Abstract
Analysis of the host response to viral infection generally has focused on the capacity of viruses to activate or repress transcription of cellular genes, and this approach is also characteristic of work on RNA viruses such as respiratory syncytial virus (RSV). In the present study, it appeared initially that RSV-driven expression of a critical immune regulator, the β-chemokine RANTES (regulated upon activation, normal T cell expressed and secreted), in primary-culture airway epithelial cells also depended on inducible gene transcription because expression was accompanied by coordinate increases in transcriptional initiation rate and gene promoter activity. However, RSV-driven increases in RANTES gene transcription and promoter activity were small and transient relative to RANTES expression, and they were no different in size and duration than for inactivated RSV that was incapable of fully inducing RANTES expression. These findings suggested that the increase in RANTES gene transcription was not sufficient for inducible expression and that critical regulatory effects occurred at a posttranscriptional level. This type of mechanism for virus-inducible expression of RANTES was established when we found that replicating (but not inactivated) RSV markedly increased RANTES mRNA half-life (from 0.8 to 6.8 h). In addition, RNase protection assays of heterologous promoter/reporter plasmids indicate that basal instability of RANTES mRNA is mediated at least in part by nucleotides 11–389 of the RANTES gene, and this region is also the target for induction by virus. The distinct pathway for production of RANTES (in combination with cytokine-dependent expression of RANTES and related immune-response genes) may more effectively coordinate immune cell interaction with epithelial barrier cells to mediate host defense.
Eukaryotic cells respond to a large number of distinct extracellular signals and environmental stresses, and the responses usually involve signal transduction pathways that lead to the activation of specific sets of genes. Thus, to define the innate host response to infection, considerable effort has focused on the transcriptional activation of cellular genes by viruses. In the case of DNA viruses (such as adenovirus and herpesviruses), viruses encode regulatory proteins that affect the activities of cellular transcription factors with consequences on target genes for cell growth, transformation, and cytokine responsiveness (1). Retroviruses influence the activation of cellular genes at least in part through regulating enhancer activity (2). In addition, RNA viruses, and in particular the Paramyxoviridae family, which includes Sendai virus, Newcastle disease virus, and respiratory syncytial virus (RSV), effectively stimulate the expression of host defense genes, especially cytokines, and the effect is attributed to viral activation of gene transcription (3). Perhaps the best-developed model is represented by induction of IFN-β gene expression based on assembly of an enhanceosome by Sendai virus (4). Similarly, increases in expression of the β-chemokine RANTES (originally designated as regulated on activation, normal T cell expressed and secreted) (5) by Newcastle disease virus also have been attributed to assembly of a more efficient transcriptional complex in a macrophage cell line (6).
In that context, we have used a natural host for respiratory viruses—primary-culture human tracheobronchial epithelial (hTBE) cells, along with endobronchial tissue and mouse models of viral bronchitis to define epithelial cell-dependent immunity to respiratory pathogens. Our work defined a subset of epithelial immune-response genes typified by intercellular adhesion molecule 1 (ICAM-1) and IFN regulatory factor 1 (IRF-1) that appears designed to coordinate several aspects of antiviral defense (7). Expression of these genes is mediated by an IFN-γ-responsive signaling pathway that requires activation of the first member (Stat1) of the signal transducer and activation of transcription (STAT) family of transcription factors to facilitate gene transcription (8–11). In the course of subsequent work with IFN-γ-stimulated cells and tissue, we also found that the pattern of expression for the cell adhesion molecule ICAM-1 needed to be coordinated with polarized secretion of an immune cell chemoattractant, i.e., the β-chemokine RANTES to appropriately direct immune cell traffic (12). In addition, we noted that the characteristics of RANTES gene expression, both in response to cytokines (13) and during airway inflammation (14–16), appeared to be distinct from other Stat1-dependent epithelial immune-response genes (i.e., ICAM-1 and IRF-1). Taken together, the findings suggested that complementary functional activities of ICAM-1 and RANTES gene products might be accompanied by distinct regulation of gene expression.
In the present experiments, we therefore examined the basis for viral induction of RANTES gene expression, and first noted that, among epithelial immune-response genes, RANTES was most sensitive to direct induction by virus (without immune cell derived IFN-γ) and induction selectively depended on viral replication. In the course of investigating the basis for virus-inducible expression of RANTES in this system, it became apparent that the standard approaches for defining gene regulation were inadequate for studying RANTES. To define the level of transcriptional activation of the RANTES gene, we resorted to RNA/RNA hybridization and quantitative PhosphorImaging to allow for improved sensitivity. This approach was necessary because of the relatively low level of transcriptional activity for the RANTES gene. In addition, we developed a modified pulse–chase method for analyzing RANTES mRNA half-life. This approach was necessary because the more traditional method using transcriptional inhibitors resulted in altering RANTES mRNA stability. By applying these combined methodologies for assessing gene activation and mRNA stability, we found that selective virus-dependent expression of RANTES is mediated by stabilization of RANTES mRNA as well as increases in promoter activity. These findings therefore provide evidence for viral induction of mammalian host gene expression at a posttranscriptional level, the molecular basis for β-chemokine regulation as distinct from other cytokines and from related cytokine-driven immune-response genes, and consequently, a further basis for coordinated regulation of innate immunity at the epithelial barrier.
MATERIALS AND METHODS
Virus Preparation.
RSV (strain A2) was obtained from American Type Culture Collection as a stock concentration of 1 × 106 TCID50/ml and was propagated and titered by using HEp-2 cells grown in Eagle’s MEM supplemented with 2.5% heat-inactivated FBS as described (17). Culture supernatant containing RSV was harvested 48–72 h after infection when >70% of the cells showed syncytia formation, rounding or detachment. Cellular debris was removed by centrifugation, and the RSV-containing supernatant (1 × 106 plaque-forming units/ml) was stored at −70°C. Inactivated RSV was prepared by exposure of an aliquot of the same RSV-containing medium to UV light (18 W) at 10 cm for 30 min at 4°C, and loss of viral replication capacity was verified by plaque assay. Thus, preparations of RSV and UV-inactivated RSV were handled identically except for UV irradiation, so medium constituents did not vary for the two preparations. Wild-type adenovirus 5 was propagated in 293 cells, purified by centrifugation through cesium chloride, and titered by plaque assay as described (11).
Airway Epithelial Cell Culture and Infection.
hTBE cells were isolated from tracheal and bronchial mucosal strips by enzymatic dissociation and cultured in Laboratory of Human Carcinogenesis (LHC)-8e medium for study up to passage 10 as described (18). Monolayers of hTBE cells were grown to subconfluence on 100-mm tissue culture plates and infected with RSV at a multiplicity of infection (MOI) of 0.02–2.0 for 1–48 h at 37°C. Control conditions included infection with inactivated RSV or with wild-type adenovirus 5 (MOI of 0.3–30) for the same time periods as well as stimulation with recombinant human IFN-γ (Genentech) at an optimal concentration (100 units/ml) and duration (24 h) for induction of RANTES gene expression (13).
Northern Blot Analysis and Immunoassay.
For all treatment conditions, cell supernatants were collected and used to determine RANTES level in triplicate by using an ELISA (Quantikine) from R&D Systems. In addition, total cellular RNA was prepared by using the RNeasy Mini kit from Qiagen and subjected to Northern blot analysis as described (11, 18) by using 32P-labeled cDNA probes for RANTES (from T. Schall, DNAX); macrophage inflammatory protein (MIP) 1α and MIP-1β (from M. Krangel, Duke University, Durham, NC); monocyte chemoattractant protein (MCP) 1 and IL-8 (from J. Oppenheim, National Cancer Institute, Bethesda, MD); MCP-3 (from G. Opdenakker, Rega Institute, Leuven, Belgium); eotaxin from A. Luster (Massachusetts General Hospital, Boston); IRF-1 (from Y. Henderson and A. Deisseroth, University of Texas, Houston); ICAM-1 (from D. Staunton, Harvard University, Boston); and β-actin in pUC9 from B. Saha (Emory University, Atlanta).
Generation of cRNAs for RANTES.
To detect RANTES transcripts most efficiently in nuclear run-on and mRNA half-life assays, we used RNA/RNA hybridization (19). Sense and antisense RANTES cRNAs were transcribed in vitro by using T3 (for sense) or T7 (for antisense) RNA polymerase (Promega) with the linearized RANTES cDNA in pBluescript as template. The transcription reaction was performed in 10 μl containing 1× transcription buffer (Promega), 10 mM DTT, 1.6 units/ml of RNasin (Promega), 0.5 mM rNTP mixture (Promega), and 0.5 μg of linearized template DNA for 1 h at 37°C. After digesting the template DNA with RQ1 DNase (Promega), RNA was purified by phenol/chloroform extraction followed by ethanol precipitation and then resuspended in RNase-free water.
Nuclear Run-on Assay.
hTBE cell monolayers (5–7 × 107 cells/condition) with and without RSV infection were scraped into PBS and then resuspended in hypotonic lysis buffer (10 mM Tris⋅HCl, pH 7.4/10 mM NaCl/3 mM MgCl2/0.5% NP-40) for 10 min at 4°C. Isolated nuclei were collected by centrifugation, washed, and resuspended in 200 μl of transcription buffer (10 mM Tris, pH 7.4/5 mM MgCl2/80 mM KCl/0.1 mM EDTA/0.5 mM DTT/35% glycerol), and transcription was initiated by adding 4 mM ATP, GTP, and CTP (Boehringer Mannheim) and 200 μCi [α-32P]UTP. After incubation for 60 min at 30°C, the 32P-labeled nuclear RNA was purified with guanidium isothiocyanate lysis and phenol-chloroform extraction (20), denatured at 80°C for 10 min, and then hybridized (1 × 107 cpm/condition) with target cRNAs (2 μg of RANTES sense and antisense cRNA) and 10 μg of linearized and denatured β-actin cDNA slotted onto nitrocellulose filters. Filters were prehybridized in 20 mM Tris⋅HCl (pH 7.4), 50% formamide, 5× SSC, 5× Denhardt’s solution, 200 μg denatured salmon sperm DNA, and 200 μg/ml of yeast tRNA at 42°C for 2 h and hybridized under the same conditions for 48–72 h. After hybridization, filters were washed in 0.5× SSC (SSC = 0.15 M NaCl and 0.015 M sodium citrate, pH 7.0)/0.1% SDS for 20 min at 25°C followed by 0.1× SSC/0.1% SDS at 65°C for 20 min before autoradiography or PhosphorImaging.
RANTES Promoter, Luciferase Reporter Plasmids, and Transactivation Assay.
RANTES 5′ flanking sequence from nucleotides −1006 to −1 (numbered from the translation start site) was generated by PCR with synthetic oligonucleotide primers (5′-AGCTTTCATATTCTGTAACT-3′ for upstream and 5′-GTACCTGTGGGAGAGGCTGT-3′ for downstream) and human genomic DNA as template. The PCR product was sequenced and was identical to one reported previously for the RANTES promoter region (21) with binding sites for Stat1 (at −303) and NF-κB (at −85, −99, −268, and −634) (22, 23). RANTES gene sequence (−1006 to −1) then was inserted upstream to the Photinus pyralis luciferase gene in the luciferase-reporter plasmid pBH-luc (24) to yield the reporter plasmid pBH-RANTES(-1006–1)-luc. Plasmid DNA was purified by two successive centrifugations through cesium chloride and then used to transfect duplicate hTBE cell monolayers at 90% confluence. Each monolayer (in a 35-mm tissue-culture plate) was treated with 1 ml of antibiotic-free LHC-8e containing 1.5 μg of pBH-RANTES(-1006–1)-luc, 5 ng of a control reporter plasmid pRL-CMV (Promega) containing the Renilla reniformis luciferase gene, and 9 μg of GeneFECTOR (VennNova, Miami, FL) for 16 h at 37°C. Cells were washed with LHC-8e containing 0.5% BSA, and then treated without virus, with RSV MOI 1.0, or with UV-treated RSV in LHC-8e for 24 h at 37°C. Cell monolayers were lysed and luciferase-reporter gene activities were determined as described (8–11). Results were normalized to R. reniformis luciferase levels, and multiple DNA transfections (at least five experiments done in duplicate) were performed for each experimental condition to define consistent results.
mRNA Half-Life Analysis.
In initial experiments, RANTES mRNA half-life was determined in uninfected or RSV-infected hTBE cells (MOI 1.0 for 24 h) by treatment with actinomycin D (5 μg/ml) or α-amanitin (2 μg/ml) for 0–12 h. For each condition, poly(A)+ RNA was isolated by using PolyA Tract (Promega) and subjected to Northern blot analysis (2 μg/condition) as described above. In addition, the half-life of RANTES mRNA was determined by a pulse–chase method modified from one described previously (25). For this assay, monolayers of hTBE cells with or without concomitant RSV infection (MOI 1.0) were first incubated (pulsed) with [5,6-3H]uridine (35 Ci/mmol, 30 μCi/ml) in LHC-8e for 24 h at 37°C to label cellular RNA. Monolayers were rinsed twice with LHC-8e containing 20 mM glucosamine and 5 mM uridine and cytidine, and the radiolabel was chased by incubation in the same medium for 0, 1, 3, or 6 h to rapidly deplete radiolabeled precursors (26). At each time point, total cellular RNA was isolated as described above, and the radiolabeled RANTES transcripts were selected out of total radiolabeled RNA (2–5 × 108 dpm for each condition) by hybridization with filters slotted with RANTES sense and antisense cRNA as described above. Filters then were washed twice with 2× standard saline phosphate/EDTA (SSPE)/0.1% SDS at 65°C for 10 min and twice with 0.2× SSPE/0.1% SDS at 65°C for 10 min, and the amount of filter-bound radioactivity was quantified by liquid scintillation counting. The level of radiolabeled RANTES transcript at each time point was calculated as the amount of radioactivity bound to antisense cRNA minus the amount bound to sense cRNA and corrected for the total amount of radioactivity in each sample. These values also were expressed as a percentage of the initial value at the start of the chase period to calculate mRNA half-life.
Heterologous Promoter/Reporter Plasmids and RNase Protection Assay.
RANTES sequences encoding the complete mRNA (nucleotides −53 to 1160) (21, 27) or with portions of the coding sequence (nucleotides 1–276) and/or 3′ untranslated region (UTR) sequence (nucleotides 277-1160) deleted or reverse-complemented were cloned into pGL3-basic downstream to a potent heterologous promoter (cytomegalovirus, CMV) and in the context of a heterologous reporter gene fragment (P. pyralis luciferase nucleotides 1341–1649) as diagrammed below (see Fig. 4). Each construct included the same endogenous RANTES polyadenylation signal. The resulting plasmids were transfected into hTBE cells along with control reporter plasmid pRL-CMV, and 42 h later the cells were treated with RSV or inactivated RSV for an additional 24 h as described above. For each condition, the level of luciferase reporter mRNA was determined by a single-tube RNase protection assay (RPA II) from Ambion (Austin, TX). In this assay, total cellular RNA from hTBE cells first was coprecipitated with an excess of 32P-labeled single-stranded RNA probe generated by in vitro transcription with T7 RNA polymerase and the linearized luciferase cDNA fragment in pBluescript as template. Next, the cellular RNA/probe mixture was treated with RNaseA and cloned RNase T1, and the RNA (containing the protected fragment bound to probe) was precipitated and subjected to gel electrophoresis in 5% polyacrylamide under nondenaturing conditions. Signal was detected by autoradiography and quantified by using a PhosphorImager system. For each condition, similar transfection/expression efficiency was verified by determination of R. reniformis luciferase levels as well as plasmid copy number (using PCR for the P. pyralis luciferase fragment). In addition, as noted above, multiple transfections (five experiments done in duplicate) were performed for each experimental condition to define consistent results. In addition, an RNA probe for glyceraldehyde-3-phosphate dehydrogenase was used to verify RNA integrity and loading.
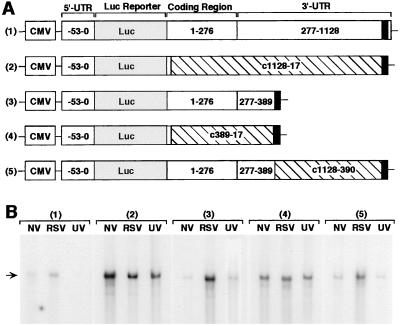
Figure 4.
Basal destabilizing and virus-induced stabilizing activity of RANTES mRNA sequence. (A) The construct designs are depicted for five reporter plasmids: 1) pGL3-CMV-RANTES(-53–1160)-luc, 2) pGL3-CMV-RANTES(-53–10/c1128–17)-luc, 3) pGL3-CMV-RANTES(-53–389)-luc, 4) pGL3-CMV-RANTES(-53–10/c389–17)-luc, and 5) pGL3-CMV-RANTES(-53–389/c1128–390)-luc. Plasmid components include: CMV promoter, P. pyralis luciferase (luc) reporter (309 bp), and RANTES 5′ UTR (nucleotide −53 to transcription start site), coding region (nucleotides 1–276), and 3′ UTR (nucleotides 277-1160) including the polyadenylation signal (nucleotides 1129–1154 represented as darkened box), and reverse-complement of RANTES sequence (denoted by c and crosshatched boxes). (B) hTBE cell monolayers were transfected with one of the six reporter plasmids and a control reporter plasmid pRL-CMV expressing R. reniformis luciferase, and 18–42 h later the transfected cells were treated with no virus (NV), RSV MOI 1.0 (RSV), or UV-inactivated RSV (UV) for an additional 24 h. For each condition, the level of luciferase reporter mRNA was determined by a single-tube RNase protection assay in which total cellular RNA (30 μg/condition) was isolated and hybridized with 32P-labeled RNA luc probe, and protection from RNase degradation was detected by 8% nondenaturing PAGE and autoradiography. Arrow indicates position of protected luciferase mRNA fragment. Transfection efficiency/expression (based on R. reniformis luciferase activity and plasmid copy number) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels (based on protection by an RNA probe for GAPDH) was similar for all treatment conditions (data not shown). Results are representative of five experiments.
RESULTS AND DISCUSSION
Selective RSV-Dependent Induction of RANTES Expression in Airway Epithelial Cells.
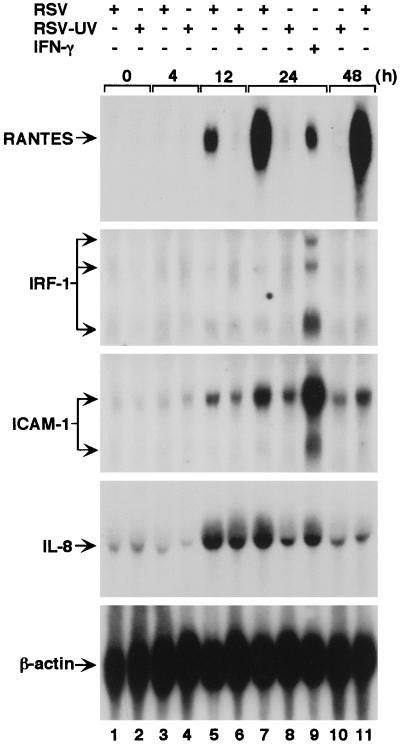
We observed time-dependent increases in RANTES mRNA and protein levels when primary-culture airway epithelial cells (hTBE cells) were infected with RSV (Fig. 1 and data not shown). The response is similar to ones observed by others for this cell type and by us and others for transformed bronchial epithelial cells, primary-culture nasal epithelial cells, and nasal washings from subjects with RSV but not non-RSV infection (refs. 28 and 29; E.S. and M.J.H., unpublished results) as well as airway epithelium in mice with paramyxoviral infection (15). Specificity of the response was demonstrated by the findings that RSV, but not adenovirus, infection was effective in stimulating RANTES expression and that expression of RANTES mRNA, but not of other β-chemokines (MIP-1α, MIP-1β, MCP-1, MCP-3, and eotaxin), was induced by infection (data not shown). In addition, related epithelial immune-response genes that (like RANTES) are regulated by IFN-γ-dependent signal transduction (8–10) were either unresponsive (for IRF-1) or less responsive (for ICAM-1) to RSV infection (Fig. 1).
Figure 1.
Time- and replication-dependent induction of RANTES expression after RSV infection in airway epithelial cells. Epithelial (hTBE) cell monolayers were uninfected (time 0) or infected with RSV or RSV rendered nonreplicative by UV irradiation (RSV-UV) at an MOI of 1.0 (or equivalent for RSV-UV) for 4–48 h, and total cellular RNA (20 μg/lane) was subjected to Northern blot analysis with RANTES, IRF-1, ICAM-1, IL-8, and β-actin cDNAs, and corresponding mRNAs were detected by autoradiography. Comparative responses to IFN-γ (100 units/ml for 24 h, lane 9) are shown as a positive control for IRF-1 expression. No signal was detected after hybridization with cDNAs for other β-chemokines: MCP-1, MCP-3, MIP-1α, or MIP-1β, or eotaxin (data not shown). Comparative responses for IL-8 mRNA served as a positive control for RSV-UV effect. Results are representative of five experiments.
RSV Replication Is Required for Induction of RANTES Expression.
The largest increase in RANTES mRNA (and protein) levels coincided with the replicative phase of RSV infection (continuing to increase through 48 h when cytopathic changes become evident), which suggested that viral replication was linked to inducible RANTES expression. To confirm that RSV induction of RANTES requires replication of the virus, hTBE cells were infected with UV-irradiated RSV that was unable to replicate. In contrast to untreated RSV, the UV-irradiated RSV exhibited a markedly decreased capacity to induce expression of RANTES mRNA and protein (Fig. 1 and data not shown), indicating that the RSV replication is essential for full induction of RANTES gene expression and excluding the possibility that viral stock solution components (RSV or non-RSV) induced the expression of RANTES in a manner described for other systems (30). This finding is also consistent with responsiveness of epithelial RANTES expression to IFN-γ but not other cytokines (because this medium does not contain IFN-γ) (13).
RSV Replication Is Not Required for Inducible Transcription of the RANTES Gene.
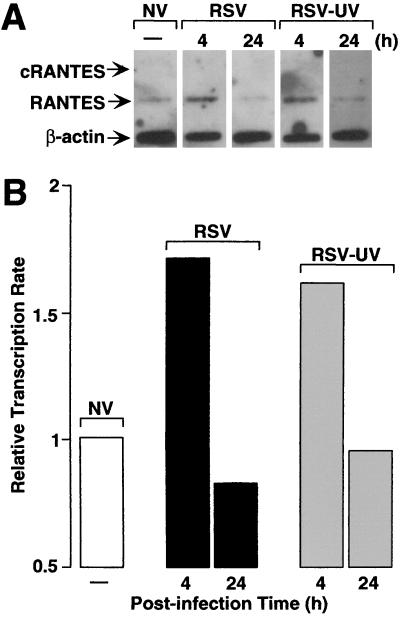
To begin to directly define the mechanism of RSV-dependent RANTES induction in airway epithelial cells, we quantified the level of nascent RANTES mRNA transcripts by nuclear run-on assay. Preliminary experiments indicated that the RANTES was transcribed at low levels compared with related immune-response genes (ICAM-1 and IRF-1) (8–11), so we increased the sensitivity of the nuclear run-on assay for RANTES by generating cRNA probes that permitted RNA/RNA hybridization (19). Under these hybridization conditions, we found low basal levels of RANTES transcripts as well as some degree of induction in response to replicating and nonreplicating RSV (Fig. 2). Transcription rate was increased approximately 2-fold after 4 h of infection either by intact or UV-irradiated RSV compared with uninfected cells and then declined to basal levels (or below basal levels) after 24 h of infection (when mRNA was actively accumulating). The increases in transcriptional initiation of the endogenous RANTES gene by replicating or UV-inactivated RSV were similar to increases in promoter activity of the 5′ flanking region of the RANTES gene fused to a luciferase reporter gene (fold increases in luciferase activities were 1.91 ± 0.08 and 1.88 ± 0.11 for RSV and RSV-UV treatment, respectively). Taken together, transcription rates for RANTES gene correlated poorly with inducible expression either in amount, duration, or requirement for viral replication, all suggesting that an additional posttranscriptional mechanism is responsible for induction of RANTES by RSV infection.
Figure 2.
RSV-dependent increases in RANTES expression detected by nuclear run-on assay. (A) RANTES sense or antisense cRNA or β-actin cDNA was bound to nitrocellulose filters and hybridized with 32P-labeled, nascent, run-off transcripts isolated from hTBE cell monolayers uninfected (NV) or infected with RSV (RSV) or UV-treated RSV (RSV-UV) for 4 h or 24 h at a MOI of 1.0. Filters then were subjected to autoradiography, and arrows indicate positions of mRNAs detected by sense RANTES cRNA (c-RANTES), antisense RANTES cRNA (RANTES), and β-actin cDNA (β-actin). (B) The same blots were subjected to quantitative PhosphorImaging, and results were expressed relative to background (value for sense RANTES cRNA) and corrected for internal standard (the level of β-actin) in each sample. Results are representative of three experiments.
Direct Analysis of RSV Effect on RANTES mRNA Half-Life.
To directly address RSV-dependent regulation of RANTES mRNA levels at a posttranscriptional level, we first attempted to determine the stability of RANTES mRNA by using the standard approach of treatment with a transcriptional inhibitor (actinomycin D) and subsequent timed determinations of the level of poly(A)+ RANTES mRNA. However, no decrease in basal or RSV-stimulated RANTES mRNA level was detected for up to 12 h after actinomycin D or α-aminitin treatment (data not shown). These findings (together with cycloheximide capacity to induce and superinduce RANTES mRNA levels; T.K. and M.J.H, unpublished results) suggest that newly transcribed mRNA(s) and subsequently translated protein(s) normally act to destabilize RANTES mRNA.
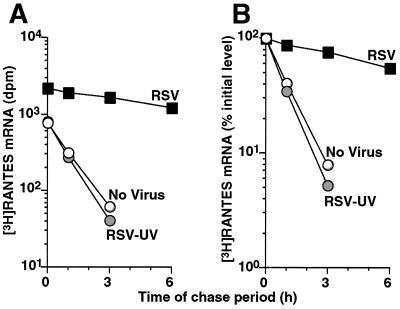
To avoid alterations of RANTES mRNA half-life by transcriptional inhibitors and to better evaluate mRNA stability under physiologic conditions, we modified previous methodology to determine RANTES mRNA half-life by using a pulse–chase method that relied on pulsed 3H-labeling of nascent RANTES transcripts followed by a chase period with timed determinations of labeled RANTES mRNA transcripts captured by RNA/RNA hybridization. By using this approach, we found that RSV prolonged RANTES mRNA half-life by 8-fold (from 0.8 h up to 6.8 h), thereby providing a mechanism for the selective effect of RSV on RANTES expression in airway epithelial cells (Fig. 3). This decay data (based on analysis of total cellular RNA) also suggests that the mature (cytosolic) RANTES mRNA (the predominant species of mRNA in this preparation) is the target of posttranscriptional regulation.
Figure 3.
Analysis of RSV-dependent increases in RANTES expression by mRNA half-life assay. Uninfected and RSV-infected (MOI 1.0 for 24 h) hTBE cell monolayers were concomitantly pulsed with 3H-uridine to label cellular RNA and then chased in medium containing unlabeled uridine and cytidine for the indicated times before isolating total cellular RNA. Radiolabeled RANTES was captured by blot hybridization with RANTES antisense cRNA. (A) The level of RANTES transcript at each time point was calculated as dpm bound to antisense cRNA minus dpm bound to sense cRNA corrected for total dpm in each sample. (B) These values were expressed as a percentage of the initial level at the start of the chase period (time 0) to calculate mRNA half-life.
Identification of RANTES mRNA Regions for Basal Destabilization and RSV-Dependent Stabilization.
To better define cis-acting elements in the RANTES mRNA that regulate basal destabilizing and/or virus-induced stabilizing functions, we next monitored the expression level of reporter plasmids containing RANTES gene sequences in the context of heterologous reporter (luciferase gene fragment) and promoter (CMV gene promoter) in airway epithelial cells. Plasmid constructs were designed to evaluate viral effect on native full-length RANTES sequence as well as sequences that were truncated or were truncated and then replaced with reversed and complementary sequence (Fig. 4). The latter strategy was designed to conserve native sequence at the DNA level (preserving DNA-protein interactions that influence gene transcription) but mutated at the mRNA level (disrupting mRNA-protein interactions that influence mRNA stability). By using this approach, RNase protection assays for reporter mRNA levels indicated that: (i) RSV infection (but not treatment with UV-inactivated RSV) increases mRNA levels when nucleotides 17–389 are included but not when this sequence is selectively mutated at the mRNA level (i.e., viral responsiveness is present in constructs 1, 3, and 5 but not in 2 and 4; Fig. 4); and (ii) basal mRNA levels increase when this same sequence (nucleotides 17–389) is selectively mutated at the mRNA level (i.e., basal levels of mRNA are increased in constructs 2 and 4 compared with 1, 3, and 5; Fig. 4). Decreased induction in this system relative to induction of endogenous mRNA detected in Northern blots (Fig. 1) likely reflects the low efficiency of transfection (only 1–2% of primary culture cells are transfected) as well as the loss of synergy with inducible activity of the endogenous promoter.
Nonetheless, because RSV (administered after plasmid transfection) does not influence plasmid transfection efficiency or transcription, the increased level of heterologous reporter mRNA provides evidence of virus-inducible mRNA stabilization conferred by RANTES mRNA sequence (nucleotides 17–389). Similarly, in conjunction with data for equivalent plasmid transfection efficiency and coexpression, the increase in basal levels of reporter mRNA also likely reflects mRNA stabilization mediated by this same sequence. Each of the constructs preserved the polyadenylation signal, indicating that the changes in mRNA levels were not influenced by this region. Instead, these results localize potential regulatory element(s) to RANTES sequence 17–389 (of the mature mRNA) that contains no previously identified consensus sites for mediating mRNA turnover in other genes (as discussed below).
Taken together, the present results indicate that posttranscriptional regulation is critical for inducible expression of immune-response genes during viral infection. In the particular case of RSV-stimulated RANTES production, the virus evokes early and transient gene transcription that is insufficient to confer gene expression but may provide additional substrate for a longer lasting and more robust increase in mRNA stability that requires viral replication and is necessary for full expression. In comparison to other types of viruses, host cells, and immune-response genes, the observed effect on mRNA stability appears distinct for RANTES. As noted above, there are many examples for viral regulation of host gene transcription, and in related examples from the Paramyxoviruses, the effect is mediated by DNA regulatory elements that bind NF-κB, IRF-1, ATF-2/c-Jun, and high mobility group protein HMG-I(Y) in the IFN-β gene or HMG-I(C) in the RANTES gene (3, 4, 6). In fact, we had assumed that NF-κB sites in the RANTES gene promoter (22, 23, 31) might be responsible for virus induction of RANTES gene expression. However, for this gene and others, the possibility of a posttranscriptional effect had not been determined directly and so was not excluded. In the case of RSV induction of RANTES expression, the degree of transcriptional activation could not possibly account for inducible gene expression, so we were prompted to define the critical role of regulation at a posttranscriptional level.
In contrast to what appear to be likely DNA-protein interactions for mediating transcriptional activation of host genes by viruses and other stimuli, little is known about viral mechanisms for controlling mRNA stability. In concomitant work on cytokine responsiveness of epithelial immune-response genes (including ICAM-1, IRF-1, and RANTES), we have noted that IFN-γ (like RSV) stimulates RANTES production via mRNA stabilization (32). It is therefore possible that RSV-driven signals for altering stability of RANTES mRNA may overlap with those for IFN-γ signal transduction. However, the available examples for IFN-γ-dependent increases in mRNA stability appear distinct from the characteristics of RANTES expression. For example, IFN-γ stabilization of ICAM-1 mRNA is mediated by a region of the translated sequence encoding the ICAM-1 cytoplasmic domain (33), but the RANTES gene (encoding a secreted protein) does not contain this sequence. Furthermore, this instability mechanism (in contrast to the one for RANTES) is uninfluenced by actinomycin D treatment. Similarly, IFN-γ up-regulates expression of the complement components C3 and C4 by stabilization of mRNA, but this system also is uninfluenced by transcriptional inhibition (34). In addition, the RANTES mRNA does not contain consensus sites for previously identified mRNA turnover elements, including AU-rich elements in other cytokine (as well as ICAM-1) mRNAs or UC-rich cleavage sites in gro α and 9E3 mRNAs (33, 35–37). Thus, the mechanism underlying basal instability as well as RSV-dependent stabilization of RANTES mRNA may be biochemically distinct from other genes.
The distinct action of RSV on RANTES production may confer a particular advantage for limiting infection in normal hosts as well as a disadvantage for subjects with pre-existing airway disease. In the case of normal hosts, the host cell apparently has engineered the direct effects of RSV to complement the stimulatory effect of IFN-γ to insure continuous expression of RANTES during infection. In fact, the concentrations of RSV causing RANTES mRNA stabilization are similar to concentrations that cause infection by inoculation of subjects (38) or those that are shed into the airway (39), suggesting that this same mechanism underlies inducible expression of RANTES found in vivo (28). The expression of RANTES in concert with other IFN-γ-inducible genes in the epithelium (notably ICAM-1) during viral infection (40) is likely to provide a more effective combination of chemoattractant and cell adhesion molecules for mediating epithelial-immune cell interaction. For example, determinations of the requirements for transepithelial T cell traffic in epithelial cell monolayers indicate a combined action of RANTES and ICAM-1 may be necessary for coordinated movement of immune cells from ablumenal to luminal surface (14). Thus, the complementary action of RANTES and ICAM-1 in coordinating immune cell movement is accompanied by complementary regulation of gene expression at transcriptional (for ICAM-1) and posttranscriptional (for RANTES) levels. The potent effects of RANTES on chemotaxis and activation of T cells (41) and other immune cells (notably eosinophils) (42) also may underlie the selective association of RSV infection with exacerbation of airway inflammatory diseases such as asthma (43). This possibility is underscored by the finding that asthma is associated with increased RANTES production (44), and that asthma exacerbation (even without viral infection) is accompanied by increased RANTES mRNA levels in airway epithelial cells (14). Thus, airway epithelial cells appear specially programmed for normal immune defense and abnormally programmed in asthma. The present results provide a molecular basis for how RANTES is overexpressed in the epithelial barrier as well as for next determining the precise RNA-protein interactions that might be targeted for modulation of β-chemokine expression and consequent immune cell activity during immunity and inflammatory disease.
Acknowledgments
We thank Douglas Dean, Michael Iademarco, and Steve Weintraub for helpful discussion and S. Brody, A. Deisseroth, M. Dustin, Y. Henderson, M. Krangel, A. Luster, G. Opdenakker, J. Oppenheim, B. Saha, T. Schall, D. Staunton, and C. Stratowa for generous gifts of reagents. This research was supported by grants from the National Institutes of Health, the Martin Schaefer Fund, and the Alan A. and Edith L. Wolff Charitable Trust.
ABBREVIATIONS
- CMV
cytomegalovirus
- hTBE cells
human tracheobronchial epithelial cells
- ICAM-1
intercellular adhesion molecule 1
- IRF-1
IFN regulatory factor 1
- LHC
Laboratory of Human Carcinogenesis
- MCP
monocyte chemoattractant protein
- MIP
macrophage inflammatory protein
- MOI
multiplicity of infection
- RANTES
regulated upon activation, normal T cell expressed and secreted
- RSV
respiratory syncytial virus
- UTR
untranslated region
References
- 1.Martin K J, Green M R. In: Transcriptional Regulation. McKnight S L, Yamamotos K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. , Part 2, pp. 695–725. [Google Scholar]
- 2.Hiscott J, Beauparlant P, Crepieux P, DeLuca C, Kwon H, Lin R, Petropoulos L. J Leuk Biol. 1997;62:82–92. doi: 10.1002/jlb.62.1.82. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka N, Taniguchi T. Adv Immunol. 1992;52:263–281. doi: 10.1016/s0065-2776(08)60877-9. [DOI] [PubMed] [Google Scholar]
- 4.Thanos D, Maniatis T. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Baruch A, Michiel D F, Oppenheim J J. J Biol Chem. 1995;270:11703–11706. doi: 10.1074/jbc.270.20.11703. [DOI] [PubMed] [Google Scholar]
- 6.Lokuta M A, Maher J, Noe K H, Pitha P M, Shin M L, Shin H S. J Biol Chem. 1996;271:13731–13738. doi: 10.1074/jbc.271.23.13731. [DOI] [PubMed] [Google Scholar]
- 7.Holtzman M J, Look D C, Sampath D, Castro M, Koga T, Walter M J. Proc Assoc Am Phys. 1998;110:1–11. [PubMed] [Google Scholar]
- 8.Look D C, Pelletier M R, Holtzman M J. J Biol Chem. 1994;269:8952–8958. [PubMed] [Google Scholar]
- 9.Look D C, Pelletier M R, Tidwell R M, Roswit W T, Holtzman M J. J Biol Chem. 1995;270:30264–30267. doi: 10.1074/jbc.270.51.30264. [DOI] [PubMed] [Google Scholar]
- 10.Walter M J, Look D C, Tidwell R M, Roswit W T, Holtzman M J. J Biol Chem. 1997;272:28582–28589. doi: 10.1074/jbc.272.45.28582. [DOI] [PubMed] [Google Scholar]
- 11.Look D C, Roswit W T, Frick A G, Gris-Alevy Y, Dickhaus D M, Walter M J, Holtzman M J. Immunity. 1998;9:871–880. doi: 10.1016/s1074-7613(00)80652-4. [DOI] [PubMed] [Google Scholar]
- 12.Taniguchi N, Mita G, Saito H, Yuo Y, Kajita T, Shida T. Allergy. 1985;40:571–573. doi: 10.1111/j.1398-9995.1985.tb00884.x. [DOI] [PubMed] [Google Scholar]
- 13.Koga T, Look D C, Taguchi M, Holtzman M J. Am J Respir Crit Care Med. 1997;155:A751. (abstr.). [Google Scholar]
- 14.Taguchi M, Sampath D, Koga T, Castro M, Look D C, Nakajima S, Holtzman M J. J Exp Med. 1998;187:1927–1940. doi: 10.1084/jem.187.12.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter M J, Kajiwara N, Sampath D, Rucker J, Holtzman M J. FASEB J. 1998;12:1453. (abstr.). [Google Scholar]
- 16.Sampath, D., Castro, M., Look, D. C. & Holtzman, M. J. (1999) J. Clin. Invest., in press. [DOI] [PMC free article] [PubMed]
- 17.Treuhaft M W, Beem M O. Infect Immun. 1982;37:439–444. doi: 10.1128/iai.37.2.439-444.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Look D C, Keller B T, Rapp S R, Holtzman M J. Am J Physiol. 1992;263:L79–L87. doi: 10.1152/ajplung.1992.263.1.L79. [DOI] [PubMed] [Google Scholar]
- 19.Flaspohler J A, Milcarek C. Biotechniques. 1992;13:68–72. [PubMed] [Google Scholar]
- 20.Srinivasula S M, Ahmad M, Ottilie S, Bullrich F, Banks S, Wang Y, Fernandes-Alnemri T, Croce C, Litwack G, Tomaselli K J, et al. J Biol Chem. 1997;272:18542–18545. doi: 10.1074/jbc.272.30.18542. [DOI] [PubMed] [Google Scholar]
- 21.Nelson P J, Kim H T, Manning W C, Goralski T J, Krensky A M. J Immunol. 1993;151:2601–2612. [PubMed] [Google Scholar]
- 22.Nelson P J, Ortiz B D, Pattison J M, Krensky A M. J Immunol. 1996;157:1139–1148. [PubMed] [Google Scholar]
- 23.Moriuchi H, Moriuchi M, Fauci A S. J Immunol. 1997;158:3483–3491. [PubMed] [Google Scholar]
- 24.Voraberger G, Schafer R, Stratowa C. J Immunol. 1991;147:2777–2786. [PubMed] [Google Scholar]
- 25.Hod Y, Hanson R W. J Biol Chem. 1988;263:7747–7752. [PubMed] [Google Scholar]
- 26.Levis R, Penman S. Cell. 1977;11:105–113. doi: 10.1016/0092-8674(77)90321-x. [DOI] [PubMed] [Google Scholar]
- 27.Schall T J, Jongstra J, Dyer B J, Jorgensen J, Clayberger C, Davis M M, Krensky A M. J Immunol. 1988;141:1018–1025. [PubMed] [Google Scholar]
- 28.Becker S, Reed W, Henderson F W, Noah T L. Am J Physiol. 1997;272:L512–L520. doi: 10.1152/ajplung.1997.272.3.L512. [DOI] [PubMed] [Google Scholar]
- 29.Saito T, Deskin R W, Casola A, Haeberle H, Olszewska B, Ernst P B, Alam R, Ogra P L, Garofalo R. J Infect Dis. 1997;175:497–504. doi: 10.1093/infdis/175.3.497. [DOI] [PubMed] [Google Scholar]
- 30.Garafolo R, Sabry M, Jamaluddin M, Yu R K, Casola A, Ogra P L, Brasier A R. J Virol. 1996;70:8773–8781. doi: 10.1128/jvi.70.12.8773-8781.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray P, Yang L, Zhang D-H, Ghosh S K, Ray A. J Biol Chem. 1997;272:20191–20197. doi: 10.1074/jbc.272.32.20191. [DOI] [PubMed] [Google Scholar]
- 32.Koga T, Look D C, Tidwell R M, Holtzman M J. Am J Respir Crit Care Med. 1998;157:A744. (abstr.). [Google Scholar]
- 33.Ohh M, Takei F. J Biol Chem. 1994;269:30117–30120. [PubMed] [Google Scholar]
- 34.Mitchell T J, Naughton M, Norsworthy P, Davies K A, Walport M J, Morley B J. J Immunol. 1996;156:4429–4434. [PubMed] [Google Scholar]
- 35.Stoeckle M Y, Hanafusa H. Mol Cell Biol. 1989;9:4738–4745. doi: 10.1128/mcb.9.11.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoeckle M Y. Nucleic Acids Res. 1992;20:1123–1127. doi: 10.1093/nar/20.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills J, Van Kirk J E, Wright P F, Chanock R M. J Immunol. 1971;107:123–130. [PubMed] [Google Scholar]
- 39.Hall C B, Douglas R G, Jr, Geiman J M. J Pediatr. 1976;89:11–15. doi: 10.1016/s0022-3476(76)80918-3. [DOI] [PubMed] [Google Scholar]
- 40.McWilliam A S, Marsh A, Holt P G. J Virol. 1997;71:226–236. doi: 10.1128/jvi.71.1.226-236.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bacon K B, Premack B A, Gardner P, Schall T J. Science. 1995;269:1727–1730. doi: 10.1126/science.7569902. [DOI] [PubMed] [Google Scholar]
- 42.Alam R, Stafford S, Forsythe P, Harrison R, Faubion D, Lett-Brown M A, Grant J A. J Immunol. 1993;150:3442–3447. [PubMed] [Google Scholar]
- 43.Pattemore P K, Johnston S L, Bardin P G. Clin Exp Allergy. 1992;22:325–336. doi: 10.1111/j.1365-2222.1992.tb03094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alam R, York J, Boyars M, Stafford S, Grant J A, Lee J, Forsythe P, Sim T, Ida N. Am J Respir Crit Care Med. 1996;153:1398–1404. doi: 10.1164/ajrccm.153.4.8616572. [DOI] [PubMed] [Google Scholar]