Abstract
The heterodimeric αβ T cell receptor (TCR) for antigen is the key determinant of T cell specificity. The structure of the TCR is very similar to that of antibodies, but the engineering of TCRs by directed evolution with combinatorial display libraries has not been accomplished to date. Here, we report that yeast surface display of a TCR was achieved only after the mutation of specific variable region residues. These residues are located in two regions of the TCR, at the interface of the α- and β-chains and in the β-chain framework region that is thought to be in proximity to the CD3 signal-transduction complex. The mutations are encoded naturally in many antibody variable regions, indicating specific functional differences that have not been appreciated between TCRs and antibodies. The identification of these residues provides an explanation for the inherent difficulties in the display of wild-type TCRs compared with antibodies. Yeast-displayed mutant TCRs bind specifically to the peptide/MHC antigen, enabling engineering of soluble T cell receptors as specific T cell antagonists. This strategy of random mutagenesis followed by selection for surface expression may be of general use in the directed evolution of other eukaryotic proteins that are refractory to display.
T cell receptors (TCRs) and antibodies have evolved to recognize different classes of ligands. Antibodies function as membrane-bound and soluble proteins that bind to soluble antigens, whereas TCRs function only as membrane-bound molecules that bind to cell-associated peptide/MHC antigens. All of the energy of the antibody:antigen interaction focuses on the foreign antigen, whereas a substantial fraction of the energy of the TCR:peptide/MHC interaction seems to be directed at the self-MHC molecule (1). In addition, antibodies can have ligand-binding affinities that are orders of magnitude higher than those of TCRs, largely because of the processes of somatic mutation and affinity maturation. In their normal cellular context, TCRs do not undergo somatic mutation and the processes of thymic selection seem to operate by maintaining a narrow window of affinities (2). The association of TCRs at the cell surface with the accessory molecules CD4 or CD8 also may influence the functional affinity of TCRs (3). Despite these differences, the three-dimensional structures of the two proteins are remarkably similar, with the hypervariable regions forming loops on a single face of the molecule that contacts the antigen (4–7).
Based on their structural similarities, it is somewhat surprising that there have been significant differences in the success of producing soluble and surface-displayed forms of the extracellular domains of TCRs and antibodies in heterologous expression systems. Many antibodies have now been expressed at high yield and solubility as either intact or Fab-fragment forms or as single-chain (sc) fragment-variable (Fv) proteins. In addition, there are numerous antigen-binding Fv fragments that have been isolated de novo and/or improved through the use of phage-display technology and, more recently, with yeast-display technology (8, 9). These expression systems for antibody fragments have been key in structural studies and in the design of diagnostic and therapeutic antibodies.
In contrast, the three-dimensional structures of a few TCR molecules were determined only after considerable effort on the expression of soluble, properly folded TCRs (10). One of the difficulties in exploring the basis of differences between Fab and TCR is that the extensive sequence diversity in antibody and TCR variable (V) regions complicates efforts to discern what features of the V regions might be important for functions other than antigen binding (e.g., V region pairing and association kinetics, stability, and folding). There have been relatively few studies that have compared the V regions of TCRs and antibodies in terms of these properties (11).
Nevertheless, the TCR from the mouse T cell clone 2C has now been expressed as an sc VαVβ (scTCR) from Escherichia coli (12), as a lipid-linked VαCαVβCβ dimer from myeloma cells (13), and as a secreted VαCαVβCβ dimer from insect cells (6). The 2C scTCR had relatively low solubility compared with most scFv, although its solubility is increased ≈10-fold by fusion at the amino terminus to thioredoxin (14). The difficulty in generating soluble, properly folded VαVβ domains has extended to other TCRs (15–17). The molecular explanation for the apparent differences between TCR and Fv in either solubility or surface-display capability has not been explored adequately. In this report, we show that the 2C scTCR can be expressed in a yeast surface-display system (8, 9) after the selection, from a random library, of specific single-site mutations at the Vα/Vβ interface or in a region of the Vβ framework suspected to interact with the CD3ɛ signal-transduction subunit. These mutations, several of which are found naturally in antibody V regions, indicate the significance of these positions in the TCR and provide a basis for further engineering of TCR-binding properties. In addition, the strategy described here that allowed display of the TCR may be of general use in the study and directed evolution of other proteins that cannot be displayed on the cell surface in their wild-type form.
MATERIALS AND METHODS
Random Mutagenesis and Expression of 2C TCR.
A library of mutant plasmids containing the 2C TCR gene (Vβ8.2-linker-Vα3.1) was generated by using the E. coli mutator strain XL1-Red (Stratagene). Plasmids were transfected into yeast, and surface-displayed TCRs were selected by using fluorescence cell sorting as described (8, 9).
Selection of Mutant TCRs by Using Flow Cytometry.
A library of yeast cells that were transfected with the mutagenized TCR plasmids was incubated with 25 μl of antibody 1B2 conjugated to biotin (20 μg/ml), washed with buffer (PBS/0.1% BSA), and incubated with streptavidin–phycoerythrin (SA-PE, 1:200, PharMingen). After washing, samples were sorted on a Coulter 753 bench sorter with an event rate of ≈4,000 cells per s. A total of 6 × 107 cells were examined during the first sorting round, and ≈5% of the population was collected. Collected cells were regrown at 30°C in selective glucose medium for ≈18–20 h, and TCR surface expression was induced at 20°C in selective galactose medium. After three rounds of sorting, collected cells were re-sorted and plated on selective medium to isolate individual clones. Flow cytometry with 1B2 conjugated to biotin and SA-PE was used to examine 20 clones further, and plasmids from 17 colonies were rescued for sequencing.
Estimation of TCR per Cell.
Yeast expressing mutant TCR (107 cells in 10 μl) were incubated with various concentrations of 125I-labeled 1B2 Fab fragments (total volume of 30 μl) on ice for 1 h in round-bottom 96-well plates. The mixture was layered over 300 μl of 80% (vol/vol) dibutyl phthalate/20% (vol/vol) olive oil in plastic tubes, and the tubes were microfuged for ≈2–3 s. Tubes were frozen on dry ice. Cell pellets containing bound 125I-labeled 1B2 Fabs were counted, and the cpm of 125I-labeled 1B2 Fab added was plotted as a function of cpm bound. To estimate the total number of receptors per yeast cell, results from the highest concentration of 125I-labeled 1B2 Fabs tested (≈70 ng/ml) were compared with results obtained by using 2C T cells, for which there are 100,000 receptors per cell (18).
Peptide/MHC Functional Binding Assay.
Soluble peptide/MHC protein (peptide/Ld) was prepared from Drosophila melanogaster cells as described by Garcia et al. (3). MHC molecules were purified by Ni-agarose (Ni2+/nitrilotriacetic acid/agarose; Qiagen, Chatsworth, CA) step-gradient affinity chromatography and ion-exchange chromatography [Mono Q in 50 mM Tris (pH 8.0), 0–500 mM NaCl gradient; Amersham Pharmacia]. Before the competition assays, nonspecific (MCMV) or specific (QL9) peptides were incubated with the soluble MHC protein/Ld preparations for ≈1 h at 4°C.
To examine binding of soluble peptide/MHC to yeast cells displaying mutant TCR, the amount of 125I-labeled 1B2 Fabs that yielded ≈400–600 cpm bound in the absence of inhibitor was used in a competitive ligand binding assay (i.e., more Fab was required for yeast that expressed lower levels of TCR to achieve this amount of bound 1B2). Yeast (107 cells in 10 μl) were incubated with 10 μl of 125I-labeled 1B2 Fabs and 10 μl of PBS (no inhibitor), MCMV/Ld, or QL9/Ld. The concentration used in these assays was ≈15 μM for both MCMV/Ld and QL9/Ld. Cell pellets were counted as described above. Inhibition by peptide/MHC was calculated by using the following equation:
Percentage of inhibition = [1 − (cpm bound in presence of inhibitor/cpm bound in absence of inhibitor)] × 100.
The approximate KD of mTCR/yeast binding to QL9/Ld was calculated by using the following equation:
KD(inhibitor) = [inhibitor]50% inhibition/[1 + (Fab)/KD(Fab)].
RESULTS
Expression and Detection of the 2C TCR on the Yeast Cell Surface.
The yeast-display system has been used to express different scFv regions as fusion proteins with a yeast-mating agglutination receptor subunit, Aga-2p (8, 9). Similarly, the gene encoding the scTCR (Vβ-linker-Vα) region from the T cell 2C was cloned into this system as a fusion with Aga-2p and a short epitope tag (HA). Yeast transformants were analyzed with a panel of anti-TCR monoclonal antibodies: KJ16, F23.1, F23.2, and 1B2. These antibodies recognize conformational epitopes of the Vβ region (KJ16, F23.1, F23.2) or the VαVβ idiotype (1B2; refs. 1 and 19). The antibody 1B2 recognizes TCR residues that are nearly identical to the TCR residues that contact the peptide/MHC (QL9/Ld or dEV8/Kb), but its affinity is over 100 times higher than the peptide/MHC interaction (1); as such, 1B2 was used as a high-affinity probe for the detection of properly folded 2C TCRs (20).
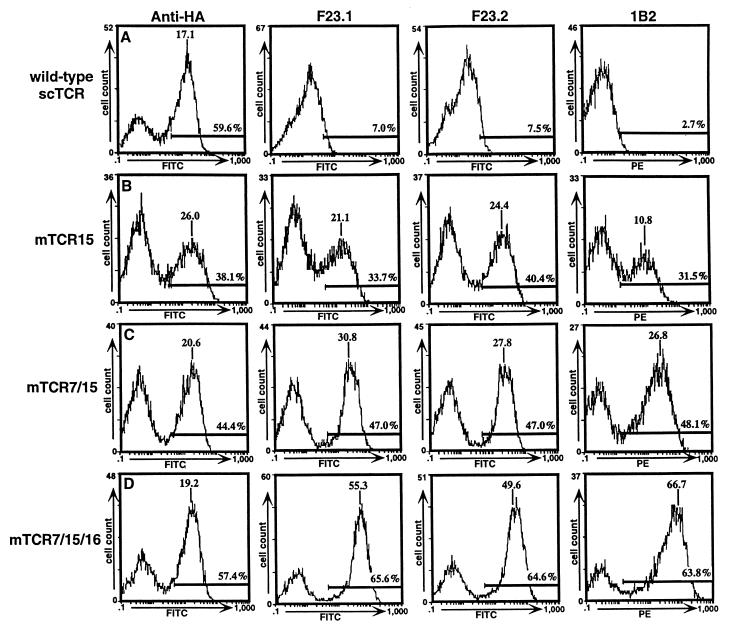
Expression of properly folded wild-type 2C scTCR on the surface of the yeast could not be detected with 1B2 (Fig. 1A). However, both the HA tag and, to a lesser degree, the KJ16, F23.1, and F23.2 epitopes were detectable on the surface (see shoulders on peaks in Fig. 1A; data not shown). Thus, properly associated wild-type VαVβ protein was not displayed at the cell surface. The eukaryotic secretory pathway retains and degrades misfolded proteins, as shown for the assembly of the TCR–CD3 complex (21). In yeast, secretion efficiency has been found to correlate with thermodynamic stability of a series of mutants of bovine pancreatic trypsin inhibitor (22). As surface-displayed proteins are transported through the secretory pathway, we reasoned that TCR mutants with enhanced stabilities might enable functional cell surface expression.
Figure 1.
Flow cytometric analysis of wild-type TCR/yeast and mutant TCR/yeast. Yeast displaying wild-type TCR (A), single-mutant mTCR15 (B), double-mutant mTCR7/15 (C), or triple-mutant mTCR7/15/16 (D) were stained with anti-HA monoclonal antibody 12CA5 (Boehringer Mannheim), anti-Vβ8 antibodies F23.1 and F23.2, and biotinylated 1B2 followed by FITC-labeled F(ab′)2 goat anti-mouse IgG or SA-PE. Labeled yeast cells were analyzed on a Coulter Epics XL flow cytometer. Peak mean fluorescence and the percentage of the population that was positive (i.e., within the region of the cursor bar) are indicated.
Selection for Increased 1B2+ Surface Levels from a Randomly Mutagenized TCR Library.
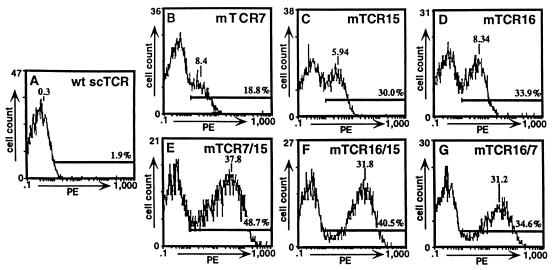
To determine whether surface expression could be achieved by single-residue changes in the Vα or Vβ domains, a randomly mutagenized library of 7 × 106 cells was screened by flow cytometric sorting with the 1B2 antibody. The Aga-2p-TCR expression plasmid was randomly mutagenized by growth in the XL1-Red mutator strain of E. coli. Yeast were selected through three rounds of flow cytometric sorting after labeling with biotinylated 1B2 followed by SA-PE. Selected cells were plated, and expanded individual colonies were analyzed by flow cytometry with the panel of anti-TCR antibodies (Figs. 1 and 2; data not shown). Individual colonies had various levels of 1B2+ immunofluorescence (Fig. 2). Among the different colonies, the fluorescence levels detected with 1B2 were directly proportional to the levels detected with the other anti-TCR antibodies (Fig. 1B for mutant 15; data not shown for other single mutants).
Figure 2.
Flow cytometric analysis of wild-type and mutant TCR/yeast after staining with antibody 1B2. Yeast displaying wild-type or mutant TCR were stained with biotinylated 1B2 and detected with SA-PE. Wild-type TCR/yeast (A); single-mutant TCR/yeast mTCR7 (B), mTCR15 (C), and mTCR16 (D); and double-mutant scTCR/yeast mTCR7/15 (E), mTCR16/15 (F), and mTCR16/7 (G).
Sequence Analysis of TCR Mutants.
Sequencing of the scTCR genes that were rescued from 17 different colonies indicated that, in every case, there were single-site mutations in either the Vα or Vβ region (data not shown). Among the 17 isolates, five unique mutations were observed: VαL43P (mTCR7, 9 isolates), VαL104P (mTCR16, 1 isolate), VβA13V (mTCR2, 4 isolates), VβG17E (mTCR15, 2 isolates), and VβT105A (mTCR3, 1 isolate).
Cell-Surface Levels of Double and Triple TCR Mutants.
To determine whether the effects of the mutations on the levels of surface scTCR were additive, three of the mutations (mTCR7, mTCR15, and mTCR16: VαL43P, VβG17E, and VαL104P) were combined into double or triple mutants. Compared with the single mutants, all combinations had increases in 1B2+ surface levels, and the triple mutant had the highest mean fluorescence level in the test (Figs. 1 C and D, and 2). We conclude that the mechanism or mechanisms involved in the increased surface expression of these mutants seem to operate independently and that the mutations can be combined to engineer TCR display libraries with enhanced surface expression levels. In addition, the observation that the increased surface levels could be transferred by site-directed mutagenesis to produce double and triple mutants suggests that the effects are caused by the single-site mutations and not by mutations that reside outside of the coding region (e.g., in the promoter region). Finally, titrations of the mutants with the 1B2 antibody indicated that there was no overall increase in the affinity of the 1B2 antibody that could account for higher levels of 1B2 binding (data not shown).
The total number of TCRs per yeast cell was estimated by using 125I-labeled 1B2 Fab fragments: ≈10,000 for the single mutants (mTCR7, mTCR15, and mTCR16), ≈30,000 for the double mutant (mTCR7/15), and ≈50,000 for the triple mutant (mTCR7/15/16). Although the number of TCRs per yeast cell is lower than the number of TCRs per 2C T cell (≈100,000), the number of TCRs per unit area is considerably higher for yeast that express the double and triple TCR mutants than for 2C T cells (i.e., the diameter of 2C T cells is 2–3 times larger; thus, they have 4- to 9-fold more surface area than the yeast cells).
Peptide/MHC Binding to TCR Mutants.
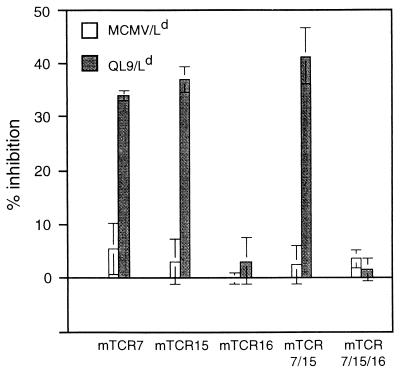
To determine whether the TCR mutants were capable of binding the peptide/MHC ligand recognized by T cell clone 2C (QL9/Ld), a sensitive competition assay was performed (18). In this assay, soluble QL9/Ld complexes were used as inhibitors of binding of 125I-labeled 1B2 Fab fragments to yeast cells that express the TCR. QL9/Ld complexes, but not an MCMV/Ld control complex, inhibited the binding of 1B2 to three of the mutants: VαL43P (mTCR7), VβG17E (mTCR15), and the mTCR7/15 double mutant (Fig. 3). However, the TCR with the VαL104P mutation (mTCR16 and the mTCR7/15/16 triple mutant), which is in CDR3, was not capable of binding QL9/Ld, at least within the detection limits of this assay. This result may not be surprising; the phenylalanine at position 100 within this CDR3 loop contributes binding energy to the QL9/Ld interaction (1), and a nonconservative CDR3 substitution such as proline could affect the conformation of the entire loop. Calculation of the approximate KD from these experiments (15 μM) indicated that the affinity was 5-fold lower than that determined by using surface plasmon resonance with E. coli-derived 2C TCR (23). This difference could be caused either by the difference in forms of the TCR analyzed (chip-immobilized sc-thioredoxin-VαVβ vs. yeast-immobilized Aga-2-VαVβ) or by minor effects of the mutations themselves.
Figure 3.
Peptide/MHC binding by surface-displayed TCR mutants. Yeast displaying mutant TCR were monitored for peptide/MHC binding in a competitive inhibition assay by using the amount of 125I-labeled 1B2 Fab fragments that yielded ≈400–600 cpm bound (in the absence of inhibitor) as the probe. The percentage of inhibition of 125I-labeled 1B2 Fab fragment binding by either QL9/Ld or the same concentration of MCMV/Ld, which is not recognized by the 2C TCR, was examined for five mutant TCR/yeast isolates, as shown. For the experiments shown, the maximum cpm bound in the absence of peptide/MHC were 570 cpm (mTCR7), 600 cpm (mTCR15), 550 cpm (mTCR16), 470 cpm (mTCR7/15), and 580 cpm (mTCR7/15/16). In the presence of excess unlabeled 1B2 as an inhibitor (i.e., nonspecific 125I-labeled 1B2 Fab binding), background cpm were 200 cpm (mTCR15), 150 cpm (mTCR16), 80 cpm (mTCR7/15), and 60 cpm (mTCR7/15/16). Results are representative of two to four experiments.
Location of Mutations in the TCR Structure.
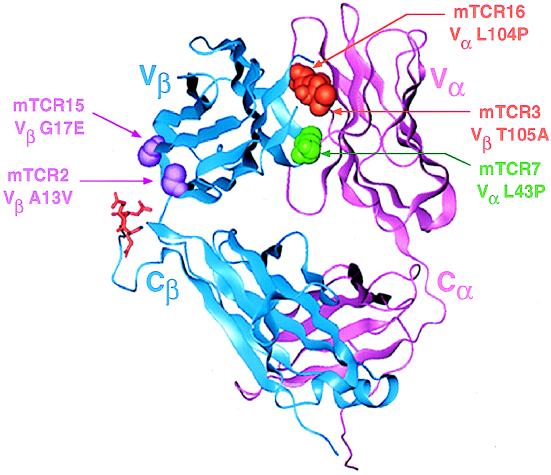
To explore the possible molecular basis for the effects of these mutations on yeast surface display, their positions in the 2C TCR structure (6, 24) were examined (Fig. 4); three of the mutations, VαL43P, VαL104P, and VβT105A, reside at the interface of the Vα and Vβ domains. The two other mutations, VβA13V and VβG17E, are located in the FR1 where in the full TCR a protruding knob in the Cβ region is in proximity to the Vβ region. The VαL104P and VβT105A interface mutations are located in the two CDR3 loops, within 5 Å of each other. The observation that the VαL104P mutation also affects peptide/MHC binding has recent precedence, as a proline substitution within the CDR3 of a different TCR also affected peptide/MHC binding (presumably caused by changing the conformation of the CDR loop; ref. 25). Interestingly, a recent alanine-scanning mutagenesis study of the 2C TCR showed that VαL104A reduced binding to QL9/Ld, whereas the VβT105A mutation yielded a slight increase in affinity for the 1B2 antibody but had no effect on QL9/Ld (1). Despite these effects on 1B2 or peptide/MHC binding, the selection of mutations only in the CDR3, which are uniquely positioned among the six CDR at the Vα/Vβ interface, suggests that they act by influencing the stability of the Vα/Vβ interaction.
Figure 4.
X-ray crystallographic structure of 2C TCR with mutated residues highlighted. The 2C TCR structure (6) is shown with residues isolated as mutants in the yeast display system highlighted. The FR2 interface mutation is colored green. The CDR3 interface mutations are colored orange. First-framework-region (FR1) mutations proximal to the Cβ “knob” (in the full TCR) are colored purple. There are three negatively charged Cβ knob residues (Glu-221, Glu-222, and Asp-223) indicated in red.
DISCUSSION
Selection of the displayed TCR mutants from a yeast library raises the question of how these specific mutations act in allowing surface expression. A clue to the mechanism comes from a comparative analysis of TCR and antibody V region sequences. The three framework mutations identified here, VβA13V, VβG17E, and VαL43P, are found naturally in many VH and VL sequences (Table 1). Valine is the most frequent residue in Vα, VH, and Vκ sequences at the FR1 site corresponding to the VβA13V mutant. Glutamic acid is the most frequent residue in Vκ and the second most common residue in Vα and Vλ at the FR1 site corresponding to the VβG17E mutant. And perhaps most compellingly, proline is the most frequent residue in Vα, Vκ, and Vλ sequences at the FR2 site corresponding to the VαL43P mutant. A leucine–proline VH/VL interaction across the Fv interface is highly conserved in antibodies (>90%; ref. 26), and the VαL43P mutation at position 43 introduces such a leucine–proline pair into the 2C scTCR interface. Thus, the added stability of this leucine–proline interaction seems to facilitate secretion and display, perhaps through improvement in Vα/Vβ (or VH/VL) association. The presence of stability-increasing mutations at the interface has implications for preferential pairing between Vα and Vβ regions, suggesting that pairing is nonrandom.
Table 1.
Frequency of residues in V regions
| Mutation | Residue | Residue and frequency (%) in region
|
||||
|---|---|---|---|---|---|---|
| Vβ | Vα | VH | Vκ | Vλ | ||
| VβA13V (mTCR2) | Ala | 8 | 4 | 2 | 41 | 22 |
| Val | 6 | 58 | 74 | 43 | 16 | |
| Most frequent | Thr, 30 | Val, 58 | Val, 74 | Val, 43 | Gly, 40 | |
| 2nd most frequent | Lys, 17 | Leu, 23 | Lys, 12 | Ala, 41 | Ala, 22 | |
| VβG17E (mTCR15) | Gly | 13 | 9 | 26 | 2 | 11 |
| Glu | 8 | 26 | 14 | 50 | 18 | |
| Most frequent | Gln, 38 | Ala, 27 | Ala, 28 | Glu, 50 | Gln, 58 | |
| 2nd most frequent | Gly, 13 | Glu, 26 | Gly, 26 | Asp, 33 | Glu, 18 | |
| VβL43P (mTCR7) | Leu | 65 | 34 | 97 | 0.3 | 14 |
| Pro | 20 | 56 | 1 | 92 | 70 | |
| Most frequent | Leu, 65 | Pro, 56 | Leu, 97 | Pro, 92 | Pro, 70 | |
| 2nd most frequent | Pro, 20 | Leu, 34 | Pro, 1 | Val, 5 | Phe, 15 | |
Comparison of amino acid frequencies in TCR (Vα and Vβ) and antibody (VH, Vκ, Vλ) variable regions. Residues found at the three framework positions indicated by mutants from the TCR/yeast display system are found naturally in antibodies. Values determined by searches of all ig and all tcr v region sequences (Kabat database, immuno.bme.nwu.edu, 11/97 data sets).
It is possible that antibody V regions evolved residues at these positions to facilitate high-level secretion in soluble form, and expression requirements for TCR Vα/Vβ as part of a membrane complex may differ. The identification of FR1 mutations VβA13V and VβG17E raises the question of how this region of the TCR might differ from antibodies. One difference between TCRs and antibodies in this region is the presence of a negatively charged knob (Fig. 4) formed by residues Glu-221, Glu-222, and Asp-223 within the Cβ (4). These residues are positioned 7–10 Å from the Vβ FR1 region that contains the VβA13V and VβG17E mutations. Interestingly, Vβ sequences from the Kabat database have a strong bias toward positive charges in this region of FR1, with a mean net charge of +1.6 for residues 8–17 (56% of Vβ sequences possess a net charge of 2 or greater). By contrast, Vα, VH, Vκ, or Vλ sequences have a mean net charge of +0.4 or lower in this FR1 region and fewer than 2% possess net charges of 2 or higher. Thus, we predict that electrostatic interactions between Vβ FR1 and the negatively charged Cβ knob contribute to stabilization of the full TCR. The absence of the Vβ/Cβ knob interaction in the scTCR design might destabilize the fold, such that introduction of a more hydrophobic buried residue (VβA13V) or a negative residue (VβG17E) could stabilize the protein.
Consistent with the possibility that the Cβ knob stabilizes the Vβ region, studies have shown that TCR constructions that contain the Cβ region are more stable than VαVβ alone (27, 28). During review of this manuscript, a study showed that phage display of a TCR was possible only when the Cβ region was included (29). We have shown that solubly expressed forms of mTCR7, mTCR15, and mTCR16 have enhanced thermal stability compared with wild-type scTCR (data not shown), suggesting that it may be possible to stabilize VαVβ pairs without including the entire Cβ domain and without affecting peptide/MHC reactivity. Finally, it is possible that the Vβ/Cβ knob interaction also may influence the signaling of a T cell through the TCR complex. This possibility is supported by a recent finding (30) that one of the two CD3ɛ subunits is associated with the TCR near the Cβ knob (called Cβ FG loop in the study). One prediction from this idea is that introduction of the Vβ FR1 mutations described here into a full TCR expressed on a T cell may affect signaling functions.
Expression of the HA tag on the surface of yeast cells that contain the wild-type TCR construct (Fig. 1A) indicated that this region of the Aga-2 fusion protein was transported correctly through the secretory pathway. Both the F23.1 and F23.2 antibodies (anti-Vβ8) also showed some reactivity with the wild-type TCR construct. In contrast, antibody 1B2, which requires both Vα and Vβ domains for reactivity, did not bind to yeast that contain the wild-type TCR plasmid. Thus, the Aga-2–HA–Vβ–linker–Vα construction showed highest reactivity with antibodies that recognize the most amino-terminal regions (Aga-2 > Vβ > Vα). Recently, we have shown that the HA-positive protein on the surface of the wild-type TCR yeast has been cleaved, accounting for its lower level of reactivity with the other antibodies (E.V.S., M.C.K., E. A. Parke, D.M.K., and K.D.W., unpublished work). Thus, less stable Vα/Vβ domains seem to be more susceptible to intracellular proteases.
The results described here suggest that the yeast surface-display system could be useful for genetic engineering of T cell receptors by combinatorial library screening. As there are a number of Vα and Vβ regions that share the particular residues identified here with the 2C TCR, the single-site changes are likely to show similar effects for other TCRs. The specific mutations thus may allow the expression of other TCR Vα/Vβ regions that have been problematic. As has been shown by using different scFv antibodies in the yeast display system, it may now be possible to isolate TCR variants with higher antigen-binding affinities. By analogy to the variety of therapeutic antibody applications resulting from phage-display technology, one might expect numerous applications that derive from the generation of stable, high-affinity T cell receptor fragments.
Finally, there are many proteins that have inherent difficulties in expression as fusion proteins on the surface of phage. As shown here, these difficulties might be overcome by combining a eukaryotic expression system with a strategy of random mutagenesis and selection to isolate mutants that are expressed at high levels on the yeast cell surface.
Acknowledgments
We thank Herman Eisen and Ian Wilson for helpful advice and discussions, Bryan Cho and Tom Manning for advice on assays and experiments, and Gary Durack of the University of Illinois Biotechnology Center for assistance with flow cytometry. M.C.K. was supported as a trainee by National Institutes of Health Grant T32 GM07283, and E.T.B. was supported by an National Science Foundation Graduate Fellowship. This work was supported by grants from the Whitaker Biomedical Engineering Foundation (to K.D.W.) and by National Institutes of Health Grants GM55767 (to D.M.K.) and AI42267 (to L.T.).
ABBREVIATIONS
- TCR
T cell receptor
- Fv
fragment variable
- sc
single-chain
- V
variable
- SA-PE
streptavidin–phycoerythrin
- FR1
first framework region
References
- 1.Manning T C, Schlueter C J, Brodnicki T C, Parke E A, Speir J A, Garcia K C, Teyton L, Wilson I A, Kranz D M. Immunity. 1998;8:413–425. doi: 10.1016/s1074-7613(00)80547-6. [DOI] [PubMed] [Google Scholar]
- 2.Alam S M, Travers P J, Wung J L, Nasholds W, Redpath S, Jameson S C, Gascoigne R J. Nature (London) 1996;381:616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- 3.Garcia K C, Scott C A, Brunmark A, Carbone F R, Peterson P A, Wilson I A, Teyton L. Nature (London) 1996;384:577–581. doi: 10.1038/384577a0. [DOI] [PubMed] [Google Scholar]
- 4.Bentley G A, Boulot G, Karjalainen K, Mariuzza R A. Science. 1995;267:1984–1987. doi: 10.1126/science.7701320. [DOI] [PubMed] [Google Scholar]
- 5.Fields B A, Ober B, Malchiodi E L, Lebedeva M I, Braden B C, Ysern X, Kim J K, Shao X, Ward E S, Mariuzza R A. Science. 1995;270:1821–1824. doi: 10.1126/science.270.5243.1821. [DOI] [PubMed] [Google Scholar]
- 6.Garcia K C, Degano M, Stanfield R L, Brunmark A, Jackson M R, Peterson P A, Teyton L, Wilson I A. Science. 1996;274:209–219. [PubMed] [Google Scholar]
- 7.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Nature (London) 1996;384:131–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 8.Boder E T, Wittrup K D. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 9.Kieke M C, Cho B K, Boder E T, Kranz D M, Wittrup K D. Protein Eng. 1997;10:1303–1310. doi: 10.1093/protein/10.11.1303. [DOI] [PubMed] [Google Scholar]
- 10.Bentley G A, Mariuzza R A. Annu Rev Immunol. 1996;14:563–590. doi: 10.1146/annurev.immunol.14.1.563. [DOI] [PubMed] [Google Scholar]
- 11.Wulfing C, Pluckthun A. J Mol Biol. 1994;242:655–669. doi: 10.1006/jmbi.1994.1615. [DOI] [PubMed] [Google Scholar]
- 12.Soo Hoo W F, Lacy M J, Denzin L K, Voss E W J, Hardman K D, Kranz D M. Proc Natl Acad Sci USA. 1992;89:4759–4763. doi: 10.1073/pnas.89.10.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slanetz A E, Bothwell A L M. Eur J Immunol. 1991;21:179–183. doi: 10.1002/eji.1830210127. [DOI] [PubMed] [Google Scholar]
- 14.Schodin B A, Schlueter C J, Kranz D M. Mol Immunol. 1996;33:819–829. doi: 10.1016/0161-5890(96)00038-7. [DOI] [PubMed] [Google Scholar]
- 15.Novotny J, Ganju R K, Smiley S T, Hussey R E, Luther M A, Recny M A, Siliciano R F, Reinherz E L. Proc Natl Acad Sci USA. 1991;88:8646–8650. doi: 10.1073/pnas.88.19.8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward E S. J Mol Biol. 1992;224:885–890. doi: 10.1016/0022-2836(92)90455-s. [DOI] [PubMed] [Google Scholar]
- 17.Ward E S. Scand J Immunol. 1991;34:215–220. doi: 10.1111/j.1365-3083.1991.tb01539.x. [DOI] [PubMed] [Google Scholar]
- 18.Sykulev Y, Brunmark A, Tsomides T J, Kageyama S, Jackson M, Peterson P A, Eisen H N. Proc Natl Acad Sci USA. 1994;91:11487–11491. doi: 10.1073/pnas.91.24.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brodnicki T C, Holman P O, Kranz D M. Mol Immunol. 1996;33:253–263. doi: 10.1016/0161-5890(95)00142-5. [DOI] [PubMed] [Google Scholar]
- 20.Schlueter C J, Schodin B A, Tetin S Y, Kranz D M. J Mol Biol. 1996;256:859–869. doi: 10.1006/jmbi.1996.0132. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Bonifacino J S, Yuan L C, Klausner R D. J Cell Biol. 1988;107:2149–2161. doi: 10.1083/jcb.107.6.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalski J M, Parekh R N, Mao J, Wittrup K D. J Biol Chem. 1998;273:19453–19458. doi: 10.1074/jbc.273.31.19453. [DOI] [PubMed] [Google Scholar]
- 23.Manning T C, Parke E A, Teyton L, Kranz D M. J Exp Med. 1999;189:461–470. doi: 10.1084/jem.189.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia K C, Degano M, Pease L R, Huang M, Peterson P, Teyton L, Wilson I A. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 25.Daniel C, Horvath S, Allen P M. Immunity. 1998;8:543–552. doi: 10.1016/s1074-7613(00)80559-2. [DOI] [PubMed] [Google Scholar]
- 26.Chothia C, Boswell D R, Lesk A M. EMBO J. 1988;7:3745–3755. doi: 10.1002/j.1460-2075.1988.tb03258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung S, Wucherpfenning K A, Friedman S M, Hafler D A, Strominger J L. Proc Natl Acad Sci USA. 1994;91:12654–12658. doi: 10.1073/pnas.91.26.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plaksin D, Polakova K, McPhie P, Margulies D H. J Immunol. 1997;158:2218–2227. [PubMed] [Google Scholar]
- 29.Weidanz J A, Card K F, Edwards A, Perlstein E, Wong H C. J Immunol Methods. 1998;221:59–76. doi: 10.1016/s0022-1759(98)00153-7. [DOI] [PubMed] [Google Scholar]
- 30.Ghendler Y, Smolyar A, Chang H C, Reinherz E L. J Exp Med. 1998;187:1529–1536. doi: 10.1084/jem.187.9.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]