Cytologists have long observed that individual eukaryotic species segregate their chromosomes in one of two apparently different ways. Monocentric chromosomes attach to microtubules at a particular region (the centromere) and move toward the pole during anaphase with the centromere leading. In contrast, holocentric chromosomes bind to microtubules along their entire length and move broadside to the pole from the metaphase plate. Holocentric chromosomes are scattered throughout the plant and animal kingdoms, and may be products of convergent evolution. Alternatively, the ancestral eukaryotic chromosome may have been holocentric, in which case the restriction of kinetic activity to a specialized region must have been an evolutionary event that occurred again and again.
Perhaps because most laboratory organisms have monocentric chromosomes, holocentric species have been regarded with a mixture of curiosity and suspicion. These attitudes have begun to change precipitously due to three major factors: the development of the holocentric nematode worm Caenorhabditis elegans as a robust molecular genetic system, the availability of extensive genome sequences for both C. elegans and monocentric species, and the harnessing of RNA interference (RNAi) as an experimental technique (Fire et al. 1998). As these tools are enabling holocentric behavior to be studied at a molecular level, we can finally explore how a chromosome function as basic and essential as microtubule attachment has assumed such distinct evolutionary forms. Three studies published in this issue, combined with other recent results, strongly suggest that many components and mechanisms underlying kinetochore function are highly conserved between holocentric and monocentric chromosomes.
Several C. elegans proteins have now been implicated in centromere function or kinetochore structure. The majority were first identified by virtue of their homology to components identified in monocentric organisms, including ZW10 (Starr et al. 1997), CENP-A (HCP-3) (Buchwitz et al. 1999), and now CENP-C (HCP-4) (see Moore and Roth 2001; Oegema et al. 2001, in this issue), Bub1, and MCAK (Oegema et al. 2001). Homologs of chromosome “passenger proteins” similar to Aurora and INCENPs have also been recognized and shown to play roles in chromosome segregation (Schumacher et al. 1998; Kaitna et al. 2000; Oegema et al. 2001).
In contrast, discovery of HCP-1 (and its partially redundant paralog HCP-2) originated with a monoclonal antibody that recognized the poleward face of mitotic chromosomes (Moore et al. 1999). HCP-1 and -2 are likely homologs of mammalian CENP-F, a kinetochore protein that is a component of the spindle assembly checkpoint (Rattner et al. 1993). Another player, HIM-10, was identified genetically through a partial loss-of-function allele that causes a nonlethal segregation defect (Hodgkin et al. 1979). In this issue, Howe et al. 2001 have now demonstrated a role in kinetochore function for HIM-10, which is homologous to Nuf2, originally identified as a spindle pole body–associated factor from budding yeast and recently shown to be centromeric in organisms from Schizosaccharomyces pombe to humans (Osborne et al. 1994; Wigge and Kilmartin 2001). Each of these C. elegans centromere or kinetochore proteins thus shares similarity with centromere-associated factors from monocentric organisms. By studying these proteins in the context of holocentric chromosomes, work in this issue contributes to our understanding of the conserved, and probably most fundamental, properties of kinetochores. A summary of available information about kinetic assembly is presented in Fig. 1 (RNAi).
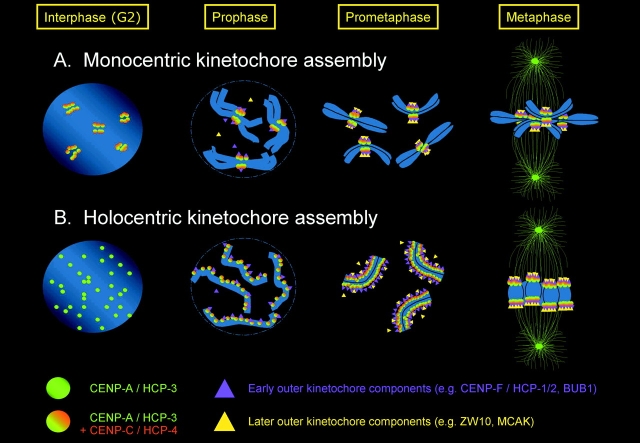
Figure 1.
Assembly of kinetochores on monocentric and holocentric chromosomes. This schematic diagram illustrates the major events of kinetochore assembly, based on information from a body of work. CENP-A/HCP-3 decorates centromeres throughout the cell cycle. In monocentric organisms (A), CENP-A staining appears as distinct foci corresponding to the number of chromosomes. These foci recruit other components, including checkpoint proteins, motors, and structural elements, during prophase to build the mitotic kinetochore. The order of assembly of many components is not yet known. In C. elegans interphase nuclei (B), HCP-3 staining reveals a multitude of tiny foci. However, by the time worm chromosomes can be seen as individual entities at prophase, a single ribbon of semicontiguous HCP-3 staining is evident along one edge of each pair of sisters. HCP-4 (CENP-C) and HCP-1 (CENP-F) appear to be recruited during prophase, as does BUB-1. MCAK loads slightly later than BUB-1. As the chromosomes congress to the metaphase plate, the single ribbon of staining splits into two ribbons that separate to opposing faces of the paired sisters. Between this stage and the alignment of the chromosomes at the metaphase plate, each ribbon of kinetochore proteins condenses dramatically to form a dot ∼0.2–0.5 μm in diameter, similar in size to a mammalian centromere. A notable difference between mono- and holocentric kinetochores is that “splitting” of moncentric kinetochores into two sister foci is already evident by G2, when centromeres appear as “double dots.” In contrast, in C. elegans there is no cytological indication of the duality of the replicated chromosome until late in mitotic prophase. Why this splitting is apparent later in worms is unknown.
Reverse Genetics in C. elegans as a Power Tool for Investigation of Kinetochore Structure and Function
Each of the papers presented here has employed the technique of RNA interference in C. elegans. This reverse genetic approach makes it technically straightforward to examine the consequences of depleting a specific gene product even when a mutant allele is unavailable. All of the conserved centromeric or kinetochore functions examined in these papers prove to be essential for the early embryonic divisions, and are thus required for viability. Nevertheless, cytological analysis of early embryonic divisions demonstrated that these factors play distinct roles in chromosome segregation.
Several different cytological approaches have been employed to probe the effects of depleting specific kinetochore components in the early embryo. Immunofluorescent detection of kinetochore components has revealed that kinetochore assembly occurs by sequential recruitment of factors. For example, normal localization of HCP-1 (CENP-F), BUB-1, and MCAK all depend on the presence of CENP-C at the kinetochore, which in turn depends on CENP-A. This is consistent with observations in mouse cells lacking CENP-A (Howman et al. 2000), suggesting monocentric kinetochores may assemble through a similar hierarchical assembly process.
Making elegant use of quantitative real-time imaging, Oegema et al. 2001 have investigated the relationship between kinetochore protein function and spindle dynamics. Among other observations, they find that embryonic mitosis does not involve an obvious anaphase A (in which chromosomes approach the poles); rather, separation of chromatids occurs through an anaphase B mechanism (in which the spindle poles move away from each other). Interestingly, they observe distinct perturbations of microtubule–chromosome interactions when they deplete kinetochore factors or ICP-1 using RNAi.
Using electron microscopy, Howe et al. 2001 have characterized kinetochore ultrastructure. They demonstrate that kinetochore morphology in C. elegans resembles what was observed in monocentric PtK cells (McEwen et al. 1998) when high-pressure freezing methods are used to fix each tissue. Moreover, they find that depletion of HIM-10 has a marked impact on both mitotic and meiotic kinetochore structure. Although HIM-10 is an essential kinetochore component, a temperature-sensitive allele of the gene enabled them to observe the effects of HIM-10 depletion in the adult germline by a postembryonic temperature shift. It might be possible to mimic this type of experiment with other kinetochore components by soaking young larvae in double-stranded RNA, which can bypass the problem of embryonic inviability due to loss of gene function (Kuroyanagi et al. 2000).
It is striking that while all of the proteins examined in these studies affect mitosis, only inactivation of the “passenger protein” ICP-1 produced a strong defect in female meiotic chromosome segregation. In ICP-1–depleted embryos, the four oocyte meiotic products fail to separate and they migrate together toward the male pronucleus. (Oegema et al. 2001). However, in HCP-3– and HCP-4–depleted animals, the oocytes appear to proceed through both meiotic divisions quite normally and to segregate polar bodies (into which three of the four female meiotic products are discarded). There are no apparent differences between the maternal and paternal pronuclei in these early embryos, as would be expected if the maternal pronucleus had just undergone an aberrant meiosis. Howe et al. 2001 have gone a step further by quantitating segregation defects after HIM-10 RNAi, and they conclude that meiotic segregation in the oocytes is not significantly impaired by loss of this factor, even though defects are observed in spermatocyte divisions, and HIM-10 is clearly present on oocyte chromosomes.
What might account for the relative insensitivity of oocyte divisions to depletion of kinetochore components essential for mitosis? To some extent, these results may reflect incomplete depletion of proteins from the oocytes under the experimental conditions used. However, meiotic divisions in the oocyte are mechanistically distinct from spermatocyte divisions and all mitotic divisions in that the spindle forms in the absence of centrioles (Albertson and Thompson 1993). This unusual division may impose fundamentally different requirements for kinetochore factors in establishing and regulating chromosome-microtubule interactions. It would be interesting to subject younger hermaphrodites to RNAi to compare the effects of depleting these factors on oocyte and spermatocyte divisions.
Implications and Outstanding Questions
The observations presented here, in conjunction with studies of centromeres in other species, have reshaped the way we might think about some of the long-standing riddles surrounding holocentric chromosome behavior. Answers to the following questions will likely have relevance for chromosome biology in all eukaryotes.
What sequence determinants underlie the positioning of holocentric centromeres?
In monocentric species, the histone H3-like protein CENP-A is specifically incorporated into nucleosomes at centromeres. In C. elegans, the observed staining pattern of anti–HCP-3 (CENP-A) antibodies as puncta within interphase nuclei and bands along mitotic chromosomes indicates that not all genomic sequences are associated with this protein, at least not in the same nucleus. At this point, we know nothing about HCP-3 binding sites. Chromatin immunoprecipitation experiments should make it possible to isolate HCP-3–associated sequences and to discern whether they have any notable features in common, or whether they are even consistent among nuclei within a given animal or among individuals within a population
Centromeric DNA sequences in S. pombe, Drosophila, and mammals appear to be neither necessary nor sufficient for centromere function (Karpen and Allshire 1997). Nevertheless, the appearance of “neocentromeres” (normally noncentromeric DNA that acquires and faithfully propagates centromere/kinetochore function) is rare. In contrast, neocentromere formation appears to be a relatively permissive event in C. elegans. Naked DNA microinjected into the gonad of adult worms can assemble into extrachromosomal multimeric arrays, which bind to factors such as HCP-3 and HIM-10 (Howe et al. 2001). Such arrays rapidly acquire the ability to segregate through mitosis and meiosis. Interestingly, there appears to be little or no requirement for authentic C. elegans sequences in this process, since even prokaryotic DNA (e.g., bacteriophage lambda or Myxococcus xanthus genomic DNA) can form functional extrachromosomal arrays when injected into C. elegans. Promiscuous centromere factor binding in worms may provide a useful experimental system for investigating how DNA sequences acquire the ability to assemble centromeres de novo.
How does mitotic chromosome segregation work when the kinetochore is distributed along each chromatid?
For sister chromatids to separate at anaphase, each of them must first attach to one of the two spindle poles. If a single kinetochore extends along the length of a flexible chromatid, how does it avoid attaching to both poles of the mitotic spindle? Recent findings have suggested a partial answer to this question. Moreover, a number of different observations, taken together, indicate that the challenge of orienting toward only one pole is shared by all kinetochores and is not specific to holocentric chromosomes.
A surprising and significant observation revealed by HCP-3 (CENP-A) or HCP-4 (CENP-C) immunostaining is that by the time chromosomes have fully congressed at the metaphase plate, the “ribbon” of centromeric/kinetochore material observed along prophase chromosomes has condensed into a dot-like structure (see, for example, Figure 3 in Moore and Roth 2001). How this happens presents a new mystery, but the appearance of a metaphase structure no larger than a typical mammalian kinetochore suggests that holocentric centromeres may not face any unique geometrical obstacles in orienting properly.
Moreover, the ability of monocentric chromatids to form merotelic (bipolar) attachments at alarmingly high frequencies has been demonstrated in insects, plants, and cultured mammalian cells (LaFountain 1985; Yu and Dawe 2000; Cimini et al. 2001). Thus, being monocentric is no guarantee that a kinetochore will only interact with microtubules emanating from a single spindle pole. All kinetochores may share common structural features that restrict their tendency to form merotelic attachments.
A further hint as to how a holocentric kinetochore can interact specifically with one pole comes from a series of mitotically stable dicentric chromosomes that have been isolated from human patients (Sullivan and Willard 1998; Higgins et al. 1999). Their two centromeres are invariably close together relative to the chromosome as a whole; the distance separating them has been measured and shown to be in the range of 2–20 Mb. This implies that mammalian centromeres can act in concert if they lie within ∼20 Mb of each other. Perhaps not coincidentally, C. elegans chromosomes range in length from ∼14 to 21 Mb. Monocentric kinetochores have been proposed to be comprised of repeating structural subunits that amalgamate to form a single unit (Zinkowski et al. 1991); the holocentric kinetochore may simply involve the coalescence of subunits that have a higher ratio of interstitial DNA sequences.
In a holocentric organism, what stabilizes the karyotype?
Chromosome rearrangements in monocentric organisms can give rise to acentric and dicentric chromosomes that usually fail to propagate. In contrast, holocentric chromosome fragments segregate normally in mitosis, which raises the question of how genome integrity is maintained in worms.
One clue to the stabilization of the karyotype of C. elegans may come from the meiotic segregation behavior of chromosome rearrangements. Experimental evidence has demonstrated that homologous chromosomal sequences will not undergo crossing-over if they become separated from a particular region near one end of the chromosome by breakage, translocation, or insertion of intervening sequences (reviewed by Zetka and Rose 1995). Even simple translocations have the effect of suppressing meiotic exchange over large regions of the genome, resulting in aberrant meiotic segregation. While chromosome rearrangements can be maintained in laboratory strains, and some even segregate efficiently in homozygotes during meiosis, all cause marked reduction of viable progeny in heterozygotes. In a wild population, this would lead to rapid loss of most types of gross rearrangements. Thus, in a holocentric organism, somatic chromosome segregation can tolerate breakage or rearrangement, but these abnormal chromosomes may be poorly transmitted to the next generation.
Despite clear evidence that chromosome rearrangements pose major problems for meiotic segregation, reassortment of the genome has apparently occurred extensively during nematode evolution (Kent and Zahler 2000). Interspecies sequence comparisons are becoming possible on a genome-wide level, and will likely offer some revelations concerning the relationship between karyotype evolution and the distribution of kinetic activity along the chromosomes.
How do holocentric chromosomes accomplish meiosis?
Over a century ago, van Beneden 1883 and Boveri 1890 established the reductional nature of meiosis through careful observation of germ cell formation in holocentric Ascaris species. Ironically, while major progress has been made in understanding reductional division in monocentric organisms, key aspects of this process remain enigmatic for holocentric chromosomes.
In all familiar organisms, reduction occurs during the first meiotic division. Homologous chromosomes segregate to opposite poles, usually after undergoing genetic exchange, and sister chromatids stay together until anaphase of meiosis II. For recombinant homologs without a defined centromeric locus, the distinction between homologue and sister becomes blurred. In C. elegans, achiasmate chromosomes such as the male X or univalents arising from defects in recombination (Lucia Wille and Diane Shakes, personal communication) segregate as a single entity during meiosis I, and sisters do not separate until meiosis II. The first division can thus be regarded as reductional since it is not equational.
A unique feature of reductional divisions in species with localized centromeres is that cohesion must be maintained differentially along the length of recombinant chromosomes. Loss of cohesion distal to the chiasmata allows recombinant homologous chromosomes to separate from each other, while sister chromatids remain associated proximal to chiasmata. Defects in this process result in meiotic nondisjunction (Dej and Orr-Weaver 2000). From model organisms, there is good evidence that a key function of the centromeric region is to serve as a zone where meiotic cohesion is maintained until anaphase II. Centromeric regions are highly enriched in molecular components that promote proper meiotic cohesion, including both novel factors such as Drosophila Mei-S332 and specialized cohesins (Kerrebrock et al. 1995; Toth et al. 2000).
The conceptual challenge posed by holocentric chromosomes is that without a single point of microtubule attachment there is no meaning to the phrase “distal to the chiasmata.” Without a defined centromeric locus, how do sisters ensure that they maintain cohesion when homologs disjoin during the first meiotic division?
This feat may be accomplished by distinct mechanisms in different holocentric organisms. In Parascaris univalens, microtubule attachment during meiosis appears to be restricted to a large heterochromatic region near one end of the lone chromosome pair, in effect making meiotic chromosomes monocentric (Goday and Pimpinelli 1989). However, in C. elegans, fluorescence microscopy reveals that HCP-3 (CENP-A) localizes throughout meiotic bivalents, and inner kinetochore components cover the entire surface (Moore et al. 1999). Howe et al. 2001 have now demonstrated by electron microscopy that a ribosome-free zone, the presumptive meiotic kinetochore, surrounds each bivalent. However, it is still not known whether microtubules actually attach all along the chromosomes during C. elegans meiosis I. These observations underscore the mysteries of how a holocentric bivalent forms a stable bipolar attachment, and how its cohesion could be regulated to promote proper meiotic segregation.
A long-standing cytological concept leads to one possible model for meiotic segregation of C. elegans chromosomes. Chiasmata in some organisms have been proposed to “terminalize” at the end of meiotic prophase, or somehow to slip along the chromosome away from the original point of strand exchange until they leave the chromosome at a telomere (discussed in Albertson et al. 1997). The idea of terminalization can be translated into contemporary terms and molecules such as separases and cohesins. For example, the act of crossing over might somehow influence the local region of the chromosome so that cohesion is removed initially at the site of chiasmata, and then is eliminated processively to the nearest end. This would be functionally equivalent to terminalization and would be consistent with observations that C. elegans achiasmate chromosomes retain sister cohesion until meiosis II. While it may seem far fetched to imagine that a chiasma can trigger loss of cohesion between itself and one end, but not the other, it is intriguing to note that on the C. elegans autosomes there is usually just a single meiotic crossover that most often occurs within the distal third of the chromosome from either end (Barnes et al. 1995). From the phenomenon of recombination interference, it is evident that crossovers must influence some aspect of chromosome structure over long distances from the point of strand exchange. In addition to preventing additional crossovers, this structural modification could hypothetically promote loss of cohesion. Local microtubule attachment might also be inhibited, consistent with the observation that the ends most distant from the chiasma usually face the poles at metaphase I (Albertson et al. 1997).
Future Prospects
Nematodes have played key roles in the history of our understanding of mitosis and meiosis. The availability of the genome sequence and new molecular tools for C. elegans has sparked a renaissance of interest in holocentric chromosomes. Evidence from work presented in this issue demonstrates that molecules implicated in centromere and kinetochore function in monocentric species play conserved roles in holocentric chromosome segregation. Beyond the conservation of the kinetochore at the level of protein components, Howe et al. 2001 have shown by electron microscopy that its ultrastructure may not be fundamentally different when it is built along an entire chromosome.
Work from Moore and Roth 2001 and Oegema et al. 2001 underscores the power of RNAi by demonstrating that construction of a mitotic kinetochore involves a hierarchical assembly of protein factors, which would have been more difficult to establish by traditional mutant analyses. These findings will clearly shape the way that kinetochores are studied in all species, and will lead to a greater understanding of the commonalities and differences among holocentric and monocentric chromosomes. In light of these results, we should sound a note of caution in interpreting phenotypes arising from loss-of-function of individual kinetochore components. If the absence of a protein affects the recruitment of downstream components in the assembly process, or results in changes in kinetochore structure, this may have indirect but profound effects on behaviors such as microtubule attachment and chromatid orientation. A complete understanding of the roles these proteins play at the centromere awaits more detailed biochemical and cell biological analyses.
Based on RNAi-induced phenotypes with conserved factors such as ZW10, HCP-3, HCP-1/2, and HIM-10, loss of function of essential mitotic centromere or kinetochore components is expected to cause early lethality in C. elegans. Systematic secondary screening by observing early events in embryonic-lethal mutants has yielded a collection of new loci that influence chromosome segregation. Ongoing screens using both traditional and reverse genetics (e.g., Gonczy et al. 1999, Gonczy et al. 2000) will likely uncover a battery of new kinetochore factors. It will be intriguing to see which, if any, of these components perform functions uniquely required by holokinetic chromosomes. If these studies are any indication, it is likely that we will see even more conservation of function at the molecular level.
Acknowledgments
I thank Beth Sullivan for insightful contributions, Gary Karpen for stimulating discussions and critical reading of the manuscript, Paul Kaufman for title suggestions, and Lucia Wille and Diane Shakes for helpful comments.
Submitted: 16 May 2001 Accepted: 17 May 2001
References
- Albertson, D.G., A.M. Rose, and A.M. Villeneuve. 1997. Chromosomes, Mitosis, and Meiosis. C. elegans II. D.L. Riddle, T. Blumenthal, B.J. Meyer, and J.R. Priess, editors. Cold Spring Harbor Laboratory Press, Plainview, NY. 47–78. [PubMed]
- Albertson D.G., Thompson J.N. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans . Chromosome Res. 1993;1:15–26. doi: 10.1007/BF00710603. [DOI] [PubMed] [Google Scholar]
- Barnes T.M., Kohara Y., Coulson A, Hekimi S. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans . Genetics. 1995;141:159–179. doi: 10.1093/genetics/141.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. Zellenstudien IIIÜber das Verhalten der chromatischen Kemsubstanz bei der Bildung der Richtungskörper und bei der Befruchtung. Jena. Zeit. Naturwiss. 1890;24:374. [Google Scholar]
- Buchwitz B.J., Ahmad K., Moore L.L., Roth M.B., Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- Cimini D., Howell B., Maddox P., Khodjakov A., Degrassi F., Salmon E.D. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 2001;153:517–528. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dej K.J., Orr-Weaver T.L. Separation anxiety at the centromere. Trends Cell Biol. 2000;10:392–399. doi: 10.1016/s0962-8924(00)01821-3. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Goday C., Pimpinelli S. Centromere organization in meiotic chromosomes of Parascaris univalens . Chromosoma. 1989;98:160–166. doi: 10.1007/BF00329679. [DOI] [PubMed] [Google Scholar]
- Gonczy P., Echeverri G., Oegema K., Coulson A., Jones S.J., Copley R.R., Duperon J., Oegema J., Brehm M., Cassin E. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Gonczy P., Schnabel H., Kaletta T., Amores A.D., Hyman T., Schnabel R. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J. Cell Biol. 1999;144:927–946. doi: 10.1083/jcb.144.5.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins A.W., Schueler M.G., Willard H.F. Chromosome engineeringgeneration of mono- and dicentric isochromosomes in a somatic cell hybrid system. Chromosoma. 1999;108:256–265. doi: 10.1007/s004120050376. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Horvitz H.R., Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans . Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M., McDonald K.L., Albertson D.G., Meyer B.J. HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J. Cell Biol. 2001;153:1227–1238. doi: 10.1083/jcb.153.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howman E.V., Fowler K.J., Newson A.J., Redward S., MacDonald A.C., Kalitsis P., Choo K.H. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitna S., Mendoza M., Jantsch-Plunger V., Glotzer M. Incenp and an Aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol. 2000;10:1172–1181. doi: 10.1016/s0960-9822(00)00721-1. [DOI] [PubMed] [Google Scholar]
- Karpen G.H., Allshire R.C. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Zahler A.M. Conservation, regulation, synteny, and introns in a large-scale C. briggsae–C. elegans genomic alignment. Genome Res. 2000;10:1115–1125. doi: 10.1101/gr.10.8.1115. [DOI] [PubMed] [Google Scholar]
- Kerrebrock A.W., Moore D.P., Wu J.S., Orr-Weaver T.L. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H., Kimura T., Wada K., Hisamoto N., Matsumoto K., Hagiwara M. SPK-1, a C. elegans SR protein kinase homologue, is essential for embryogenesis and required for germline development. Mech. Dev. 2000;99:51–64. doi: 10.1016/s0925-4773(00)00477-9. [DOI] [PubMed] [Google Scholar]
- LaFountain J.R., Jr. Malorientation in half-bivalents at anaphase in crane fly spermatocytes following Colcemid treatment. Chromosoma. 1985;91:337–346. doi: 10.1007/BF00291005. [DOI] [PubMed] [Google Scholar]
- McEwen B.F., Hsieh C.E., Mattheyses A.L., Rieder C.L. A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma. 1998;107:366–375. doi: 10.1007/s004120050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L.L., Morrison M., Roth M.B. HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans . J. Cell Biol. 1999;147:471–480. doi: 10.1083/jcb.147.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L.L., Roth M.B. HCP-4, a CENP-C–like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 2001;153:1199–1207. doi: 10.1083/jcb.153.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A.A. Functional analysis of kinetochore assembly in Caenorhabditis elegans . J. Cell Biol. 2001;153:1209–1225. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne M.A., Schlenstedt G., Jinks T., Silver P.A. Nuf2, a spindle pole body-associated protein required for nuclear division in yeast. J. Cell Biol. 1994;125:853–866. doi: 10.1083/jcb.125.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner J.B., Rao A., Fritzler M.J., Valencia D.W., Yen T.J. CENP-F is a >400 kDa kinetochore protein that exhibits a cell-cycle dependent localization. Cell Motil. Cytoskelet. 1993;26:214–226. doi: 10.1002/cm.970260305. [DOI] [PubMed] [Google Scholar]
- Schumacher J.M., Golden A., Donovan P.J. AIR-2an Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 1998;143:1635–1646. doi: 10.1083/jcb.143.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Williams B.C., Li Z., Etemad-Moghadam B., Dawe R.K., Goldberg M.L. Conservation of the centromere/kinetochore protein ZW10. J. Cell Biol. 1997;138:1289–1301. doi: 10.1083/jcb.138.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.A., Willard H.F. Stable dicentric X chromosomes with two functional centromeres. Nat. Genet. 1998;20:227–228. doi: 10.1038/3024. [DOI] [PubMed] [Google Scholar]
- Toth A., Rabitsch K.P., Galova M., Schleiffer A., Buonomo S.B., Nasmyth K. Functional genomics identifies monopolina kinetochore protein required for segregation of homologs during meiosis I. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- van Beneden E. Récherches sur la maturation de l'oeuf, la fecundation et al division cellulaire. Arch. Biol. 1883;4:265–640. [Google Scholar]
- Wigge P.A., Kilmartin J.V. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 2001;152:349–360. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.G., Dawe R.K. Functional redundancy in the maize meiotic kinetochore. J. Cell Biol. 2000;151:131–142. doi: 10.1083/jcb.151.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka M., Rose A. The genetics of meiosis in Caenorhabditis elegans . Trends Genet. 1995;11:27–31. doi: 10.1016/s0168-9525(00)88983-0. [DOI] [PubMed] [Google Scholar]
- Zinkowski R.P., Meyne J., Brinkley B.R. The centromere-kinetochore complexa repeat subunit model. J. Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]