Abstract
Tom40 is the main component of the preprotein translocase of the outer membrane of mitochondria (TOM complex). We have isolated Tom40 of Neurospora crassa by removing the receptor Tom22 and the small Tom components Tom6 and Tom7 from the purified TOM core complex. Tom40 is organized in a high molecular mass complex of ∼350 kD. It forms a high conductance channel. Mitochondrial presequence peptides interact specifically with Tom40 reconstituted into planar lipid membranes and decrease the ion flow through the pores in a voltage-dependent manner. The secondary structure of Tom40 comprises ∼31% β-sheet, 22% α-helix, and 47% remaining structure as determined by circular dichroism measurements and Fourier transform infrared spectroscopy. Electron microscopy of purified Tom40 revealed particles primarily with one center of stain accumulation. They presumably represent an open pore with a diameter of ∼2.5 nm, similar to the pores found in the TOM complex. Thus, Tom40 is the core element of the TOM translocase; it forms the protein-conducting channel in an oligomeric assembly.
Keywords: TOM complex, Tom40, mitochondria, protein translocation channel, protein targeting
Introduction
Transport of nuclear-encoded proteins into mitochondria is mediated by distinct multisubunit translocation machineries located in the outer and inner membranes of mitochondria (Schatz and Dobberstein 1996; Neupert 1997; Voos et al. 1999). The translocase of the outer mitochondrial membrane (TOM complex) facilitates the recognition of preproteins, their transfer through the membrane, and the insertion of resident outer membrane proteins. Two translocation machineries in the inner mitochondrial membrane (TIM complexes), which are specific for different subsets of preproteins, mediate the transfer of preproteins across or into the inner membrane.
Biochemical and biophysical characterization of the TOM complex of Neurospora crassa and Saccharomyces cerevisiae revealed Tom40 as the key structural component of the protein-conducting channel in the mitochondrial outer membrane (Hill et al. 1998; Künkele et al. 1998a,Künkele et al. 1998b). Tom40 is an integral membrane protein that is essential for viability in yeast and Neurospora (Vestweber et al. 1989; Kiebler et al. 1990). Multiple copies of this protein are organized in the TOM core complex together with up to three small membrane-embedded subunits, Tom5 (Dietmeier et al. 1997), Tom6 (Kassenbrock et al. 1993; Alconada et al. 1995; Cao and Douglas 1995), and Tom7 (Hönlinger et al. 1996) and Tom22 (Kiebler et al. 1993; Lithgow et al. 1994; Hönlinger et al. 1995; Nakai and Endo 1995), a subunit with hydrophilic domains exposed to both sides of the outer membrane. The TOM holo complex in addition comprises the receptors Tom20 (Söllner et al. 1989; Ramage et al. 1993) and Tom70 (Hines et al. 1990; Söllner et al. 1990), single-spanning membrane proteins with hydrophilic domains exposed to the cytosol. They are only loosely attached to the TOM core complex (Dekker et al. 1998; Ahting et al. 1999).
The recent isolation and purification of the TOM holo complex of N. crassa has provided information about its composition, structure, and function as a protein-conducting channel. EM revealed particles, the majority of which contained two and three centers of stain accumulation (Künkele et al. 1998a). Electron tomography and three-dimensional image reconstruction of the TOM core complex yielded a map with two channels traversing the complex. Removal by proteolysis of the cytosolic or the intermembrane space domains of Tom22 did not interfere with the structural integrity of the complex of Neurospora (Ahting et al. 1999). On the other hand, in S. cerevisiae Tom22 was proposed to be crucial for the high level organization of the complex (van Wilpe et al. 1999).
Here, we report on the isolation and biochemical and biophysical characterization of a TOM subcomplex, consisting exclusively of Tom40 of N. crassa. Using circular dichroism (CD) and attenuated total reflection Fourier transform infrared spectroscopy (ATR-FTIR), we analyze the secondary structure of the protein and compare it with those of purified TOM core complex and the voltage-dependent anion channel (VDAC), the mitochondrial porin. The results provide insights into the secondary structure of Tom40, revealing both β-sheet and α-helical elements. Electrophysiological measurements suggest Tom40 to be the essential element in the formation of the pore. Analysis of purified Tom40 by EM and image analysis reveals particles, most of which contain one center of stain accumulation. The results indicate that Tom6, Tom7, and Tom22 are important for the stability of the TOM complex but play only a relatively minor structural role in the formation of the protein translocation channel.
Materials and Methods
Cell Growth and Isolation of Mitochondria
TOM core complex was isolated and purified from mitochondrial membranes of an N. crassa strain (GR-107) containing a hexahistidinyl-tagged form of Tom22 as described previously (Künkele et al. 1998a; Ahting et al. 1999). Cells were grown overnight under vigorous aeration in 100 liters Vogel's minimal medium supplemented with 1.3 mM histidine and 2% (wt/vol) sucrose at 27°C. The cultures were inoculated with 109 conidia per liter and grown under bright illumination. Hyphae were harvested by centrifugation, cooled on ice, and homogenized in a Waring blender with quartz sand in SEM buffer (0.2 M sucrose, 1 mM EDTA, 10 mM MOPS, 1 mM PMSF). After passing the cells through a corundum mill, sand and cellular debris were removed by two centrifugation steps for 5 min at 3,000 g. Mitochondria were sedimented by centrifugation for 50 min at 17,700 g, washed by resuspending them in SM buffer (0.2 M sucrose, 10 mM MOPS, 1 mM PMSF, complete protease inhibitor cocktail; Roche). After a second centrifugation step for 50 min at 17,700 g, mitochondria were resuspended in SM buffer at a protein concentration of ∼50 mg/ml and stored in aliquots at −20°C.
Isolation and Purification of TOM Core Complex
TOM core complex was purified as described previously (Ahting et al. 1999) with minor modifications. In brief, isolated mitochondria were solubilized in 50 mM potassium acetate, 10 mM MOPS, pH 7.0, 20% glycerol, and 1% (wt/vol) n-dodecyl β-d-maltoside (DDM; Anatrace, Inc.) in the presence of 1 mM PMSF and a cocktail of protease inhibitors at a protein concentration of 10 mg/ml for 30 min at 4°C. Insoluble material was removed by centrifugation, and the clarified extract was loaded onto a nickel nitrilotriacetic acid agarose column (Ni-NTA; QIAGEN) using 4 ml resin per 1 g of total mitochondrial protein. The column was washed with 20 column volumes of a buffer containing 50 mM potassium acetate, 10 mM MOPS, pH 7.0, 20% glycerol, 0.1% DDM, and 40 mM imidazole. Specifically bound material was eluted with 300 mM imidazole in the same buffer. Fractions containing TOM core complex were pooled and loaded onto a Resource Q anion exchange column (Amersham Pharmacia Biotech) equilibrated with 50 mM potassium acetate, 10 mM MOPS, pH 7.0, 20% glycerol, and 0.1% DDM. The complex was eluted by a linear 0–500 mM KCl gradient in the same buffer. Stock solutions of purified TOM core complex were stored at a protein concentration of ∼5 mg/ml at 4°C.
Preparation of Tom40
For the isolation of Tom40, purified TOM core complex (1–5 mg) was reloaded onto an Ni-NTA affinity column (1 ml resin) equilibrated with 50 mM potassium acetate, 10 mM MOPS, pH 7.0, 10% glycerol, and 0.1% (wt/vol) DDM at 4°C. The column was washed with two column volumes of equilibration buffer. Tom40 and Tom7 were eluted with 3% (wt/vol) n-octyl β-d-glucopyranoside (OG; Fluka) in the same buffer using a column flow rate of 0.05 ml/min. 10 column fractions of a volume of 1 ml were collected. Specifically bound material was eluted with 300 mM imidazole in the same buffer containing 1% (wt/vol) OG. To prevent Tom40 from aggregation, the fractions containing Tom40 were supplemented with 0.5% (wt/vol) DDM (final concentration) and subsequently loaded onto a sucrose gradient (7–35%) containing 50 mM potassium acetate, 10 mM MOPS, pH 7.0, and 0.5% (wt/vol) DDM and centrifuged overnight to replace OG with DDM. Alternatively, detergent exchange was performed by anion exchange chromatography using a Resource Q column (Amersham Pharmacia Biotech). Isolated Tom40 was stored in 0.5% DDM at a protein concentration of 0.8–1 mg/ml at 4°C. The purity of protein was assessed by denaturing gel electrophoresis (Laemmli 1970).
Purification of Mitochondrial Porin
Mitochondrial porin (VDAC) was isolated from N. crassa according to Freitag et al. 1982. In brief, mitochondrial outer membrane vesicles (1 mg protein per ml) were prepared as described by Künkele et al. 1998a and solubilized by incubation in 1% DDM, 50 mM potassium acetate, pH 7.0, 10 mM MOPS, 20% glycerol, and 1 mM PMSF at 4°C for 30 min. Insoluble material was removed by centrifugation at 226,200 g, and the supernatant was passed over a Resource Q anion exchange column (1 ml resin; Amersham Pharmacia Biotech). Mitochondrial porin was recovered from the flow through fraction and stored at 4°C at a protein concentration of ∼3 mg/ml.
Size Exclusion Chromatography
For determination of the molecular mass of Tom40, 100 μg of protein was loaded onto a Superose 6 size exclusion column (Amersham Pharmacia Biotech) equilibrated with 50 mM potassium acetate, 10 mM MOPS, pH 7.0, 10% glycerol, and 0.1% DDM at 4°C. Protein was eluted at a flow rate of 0.5 ml/min. Control experiments were carried out using purified TOM core complex. The molecular masses of Tom40 complexes were estimated using thyroglobulin (669 kD), apoferritin (443 kD), alcohol dehydrogenase (155 kD), and carboanhydrase (29 kD) as protein standards.
CD Spectroscopy
CD measurements were performed using a Jasco J-715 spectrometer in quartz cuvettes of 0.1-cm path length. Spectra were recorded at 4°C from 198 to 250 nm with a resolution of 0.1 nm and an acquisition time of 50 nm/min. Final CD spectra were obtained by averaging 10 consecutive scans and corrected for background by subtraction of spectra of protein-free samples recorded under the same conditions. Mean ellipticities per residue ΘR were calculated based on the molar protein concentration and the amino acid composition (Tom40 and mitochondrial porin) or based on the absolute protein concentration and a mean residue molecular weight of 113 (TOM core complex). The concentrations of Tom40 and mitochondrial porin were determined by ultraviolet absorbance spectroscopy after unfolding of protein in 7.2 M urea and using extinction coefficients εW,280nm ∼ 5,600 M−1cm−1 for tryptophan and εY,280nm ∼ 1,200 M−1cm−1 for tyrosine (Pace et al. 1995). Secondary structure predictions were based on algorithms by Sreerama and Woody 1993 using the Dicroprot version 2.5 software package by G. Deleage (CNRS).
ATR-FTIR
ATR-FTIR was performed using a Nicolet 740 FT spectrometer. The spectrometer was purged continuously with nitrogen gas to remove water vapor. The internal reflection element was a germanium crystal (ATR plate) with an aperture angle of 45°. Purified TOM core complex, Tom40, or mitochondrial porin was dialyzed against 5 mM phosphate buffer at 4°C. 50–100 μg of protein was applied onto one side of the glow-discharged ATR plate and taken to dryness under a stream of nitrogen. The ATR plate was sealed in a home built sample holder. Infrared (IR) spectra were recorded from 600 to 4,000 cm−1 at a resolution of better than 2 cm−1. The data are means of 1,024 scans. To differentiate α-helical components from random coil, the sample compartment was flushed with D2O-saturated nitrogen for 120 min at room temperature. This shifted the absorption peak corresponding to the random coil structure elements from ∼1,655 to ∼1,642 cm−1. The exchange of hydrogen with deuterium was monitored by repeated measurements and judged by decrease of the amide band II centered around 1,530 cm−1. The content of β-sheet, random coil, and α-helical secondary structure elements was estimated by analyzing the amide I region between 1,600 and 1,700 cm−1 using Fourier self deconvolution according to Kauppinen et al. 1981 for determining the position of absorption bands, and constrained band fitting to original spectra essentially by following the approach described by Byler and Susi 1986. Spectra of mitochondrial porin revealed a significant contribution of residual lipid, indicated by carbonyl ester vibrations ∼1,730 cm−1. These absorptions were fitted and subtracted from porin spectra before further quantitative analysis.
Electrophysiological Procedures
Conductance measurements of Tom40 in planar black lipid membranes were carried out as described previously (Benz et al. 1978; Künkele et al. 1998a). Membranes were formed from a 1% (wt/vol) solution of diphytanoyl phosphatidyl choline (Avanti Polar Lipids) in n-decane/butanol (9:1 vol/vol) across circular holes (surface area ∼0.1 mm2) in the wall of a Teflon cell separating two aqueous compartments of 5 ml each. The aqueous solutions contained 1 M KCl, 5 mM Hepes-KOH, pH 7.0 (σ0 = 96.7 mS cm−1). To improve the insertion of protein into the lipid membranes, purified TOM core complex and Tom40 were mixed with an aqueous suspension of ergosterol (Fluka) before addition to the aqueous phase bathing the black lipid membrane. Membrane currents were measured at a membrane potential of +20 mV with a pair of Ag/AgCl electrodes (Metrohm) using a 428-current amplifier (Keithley Instruments, Inc.). Amplified signals were monitored with an analogue/digital storage oscilloscope (HM 407; Hameg) and recorded with a strip chart recorder. Single channel analysis was carried out according to previously described methods (Künkele et al. 1998b). Voltages are given as Vcis−Vtrans.
EM
Purified Tom40 (∼0.1 mg protein/ml) was adsorbed to glow-discharged carbon-coated grids (Cu, 600 mesh) for 30 s. The grids were washed twice with deionized water, blotted with filter paper, and stained with 2% (wt/vol) uranyl acetate for 30–60 s. EM images of Tom40 were recorded using a Philips CM 12 electron transmission microscope equipped with a VIPS computer (TVIPS) and a large area CCD camera (Photometrics). The microscope was operated at 120 kV. Images were taken at an underfocus of ∼1.5 μm, a nominal EM magnification of 35.000× and a postmagnification factor of 1.934 on the CCD camera. This corresponded to a pixel size of 0.355 nm at the specimen.
Single particle image processing was carried out on a Silicon graphics workstation using the EM software (Hegerl 1996). Images were low-pass filtered to the first zero of the electron microscope transfer function corresponding to a cutoff frequency of ∼2.3 nm−1. A total of 1,550 particles were manually marked in the digitized images. After extraction of frames with 64 × 64 pixels, images were subjected to multireference alignment using synthetic model images with one, two, three, and four pores as first references (Frank et al. 1981). Particle images of the two most prominent classes were resubjected to separate alignment, multivariate statistical analysis (Frank and van Heel 1982), and averaging. 20 eigenimages of each class were calculated. Each data set was subsequently split into 20 groups using the 6 most significant eigenimages.
Results
Isolation of Neurospora Tom40
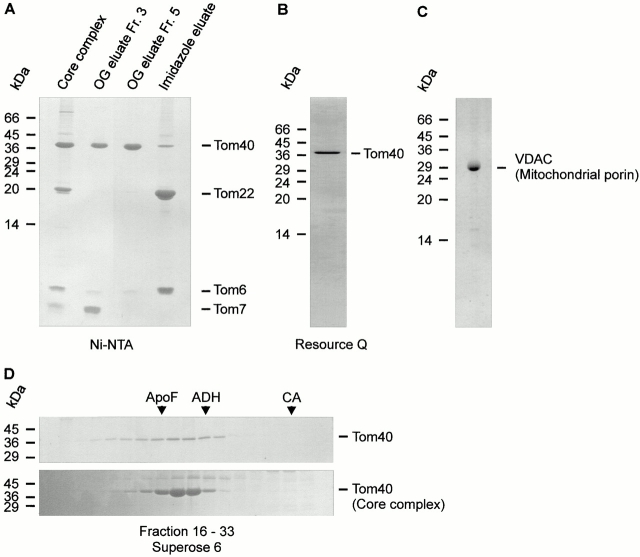
Incubation of purified TOM core complex with OG at concentrations >3% (wt/vol) led to the dissociation of the core complex into the individual components Tom40, Tom22, Tom7, and Tom6. This was exploited to isolate Tom40. Purified TOM core complex containing a Tom22 with an oligohistidinyl tag at its COOH terminus was bound to an Ni-NTA affinity column in 0.1% (wt/vol) DDM. Fig. 1 A shows the main fractions analyzed for polypeptide composition after elution of proteins successively with OG and imidazole. The initial OG fractions contained nearly all of Tom40 and Tom7, then virtually pure Tom40 was eluted. Tom22 and Tom6 eluted upon inclusion of imidazole into the buffer. A component corresponding to yeast Tom5 was not detected in the TOM core complex (Dembowski et al. 2001) or in Tom40 preparations by Coomassie blue or silver staining. To stabilize Tom40 in DDM buffer and to prepare Tom40 without contamination by Tom7, fractions obtained by elution with OG were pooled and subjected to anion exchange chromatography (Fig. 1 B). Size exclusion chromatography of purified Tom40 exhibited a peak in the high molecular mass range corresponding to ∼350 kD (Fig. 1 D), indicating that Tom40 is organized in a high molecular mass complex similar to the TOM core complex.
Figure 1.
Purification of Tom40. (A) Purified TOM core complex carrying Tom22 with a hexahistidinyl tag was solubilized in 0.1% DDM and bound to an Ni-NTA affinity column. Tom40 and Tom7 were eluted with 3% (wt/vol) OG; Tom22 and Tom6 were eluted with 300 mM imidazole. Aliquots of the resulting column fractions were analyzed by high Tris/urea SDS-PAGE and staining with Coomassie blue. (Lane 1) TOM core complex; (lanes 2 and 3) column fractions 3 and 5 of the OG eluate; (lane 4) column fraction 3 of the imidazole eluate. (B) Fractions containing Tom40 were pooled and further purified by anion exchange chromatography on a Resource Q column equilibrated with 0.5% DDM. The peak fraction of the column was analyzed by high Tris/urea SDS-PAGE and Coomassie staining. Tom7 and residual amounts of Tom6 were removed completely from Tom40 after passage over the anion exchange column. (C) SDS-PAGE of VDAC isolated from N. crassa mitochondria. The gel was stained with Coomassie blue. (D) Size exclusion chromatography of isolated Tom40. Tom40 purified by anion exchange chromatography was subjected to gel filtration on a Superose 6 column equilibrated with 0.1% DDM. Column fractions (16–33) were analyzed by SDS-PAGE and staining with Coomassie (top). For comparison, gel filtration analysis of purified TOM core complex is shown at the bottom. Protein M r standards: ApoF, apoferritin (M r 443,000); ADH, alcohol dehydrogenase (M r 155,000); CA, carboanhydrase (M r 29,000).
Structure of Isolated Tom40
Does the structure of purified Tom40 resemble that of the TOM complex? EM images of negatively stained Tom40 particles were diverse (Fig. 2 A). Nevertheless, there were several molecules with one and two stain-filled openings or pores. A total of 1,550 projections were extracted. Multireference alignment procedures (Frank et al. 1981) using synthetic reference images corresponding to particles with one, two, three, and four pores were used to eliminate poorly defined images of Tom40 and to separate the main groups of Tom40 particle images. The class averages contained predominantly one and two pores. Using multivariate statistical analysis (Frank and van Heel 1982), the data of the most prominent one and two pore classes were each broken up into 20 groups using the 6 most significant eigenimages. The group averages are shown in Fig. 2B and Fig. C. Compared with TOM core complex (Ahting et al. 1999), preparations of Tom40 revealed mainly one ring structures (n = 560). Particles with two centers of stain accumulation (n = 114) were present but much less frequent (∼7%) than one pore particles (∼36%). This suggests that the components Tom22, Tom7, and Tom6 are not necessary for the formation of the two pore form of the TOM complex but help to stabilize this form. The pore size of the Tom40 particles ranged between 2 and 3 nm and was comparable with that of the TOM core complex and the TOM holo complex (Ahting et al. 1999).
Figure 2.
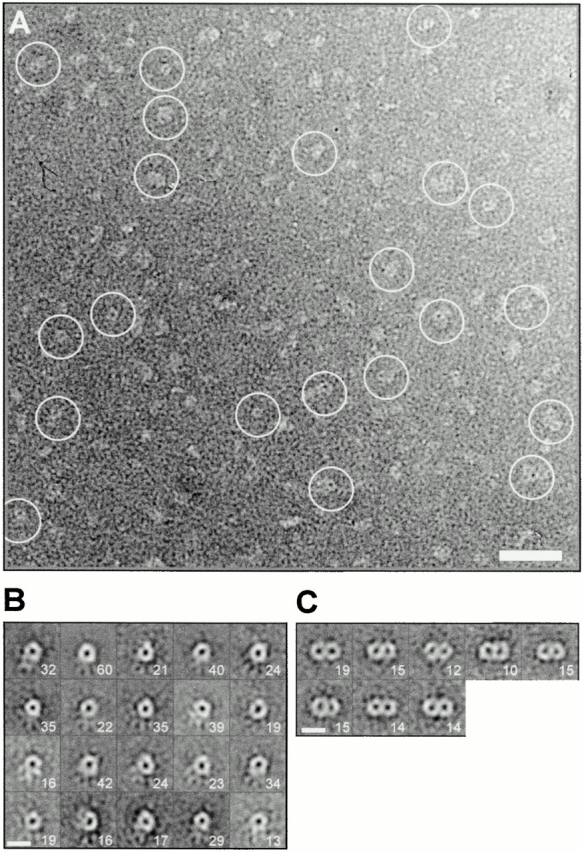
EM and projection map of purified Tom40. (A) Survey view of negatively stained Tom40 particles. The image was filtered to the first zero of the electron microscope transfer function. (B and C) Statistical analysis of Tom40 particles. From electron micrographs a total of 1,550 Tom40 particles were extracted and subjected to multireference alignment. Based on eigenimage analysis, the data sets of the two most prominent groups were split into 20 classes. (B) Class averages of one pore particle images. (C) Group averages of the two pore classes. The numbers given for a specific class average represent the number of merged particle images. Group averages containing <10 particle images were omitted. Bars: (A) 20 nm; (B and C) 10 nm.
Secondary Structure of Tom40
The secondary structure of Tom40 in detergent solution was analyzed by two different methods. CD and IR spectra of purified Tom40 were recorded and compared with those of TOM core complex and VDAC, the mitochondrial porin (Fig. 3 and Fig. 4). Table summarizes the characteristics of the CD spectra in terms of the minimum value (λmin), the wavelength at which the ellipticity equals zero (λcrossover), and the spectral deconvolution results. Tom40 revealed a spectrum with a crossover of the baseline at 202 nm and a minimum at 216.5 nm (Fig. 3 A). At wavelengths >245 nm, the CD spectrum approached ellipticity values close to zero, indicating that the Tom40 preparation was virtually free of higher order aggregates, which would cause light scattering effects and interfere with the interpretation of the data. CD measurements of the TOM core complex yielded a spectrum with additional local minima at 208 and 222 nm, which are characteristic of α-helical structure (Fig. 3 B). Both spectra were markedly different from that of mitochondrial porin (Fig. 1 C), which yielded a spectrum with a large positive ellipticity <206 nm and a less intense minimum centered at 216 nm (Fig. 3 C). The content of α-helix of Tom40 was higher than proposed previously on the basis of structure predictions (Court et al. 1995; Mannella et al. 1996).
Figure 3.
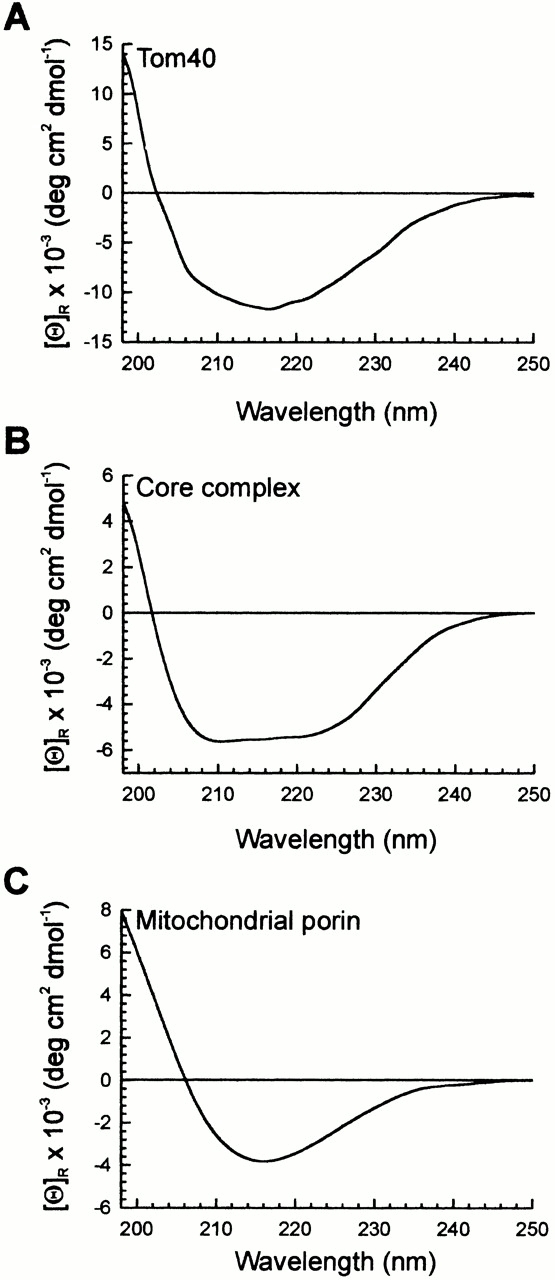
Far ultraviolet CD spectra of Tom40, TOM core complex, and mitochondrial porin isolated from N. crassa mitochondria. (A) Tom40; (B) TOM core complex; and (C) porin. Protein was solubilized in 0.1% DDM, 50 mM potassium-acetate/MOPS, pH 7.0, 10% glycerol. 10 scans were accumulated at 4°C. The protein concentrations of Tom40 (2.5 μM) and mitochondrial porin (6.9 μM) relevant for computing the molar ellipiticities ΘR were determined by UV absorbance spectroscopy. The protein concentration of the TOM core complex was determined using a colorimetric assay.
Figure 4.
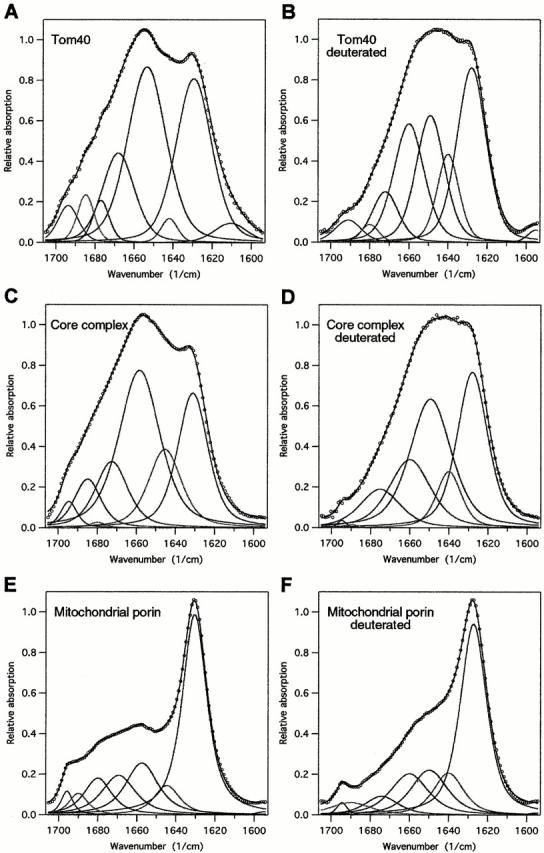
Original and deconvoluted FTIR spectra of Tom40, TOM core complex, and mitochondrial porin. The spectra of Tom40 (A), TOM core complex (C), and porin (E) were recorded on thin films on Ge crystals applying the ATR approach. After the end of measurements, the films were exposed to D2O-saturated nitrogen gas for 120 min, and spectra of deuterated protein were recorded (B, D, and F) in order to separate contributions of α-helix from random coil components. Spectral bands assigned to α-helix structure are centered at ∼1,650 cm−1, random components at 1,645–1,640 cm−1, and β-sheet at 1,630–1,625 cm−1. The shoulder at 1,695 cm−1 indicates antiparallel β-sheet with particularly short turns. For all spectra, the baseline and residual water vapor components were subtracted if necessary.
Table 1.
Comparison of the CD Spectrum of Tom40 with that of TOM Core Complex and Mitochondrial Porin
| Protein | λ crossover | λ min | ΘR min | α-Helix | β-Sheet + turn | Other |
|---|---|---|---|---|---|---|
| nm | 10−3 deg cm2 dmol−1 | % | % | % | ||
| Tom40 | 202 | 216.5 | −11.7 | 32 | 22 | 46 |
| TOM core complex | 202 | 210 and 226 | −5.6 | ND | ND | ND |
| VDAC (mitochondrial porin) | 206 | 216.0 | −3.4 | 15 | 46 | 34 |
The secondary structure estimates were computed from CD spectra of Tom40 and mitochondrial porin of N. crassa in 0.1% DDM as described in Materials and Methods. The content of α-helix and β-sheet was estimated for Tom40 and mitochondrial porin only, since the protein concentration of the TOM core complex could not be determined with the required accuracy.
The IR spectra of Tom40 and TOM core complex revealed peak signals in the amide I vibrational frequency region centered between 1,629 and 1,680 cm−1, which are characteristic of α-helical, random coil, and β-sheet structure elements (Fig. 4, A–D). Evaluation of this frequency region by Fourier self deconvolution and curve fitting revealed for Tom40 and the TOM core complex a content of β-sheet structure of ∼31 and 30%, respectively (Table ). This was significantly less than that of mitochondrial porin of Neurospora (Fig. 4 E). The shape of the amide I band of porin was typical of proteins with a high content of antiparallel β-sheet. Also for Tom40, the spectral component at 1,695 cm−1 indicated the existence of antiparallel β-strands. Analysis of the spectra of deuterated proteins (Fig. 4B and Fig. D) resulted in estimates of ∼22 and 33% α-helical structure for Tom40 and the TOM core complex. In agreement with the CD measurements, the content of α-helical structure of Tom40 was again larger than that of mitochondrial porin (Fig. 4 F). The amount of β-sheet structure of Tom40 based on CD was less than that calculated from IR spectroscopy. It should be noted that the prediction of β-strand structures by CD measurements tends generally to underestimate β-sheet relative to α-helical structures.
Table 2.
Secondary Structure of Tom40, TOM Core Complex, and Mitochondrial Porin Estimated from IR Spectra
| Protein | H2O/D2O | α-Helix | β-Sheet | Other | Position of the β-signal |
|---|---|---|---|---|---|
| % | % | % | cm−1 | ||
| Tom40 | H2O | 31 | 1,629.4 | ||
| Tom40 | D2O | 22 | 31 | 47 | 1,628.0 |
| TOM core complex | H2O | 24 | 1,631.1 | ||
| TOM core complex | D2O | 33 | 30 | 37 | 1,628.0 |
| VDAC (mitochondrial porin) | H2O | 48 | 1,630.5 | ||
| VDAC (mitochondrial porin) | D2O | 15 | 48 | 37 | 1,627.3 |
Channel Properties of Tom40
To test whether isolated Tom40 is functional and forms pores, we analyzed its channel-forming activity after reconstitution into planar lipid membranes. Fig. 5 A shows a current trace obtained for purified Neurospora Tom40 recorded in 1 M KCl at a membrane potential of +20 mV. The single channel distribution of 114 insertion events showed a characteristic maximum at ∼2.8 nS (Fig. 5 B). The mean conductance of isolated Tom40 insertions was comparable to those of the TOM holo complex (Künkele et al. 1998a), the TOM core complex, and protease-treated core complex lacking the hydrophilic domains of the receptor component Tom22 (Ahting et al. 1999).
Figure 5.
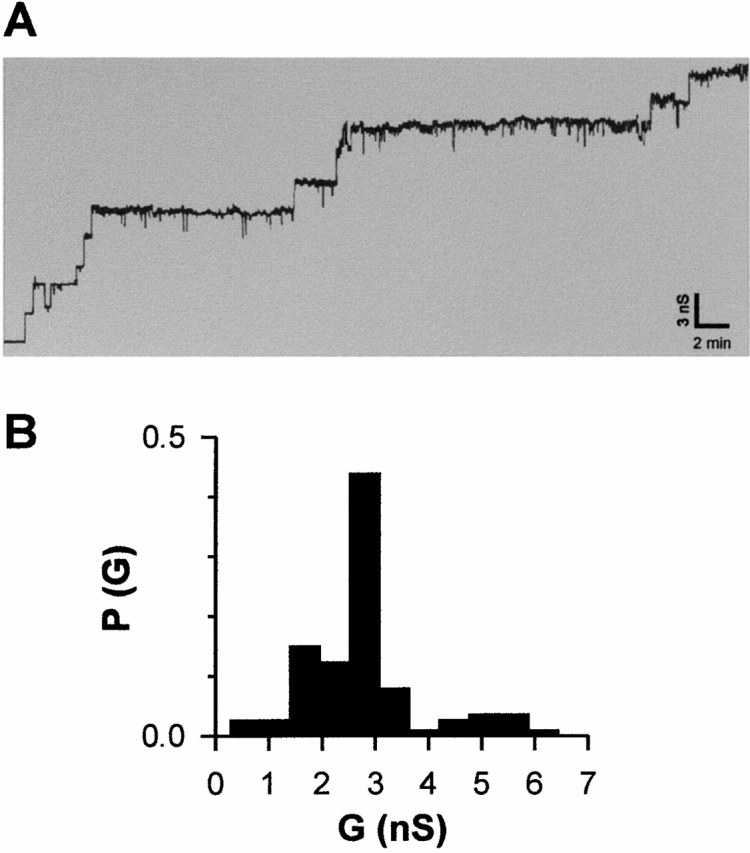
Single-channel recording of isolated Tom40. (A) Purified Tom40 (∼4 μg/ml final protein concentration) was added to both sides of a black lipid membrane formed by diphytanoyl phosphatidyl choline/n-decane/butanol, and single channel conductances were measured in the presence of a membrane potential of +20 mV. (B) Histogram of channel conductances. P(G) is the probability that a given conductance increment G is observed. A total of n = 114 conductance increments were analyzed. The aqueous phase contained 1 M KCl, 5 mM Hepes, pH 7.0.
Tom40 and TOM core complex channels were further characterized in single channel records (Fig. 6). They were either directly integrated into the bilayer by adding detergent-solubilized protein to the bath solution or they were reconstituted into proteoliposomes, which were subsequently fused to lipid bilayers. The channels described previously for the core complex showed cation selectivity and were characterized by three main conductance levels separated from the fully open state (1,100 pS in 150 mM KCl) by two identical jumps of 440 pS (Fig. 6 A). At ∼0 mV, they were fully open, and they closed with slow kinetics at potentials of either polarity. In addition, a fast flicker between the three main conductance levels occurred only at voltages of one polarity around 70 mV (Fig. 6 A). These characteristics are similar to those of the dimeric peptide-sensitive channel (PSC) identified in outer membrane and holo complex fractions of Neurospora (Künkele et al. 1998b). The channels most often found in the Tom40 fraction had similar selectivity and voltage-dependence properties, but their maximum conductance (550 pS in 150 mM KCl) was half that of the channels described above. They exhibited only two main conductance levels separated by jumps of 440 pS and a fast flicker at voltages of one polarity (Fig. 6 B). They are thus similar to the monomeric form of the Neurospora PSC described previously (Künkele et al. 1998b). This form was also found in the core complex fraction but with a lower probability than the dimeric form. These results indicate that Tom40 can form the protein translocation channel of the mitochondrial outer membrane.
Figure 6.
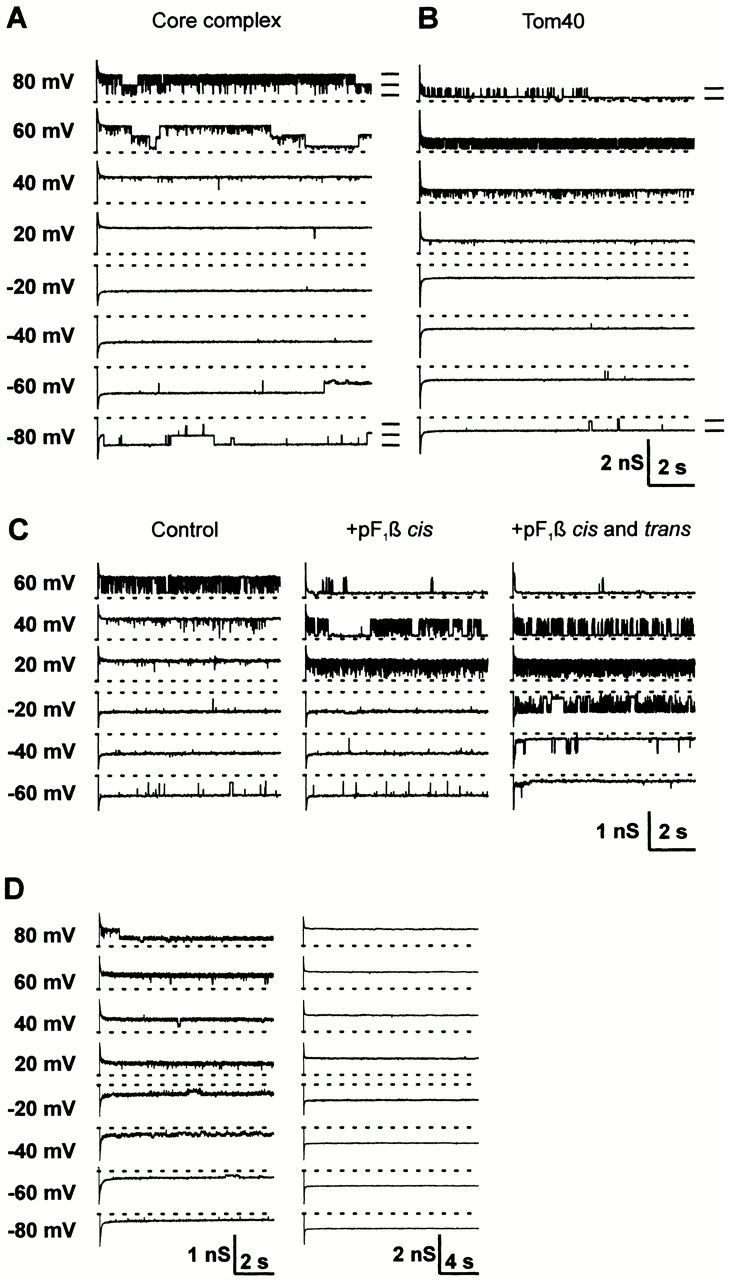
Properties of single channels of Tom40 and purified TOM core complex. Samples of current traces of channels of TOM core complex (A) and purified Tom40 (B). Currents were recorded after voltage jumps from 0 mV to the voltages indicated on the left of the traces. The main conductance levels are indicated on the right of the traces recorded at +80 and −80 mV. (C) Blockade of a channel from purified Tom40 by a mitochondrial presequence peptide. Currents were recorded after voltage jumps from 0 mV to the voltages indicated on the left of the traces. Left, control, before peptide addition; middle, after addition to the cis (cytosolic) side of a peptide corresponding to the first 32 residues of yeast pF1β (final concentration 0.5 μM); right, after further addition of the same peptide to the trans (intermembrane space) side (final concentration 1 μM). The orientation of the channels in the bilayer was determined from the polarity of the membrane potential at which the characteristic flicker occurred (Künkele et al. 1998b). (D) Properties of nontypical channels formed by purified Tom40. Samples of current traces recorded after voltage jumps from 0 mV to the voltages indicated on the left of the traces. The first type (right) is voltage dependent and exhibits rectification. The second type (right) does not rectify and is not voltage dependent. Both types are devoid of the characteristic flicker. The dashed lines represent the 0 pA levels. For all records, the cis and trans compartments contained 150 mM KCl, 10 mM Hepes, pH 7.0. Data were sampled at 400 Hz and filtered at 200 Hz.
The channel formed by purified Tom40 is blocked by mitochondrial presequences. Purified Tom40 was inserted into bilayers, and single channels were recorded at different voltages before and after addition of a synthetic peptide corresponding to the first 32 residues of the precursor of S. cerevisiae F1-ATPase β-subunit (pF1β) to one or both compartments (Fig. 6 C). pF1β reduced the channel open state probability in a voltage-dependent manner. Control peptides had no effect on the channel activities (data not shown). The channels formed by the TOM core complex were blocked in a similar manner by pF1β (data not shown). This blockade has been reported previously also for TOM holo complex channels (Künkele et al. 1998b).
When Tom40-containing proteoliposomes were fused to bilayers, not only monomeric PSC-type channels were observed but, with a frequency of <25% of all channels recorded, two other types of channels were observed (Fig. 6 D), which were not seen using holo or core complex proteoliposomes. Like PSCs, both types had cationic selectivity, but their maximum conductance and voltage dependence were different. The first type was characterized by multiple conductance levels and rectification. The second type was a pore devoid of voltage dependence. Differing from the PSCs, the two types of channels were not blocked by pF1β at the concentration of 1 μM (data not shown). A likely explanation for the occurrence of these unusual channels is the relative instability of Tom40 compared with the very stable TOM core complex; this could lead to the formation of nonnative Tom40 channels and nontypical Tom40 particles in the electron microscopic pictures in the purified Tom40 preparations (see Fig. 2).
Discussion
We have isolated Tom40 from purified TOM core complex of N. crassa and analyzed the pore-forming activity of this protein. Channels of Tom40 were recorded, which were very similar to those found with both TOM core complex and TOM holo complex. This supports the view that Tom40 is the central constituent of the protein-conducting channel of the TOM complex. The characteristic gating properties of Tom40, TOM core complex, and TOM holo complex point to common elements in the protein translocation channel that sense the electric field in planar lipid membranes. On the other hand, channels exhibiting different properties were recorded together with classical PSCs (Thieffry et al. 1992), indicating that removal of the protein environment weakens the channel stability. This leads to structural changes, which however do not preclude the ability of Tom40 to form, albeit different, pores.
Analysis of Tom40 preparations was reported previously for yeast Tom40 obtained by expression in Escherichia coli and refolding from urea-solubilized inclusion bodies (Hill et al. 1998). These preparations are different in several aspects from the Tom40 purified from TOM complex under nondenaturing conditions described here.
The maximum conductance level of recombinant Tom40 was reported to be ∼360 pS in 250 mM KCl (Hill et al. 1998). This is a rather low conductance that does not fit to previous measurements of TOM complex channels in planar lipid membranes. The conductance of single channels (the major form found in the present study, which corresponds to one pore particle) is ∼550 pS in 150 mM KCl, corresponding to ∼900 pS in 250 mM KCl both in TOM40 (this study) and holo complex fractions (Künkele et al. 1998a,Künkele et al. 1998b). Similar values were observed previously using different techniques for S. cerevisiae and adrenal cortex channels (Thieffry et al. 1992; Bathori et al. 1996).
Also, the spectral properties of purified Neurospora Tom40 were different from those of recombinant Tom40. The CD spectra of recombinant yeast Tom40 showed crossovers of the ellipticity to the baseline at ∼217 nm and a broad minimum >230 nm (Hill et al. 1998). Again, this may suggest that the folding of refolded recombinant membrane protein Tom40 differs considerably from that of Tom40 isolated from mitochondria. We presume that gentle isolation of Tom40 from the native complex conserves the basic structure and function of the channel, whereas expression in E. coli as inclusion bodies and renaturation from 8 M urea may not produce the correct higher order structure.
Tom40 was predicted to traverse the mitochondrial outer membrane as a series of antiparallel β-strands that form a β-barrel (Court et al. 1995; Mannella et al. 1996). A novel multiple alignment algorithm, called the Gibbs sampling algorithm, was used previously to detect motif-encoding regions in sequences of bacterial outer membrane proteins that correspond to transmembrane β-strands in bacterial porins (Neuwald et al. 1995). This bacterial motif has been used to screen sets of mitochondrial membrane protein sequences. Matches occurred in two proteins: mitochondrial porin and the outer membrane protein import pore Tom40. This suggested a structural relatedness between Tom40 and the bacterial and mitochondrial pore proteins (Mannella et al. 1996). CD measurements of bacterially expressed Tom40 of S. cerevisiae were suggested to indicate a predominance of >60% β-sheet (Hill et al. 1998) and high structural similarity to members of the porin membrane protein family.
We have performed CD and FTIR spectroscopy measurements with purified Neurospora Tom40. A maximum of 31% of Neurospora Tom40 was found to adopt β-sheet topology, whereas the calculated helix content is ≥22%. Surprisingly, the content of β-sheet is markedly less than that of mitochondrial porin, which in agreement with previous studies (Shao et al. 1996; Koppel et al. 1998) revealed predominantly β-sheet structure (48% β-sheet, 15% α-helix).
An important question is whether a single Tom40 protomer can form a protein translocation pore. Our data predict that ∼108 amino acid residues of Tom40 may be organized in β secondary structure elements. The mean radius of a regular β-barrel can be computed according to the number of β-strands and the shear number (Murzin et al. 1994). Given an inner diameter of 2.5 nm of the barrel, implying a larger diameter of the barrel backbone, and applying common values for the shear number (Murzin et al. 1994), >14 β-strands are necessary to fulfill the structural requirements. This denotes that the β-strands of Tom40 consist of less than seven to eight amino acid residues on average and cannot be expected to span the hydrophobic region of the membrane, if the shear number is close to or greater than the number of β-strands. In fact, the average of β-strands in bacterial outer membrane proteins are made up by ∼12 amino acid residues (Buchanan 1999; Koebnik et al. 2000; Schulz 2000). If this would also apply to Tom40, our data suggest the existence of only 8–10 β-strands in Tom40. This number of strands can hardly be expected to form stain-filled and open channels of the observed size.
On the basis of these considerations, we speculate that a β-strand solvent-accessible pore with a diameter of 2.5 nm could only be formed by β-barrel structures contributed by more than one Tom40 protomer. These protomers could be imagined to assemble in a way similar to staphylococcal α-hemolysin (Song et al. 1996) or the bacterial multidrug efflux and protein export channel TolC (Koronakis et al. 2000). However, to fully understand the structural basis of pore formation, further studies addressing the oligomeric state of Tom40 will be required. It seems possible that different oligomeric structures of Tom40 exist, similar to the bacterial protein translocase SecYEG, which forms primarily dimeric but also tetrameric structures. Formation of the tetramer is induced by SecA (Manting et al. 2000).
EM and image analysis of Tom40 revealed mainly single ring structures, whereas the TOM core complex consists predominantly of double rings. The core complex is composed of about eight Tom40 molecules (Ahting et al. 1999). From the size of the Tom40 complex as indicated by gel filtration analysis, the stoichiometry cannot be determined with certainty. Higher resolution images of the Tom40 complex are required to resolve structural symmetries.
It seems clear from the findings reported here that oligomeric Tom40 forms the basic structure of the TOM complex. At the same time, our results demonstrate that the other constituents of the core complex Tom22, Tom7, and Tom6 have important functions in generating a stable two pore channel. This agrees well with genetic experiments in which the genes for these components were deleted (Hönlinger et al. 1996; van Wilpe et al. 1999).
Acknowledgments
We thank W. Baumeister for continuous support. We are also indebted to E. Weyer for her assistance with the CD spectroscopy and U. Staudinger and M. Braun for excellent technical assistance in the isolation of mitochondria and TOM complex.
This research was supported by the Sonderforschungsbereich 184 of the Deutsche Forschungsgemeinschaft (to S. Nussberger and W. Neupert) and the Münchner Medizinische Wochenschrift (to S. Nussberger).
Footnotes
Abbreviations used in this paper: ATR, attenuated total reflection; CD, circular dichroism; DDM, n-dodecyl β-d-maltoside; FTIR, Fourier transform IR spectroscopy; IR, infrared; Ni-NTA, nickel nitrilotriacetic acid agarose; OG, n-octyl β,d-glucopyranoside; PSC, peptide-sensitive channel; TOM, translocase of the outer mitochondrial membrane; VDAC, voltage-dependent anion channel.
References
- Ahting U., Thun C., Hegerl R., Typke D., Nargang F.E., Neupert W., Nussberger S. The TOM core complexthe general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 1999;147:959–968. doi: 10.1083/jcb.147.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alconada A., Kubrich M., Moczko M., Honlinger A., Pfanner N. The mitochondrial receptor complexthe small subunit Mom8b/Isp6 supports association of receptors with the general insertion pore and transfer of preproteins. Mol. Cell. Biol. 1995;15:6196–6205. doi: 10.1128/mcb.15.11.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathori G., Szabo I., Wolff D., Zoratti M. The high-conductance channels of yeast mitochondrial outer membranesa planar bilayer study. J. Bioenerg. Biomembr. 1996;28:191–198. doi: 10.1007/BF02110650. [DOI] [PubMed] [Google Scholar]
- Benz R., Janko K., Boos W., Läuger P. Formation of large, ion-permeable membrane channels by the matrix protein (porin) of Escherichia coli . Biochim. Biophys. Acta. 1978;511:305–319. doi: 10.1016/0005-2736(78)90269-9. [DOI] [PubMed] [Google Scholar]
- Buchanan S.K. Beta-barrel proteins from bacterial outer membranesstructure, function and refolding. Curr. Opin. Struct. Biol. 1999;9:455–461. doi: 10.1016/S0959-440X(99)80064-5. [DOI] [PubMed] [Google Scholar]
- Byler D.M., Susi H. Examination of the secondary structure of proteins by deconvoluted FTIR spectra. Biopolymers. 1986;25:469–487. doi: 10.1002/bip.360250307. [DOI] [PubMed] [Google Scholar]
- Cao W., Douglas M.G. Biogenesis of ISP6, a small carboxyl-terminal anchored protein of the receptor complex of the mitochondrial outer membrane. J. Biol. Chem. 1995;270:5674–5679. doi: 10.1074/jbc.270.10.5674. [DOI] [PubMed] [Google Scholar]
- Court D.A., Lill R., Neupert W. The protein import apparatus of the mitochondrial outer membrane. Can. J. Bot. 1995;73:193–197. [Google Scholar]
- Dekker P.J.T., Ryan M.T., Brix J., Müller H., Hönlinger A., Pfanner N. Preprotein translocase of the outer mitochondrial membranemolecular dissection and assembly of the general import pore complex. Mol. Cell. Biol. 1998;18:6515–6524. doi: 10.1128/mcb.18.11.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembowski M., Künkele K.P., Nargang F.E., Neupert W., Rapaport D. Assembly of Tom6 and Tom7 into the TOM core complex of Neurospora crassa . J. Biol. Chem. 2001;276:17679–17685. doi: 10.1074/jbc.M009653200. [DOI] [PubMed] [Google Scholar]
- Dietmeier K., Hönlinger A., Bömer U., Dekker P.J.T., Eckerskorn C., Lottspeich F., Kübrich M., Pfanner N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 1997;388:195–200. doi: 10.1038/40663. [DOI] [PubMed] [Google Scholar]
- Frank J., van Heel M. Correspondence analysis of aligned images of biological particles. J. Mol. Biol. 1982;161:134–137. doi: 10.1016/0022-2836(82)90282-0. [DOI] [PubMed] [Google Scholar]
- Frank J., Verschoor A., Boublik M. Computer averaging of electron micrographs of 40S ribosomal subunits. Science. 1981;214:1353–1355. doi: 10.1126/science.7313694. [DOI] [PubMed] [Google Scholar]
- Freitag H., Genchi G., Benz R., Palmieri F., Neupert W. Isolation of mitochondrial porin from Neurospora crassa . FEBS Lett. 1982;145:72–76. doi: 10.1016/0014-5793(82)81209-x. [DOI] [PubMed] [Google Scholar]
- Hegerl R. The EM program packagea platform for image processing in biological electron microscopy. J. Struct. Biol. 1996;116:30–34. doi: 10.1006/jsbi.1996.0006. [DOI] [PubMed] [Google Scholar]
- Hill K., Model K., Ryan M.T., Dietmeier K., Martin F., Wagner R., Pfanner N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 1998;395:516–521. doi: 10.1038/26780. [DOI] [PubMed] [Google Scholar]
- Hines V., Brandt A., Griffiths G., Horstmann H., Brutsch H., Schatz G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 1990;9:3191–3200. doi: 10.1002/j.1460-2075.1990.tb07517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönlinger A., Kübrich M., Moczko M., Gärtner F., Mallet L., Bussereau F., Eckerskorn C., Lottspeich F., Dietmeier K., Jacquet M., Pfanner N. The mitochondrial receptor complexMom22 is essential for cell viability and directly interacts with preproteins. Mol. Cell. Biol. 1995;15:3382–3389. doi: 10.1128/mcb.15.6.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönlinger A., Bömer U., Alconada A., Eckerskorn C., Lottspeich F., Dietmeier K., Pfanner N. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. EMBO J. 1996;15:2125–2137. [PMC free article] [PubMed] [Google Scholar]
- Kassenbrock C.K., Cao W., Douglas M.G. Genetic and biochemical characterization of ISP6, a small mitochondrial outer membrane protein associated with the protein translocation complex. EMBO J. 1993;12:3023–3034. doi: 10.1002/j.1460-2075.1993.tb05971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppinen J.K., Moffat D.J., Mantsch H.H., Cameron D.G. Fourier-self-deconvolutiona method for resolving intrinsically overlapped bands. Appl. Spectroscopy. 1981;35:271–276. [Google Scholar]
- Kiebler M., Pfaller R., Sollner T., Griffiths G., Horstmann H., Pfanner N., Neupert W. Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature. 1990;348:610–616. doi: 10.1038/348610a0. [DOI] [PubMed] [Google Scholar]
- Kiebler M., Keil P., Schneider H., van der Klei I.J., Pfanner N., Neupert W. The mitochondrial receptor complexa central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell. 1993;74:483–492. doi: 10.1016/0092-8674(93)80050-o. [DOI] [PubMed] [Google Scholar]
- Koebnik R., Locher K., Van Gelder P. Structure and function of bacterial outer membrane proteinsbarrels in a nutshell. Mol. Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- Koppel D.A., Kinnally K.W., Masters P., Forte M., Blachly-Dyson E., Mannella C.A. Bacterial expression and characterization of the mitochondrial outer membrane channel. Effects of N-terminal modifications. J. Biol. Chem. 1998;273:13794–13800. doi: 10.1074/jbc.273.22.13794. [DOI] [PubMed] [Google Scholar]
- Koronakis V., Sharff A., Koronakis E., Luisi B., Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- Künkele K.P., Heins S., Dembowski M., Nargang F.E., Benz R., Thieffry M., Walz J., Lill R., Nussberger S., Neupert W. The preprotein translocation channel of the outer membrane of mitochondria Cell. 93 1998. 1009 1019a [DOI] [PubMed] [Google Scholar]
- Künkele K.P., Juin P., Pompa C., Nargang F.E., Henry J.P., Neupert W., Lill R., Thieffry M. The isolated complex of the translocase of the outer membrane of mitochondria. Characterization of the cation-selective and voltage-gated preprotein-conducting pore J. Biol. Chem 273 1998. 31032 31039b [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lithgow T., Junne T., Suda K., Gratzer S., Schatz G. The mitochondrial outer membrane protein Mas22p is essential for protein import and viability of yeast. Proc. Natl. Acad. Sci. USA. 1994;91:11973–11977. doi: 10.1073/pnas.91.25.11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannella C.A., Neuwald A.F., Lawrence C.E. Detection of likely transmembrane beta strand regions in sequences of mitochondrial pore proteins using the Gibbs sampler. J. Bioenerg. Biomembr. 1996;28:163–169. doi: 10.1007/BF02110647. [DOI] [PubMed] [Google Scholar]
- Manting E.H., van Der Does C., Remigy H., Engel A., Driessen A.J. SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J. 2000;19:852–861. doi: 10.1093/emboj/19.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A.G., Lesk A.M., Chothia C. Principles determining the structure of beta-sheet barrels in proteins. I. A theoretical analysis. J. Mol. Biol. 1994;236:1369–1381. doi: 10.1016/0022-2836(94)90064-7. [DOI] [PubMed] [Google Scholar]
- Nakai M., Endo T. Identification of yeast MAS17 encoding the functional counterpart of the mitochondrial receptor complex protein MOM22 of Neurospora crassa . FEBS Lett. 1995;357:202–206. doi: 10.1016/0014-5793(94)01362-5. [DOI] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Neuwald A.F., Liu J.S., Lawrence C.E. Gibbs motif samplingdetection of bacterial outer membrane protein repeats. Protein Sci. 1995;4:1618–1632. doi: 10.1002/pro.5560040820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace C.N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage L., Junne T., Hahne K., Lithgow T., Schatz G. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO J. 1993;12:4115–4123. doi: 10.1002/j.1460-2075.1993.tb06095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz G., Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Schulz G.E. Beta-barrel membrane proteins. Curr. Opin. Struct. Biol. 2000;10:443–447. doi: 10.1016/s0959-440x(00)00120-2. [DOI] [PubMed] [Google Scholar]
- Shao L., Kinnally K., Mannella C. Circular dichroism studies of the mitochondrial channel, VDAC, from Neurospora crassa . Biophys. J. 1996;71:778–786. doi: 10.1016/S0006-3495(96)79277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T., Griffiths G., Pfaller R., Pfanner N., Neupert W. MOM19, an import receptor for mitochondrial precursor proteins. Cell. 1989;59:1061–1070. doi: 10.1016/0092-8674(89)90762-9. [DOI] [PubMed] [Google Scholar]
- Söllner T., Pfaller R., Griffiths G., Pfanner N., Neupert W. A mitochondrial import receptor for the ADP/ATP carrier. Cell. 1990;62:107–115. doi: 10.1016/0092-8674(90)90244-9. [DOI] [PubMed] [Google Scholar]
- Song L., Hobaugh M.R., Shustak C., Cheley S., Bayley H., Gouaux J.E. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- Sreerama N., Woody R.W. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- Thieffry M., Neyton J., Pelleschi M., Fevre F., Henry J.P. Properties of the mitochondrial peptide-sensitive cationic channel studied in planar bilayers and patches of giant liposomes. Biophys. J. 1992;63:333–339. doi: 10.1016/S0006-3495(92)81626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wilpe S., Ryan M.T., Hill K., Maarse A.C., Meisinger C., Brix J., Dekker P.J., Moczko M., Wagner R., Meijer M. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature. 1999;401:485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- Vestweber D., Brunner J., Baker A., Schatz G. A 42K outer-membrane protein is a component of the yeast mitochondrial protein import site. Nature. 1989;341:205–209. doi: 10.1038/341205a0. [DOI] [PubMed] [Google Scholar]
- Voos W., Martin H., Krimmer T., Pfanner N. Mechanisms of protein translocation into mitochondria. Biochim. Biophys. Acta. 1999;1422:235–254. doi: 10.1016/s0304-4157(99)00007-6. [DOI] [PubMed] [Google Scholar]