack of SP CD8+ thymocytes in NP34tg mice suggested that there was no increase in thymic positive selection in these animals (Fig. 1A). However, small numbers of DP cells and SP CD8+ T could sometimes be observed in the spleens of NP34tg animals over 10 weeks of age (Fig. 1A). It was noted in NP68tg animals that the level of CD3 expression on CD8+ IELs was lower than on IELs from Tap-1+/+ mice, while CD8− IELs from NP68tg mice and CD8+ IELs from NP34tg mice showed intermediate expression (Fig. 1C).
Different Numbers and Phenotypes of Mucosal T Cells Develop in NPtg Mice.
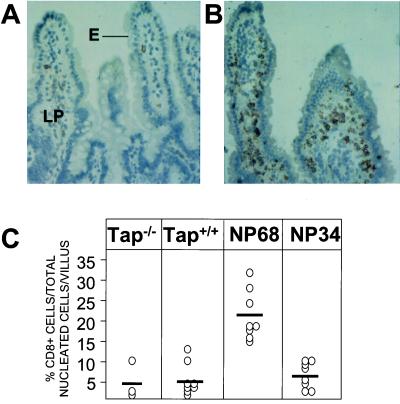
To determine the distribution of T cells within the intestinal mucosa, immunohistochemical staining for CD8 expression was performed on small bowel tissue from different lines of F5 transgenic mice. As shown in Fig. 2B, the accumulation of CD8+ cells in the intestine of NP68tg mice was particularly striking when compared with the number of cells staining for CD8 in Tap-1−/− animals (Fig. 2A). These cells not only were detected in the epithelium but also were prominent in the LP. To quantify the intestinal T cells in the different lines of mice, the number of CD8+ cells staining in tissue sections was determined as a percentage of nucleated cells per villus in each line of mice. As shown in Fig. 2C, as many as 4 times the number of mucosal CD8+ T cells were present in the intestine of NP68tg mice compared with intestinal tissue from NP34tg or Tap-1+/+ animals. These results demonstrate that compared with endogenous ligands and the weaker affinity NP34 peptide, the NP68 peptide induced significant selection and expansion of antigen-specific T cells within the intestinal mucosa.
Figure 2.
Immunohistochemical analysis of CD8α expression by mucosal T cells. Sections of small intestine from Tap-1−/− (A) or NP68tg (B) mice were analyzed by immunohistochemical staining for CD8α chain expression. (Original magnification 200×.) Large numbers of CD8α staining cells were detected in both the LP and epithelial (E) compartments of NP68tg, but not Tap-1−/− mice. (C) Total CD8α+ cell counts. Sections from small bowel of different lines of mice were stained for CD8α expression as described above. Positive-staining cells in the mucosal LP and epithelium were then counted as total nucleated cells per villus (epithelial cells and lymphocytes) in 12 different objective fields. Each symbol represents the mean score for an individual mouse.
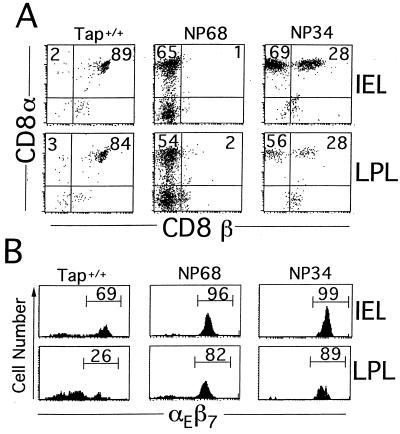
Possible phenotypic differences between intestinal T cells selected on alternative ligands were next explored by using flow cytometry of IELs and LPLs. Under normal circumstances murine CD8αα cells are restricted to the epithelial layer of the intestine, whereas CD8αβ cells reside in both compartments. Indeed, in F5+Tap-1+/+ mice almost all (95–100%) CD8+ IELs and LPLs expressed both CD8α and β chains (Fig. 3A). In F5+Tap-1−/−NP68tg mice, only CD8αα and CD8− T cells developed that, surprisingly, occupied both the IE and the LP compartments. The NP34 transgene induced the appearance of both CD8αα and CD8αβ F5 TCR+ T cells in the IE and LP compartments (Fig. 3A). The fact that CD8αα+ cells were present in the LP of NPtg mice is somewhat unexpected in light of previous studies showing these cells to be generally restricted to the mucosal epithelium. In normal mice IELs can be distinguished from LPLs and peripheral blood lymphocytes by their relatively homogeneous expression of the mucosal integrin αEβ7 (31, 32). Examination of F5 TCR+ CD8+ IELs and LPLs from NP68tg and NP34tg mice revealed a pattern of relatively uniform αEβ7 expression on both populations of cells. In contrast, those from Tap-1+/+ mice were more heterogeneous in expression, with a minority of LPLs expressing high levels of this marker (Fig. 3B).
Figure 3.
Phenotype of T cells from LP and IE compartments. (A) IELs and LPLs were stained for CD8α and CD8β chain expression after analytical gating on CD3+ cells in FCS vs. CD3 plots. In Tap-1+/+ mice the majority of F5 TCR+ cells expressed both CD8α and CD8β chains. In NP68tg animals very few CD8β-expressing T cells were present, with the majority expressing CD8α. IELs and LPLs from NP34tg mice comprised both CD8αβ and CD8αα T cells. (B) Integrin αEβ7 expression on IELs and LPLs is shown in histogram plots after analytical gating on CD3+ cells.
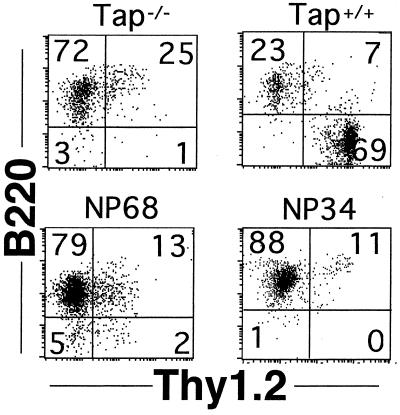
In wild-type mice, IELs also show varied patterns of Thy-1 and B220 expression that are thought to reflect particular maturational or functional states (7, 33). Examination of these markers revealed that IELs from Tap-1+/+ mice were predominantly Thy-1+B220−, whereas in NP68tg and NP34tg mice the reverse phenotype prevailed (Fig. 4). Taken together, these findings indicate that expression of NP peptides in Tap-1−/− mice results in the development of CD8αα and CD8αβ IEL sub-sets that are distinct from those in Tap-1+/+ mice.
Figure 4.
B220 and Thy-1 expression on NPtg mice. CD8+ IELs from NPtg mice, stained for B220 and Thy-1, are compared with those from Tap-1+/+ animals by using two-color dot plots of cells gated for CD8α expression.
NP68 and NP34 Have Partially Opposing Effects on IEL and LPL Development.
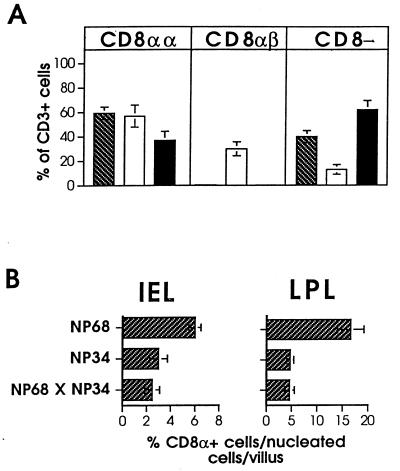
In the F5 TCR transgenic system, NP68 and NP34 possess functionally distinct properties. While NP68 induces CTL activation and the strong deletion of F5 TCR-expressing thymocytes, NP34 partially inhibits these effects (22). In light of these findings, we sought to determine what effects co-expression of the NP34 and NP68 transgenes might have on IEL development. Fig. 5A compares the representation of the different subsets of F5 TCR+ IELs in mice expressing the single NP transgenes with mice co-expressing both. Most notably, CD8αβ IELs were absent from NP68tg × NP34tg mice, providing evidence for the inhibition of CD8αβ IEL development by the NP68 peptide. In addition, there was a measurable reduction in the frequency of CD8αα cells in double NPtg mice relative to NP68tg animals. This decrease was also apparent in IELs and LPLs detected in immunostained small bowel sections of these animals (Fig. 5B). These data reveal that although the NP68 and NP34 ligands could themselves induce development of intestinal T cells, they also possessed a capacity to interfere with the selection of CD8αβ or CD8αα subsets, respectively.
Figure 5.
Co-expression of NP peptides leads to deletion and inhibition of selection. (A) The mean representations of each of the subsets of IEL are shown after FACS analysis of IELs from NP68tg (hatched bars, n = 8), NP34tg (open bars, n = 8), and NP68tg × NP34tg mice (filled bars, n = 5). Results are shown ±SEM. (B) Percentage of CD8α+ T cells in sections of small bowel from each line of NPtg mice. Bars represent mean percentages ± SEM.
DISCUSSION
The experiments presented in this paper demonstrate the capacity of well-defined peptide/MHC ligands to support the development of CD8+ T cells expressing the F5 TCR within the mucosa of the small intestine. In contrast to similar studies conducted previously, we have minimized the potential influence that endogenous ligands might have in selection of IELs by the nominal antigen (15–18). Our data thus reveal that solitary peptide/MHC ligands are sufficient to select intestinal T cells in the absence of endogenous peptide/MHC complexes. A second important aspect of our findings is that two peptides with different affinities for the F5 TCR influenced T cell development in different ways. Thus, large numbers of CD8αα and CD8− F5 TCR+ cells accumulated in the presence of the high-affinity (NP68) peptide, whereas development of CD8αβ F5 TCR+ cells was inhibited. In the presence of the lower-affinity peptide (NP34), smaller numbers of cells comprising both CD8αα and CD8αβ subsets developed. During intrathymic T cell development, the same peptides either induced negative selection (NP68) or failed to positively select CD8αβ thymocytes (NP34). By comparison, CD8αβ T cells developed in both the thymus and the intestine of F5+Tap-1+/+ mice, but CD8αα IELs were scarce.
These observations and findings from other TCR transgenic systems suggest that the development of CD8αβ IELs is promoted by low-affinity TCR/peptide–MHC interactions, whereas their development is inhibited by high-affinity interactions. For example, it was shown that CD8αβ IELs carrying the 2C TCR developed in mice expressing the low-affinity ligand H-2Kb, whereas these cells were deleted in the presence of H-2Ld, a high-affinity ligand. This is not a strict rule, as is exemplified in mice transgenic for the H-Y TCR, which specifically recognizes the male H-Y antigen presented by H-2Db. In this transgenic TCR system, the development of CD8αβ IELs was severely limited in TCR transgenic females carrying the low-affinity ligand, whereas in males in which the high-affinity ligand is expressed, CD8αβ IELs were present (17). It is possible that the discrepancies between CD8αβ IEL development in the various models are partially caused by differences in the expression levels of the selecting ligands in the intestine.
The finding that CD8αβ IEL development was independent of the presence of CD8αβ T cells in the thymus, spleen, or lymph nodes of NP34tg mice suggests that these CD8αβ IELs were not derived from mature peripheral CD8αβ cells. Furthermore, CD8αβ IEL in NP34tg mice had several characteristics similar to those of CD8αα IELs, such as expression of αEβ7 and B220 and lack of Thy-1. Together, these observations imply that CD8αβ IELs may have developed locally within the intestinal compartment in a fashion similar to their CD8αα counterparts. However, several studies have shown that CD8αβ IELs do not mature efficiently in the absence of a thymus (4), making it seem likely that the CD8αβ IELs found in NP34tg mice developed from thymus-derived precursors or were dependent on certain thymic cytokines. The CD8αβ IELs that developed in F5+Tap-1+/+ mice were Thy-1+, αEβ7−, and B220−, a phenotype closer to that of thymus-derived CD8 T cells. It is possible, therefore, that different subsets of CD8αβ IEL exist that follow alternative routes of maturation.
Our data show that class I-restricted CD8αα IELs and LPLs are selected more efficiently in the presence of a high-affinity ligand than in the presence of a low-affinity ligand. This observation is in line with studies in which other TCR transgenic mouse models were employed. It is surprising, though, that large numbers of CD8αα T cells were present in the LP of both NP34tg and NP68tg mice. This contrasts with previous reports showing that CD8αα T cells reside principally within the mucosal epithelium (1–7). The expression of αEβ7 by both IELs and LPLs suggests that they formed part of an equivalent population, despite their distinct localization. Thus, it seems plausible that CD8αα LPLs in peptide transgenic mice were derived from T cells that had been selected in the epithelium and had, in the absence of other lymphoid cells, emigrated to expand within the LP.
In animals cotransgenic for NP34 and NP68, the prevailing phenotype appeared to reflect the contributions of both peptides. First, CD8αβ selected by NP34 were specifically prevented from developing by the presence of NP68. At the same time, there was a decrease of CD8αα cells relative to NP68 single transgenic mice, suggesting that the presence of the NP34 transgene somehow inhibited the actions of the NP68 transgene. As NP34 has been described to act as an antagonist for the F5 TCR during cytotoxicity or T cell selection, it is possible that NP34 is also capable of antagonizing the effect of NP68 in inducing maturation of F5 TCR+CD8αα IELs. For NP34 to act effectively as an antagonist, however, it has to be present at significantly higher concentrations than NP68. This is likely to be the case in the double transgenic mice, as expression of the NP34 transgene was easily detected by Northern blot analysis, whereas expression of the NP68 transgene could be detected only by reverse transcriptase–PCR.
What might be the molecular entities responsible for the differential sensitivities of intestinal CD8αα and CD8αβ T cells for ligands that induce their maturation or elimination? First, the CD8β chain has been shown to increase the sensitivity of the TCR by enhancing recruitment of the protein-tyrosine kinase Lck to the CD8α chain (34). Consequently, mice that are deficient for the CD8β chain show reduced negative selection of thymocytes carrying a self-reactive TCR (35, 36). By analogy, CD8αβ IELs are expected to undergo negative selection at lower avidity TCR-peptide/MHC interactions than CD8αα IELs.
A second influence may lie in the fact that CD8αα, but not CD8αβ, IELs have the capacity to utilize the FcɛRγI chain as a surrogate or partner signaling molecule for CD3ζ (37–39). Several studies have shown that the protein-tyrosine kinases that are involved in signal transduction mediated by the TCR have distinct preferences for FcɛRγI or CD3ζ. Thus, while CD3ζ can be phosphorylated by either Lck or Fyn, FcɛRγI is preferentially phosphorylated by Lck. Upon phosphorylation, CD3ζ associates with Zap-70, whereas FcɛRγI associates with Syk. As a result, signals received by CD8αα IELs are likely to be different from those received by CD8αβ IELs and peripheral T cells. In agreement with the idea that distinct signaling cascades are used by different IEL subsets is the finding that the maturation of CD8αα IELs is severely impaired in Lck−/− mice, whereas that of CD8αβ IELs is only partially affected (40). Similar to the situation in early thymocyte development, Fyn can partially substitute for Lck during CD8αβ IEL development, as can be concluded from the absence of CD8αβ IELs in Lck/Fyn double mutants (40). As CD8αα IELs express TCR complexes containing FcɛRγI, Fyn may be less efficient at replacing Lck because of the reduced usage of its preferred substrate CD3ζ.
A third effect on mucosal CD8αα T lymphocyte development may be that these cells often fail to express specific costimulatory molecules normally expressed by mature T cells, such as CD2, CD5, CD28, and Thy-1. Last, CD8αα IELs express higher levels of Bcl-2 than do CD8αβ IELs, which undoubtedly contributes to the observed resistance of these cells to apoptosis (41).
In conclusion, the data presented here show that the development of CD8αβ and CD8αα IEL can be mediated by single-peptide MHC ligands, and that this can occur in the absence of intrathymic positive selection. Whereas lower-affinity ligands stimulate limited CD8αα and/or CD8αβ IEL development, high-affinity ligands delete CD8αβ IELs but efficiently drive the maturation of CD8αα IELs. Our data also suggest that the latter process can be counteracted by the presence of antagonist peptides. These findings illuminate the processes that influence class I-restricted T cell selection in the intestine and may help to clarify the function of mucosal T lymphocytes.
Acknowledgments
We thank Dr. D. Kioussis for generously providing the F5 transgenic mice and Drs. H. Pircher and R. Zinkernagel for providing plasmid pHSE3′. We are grateful to Drs. W. Haas, D. Doherty, J. Chen, and A. Smith for critical reading of the manuscript. This work was supported by grants from the Crohn’s and Colitis Foundation of America (C.T.), National Institutes of Health Grants P30DK433-51 and DK47677-01 (A.K.B.), and National Institutes of Health Grant CA53874 and a gift from the Chiba Prefectural Government of Japan (S.T.). C.N.L. received a postdoctoral fellowship from the Cancer Research Institute. S.J.S. and Y.P.d.J. were supported by fellowships from the Crohn’s and Colitis Foundation of America.
ABBREVIATIONS
- IE
intraepithelium
- IEL
IE lymphocyte
- TCR
T cell antigen receptor
- LP
lamina propria
- LPL
LP lymphocyte
- Tap
transporter associated with antigen processing
- NP
nucleoprotein
- NPtg
NP-transgenic
- SP
single-positive
- DP
double-positive
References
- 1.Rocha B, Guy-Grand D, Vassalli P. Curr Opin Immunol. 1995;7:235–242. doi: 10.1016/0952-7915(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 2.Rocha B, Vassalli P, Guy-Grand D. Immunol Today. 1992;13:449–454. doi: 10.1016/0167-5699(92)90074-H. [DOI] [PubMed] [Google Scholar]
- 3.Poussier P, Julius M. Annu Rev Immunol. 1994;12:521–553. doi: 10.1146/annurev.iy.12.040194.002513. [DOI] [PubMed] [Google Scholar]
- 4.Lefrancois L, Puddington L. Immunol Today. 1995;16:449–454. doi: 10.1016/0167-5699(95)80065-4. [DOI] [PubMed] [Google Scholar]
- 5.Mosley L, Klein J. J Exp Med. 1992;176:1365–1373. doi: 10.1084/jem.176.5.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein J R. Semin Immunol. 1995;7:291–297. doi: 10.1016/1044-5323(95)90010-1. [DOI] [PubMed] [Google Scholar]
- 7.Rocha B, Vassalli P, Guy-Grand D. J Exp Med. 1994;180:681–686. doi: 10.1084/jem.180.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanamori Y, Ishimaru K, Nanno M, Maki K, Kikuta K, Nariuchi H, Ishikawa H. J Exp Med. 1996;280:1449–1459. doi: 10.1084/jem.184.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito H, Kanamori Y, Takemori T, Nariuchi H, Kubota E, Takahashi-Iwanaga H, Iwanaga T, Ishikawa H. Science. 1998;280:275–278. doi: 10.1126/science.280.5361.275. [DOI] [PubMed] [Google Scholar]
- 10.Van Kaer L, Ashton-Rickardt P G, Ploegh H L, Tonegawa S. Cell. 1992;71:1205–1214. doi: 10.1016/s0092-8674(05)80068-6. [DOI] [PubMed] [Google Scholar]
- 11.Sydora B, Brossay L, Hagenbaugh A, Kronenberg M, Cheroutre H. J Immunol. 1996;156:4209–4216. [PubMed] [Google Scholar]
- 12.Fujiura Y, Kawaguchi M, Kondo Y, Obana S, Yamamoto H, Nanno M, Ishikawa H. J Immunol. 1996;156:2710–2715. [PubMed] [Google Scholar]
- 13.Fuller B, Lefrancois L. J Immunol. 1995;155:2808–2811. [PubMed] [Google Scholar]
- 14.Rocha B, Vassalli P, Grand D G. J Exp Med. 1991;173:483–486. doi: 10.1084/jem.173.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poussier P, Teh H S, Julius M. J Exp Med. 1993;178:1947–1957. doi: 10.1084/jem.178.6.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rocha B, von Boehmer H, Guy-Grand D. Proc Natl Acad Sci USA. 1992;89:5336–5340. doi: 10.1073/pnas.89.12.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cruz D, Sydora B C, Hetzel K, Yakoub G, Kronenberg M, Cheroutre H. J Exp Med. 1998;188:255–265. doi: 10.1084/jem.188.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guehler S R, Bluestone J A, Barrett T A. J Exp Med. 1996;184:493–503. doi: 10.1084/jem.184.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mamalaki C, Norton T, Tanaka Y, Townsend A, Chandler P, Simpson E, Kioussis D. Proc Natl Acad Sci USA. 1992;89:11342–11346. doi: 10.1073/pnas.89.23.11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mamalaki C, Tanaka Y, Corbella P, Townsend A, Chandler P, Simpson E, Kioussis D. Int Immunol. 1993;5:1285–1292. doi: 10.1093/intimm/5.10.1285. [DOI] [PubMed] [Google Scholar]
- 21.Levelt C N, Mizoguchi E, Huang X, Zacks R, Bhan A K, Tonegawa S. Proc Natl Acad Sci USA. 1998;95:14349–14354. doi: 10.1073/pnas.95.24.14349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams O, Tanaka Y, Bix M, Murdjeva M, Littman D, Kioussis D. Eur J Immunol. 1996;26:532–538. doi: 10.1002/eji.1830260305. [DOI] [PubMed] [Google Scholar]
- 23.Snyder H, Yewdell L J, Bennink J R. J Exp Med. 1994;180:2389–2394. doi: 10.1084/jem.180.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliott T, Willis A, Cerundolo V, Townsend A. J Exp Med. 1995;181:1481–1491. doi: 10.1084/jem.181.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mombaerts P, Iacomini J, Johnson R, Herrup K, Tonegawa S, Papaioannou V E. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 26.Simpson S J, Hollander G, She J, Levelt C, Huang M, Terhorst C. Int Immunol. 1995;7:287–293. doi: 10.1093/intimm/7.2.287. [DOI] [PubMed] [Google Scholar]
- 27.Mizoguchi E, Mizoguchi A, Bhan A K. Lab Invest. 1997;76:385–397. [PubMed] [Google Scholar]
- 28.Lin T, Matsuzaki G, Yoshida N, Kobayashi N, Kenai H, Omoto K, Nomoto K. Eur J Immunol. 1994;24:1080–1087. doi: 10.1002/eji.1830240511. [DOI] [PubMed] [Google Scholar]
- 29.She J, Simpson S J, Hollander G, Liu C-P, Allen D, van Houten N, Wang B, Terhorst C. J Immunol. 1997;158:4678–4687. [PubMed] [Google Scholar]
- 30.Teh H-S, Kishi H, Scott B, von Boehmer H. J Exp Med. 1989;169:795–806. doi: 10.1084/jem.169.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilshaw P J, Murant S J. Eur J Immunol. 1991;21:2591–2597. doi: 10.1002/eji.1830211041. [DOI] [PubMed] [Google Scholar]
- 32.Lefrancois L, Barrett T A, Havran W L, Puddington L. Eur J Immunol. 1994;24:635–640. doi: 10.1002/eji.1830240322. [DOI] [PubMed] [Google Scholar]
- 33.Ibraghiomov A R, Lynch R. J Exp Med. 1994;180:433–444. doi: 10.1084/jem.180.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irie H Y, Ravicandran K S, Burakoff S. J Exp Med. 1995;181:1267–1273. doi: 10.1084/jem.181.4.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fung-Leung W P, Kundig T M, Ngo K, Panakos J, De Sousa-Hitzler J, Wang E, Ohashi P S, Mak T W, Lau C Y. J Exp Med. 1994;180:959–967. doi: 10.1084/jem.180.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crooks M E, Littman D R. Immunity. 1994;1:277–285. doi: 10.1016/1074-7613(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 37.Guy-Grand D, Rocha B, Mintz P, Malassis-Seris M, Selza F, Malissen B, Vassalli P. J Exp Med. 1994;180:673–679. doi: 10.1084/jem.180.2.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C-P, Ueda R, She J, Sancho J, Wang B, Weddell G, Loring J, Kurahara C, Dudley E C, Hayday A, Terhorst C, Huang M. EMBO J. 1993;12:4863–4875. doi: 10.1002/j.1460-2075.1993.tb06176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malissen M, Gillet A, Rocha B, Trucy J, Vivier E, Boyer C, Kontgen F, Brun N, Mazza G, Spanopoulou E. EMBO J. 1993;12:4347–4355. doi: 10.1002/j.1460-2075.1993.tb06119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Page S, van Oers N S, Perlmutter R M, Weiss A, Pullen A M. Eur J Immunol. 1997;27:554–562. doi: 10.1002/eji.1830270229. [DOI] [PubMed] [Google Scholar]
- 41.Van Houten N, Blake S, Li E, Hallam T, Chilton D, Gourley W, Boise L, Thompson C, Thompson E. Int Immunol. 1997;9:945–953. doi: 10.1093/intimm/9.7.945. [DOI] [PubMed] [Google Scholar]