Abstract
In addition to their role in peptide antigen presentation, class I MHC proteins also play a critical role in inhibiting natural killer (NK) cytotoxicity through interaction with NK inhibitory receptors. Thus, NK cells are cytotoxic to virus-infected and tumor cells that have lost class I MHC protein expression. However, the nature of the receptors involved in the triggering of lysis of target cells is poorly understood. CD16 (Fcγ receptor III) has been described as a receptor expressed on NK cells that facilitates antibody-dependent cellular cytotoxicity (ADCC) by binding to the Fc portion of various antibodies. However, we show here that CD16 has a broader function and is directly involved in the lysis of some virus-infected cells and tumor cells, independent of antibody binding. The presence of a putative CD16 ligand on appropriate target cells has also been demonstrated by the use of a CD16-Ig fusion protein.
Keywords: Fcγ receptor III, innate immunity, Ig superfamily
Natural killer (NK) cells are major components of the cellular mechanism by which an immune response leads to the destruction of foreign or infected tissue (1). In contrast to cytotoxic T lymphocytes (CTL), which are triggered by ligation to class I MHC molecules complexed with an appropriate specific peptide, one well defined function of NK cells is the lysis of target cells deficient in expression of class I MHC proteins. In this manner NK cells carry out immunosurveillance for “missing self” (2), rather than for direct detection of foreign antigens.
Recognition of polymorphic determinants on HLA molecules by human NK cells is mediated by two types of class I MHC-binding inhibitory receptors: the Ig superfamily of inhibitory receptors, which includes both the NKIR proteins (3–5) and the ILT-2 protein (6), whose ligands are various HLA-A, -B, and -C proteins, and the lectin-like CD94/NKG2 complex, which delivers an inhibitory signal upon binding the HLA-E protein (7–9). This variety of class I MHC protein-specific receptors illustrates the importance of these molecules in modulating NK function. On the other hand, thus far only limited information has been available on the lysis receptor(s) involved in triggering NK cell cytotoxicity against target cells. Recently however, a triggering receptor NKp46 was cloned and shown to be involved in lysis of some tumor cells (10).
CD16, a molecule of the Ig superfamily known to be involved in antibody-dependent cellular cytotoxicity (ADCC), is the best-characterized membrane receptor responsible for triggering of lysis by NK cells. CD16, the low-affinity receptor for the Fc portion of some IgGs, is associated with CD3ζ or Fcɛ receptor I (FcɛRI) γ chains (11), which participate in signal transduction (12). Cross-linking of CD16 on NK cells resulted in increased intracellular Ca2+ levels and a cascade of biochemical events similar to those activated by the T cell receptor (13). The work presented here demonstrates an additional role for CD16 on human NK cells as a lysis receptor that mediates the direct killing of some virus-infected and tumor cells, independent of antibody ligation.
MATERIALS AND METHODS
MAbs.
The hybridoma-producing mAb 368 (14) was kindly given by Jay Unkeless (Mt. Sinai School of Medicine, New York). The F(ab′)2 fragment of the anti-CD16 mAb 3G8 (14) was purchased from Medarex (Annandale, NJ). The anti-CD99 mAb 12E7 (15), used as a control, was a kind gift from A. Bernard (Hôpital de l’Archet, Nice, France). The Fab fragment of mAb 12E7 was generated by using the ImmunoPure Fab Kit (Pierce).
Soluble CD16 and CD99 Fusion Proteins and Direct Binding Assays for the CD16 Ligand.
The sequences encoding the extracellular portion of either CD16 or CD99 proteins were amplified by PCR from cDNA isolated from NK clones. The CD99-specific primers were 5′ primer (including a HindIII site and Kozak sequence), 5′-CCCAAGCTTGGGGCCGCCACCATGGCCCGCGGGGCTGCGCTG-3′, 3′ primer (including the BamHI site), 5′-GGGATCCGCGTCGGCCTCTTCCCCTTCTTT-3′. The CD16-specific primers were 5′ primer (including a HindIII site and Kozak sequence), 5′-CCCAAGCTTGGGGCCGCCACCATGTGGCAGCTGCTCCTCCCAACT-3′, 3′ primer (including the BamHI site), 5′-GGGATCCCCAGGTGGAAAGAATGATGAGAT-3′. These PCR-generated fragments were cloned into a mammalian expression vector containing the Fc portion of human IgG1 (16) (a kind gift from B. Seed, Massachusetts General Hospital Cancer Center, Charlestown, MA). Sequencing of the constructs revealed that both CD16-Ig and CD99-Ig cDNA were in frame with the human Fc genomic DNA and were identical to the reported sequences. COS-7 cells were transiently transfected with the plasmids containing either the CD16 or CD99 cDNAs, and supernatants were collected and purified on a Poros 20 protein G column in the High Pressure Perfusion Chromatography Station, BioCAD (PerSeptive Biosystems). SDS/PAGE analysis revealed that both Ig fusion proteins were approximately 95% pure and of the proper molecular mass (approximately 65 kDa for CD16-Ig and 55 kDa for CD99-Ig, under reducing conditions). Furthermore, both Ig-fusion proteins could be detected by ELISAs using specific mAbs: 3G8 and B73.1 (17) for CD16-Ig and 12E7 and 0662-E3 (15) for CD99-Ig. To assay for the CD16 ligand, various cells were incubated with 40 μg/ml either CD16-Ig or CD99-Ig fusion protein as a control for 1 hr on ice. The cells were washed and incubated with Fcγ-fragment-specific (minimal cross-reaction to bovine, horse, and mouse serum proteins), phycoerythrin (PE)-conjugated affinity-purified F(ab′)2 fragment of goat anti-human IgG (Jackson ImmunoResearch) for 30 min and analyzed by flow cytometry with a FACScan (Becton Dickinson).
Cytolytic Assays.
The cytolytic activity of NK lines and clones against the various targets was assayed in 5-hr 35S-release assays as previously described (18). In experiments in which mAbs were included, these were added to be at a final concentration of 40 μg/ml. In all presented experiments the spontaneous release was less than 25% of maximal release. The range of the triplicates was under 5% of their mean.
RESULTS AND DISCUSSION
CD16: A Receptor for Lysis of Some Targets by Human NK Cells.
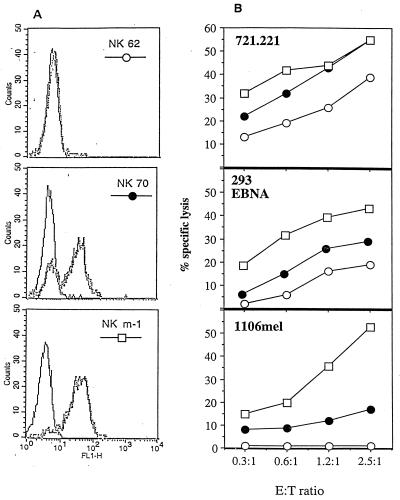
To determine whether the CD16 protein might be involved in direct cellular lysis by NK cells, initially three NK lines [cultured as previously described (18)] that differed in the level of CD16 expression were selected (Fig. 1A). NK line 62 did not express CD16, about 60% of the cells in the NK line 70 expressed CD16, and all the cells in NK line m-1 expressed CD16. The CD16-negative NK line 62 moderately lysed 721.221 target cells (38% lysis at an E:T ratio of 2.5:1), weakly lysed 293 EBNA cells (17% lysis), and did not lyse the melanoma cell line 1106 mel at all (Fig. 1B). Strong (50%), moderate (25%), or weak (15%) lysis of all three targets was observed with the NK line 70, in which only 2/3 of the cells expressed the CD16 proteins. In contrast, all target cells were efficiently lysed by the CD16 expressor line NK m-1 (43–55% lysis). The level of CD16 expression was further demonstrated to correlate with efficient NK killing of 1106 mel target cells by using an additional 16 NK lines sorted to either 3G8 dim or 3G8 bright (Table 1).
Figure 1.
Relationship between CD16 expression on NK cell lines and lysis of various target cells. (A) FACS analysis of NK lines (NK 62, NK 70, and NK m-1) stained with FITC-labeled anti-CD16 mAb 3G8 (dotted line), overlaid on a control FITC-labeled IgG1 mAb (bold line). FL1-H, fluorescence (FITC). (B) Killing of different target cells by various NK cell lines. E:T ratio, ratio of effector to target cells.
Table 1.
Killing of 1106 mel cells correlated with high levels of CD16 expression on NK cells (measured with FITC-mAb 3G8 using FITC-IgG1 as a control)
| Line | MFI
|
% lysis of 1106 mel | |
|---|---|---|---|
| Control | CD16 | ||
| 3G8 dim | |||
| E4 | 10 | 17 | 6 |
| C1 | 11 | 25 | 5 |
| C10 | 10 | 33 | 3 |
| C11 | 10 | 36 | 2 |
| G11 | 12 | 19 | 3 |
| D12 | 9 | 41 | 3 |
| D3 | 10 | 33 | 2 |
| H1 | 11 | 28 | 12 |
| Mean | 10.3 | 29 | 4.5 |
| 3G8 bright | |||
| 17 | 10 | 184 | 34 |
| 24 | 11 | 207 | 28 |
| 3–2 | 12 | 199 | 29 |
| 4–2 | 11 | 296 | 37 |
| 30 | 12 | 261 | 32 |
| 32 | 10 | 171 | 31 |
| 33 | 9 | 184 | 42 |
| 334 | 9 | 314 | 28 |
| Mean | 10.5 | 227 | 32.6 |
NK cells lines were sorted (30 cells per well) to be either 3G8 dim (upper half of the table) or 3G8 bright (lower half of the table). Median fluorescence intensity (MFI) was determined by using FACS analysis. Assays were performed at an E:T ratio of 2.5:1. The table shows one representative experiment out of three performed.
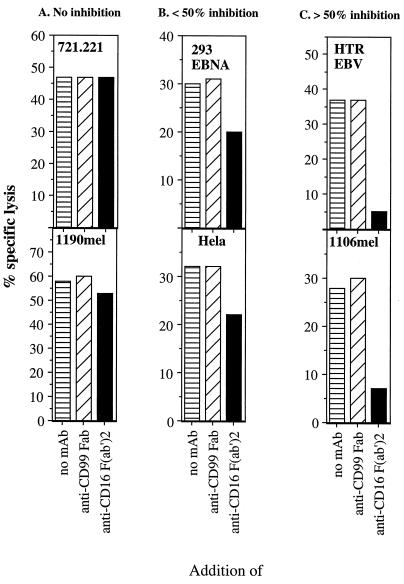
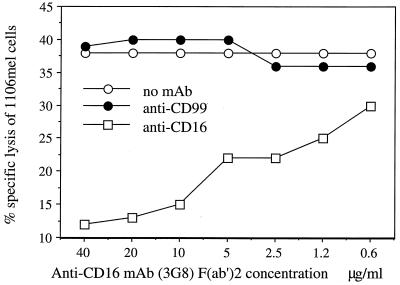
A direct demonstration of the role of CD16 as a lysis receptor was obtained by blocking CD16 protein with the F(ab′)2 fragment of the high-affinity mAb 3G8. On the basis of the results of these experiments, three different groups of target cells could be identified. In group A addition of the F(ab′)2 fragment of mAb 3G8 had no effect on lysis of either the 721.221 B cell line or the 1190 mel melanoma line (Fig. 2A). Among other NK targets whose lysis could not be blocked were the B cell line Raji, the erythroleukemia line K562, the T cell line CEM, and the mouse BW and YAC-1 cell lines (data not shown). In group B, addition of the F(ab′)2 fragment of mAb 3G8 resulted in a moderate inhibition of lysis (less than 50%). This group included the cervical carcinoma cell line HeLa, the adenovirus-transformed kidney cell line 293 EBNA (Fig. 2B), and the monkey cell line COS-7 (data not shown). In group C, addition of the F(ab′)2 fragment of mAb 3G8 resulted in efficient blocking of lysis (more than 70%). This group included the 1106 mel melanoma line and the class I-positive, EBV-transformed B cells from healthy donors HTR (Fig. 2C) and 1612 that are effectively lysed by only a minority of NK clones (data not shown). No blocking of lysis was observed with the Fab fragments of an isotype-matched control anti-CD99 mAb 12E7 (Fig. 2) or with the intact anti-CD99 or anti-CD7 mAb (data not shown). Blocking of lysis of 1106 mel cells was proportional to the amount of the F(ab′)2 fragment added (Fig. 3). Thus, blocking by the F(ab′)2 fragment of the high-affinity anti-CD16 mAb 3G8 shows that CD16 can directly participate in the killing of some targets by NK cells. NK cells are likely to have multiple receptors capable of mediating direct cellular cytotoxicity. The lysis of targets that could not be blocked by the F(ab′)2 fragments of mAb 3G8 must therefore employ a different NK lysis receptor. Indeed, lysis of the BW, YAC-1, and 721.221 cells that was not affected by the addition of the F(ab′)2 fragments mAb 3G8 (Fig. 2A and data not shown) was shown to be blocked with the mAb against the NKp46 receptor (10). Both CD16 and NKp46 proteins have two extracellular C-2 set Ig domains as well as transmembrane and cytoplasmic domains (10, 19) and were found to be associated with CD3ζ (10, 11). However, whereas a comparison of CD16 and NKp46 proteins revealed 19.4% identity and 32.4% similarity when conserved substitutions were included, the similarities were due primarily to motifs conserved among all C-2 set Ig domains. Furthermore, analysis of the CD16 and NKp46 sequences by using gene trees suggested that they are divergent (data not shown). Thus, while CD16 and NKp46 may be functionally similar, they are relatively distantly related and in fact are encoded on different chromosomes.
Figure 2.
F(ab′)2 of anti-CD16 mAb 3G8 blocks lysis of some target cells. NK clone 721 (CD56+, CD16+, NKIR2+ NKIR1−, NKB1−, CD94+) was incubated with 40 μg/ml of the F(ab′)2 fragment of either the anti-CD16 mAb 3G8 or the anti-CD99 mAb 12E7 for 1 hr on ice. The NK clone 721 incubated with no mAb was used as a control. Cells were washed and incubated with the 35S-labeled target cells at an E:T ratio of 5:1 in a 5-hr killing assay. The figure shows one representative experiment of three performed. Similar results were obtained with various E:T ratios and with 10 other NK clones and 10 other NK lines from different donors.
Figure 3.
Blocking the lysis of 1106 mel target cells is proportional to the amount of mAb added. NK clone 10 (CD56+, CD16+, NKIR2+, NKIR1−, NKB1−, CD94+) was incubated with various concentrations of F(ab′)2 fragment of either the anti-CD16 mAb 3G8 or the anti-CD99 mAb 12E7 for 1 hr on ice. The NK clone 10 incubated with no mAb was used as a control. Cells were washed and incubated with the 35S-labeled target cells at an E:T ratio of 5:1 in a 5-hr killing assay. The figure shows one representative experiment of three performed.
Lysis of Targets by the NK92 Tumor Line.
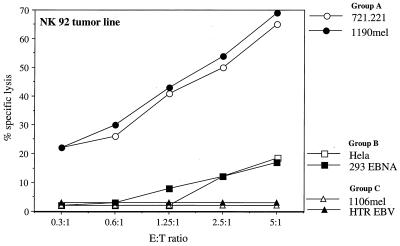
To further corroborate the role of the CD16 protein as a lysis receptor on NK cells, lysis of the various target cells was tested with the CD16-negative NK92 tumor line (20). 721.221 and 1190 mel cells, whose lysis could not be blocked by the addition of the F(ab′)2 fragments of mAb 3G8 (Fig. 2A), were efficiently lysed by the CD16-negative NK92 tumor line (Fig. 4). The lysis of HeLa and of 293 EBNA cells line was moderately blocked by the addition of the F(ab′)2 fragments of mAb 3G8 (Fig. 2B), and these cells were moderately lysed by the NK92 tumor line (Fig. 4). Finally, HTR EBV and the 1106 mel lines, whose lysis was effectively blocked by the addition of the F(ab′)2 fragments mAb 3G8 (Figs. 2C and 3), could not be killed by the NK92 tumor line (Fig. 4). Interestingly, however, some of the targets that belong to group B, such as COS-7 cells, were completely resistant to the NK92-mediated killing (data not shown).
Figure 4.
Killing pattern of the NK92 tumor clone. The NK92 tumor line (CD56+, CD16−, NKIR2−, NKIR1−, NKB1−, CD94−) was incubated with various 35S-labeled target cells for 5 hr at different E:T ratios. The figure shows one representative experiment of three performed.
Thus, these data suggest the existence of multiple lysis receptors and/or multiple mechanisms of killing of different targets by NK clones and, therefore, multiple ligands on target cells. This situation parallels the finding of multiple mechanisms for killing by T cells—i.e., the ligation of the T cell receptor by the MHC/peptide complex to which the ligation of CD16 by the CD16 ligand might be analogous, Fas ligand (whose expression induces killing of cells expressing Fas), secretion of tumor necrosis factor (TNF) α (through engagement of TNF receptors) (reviewed in refs. 21 and 22) and CD40 ligand (through engagement of CD40 on target cells) (23).
Detection of a CD16 Ligand.
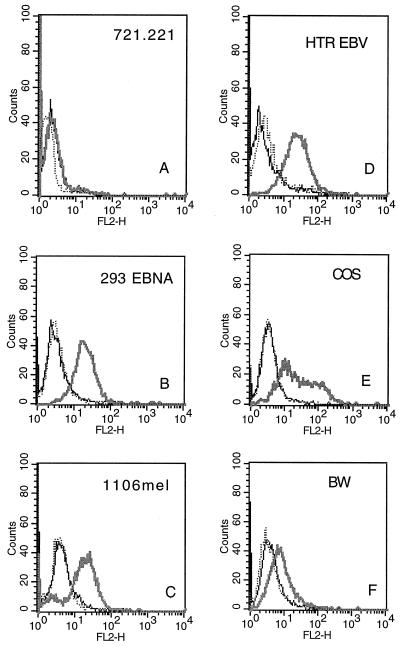
To demonstrate direct binding between the CD16 protein and the appropriate target cells, the cDNA encoding the extracellular domain of CD16 was fused to the genomic DNA segments encoding human IgG1 according to the method previously described (16). cDNA encoding the extracellular domain of CD99 fused to the human IgG1 DNA was used as control. These constructs were transiently transfected into COS-7 cells, and the secreted fusion proteins were purified on a protein G column using the BioCAD. The Ig fusion proteins were incubated with the various targets and were analyzed for binding by indirect staining using the F(ab′)2 fragments and phycoerythrin-labeled goat anti-human Fc as a secondary mAb. Strikingly, CD16-Ig specifically bound all target cells whose NK cell-mediated killing was blocked by the F(ab′)2 fragments of anti-CD16 mAb—e.g., the 293 EBNA cells from group B and the 1106 mel and HTR EBV cells from group C (Fig. 5 B–D). The CD16 binding was specific, since no binding could be detected with another Ig-fusion protein, CD99-Ig. In contrast, little or no binding was observed to group A target cells that were not blocked [i.e., 721.221 or the mouse BW cells (Fig. 5 A and F)]. These data provide evidence for the existence of an unknown ligand(s) for CD16 present on these cells. The most efficient binding of the CD16-Ig fusion protein was observed with the 293 EBNA cells and Epstein–Barr virus (EBV)-transformed B cell lines HTR (Fig. 5B and D). Both cell lines express class I MHC protein and both are protected from lysis by the vast majority of NK clones tested (data not shown). Therefore, expression of the putative CD16 ligand on these cells, even at levels as high as observed with the EBV-transformed HTR line, cannot overcome the inhibition mediated by the class I MHC proteins.
Figure 5.
Staining of various target cells with the CD16-Ig fusion protein. Various target cells were incubated with 50 μg/ml of either the CD16-Ig (thick line) or the CD99-Ig (dotted line) fusion proteins for 1 hr on ice and analyzed by flow cytometry. Controls were the same cells incubated with the secondary mAb alone (thin line). The figure shows one representative experiment of five performed.
The presence of a CD16 ligand on COS-7 cells (Fig. 5E) suggests that the ligand might be conserved among some primates. In contrast, little or no binding of CD16-Ig was observed on any of the mouse cells tested such as BW (Fig. 5F) or YAC-1 (data not shown). This result is most likely to be caused by the use of human CD16-Ig to identify the mouse ligand or the limited number of mouse targets tested. Alternatively, mouse NK cells might not use CD16 as a lysis receptor for cellular cytotoxicity. In support of the latter explanation, no difference in the killing of BW, YAC-1, and EL-4 cells was observed between NK cells derived from normal mice or NK cells derived from knockout mice generated by a targeted insertion into the γ subunit, which is common between the IgG Fc receptors (24). The γ subunit is required for the cell surface expression and signal transduction of the high-affinity IgE receptor FcɛRI, the high-affinity IgG receptor FcγRI, and the low-affinity CD16 receptor FcγRIII. Other important differences have been observed between mice and humans in the mechanism of regulation of NK cell cytotoxicity. For example, mouse NK cells recognize class I MHC proteins mainly by means of the C-type lectin family of receptors—e.g., Ly49. In contrast, human NK cells recognize class I MHC proteins primarily by means of the Ig superfamily of receptors—e.g., NKAT or ILT proteins (25).
In summary, the CD16 receptor is shown here to be directly involved in the lytic process mediated by human NK cells, in addition to its role in antibody-dependent cellular cytotoxicity (ADCC). The failure of a target cell to express the appropriate ligand for either CD16 or other NK lysis receptors would be a mechanism by which cells could escape from NK cell-mediated cytotoxicity. The nature of the ligand for CD16 that was detected on target cells by the CD16-Ig fusion protein remains unknown. It may be one or several of the known Ig superfamily members ubiquitously expressed on cells, since its ligand in ADCC is the Fc domains of Ig. The current hypothesis to explain the function of NK cells suggests that NK cells carry out immunosurveillance for “missing self” (2) rather than for direct detection of foreign antigens. However, the results presented here and elsewhere (10) suggest that expression of the CD16 ligand and of other NK lysis ligands on target cells lends a degree of specificity to the killing mediated by NK cells.
Acknowledgments
We are very grateful to Dr. G. Trinchieri for helpful suggestions. We thank Dr. H. T. Reyburn for his help in the construction of the Ig-fusion plasmids. This work was supported by National Institutes of Health research grants (R35CA47554 and NO1AI45198). O.M. is supported by the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation Fellowship Grant DRG 1454, and D.M.D. is supported by a fellowship from the Irvington Institute.
ABBREVIATIONS
- NK
natural killer
- FcR
Fc receptor
- E:T ratio
ratio of effector to target cells
Note Added in Proof
After this manuscript was submitted, the cloning of a third putative lysis receptor, NKp44, was reported (26, 27). NKp44, NKp46, and CD16 are all members of the Ig superfamily. Pairwise sequence comparisons suggest no significant homology among them, and they are each encoded on a different chromosome.
References
- 1.Trinchieri G. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ljunggren H-G, Karre K. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 3.Colonna M, Samaridis J. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 4.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati M S, Vitale M, Bottino C, Moretta L, Moretta A, Long E O. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 5.D’Andre A, Chang C, Franz-Bacon K, McClanahan T, Phillips J H, Lanier L L. J Immunol. 1995;155:2306–2310. [PubMed] [Google Scholar]
- 6.Colonna M, Navarro F, Bellon T, Liano M, Garcia P, Samaradis J, Angman L, Cella M, Lopez-Botet M. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrego F, Ulbrecht M, Weiss E H, Coligan J E, Brooks A G. J Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braud V M, Allan D S, O’Callaghan C A, Soderstrom K, D’Andrea A, Ogg G S, Lazetic S, Young N T, Bell J I, Phillips J H, Lanier L L, McMichael A. Nature (London) 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 9.Lee N, Liano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, Geraghty D E. Proc Natl Acad Sci USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pessino A, Sivori S, Bottino C, Malaspina A, Morelli L, Moretta L, Biassoni R, Moretta L. J Exp Med. 1998;188:953–960. doi: 10.1084/jem.188.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanier L L, Yu G, Phillips J H. J Immunol. 1991;146:1571–1576. [PubMed] [Google Scholar]
- 12.Vivier E, Morin P, O’Brien C, Druker B, Schlossman S F, Anderson P A. J Immunol. 1991;146:206–210. [PubMed] [Google Scholar]
- 13.Aramburu J, Azzoni L, Rao A, Perussia B. J Exp Med. 1995;182:801–810. doi: 10.1084/jem.182.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleit H B, Wright S D, Unkeless J. Proc Natl Acad Sci USA. 1982;79:3275–3279. doi: 10.1073/pnas.79.10.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernard G, Zoccola D, Deckert M, Breittmayer J P, Aussel C, Bernard A. J Immunol. 1995;154:26–32. [PubMed] [Google Scholar]
- 16.Aruffo A, Stamenkovic I, Melinic M, Underhill C B, Seed B. Cell. 1990;61:1303–1313. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- 17.Perussia B, Starr S, Abraham S, Fanning V, Trinchieri G. J Immunol. 1983;130:2133–2141. [PubMed] [Google Scholar]
- 18.Mandelboim O, Reyburn H T, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, Strominger J L. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamm A, Reinhold S E. J Immunol. 1996;157:1567–1581. [Google Scholar]
- 20.Goodman D J, Von Albertini M, Willson A, Millan M T, Bach F H. Transplantation. 1996;61:763–771. doi: 10.1097/00007890-199603150-00016. [DOI] [PubMed] [Google Scholar]
- 21.Nagata S, Golstein P. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 22.Depraetere V, Golstein P. Semin Immunol. 1997;9:1–10. doi: 10.1006/smim.1997.0062. [DOI] [PubMed] [Google Scholar]
- 23.Carbone E, Ruggiero G, Terrazzano G, Palomba C, Manzo C, Fontana S, Spits H, Karre K, Zappacosta S. J Exp Med. 1997;185:2053–2060. doi: 10.1084/jem.185.12.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takai T, Li M, Sylvestre D L, Clynes R, Ravetch J L. Cell. 1994;76:519–529. doi: 10.1016/0092-8674(94)90115-5. [DOI] [PubMed] [Google Scholar]
- 25.Parham, P., ed. (1997) Immunol. Rev.155, 5–220.
- 26.Vitale M, Bottino C, Sivori S, Sanseverino L, Castriconi R, Marcenaro E, Augugliaro R, Moretta L, Moretta A. J Exp Med. 1998;187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantoni C, Bottino C, Vitale M, Pessino A, Augugliaro R, Malaspina A, Parolini S, Moretta L, Moretta A, Biassoni R. J Exp Med. 1999;189:787–795. doi: 10.1084/jem.189.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]