Abstract
The spindle checkpoint inhibits the metaphase to anaphase transition until all the chromosomes are properly attached to the mitotic spindle. We have isolated a Xenopus homologue of the spindle checkpoint component Bub1, and investigated its role in the spindle checkpoint in Xenopus egg extracts. Antibodies raised against Bub1 recognize a 150-kD phosphoprotein at both interphase and mitosis, but the molecular mass is reduced to 140 upon dephosphorylation in vitro. Bub1 is essential for the establishment and maintenance of the checkpoint and is localized to kinetochores, similar to the spindle checkpoint complex Mad1–Mad2. However, Bub1 differs from Mad1–Mad2 in that Bub1 remains on kinetochores that have attached to microtubules; the protein eventually dissociates from the kinetochore during anaphase. Immunodepletion of Bub1 abolishes the spindle checkpoint and the kinetochore binding of the checkpoint proteins Mad1, Mad2, Bub3, and CENP-E. Interestingly, reintroducing either wild-type or kinase-deficient Bub1 protein restores the checkpoint and the kinetochore localization of these proteins. Our studies demonstrate that Bub1 plays a central role in triggering the spindle checkpoint signal from the kinetochore, and that its kinase activity is not necessary for the spindle checkpoint in Xenopus egg extracts.
Keywords: spindle checkpoint, Xenopus, kinetochore, Bub1, Mad1
Introduction
During each cell division cycle, the newly duplicated chromosomes must be distributed evenly into the new cells so that each cell receives exactly one copy of the chromosomes. Mistakes in this process result in aneuploidy that is manifested in genetic disorders and cancers. Accurate sister chromatid segregation relies on the attachment and alignment of chromosomes on the mitotic spindle. The presence of a damaged mitotic spindle or misaligned chromosomes triggers the spindle checkpoint, leading to inhibition of anaphase onset (for review see Amon 1999). Kinetochores play an important role in relaying the checkpoint signal to the cell. Unattached kinetochores (Rieder et al. 1995) or a lack of tension on kinetochores (Li and Nicklas 1995) produces a checkpoint signal.
Genes involved in the spindle checkpoint were first isolated from the budding yeast Saccharomyces cerevisiae, and they include MAD 1–3 (mitotic arrest deficient) (Li and Murray 1991), BUB 1–3 (budding uninhibited by benzimidazole) (Hoyt et al. 1991), and MPS1 (monopolar spindle) (Weiss and Winey 1996). When treated with microtubule depolymerizing agents, mad or bub mutants fail to arrest in mitosis and enter anaphase without a functional spindle, leading to chromosome loss and rapid cell death. The mutant phenotypes suggest that these genes play an important role in controlling the metaphase to anaphase transition.
Checkpoint proteins have been isolated and characterized in yeast and higher eukaryotes. Some of these checkpoint components localize to kinetochores in metazoa and are likely involved in generating the checkpoint signal; these components include Mad1 (Chen et al. 1998; Jin et al. 1998), Mad2 (Chen et al. 1996; Li and Benezra 1996), Bub1 (Taylor and McKeon 1997), and Bub3 (Taylor et al. 1998). Mps1 and Bub1 are protein kinases (Roberts et al. 1994; Weiss and Winey 1996) that likely act at an early stage in establishing the checkpoint (Hardwick and Murray 1995; Hardwick et al. 1996; Farr and Hoyt 1998). Mps1 is thought to phosphorylate Mad1 in a Mad2-, Bub1-, and Bub3-dependent manner (Chen et al. 1999). Mad1 interacts with several checkpoint proteins, forming a complex with Mad2 in Xenopus, and recruiting Mad2 to unattached kinetochores (Chen et al. 1998). Both Mad1 and Mad2 dissociate from kinetochores that have stably bound to spindle microtubules (Chen et al. 1996, Chen et al. 1998; Waters et al. 1998). In addition to Mad2, yeast Mad1 associates with Bub1 and Bub3 when the spindle checkpoint is active (Brady and Hardwick 2000). The ability of Mad1 to interact with Mad2, Bub1, and Bub3 is important for its checkpoint function, because mutants that fail to bind these proteins are checkpoint deficient (Chen et al. 1999; Brady and Hardwick 2000). Bub3 forms a complex with Bub1, an interaction that is essential for Bub1 to localize to kinetochores (Taylor et al. 1998). Bub3 can also associate with a Bub1-related kinase (BubR1) (Jablonski et al. 1998; Taylor et al. 1998). The Bub1–Bub3 complex remains on kinetochores until early anaphase (Jablonski et al. 1998; Basu et al. 1999).
Kinetochore component CENP-E, a plus end–directed motor (Wood et al. 1997), is also important for the spindle checkpoint in Xenopus egg extract (Abrieu et al. 2000). The protein is involved in tethering microtubules to the kinetochore, because inhibition of CENP-E function leads to a failure to establish chromosome alignment at metaphase (Schaar et al. 1997; Wood et al. 1997; Yao et al. 2000). It also plays a role in the spindle checkpoint because immunodepletion of CENP-E, or the addition of a neutralizing anti–CENP-E antibody to Xenopus egg extract, abolishes the spindle checkpoint and prevents Mad1–Mad2 from binding to kinetochores (Abrieu et al. 2000). An association between CENP-E and BubR1 has also been demonstrated in human cells (Chan et al. 1998). Furthermore, CENP-E is phosphorylated by mitogen-activated protein (MAP) kinase in vitro, and preferentially associates with active MAP kinase in mammalian cells during mitosis (Zecevic et al. 1998). Active MAP kinase is also enriched on kinetochores of misaligned chromosomes, and disappears from kinetochores after midanaphase (Shapiro et al. 1998; Zecevic et al. 1998). MAP kinase is important for the spindle checkpoint in Xenopus egg extract and in Xenopus tissue culture cells (Minshull et al. 1994; Takenaka et al. 1997; Wang et al. 1997). Together, these studies suggest that a complex network of checkpoint components controls the generation of the checkpoint signal at the kinetochore.
The downstream target of the spindle checkpoint is the anaphase-promoting complex (APC), the ubiquitin protein ligase involved in ubiquitination and degradation of the anaphase inhibitor Pds1 and cyclin B (for review see Page and Hieter 1999). Degradation of Pds1 and cyclin B triggers anaphase and exit from mitosis, respectively. The specificity of the APC to different substrates is conferred by its associated specificity factor/activator (Visintin et al. 1997). APC bound with Cdc20 targets Pds1, whereas the APC–Cdh1 complex recognizes cyclin B. When the spindle checkpoint is activated, Mad2 binds and inhibits Cdc20 (Fang et al. 1998; Hwang et al. 1998; Kim et al. 1998), thus preventing Pds1 degradation and sister chromatid segregation. Human Bub1 (Chan et al. 1999) and BubR1 (Wu et al. 2000) have also been found to associate with the APC. However, their effect on the APC has not been determined.
Vertebrate cells require the spindle checkpoint for proper timing of anaphase, even when the spindle assembly is intact (Gorbsky et al. 1998; Taylor et al. 1998). Mice deficient in the checkpoint genes MAD2 (Dobles et al. 2000) or BUB3 (Kalitsis et al. 2000) die early during embryogenesis, indicating the importance of the spindle checkpoint in normal cell division. The control of accurate chromosome segregation by the spindle checkpoint implies that mutations in the checkpoint genes may contribute to aneuploidy, often associated with human cancers. In fact, mutations in human Bub1 have been found in the subtype of colorectal cancer that exhibits chromosome instability (Cahill et al. 1998). Despite the importance of Bub1 in the spindle checkpoint, its molecular mechanism remains to be elucidated. We have now isolated a Xenopus homologue of Bub1 and used Xenopus egg extracts to investigate the question of how Bub1 might interact with other checkpoint components, Mad1–Mad2, Bub3, and CENP-E, to trigger the spindle checkpoint signal. We demonstrate that Bub1 is required for kinetochore binding of Mad1–Mad2, Bub3, and CENP-E.
Materials and Methods
Isolation of Xenopus BUB1 and BUB3 cDNAs
Xenopus BUB1 cDNA was isolated using a procedure similar to that described for Xenopus MAD1 (Chen et al. 1998). Specifically, degenerate oligonucleotides were designed corresponding to the sequences WEFYIG (amino acids 829–834 in human Bub1) and IIHGDIKPDN (amino acids 913–922 in human Bub1), which are highly conserved among all known Bub1 homologues. These oligonucleotides were used as primers in PCRs to amplify a corresponding Xenopus sequence. The template for the PCR reaction was generated by reverse transcription from Xenopus egg total mRNA, with oligo-dT as a primer. A 282–base pair PCR product was cloned into TOPO™ vector (Invitrogen) and sequenced to verify its authenticity. This sequence was then used to generate a digoxigenin-labeled probe (Boehringer) for screening a Xenopus ovary cDNA library as described (Chen et al. 1998). Two independent cDNA clones of 3.1 and 2.8 kb were isolated and sequenced on both strands. The sequences revealed that both clones were partial sequences derived from the same gene. An additional sequence at the 5′ end was isolated using SMART™ RACE cDNA Amplification kit (CLONTECH Laboratories, Inc.).
To isolate the known Xenopus BUB3 cDNA (Goto and Kinoshita 1999) (GenBank/EMBL/DDBJ accession no. AB018419), oligonucleotide primers were made corresponding to the NH2- and COOH-terminal sequences MNTQTDM and DAETKPK, respectively. The recognition sequence for SalI was added at the 5′ end of the oligonucleotides for cloning. Using these oligonucleotides as primers, the BUB3 sequence was amplified by PCR from a Xenopus egg cDNA library and verified by sequencing.
Preparation of Recombinant Bub1 and Bub3 Proteins and Generation of Anti-Bub1 and Anti-Bub3 Antibodies
To generate recombinant Bub1 protein, a restriction fragment encoding amino acids 274–467 was subcloned into pQE10 (QIAGEN) at the cognate sites. To prepare recombinant Bub3, the full-length Bub3 coding sequence was cloned into pQE9 (QIAGEN) at the SalI site. The proteins were expressed in Escherichia coli with a hexahistidine tag at the NH2 terminus. Bub1274–467 was produced as a soluble protein, and purified as described for Xenopus Mad2 protein (Chen et al. 1996). Xenopus Bub3 protein was insoluble, and the denatured protein was purified from the inclusion bodies as described for Mad1 (Chen et al. 1998). The purified proteins were used to raise antibodies in rabbits (Covance Research Products, Inc.) and to affinity purify the antibodies as described (Chen et al. 1996).
Preparation of Egg Extracts and Activation of the Spindle Checkpoint
Cytostatic factor (CSF)-arrested egg extracts and demembranated frog sperm nuclei were prepared as described (Murray 1991). Interphase extracts were prepared by adding calcium chloride to the CSF-arrested extracts to a final concentration of 0.5 mM from a 10 mM stock, and the extracts were incubated at 23°C for 1 h. The protein synthesis inhibitor cycloheximide was added to 100 μg/ml during the last 30 min of incubation to prevent extracts from reentering mitosis. Activation of the spindle checkpoint was performed as described (Minshull et al. 1994).
To determine the effect of antibodies on checkpoint establishment, CSF-arrested extracts were first incubated with antibodies for 30 min on ice before the addition of sperm nuclei and nocodazole. To determine the effect on checkpoint maintenance, CSF-arrested extracts were incubated with sperm nuclei and nocodazole at 23°C for 30 min to activate the checkpoint, followed by the addition of antibodies and incubation for another 30 min at 23°C. To determine whether the checkpoint was active, calcium chloride was added to the extracts to a final concentration of 0.5 mM. Samples were taken every 15 min for histone H1 kinase activity measurement, and for examination of chromosomal morphology under a microscope (Chen et al. 1996).
Immunoprecipitations and Bub1 Autophosphorylation
To immunoprecipitate Bub1 or Bub3, 10 μl of Affi-prep protein A support beads (Bio-Rad Laboratories) were preincubated with 1 mg/ml BSA in PBS (2.7 mM potassium chloride, 137 mM sodium chloride, 1.5 mM potassium phosphate, 4.3 mM sodium phosphate, pH 7.2) for 30 min at 4°C. Beads were washed twice with 1 ml of extract buffer (XB: 10 mM Hepes, pH 7.8, 50 mM sucrose, 100 mM potassium chloride, 10 mM magnesium chloride, 1 mM calcium chloride) containing 5 mM EGTA, additional 1 mM magnesium chloride, and 10 μg/ml each of leupeptin, pepstatin, and chymostatin (LPC). After incubating with affinity-purified anti-Bub1 or anti-Bub3 antibody, or control IgG for 1 h at 4°C, the beads were washed four times with 1 ml XB plus 5 mM EGTA, 1 mM magnesium chloride, and LPC. 20 μl of CSF-arrested egg extracts was added to the antibody-coated beads and incubated at 4°C for 1 h. The beads were isolated by centrifugation at 16,000 g for 2 min, and washed four times with XB plus LPC. Proteins bound to the beads were solubilized in SDS-PAGE sample buffer and resolved by SDS-PAGE. Immunoblot analysis was performed as described (Chen et al. 1996).
For Bub1 autophosphorylation, Bub1 immunoprecipitates were washed twice with lysis buffer (10 mM K2HPO4, pH 7.2, 50 mM β-glycerophosphate, 1 mM EDTA, 5 mM EGTA, 1 mM MgCl2, 1 mM sodium vanadate, 2 mM DTT, 0.5% Triton X-100, 1 mM PMSF, and LPC) containing 0.5 M NaCl, twice with lysis buffer and once with kinase buffer (20 mM Hepes, 10 mM MgCl2, 0.1 mg/ml BSA, 3 mM β-mercaptoethanol). The immunoprecipitates were resuspended in 10 μl of kinase buffer containing 100 μM ATP and 10 μCi [γ-32P]ATP, and incubated at 30°C for 15 min. The reactions were terminated by the addition of SDS-PAGE sample buffer and heated at 95°C for 3 min. To ensure that the phosphorylated protein is indeed Bub1, anti-Bub1 antibody was used to immunoprecipitate Bub1 again from the kinase reactions after diluting SDS to 0.1% with lysis buffer. The immuoprecipitates were resolved by SDS-PAGE, and analyzed by autoradiography.
Lambda Protein Phosphatase Treatment
Bub1 or Bub3 was immunoprecipitated as described above. To preserve any phosphorylation, the immunoprecipitates were washed three times with lysis buffer, twice with XB plus 5 mM EGTA, 1 mM MgCl2, and LPC to remove residual phosphatase inhibitors in the lysis buffer, and once with lambda protein phosphatase (LPP) reaction buffer (50 mM Tris, pH 8.0, 2 mM MnCl2, 1 mg/ml acetylated BSA). The immunoprecipitates were incubated for 30 min at 30°C with 40 U of Lambda phosphatase (New England Biolabs, Inc.) in 50 μl of LPP reaction buffer. Control reactions were performed under the same conditions in 50 μl LPP reaction buffer with or without 2 mM zinc chloride, 1 mM sodium vanadate, and 50 mM sodium fluoride as phosphatase inhibitors.
Immunodepletion
Bub1 was depleted from CSF-arrested egg extract by two sequential immunoprecipitations. Two sets of Affi-prep protein A support beads were prepared as described above for immunoprecipitation. After the final wash, as much residual buffer as possible was removed. Beads were rinsed quickly with 20 μl of CSF-arrested egg extract, spun for 1 min at 4°C at 16,000 g, and the supernatant was discarded. 20 μl extract was then added to the antibody-coated beads and incubated on a rotator at 4°C for 30 min. The beads were then pelleted, and the supernatant was transferred to a new tube containing the second set of antibody-coated beads washed as above. The extract was incubated with beads for another 30 min at 4°C. At the end of the incubation, the beads were pelleted at 4°C and the supernatant was removed for experiments. Immunodepletion of Mad1 was performed as described (Chen et al. 1998).
Biotinylation of Antibodies
Affinity-purified anti-Bub1 antibodies were dialyzed against 50 mM NaHCO3, 50 mM NaCl, pH 8.5. NHS-LC-biotin (Pierce Chemical Co.), freshly prepared in water, was then added to a final concentration of 1 mg/ml and the antibodies were incubated on ice for 1.5 h. To quench the reaction, a solution containing 1 M glycine, 1 M Tris, pH 6.8, was added to a final concentration of 50 mM, and the antibodies were dialyzed against PBS plus 0.05% sodium azide to remove unreacted biotin.
Immunofluorescencent Staining of Chromosomes Assembled in Egg Extracts
For immunofluorescent staining of unreplicated chromosomes, 20 μl of egg extract was incubated with sperm nuclei (1,000/μl of extract) at 23°C for 10 min, followed by addition of nocodazole to 10 μg/ml, and incubation for another 20 min at 23°C to disrupt microtubules. The replicated chromosomes and the in vitro metaphase to anaphase transition were performed as described (Chen et al. 1998). The chromosomes were isolated and processed for immunofluorescent staining as described (Chen et al. 1998). The anti–CENP-E antibody was provided by Dr. T. Yen (Fox Chase Cancer Center, Philadelphia, PA). For results shown in Fig. 8, the coverslips were first incubated with rabbit antibodies against Mad1, Mad2, Bub3, or CENP-E, followed by fluorescein-conjugated anti–rabbit IgG (Jackson ImmunoResearch Laboratories). To detect Bub1 in the same samples, the coverslips were then incubated with biotinylated anti-Bub1 antibody, followed by Texas red–conjugated streptavidin (Jackson ImmunoResearch Laboratories). Images were taken using a charge-coupled device camera (MicroMAX-5MHz; Princeton Instruments, Inc.) attached to a fluorescence microscope (model E800; Nikon). Images were collected and processed with the Metamorph Imaging System (v4.0; Universal Imaging Corp.) and converted to Adobe Photoshop® format. Immunofluorescent staining from samples containing a low nuclear density (1,000/μl of extract) is the same as the high density (9,000/μl of extract) that is required for activation of the spindle checkpoint.
Figure 8.
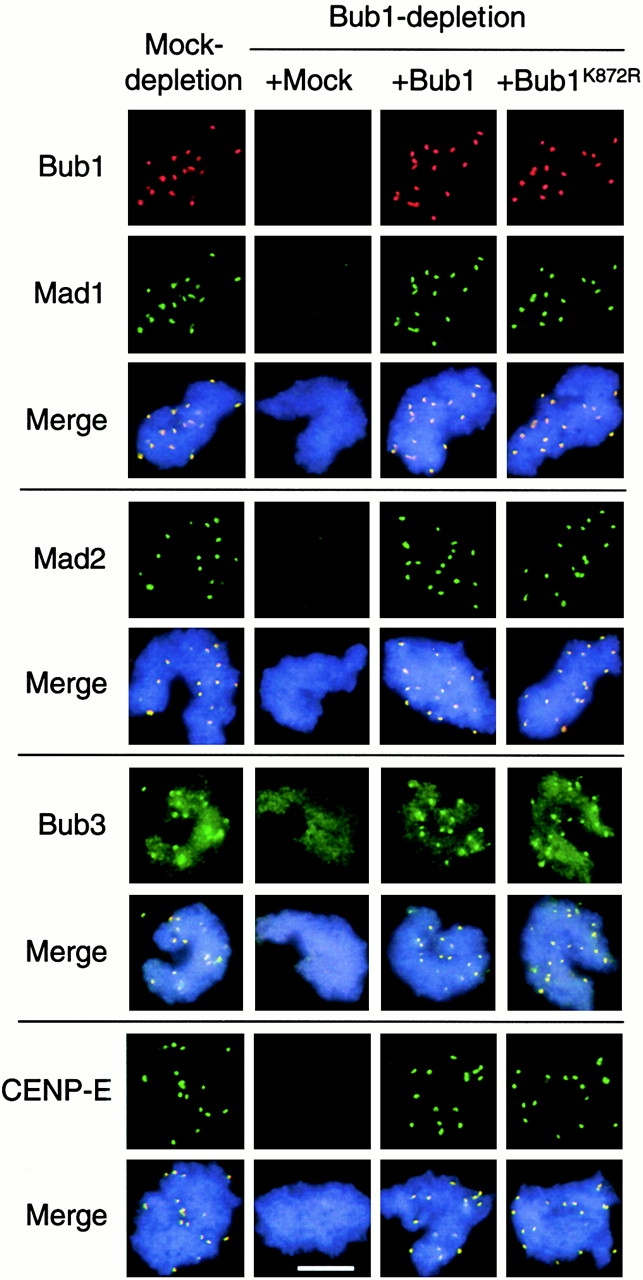
Bub1 and Bub1K872R restore kinetochore binding of Mad1, Mad2, Bub3, and CENP-E in Bub1-depleted extract. Sperm nuclei and nocodazole were incubated with mock-depleted extract or Bub1-depleted extract supplemented with mock, Bub1, or Bub1K872R translation as indicated on the top. Isolated chromosomes were incubated with rabbit antibodies against Mad1, Mad2, Bub3, or CENP-E as indicated on the left, followed by fluorescein-conjugated anti–rabbit antibody. Bub1 was then detected with biotinylated anti-Bub1 antibody and Texas red–conjugated streptavidin. For space conservation, pictures of Bub1 are shown only once for each type of extract. The merge pictures contain all three fluorochromes. Bar, 10 μm.
Generation of Kinase-deficient Bub1 and Translation of Bub1 in Egg Extracts
The kinase-deficient mutant of Bub1 was generated by overlapping PCR mutagenesis in order to change lysine 872 to arginine in the kinase domain of Bub1. To produce proteins in egg extracts, the coding region of Bub1 or Bub1K872R was cloned into a modified pGEM transcription vector that contains 5′ and 3′ untranslated regions of Xenopus cyclin B1, and the Kozak consensus sequence (ACCATGG) to facilitate translation in egg extracts. The plasmids were first cleaved with XmnI that is located 3′ to the polyadenylation signal. The linearized DNA was used to produce transcripts using the mMESSAGE mMACHINE T7 transcription kit (Ambion). Typically, we reconstitute purified transcripts from a 20-μl transcription reaction with 5 μl ribonuclease-free water. This yields transcripts of ∼4 mg/ml. To remove secondary structures, the transcripts were heated to 65°C for 3 min and left on ice immediately before use. Translation reaction was performed in CSF-arrested extracts immunodepleted of the endogenous Bub1. 21 μl of the extract was first incubated with 0.5 μl of recombinant RNasin (Promega) to 0.8 U/μl for 15 min on ice. The RNasin-treated extracts were then supplemented with 2.5 μl of rabbit reticulocyte lysate (Ambion), then incubated with 1 μl of transcript at 23°C for 4–5 h, with gentle mixing every 15 min. Mock translations were performed in parallel with ribonuclease-free water in place of transcripts. Aliquots of the translations were frozen in liquid nitrogen and stored at −80°C before use. Translation products were confirmed by immunoblot. Bub1 produced in the standard reactions was usually 10–20-fold over the endogenous levels.
Results
Characterization of Xenopus Bub1
We isolated a Xenopus BUB1 cDNA (GenBank/EMBL/DDBJ accession no. AF348328) that predicts a protein of 1,137 amino acids. An alignment between human, mouse, and budding yeast Bub1 revealed that this clone encodes a Xenopus Bub1 homologue rather than a Bub1-related protein (BubR1), because it has significant overall identity with human (37.2%) and mouse (37.8%) Bub1, and 13.2% identity with yeast Bub1. When compared with BubR1, this sequence shows only 15.6 and 13.2% overall identity with the human and mouse proteins, respectively. The identity is even less in the regions that are specific to BubR1.
By immunoblot analysis, the anti-Bub1 antibody generated against amino acids 274–467 of Bub1 specifically recognized a protein of ∼150 kD in mitotic frog egg extracts (Fig. 1 A, lane 1). Antibodies made against a known Xenopus Bub3 sequence (Goto and Kinoshita 1999) specifically recognized a 45-kD doublet (Fig. 1 A, lane 7). Immunoprecipitation with anti-Bub1 antibodies isolated the 150-kD protein (Fig. 1 A, lane 4) that was also present in the anti-Bub3 immunoprecipitate (Fig. 1 A, lane 6). Similarly, both 45-kD polypeptides that were recognized by the anti-Bub3 antibodies were present in the anti-Bub1 immunoprecipitates (Fig. 1 A, lane 10). Interestingly, only the slower-migrating polypeptide in the 45-kD doublet was immunoprecipitated by the anti-Bub3 antibodies (Fig. 1 A, lane 12). It appears that the anti-Bub3 antibodies can recognize both polypeptides that have been denatured, as in the case of immunoblot analysis (Fig. 1 A, lanes 7 and 10). However, the antibodies cannot recognize the faster migrating species under native conditions, as in the case of immunoprecipitation (Fig. 1 A, lane 12). We do not know whether these two polypeptides represent two closely related gene products, or whether they are isoforms of the same gene product. Nevertheless, these results demonstrate that Xenopus Bub1 and Bub3 associate with each other, and that the Bub1 we have isolated is an authentic Bub1 homologue.
Figure 1.
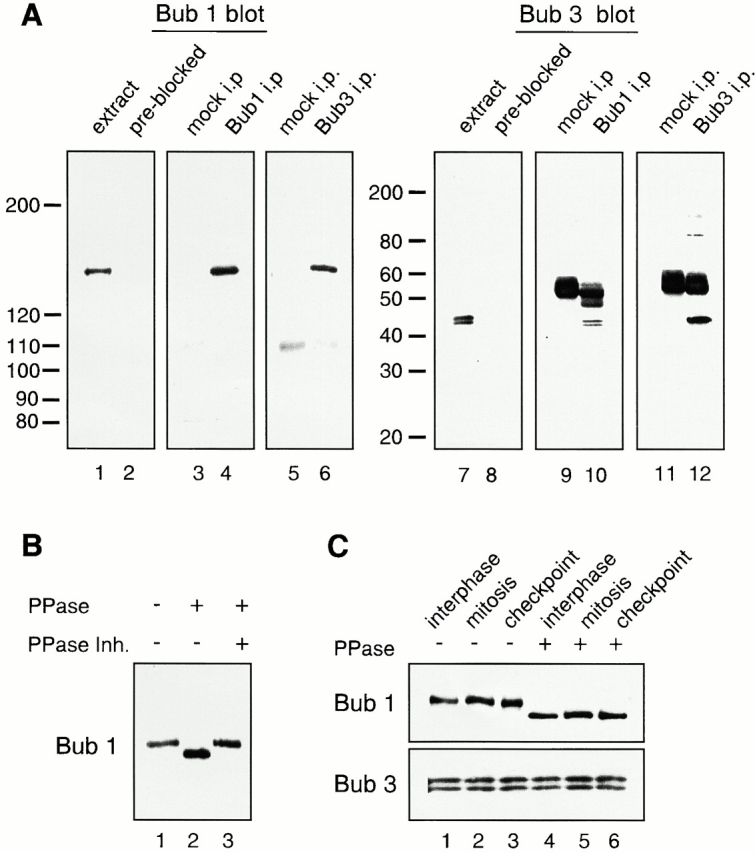
Analysis of Bub1 and Bub3 from Xenopus egg extracts. (A) Characterization of affinity-purified anti-Bub1 and anti-Bub3 antibodies. CSF-arrested egg extracts (lanes 1, 2, 7, and 8), or immunoprecipitates generated with control IgG (lanes 3, 5, 9, and 11), with anti-Bub1 (lanes 4 and 10) or anti-Bub3 antibodies (lanes 6 and 12) were probed with affinity-purified anti-Bub1 antibodies (lanes 1, 3–6), anti-Bub1 antibodies preblocked with recombinant Bub1 protein (lane 2), anti-Bub3 antibodies (lanes 7, 9–12), or anti-Bub3 preblocked with recombinant Bub3 protein (lane 8). The migration of molecular weight standards is indicated on the left. The prominent 55-kD protein is the IgG heavy chain. (B) Bub1 is a phosphoprotein. Bub1 was immunoprecipitated from CSF-arrested extract and treated with LPP (PPase) in the presence or absence of phosphatase inhibitors (PPase Inh.) as indicated on the top. (C) Bub1 is phosphorylated at interphase and mitosis, and under the checkpoint-active condition. Bub1 and Bub3 were coimmunoprecipitated with anti-Bub1 antibodies from interphase (lanes 1 and 4), CSF-arrested (lanes 2 and 5, mitosis), or checkpoint-active extracts (lanes 3 and 6, checkpoint), and left untreated (lanes 1–3) or treated with LPP (lanes 4–6). The upper panel was probed with anti-Bub1 antibody and the lower panel was probed with anti-Bub3 antibody.
Xenopus Bub1 Is a Phosphoprotein
Bub1 is a serine–threonine protein kinase that can phosphorylate itself (Roberts et al. 1994), Bub3 (Roberts et al. 1994), Mad1 (Seeley et al. 1999), and the tumor suppressor adenomatous polyposis coli (Kaplan et al. 2001), in vitro. Phosphorylated Bub1 shows a slower mobility on SDS-PAGE (Roberts et al. 1994; Chan et al. 1998). The protein recognized by our anti-Bub1 antibodies is substantially larger than its predicted molecular mass of 131 kD. Similarly, Bub3 is also larger than its predicted molecular mass of 37 kD. To determine if the apparent large size of Bub1 and Bub3 on SDS-PAGE was due to phosphorylation, we treated the Bub1 immunoprecipitate prepared from mitotic extract with the LPP. Fig. 1 B showed that the molecular mass of Bub1 was reduced to 140 kD upon phosphatase treatment (lane 2), indicating that Bub1 is phosphorylated during mitosis.
Next, we determined whether phosphorylation of Bub1 was regulated during the cell cycle. Bub1 immunoprecipitated from interphase, mitotic, or spindle checkpoint–active extracts showed a similar mobility on SDS-PAGE (Fig. 1 C, top, lanes 1–3). Phosphatase treatment of these immunoprecipitates showed a similar increase of mobility (Fig. 1 C, top, lanes 4–6). Thus, Bub1 is phosphorylated at both interphase and mitosis, and under the checkpoint-active condition. On the other hand, Bub3 that has been coimmunoprecipitated with Bub1 and similarly treated did not show any change in mobility (Fig. 1 C, bottom).
Bub1 Is Necessary for the Spindle Checkpoint
The spindle checkpoint can be reproduced in Xenopus egg extracts. The mature eggs are arrested at metaphase of the second meiotic division by the CSF. Fertilization of eggs triggers a calcium transient, leading to completion of meiosis and commencement of mitotic divisions. CSF-arrested extract, the cytoplasmic extracts prepared from eggs, can reproduce the spindle checkpoint upon incubation with nocodazole and a high concentration of sperm nuclei (Minshull et al. 1994). Once the spindle checkpoint is activated in the extracts, addition of calcium is unable to induce the mitotic exit. The mitotic arrest can be determined by examining nuclear morphology and by measuring Cdc2-associated histone H1 kinase activity. First, we asked if anti-Bub1 antibodies have an effect on the spindle checkpoint. In the absence of antibodies, the extract maintained a high level of H1 kinase activity and the chromosomes remained condensed for at least 60 min after the addition of calcium (Fig. 2 A, top), indicative of checkpoint activation. When the extract was preincubated with anti-Bub1 antibodies before adding sperm nuclei and nocodazole, calcium induced inactivation of the H1 kinase within 15 min, and the chromosomes assembled into interphase nuclei within 60 min (Fig. 2 A, middle). The failure to sustain mitotic arrest indicated that the antibody interfered with the checkpoint. This effect was specific to Bub1, as antibody preblocked with recombinant Bub1 had no effect on the checkpoint (Fig. 2 A, bottom). Anti-Bub1 antibody also abolished the checkpoint when added after the checkpoint had been established with sperm nuclei and nocodazole (Fig. 2 B). This effect is similar to that of anti-Mad1 and anti-Mad2 antibodies, which interfere with both establishment and maintenance of the checkpoint (Fig. 2 B, and Chen et al. 1996, Chen et al. 1998).
Figure 2.
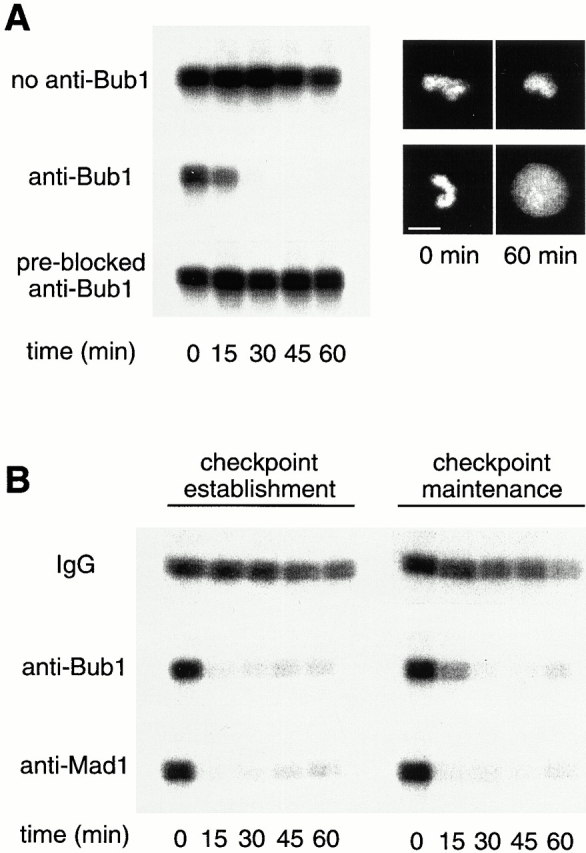
Bub1 is necessary for the establishment and maintenance of the spindle checkpoint in frog egg extract. (A) Anti-Bub1 antibodies abolish the spindle checkpoint. CSF-arrested extracts were incubated for 30 min on ice without addition of anti-Bub1 antibody (top), with anti-Bub1 antibodies (middle), or with anti-Bub1 antibodies preblocked with recombinant Bub1 protein (bottom). Sperm nuclei and nuclei were added for 20 min, followed by the addition of calcium chloride to overcome the metaphase arrest. Samples were taken immediately before (t = 0) the addition of calcium and every 15 min thereafter, and histone H1 kinase activity was determined. Autoradiograms of histone H1 kinase assay as well as photographs of chromosomes at times indicated at bottom are presented. (B) Anti-Bub1 antibodies abolish the establishment and maintenance of the spindle checkpoint. For the effect on checkpoint establishment, CSF-arrested extracts were preincubated with control IgG (top), anti-Bub1 (middle), or anti-Mad1 antibodies (bottom) as described in A. For the effect on checkpoint maintenance, checkpoint was first established in extracts with sperm nuclei and nocodazole, followed by the addition of antibodies. Samples were taken for histone H1 kinase assay as in A. The decline in H1 kinase activity was accompanied by decondensation of chromosomes (data not shown). Bar, 10 μm.
Xenopus Bub1 Localizes to Kinetochores
We performed immunofluorescent staining to determine if Xenopus Bub1 associates with kinetochores like its homologues in other species. Sperm nuclei were first allowed to replicate in interphase extract, followed by addition of CSF-arrested extracts to induce mitosis and to assemble metaphase structures. Unattached kinetochores were generated by the addition of nocodazole to disrupt microtubules. In nocodazole-treated samples, anti-Bub1 antibody showed a punctate staining that colocalized with the anti-Mad1 antibody staining (Fig. 3 A, middle), indicating that Bub1 associates with unattached kinetochores. Without nocodazole treatment, the Bub1 kinetochore staining was reduced, whereas Mad1 was absent from kinetochores (Fig. 3 A, bottom, and Chen et al. 1998). The kinetochore staining is specific to Bub1, because antibody preblocked with recombinant Bub1 did not stain kinetochores (Fig. 3 A, top). Immunofluorescent staining in unreplicated chromosomes showed a similar pattern (our unpublished results). Thus, unlike Mad1 or Mad2, microtubule attachment to kinetochores is not sufficient to dissociate Bub1 from kinetochores.
Figure 3.
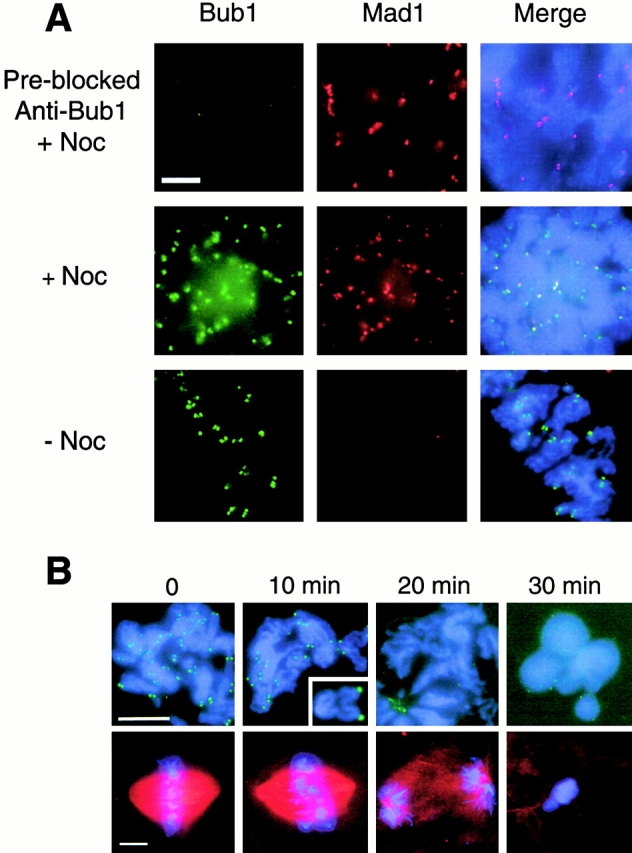
Kinetochore binding of Bub1 during the metaphase to anaphase transition. (A) Immunofluorescent staining of Bub1 and Mad1 in replicated sperm chromosomes in the presence (+Noc) or absence (−Noc) of nocodazole. Chromosomes were isolated and stained with rabbit anti-Bub1 and mouse anti-Mad1 antibodies. Fluorescein-conjugated anti–rabbit and Texas red–conjugated anti–mouse IgG antibodies were used as secondary antibodies. The DNA was stained with the DNA-binding dye Hoechst 33258. The merges of all three fluorochromes are also shown (Merge). All pictures were taken using the same magnification. (B) Bub1 dissociates from kinetochores during anaphase. Metaphase chromosomes were assembled with the addition of rhodamine-conjugated tubulin to visualize the mitotic spindle. Synchronous anaphase was induced by adding calcium chloride to inactivate the CSF activity, and samples were taken at times indicated on top. Chromosomes were isolated and stained with anti-Bub1 antibody and with Hoechst 33258 (top). A second aliquot of the sample was fixed directly with a solution containing formaldehyde and Hoechst 33258 to visualize the mitotic spindle and chromosomes (bottom). Bub1 dissociates from kinetochores after sister chromatids are well separated by 20 min after calcium addition. The inset shows an enlargement of a pair of sister chromatids that have clearly separated and still retain Bub1. Bars, 10 μm.
The in vitro metaphase established from replicated nuclei enters anaphase upon addition of calcium to inactivate the CSF activity. Anaphase began within 10 min of calcium addition, as evidenced by the increase in the distance between sister kinetochores compared with those at meta-phase (Fig. 3 B). At this stage, Bub1 still associated with kinetochores (Fig. 3 B). By 20 min, sister chromatids were well separated and kinetochores were not stained with anti-Bub1 antibody (Fig. 3 B), showing that Bub1 dissociated from kinetochores during anaphase.
Bub1 Is Required for Kinetochore Localization of Mad1, Mad2, Bub3, and CENP-E
Localization of spindle checkpoint proteins Bub1, Bub3, Mad1, Mad2, and CENP-E to kinetochores prompted us to ask whether there is any dependency between these checkpoint proteins. We investigated this question by using anti-Bub1 or anti-Mad1 antibody to remove Bub1 or Mad1, respectively, from egg extracts. Immunoblot analysis showed that immunodepletion with anti-Bub1 antibodies removed >95% of Bub1, without affecting the bulk of Mad1, Mad2, CENP-E, or Bub3 (Fig. 4, lane 6). Similarly, depletion with anti-Mad1 antibodies removed >95% of Mad1 and a fraction of Mad2, without significantly reducing Bub1, Bub3, or CENP-E (Fig. 4, lane 7). Consistent with this result, immunoblot analysis of the Bub1 immunoprecipitate detected only a small amount of Mad1 besides Bub1 and Bub3 (Fig. 4, lanes 9 and 12). A low level of Bub1 was also present in Mad1 immunoprecipitate, whereas Mad2 and CENP-E were undetectable (Fig. 4, lanes 10 and 13).
Figure 4.
Immunoblot analysis of Bub1, Bub3, Mad1, Mad2, and CENP-E in extracts depleted of Bub1 or Mad1, and in Bub1 or Mad1 immunoprecipitates. The blot contains a serial dilution of CSF-arrested extract (100, 50, 20, and 10%, lanes 1–4), mock-depleted (lane 5), Bub1-depleted (lane 6), Mad1-depleted (lane 7) extracts, or immunoprecipitates generated with control IgG (Mock IP, lanes 8 and 11), anti-Bub1 (Bub1 IP, lanes 9 and 12), or anti-Mad1 antibodies (Mad1 IP, lanes 10 and 13). The blots were probed with antibodies against CENP-E, Bub1, Mad1, Bub3, or Mad2 as indicated on the left. The blots shown in lanes 11–13 are longer exposure of the same blots shown in lanes 8–10. Immunodepletion of Bub1 generally removed 10–20% of Bub3 and depletion of Mad1 removed 20–40% of Mad2 as determined by NIH Image software.
In Bub1-depleted extract, kinetochore staining of Bub1, Bub3, Mad1, Mad2, and CENP-E was diminished (Fig. 5), indicating that Bub1 is required for these checkpoint proteins to bind to kinetochores. Besides kinetochore staining, anti-Bub3 antibody gave an overall chromosomal staining that was not affected by the Bub1 depletion (Fig. 5). Anti-Bub3 antibodies preblocked with recombinant Bub3 protein did not stain kinetochores (data not shown), indicating its specificity to Bub3. Immunodepletion of Mad1 abolished kinetochore localization of Mad2, whereas the levels of Bub1 and Bub3 at kinetochores were the same as in mock-depleted extract (Fig. 5). Thus, kinetochore binding of Bub1–Bub3 complex is independent of Mad1–Mad2. Interestingly, CENP-E staining at kinetochores was reduced in Mad1-depleted extract (Fig. 5).
Figure 5.
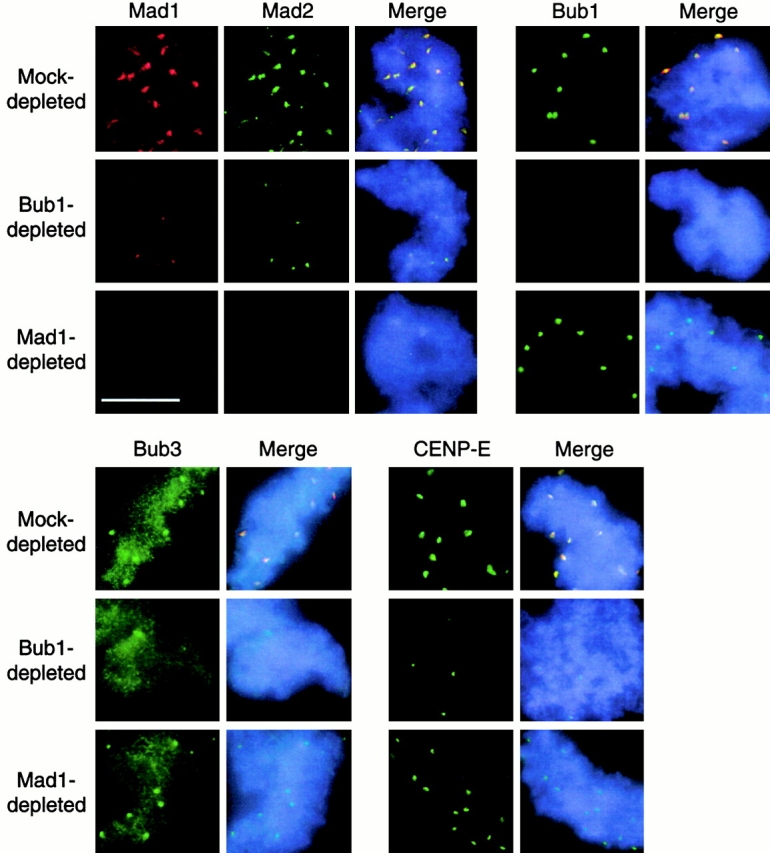
Dependency of kinetochore binding between various checkpoint proteins. Mitotic chromosomes were assembled in mock-depleted, Bub1-depleted, or Mad1-depleted extracts in the presence of nocodazole. The isolated chromosomes were then stained with mouse anti-Mad1 antibody along with rabbit anti-Mad2, anti-Bub1, anti-Bub3, or anti–CENP-E antibodies. The use of secondary antibodies and DNA staining were as described in the legend to Fig. 3. For space conservation, pictures of Mad1 staining are shown only once for each type of extract. Photographs for each antibody staining were taken for the same exposure time to reflect a difference in the staining level. Bar, 10 μm.
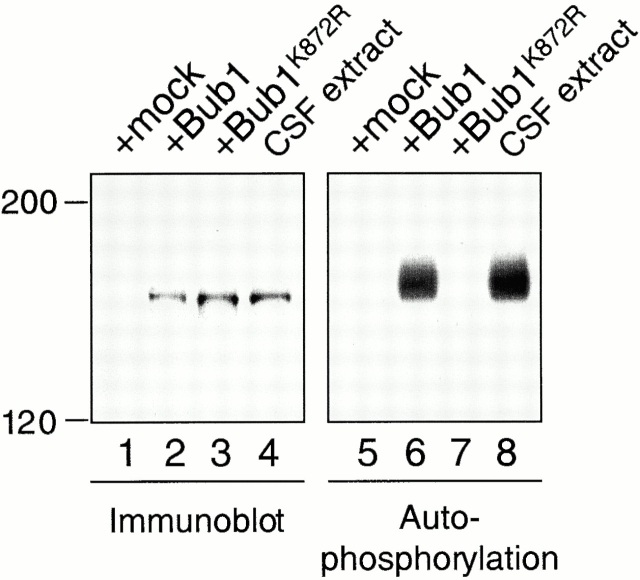
To determine if the kinase activity of Bub1 is necessary for targeting other checkpoint proteins to kinetochores, we generated a Bub1 mutant (Bub1K872R) by changing the highly conserved lysine residue in the kinase domain (amino acid 872) to arginine. The RNAs corresponding to either the wild-type Bub1 or the Bub1K872R mutant were synthesized by in vitro transcription. To produce proteins directly in the physiological condition, the transcripts were then added to egg extracts depleted for the endogenous Bub1. Immunoprecipitation of the translated proteins showed that the wild-type protein underwent autophosphorylation to a similar extent as the endogenous Bub1, whereas Bub1K872R failed to phosphorylate itself (Fig. 6). This demonstrates that the translated Bub1 behaves like the endogenous protein, and that Bub1K872R is kinase deficient.
Figure 6.
Bub1K872R is kinase deficient. Bub1 protein in various extracts was analyzed by immunoblot (lanes 1–4), or immunoprecipitated for an in vitro autophosphorylation reaction (lanes 5–8). Bub1-depleted extract was supplemented with mock translation (lanes 1 and 5), Bub1 translation (lanes 2 and 6), or Bub1K872R translation (lanes 3 and 7). Lanes 4 and 8, CSF-arrested extract. The migration of molecular weight standards is indicated on the left.
The translated Bub1 and Bub1K872R were tested for their checkpoint function in extracts depleted for the endogenous Bub1. Without calcium addition, mock- and Bub1-depleted extracts contained a similar level of H1 kinase activity, indicating that the extracts were still arrested by the CSF and that Bub1 is not necessary for maintaining the CSF-induced arrest (Fig. 7 A). However, Bub1-depleted extract failed to support the spindle checkpoint, because H1 kinase activity declined after calcium addition even in the presence of nocodazole and a high density of nuclei (Fig. 7 A). The checkpoint is restored when the wild-type Bub1 translation was added back to the depleted extract (Fig. 7 A). Interestingly, addition of Bub1K872R also sustained the H1 kinase activity (Fig. 7 A) and condensed chromosomes (data not shown), indicating that the kinase-deficient protein still retained the checkpoint function. Consistent with the H1 kinase assay, addition of either Bub1 or Bub1K872R allowed other checkpoint proteins to bind kinetochores in extracts depleted for endogenous Bub1 (Fig. 8).
Figure 7.
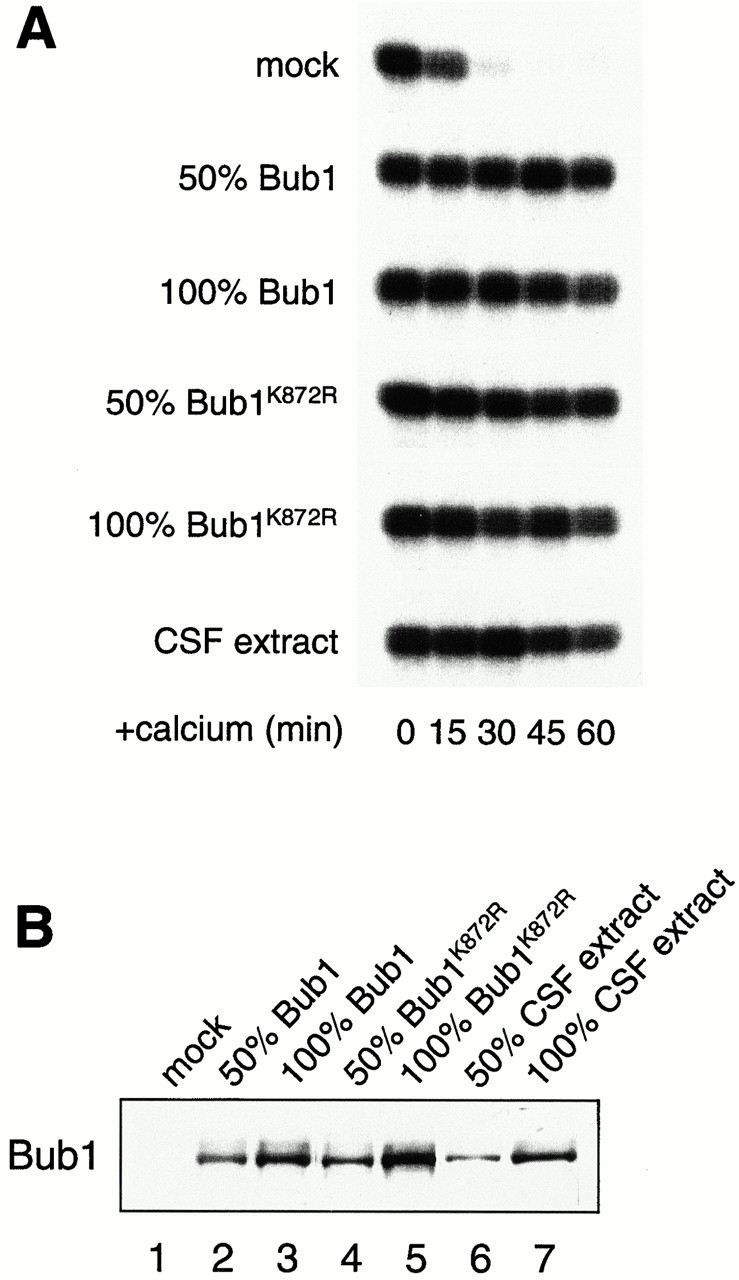
Bub1K872R is functional in the spindle checkpoint. (A) Bub1-depleted extracts containing mock, Bub1, or Bub1K872R translation as indicated on the left were incubated with sperm nuclei and nocodazole. Samples were taken for histone H1 kinase measurement as described in the legend to Fig. 2. The level of added protein compared with the endogenous protein is indicated on the left. Both Bub1 and Bub1K872R restore the checkpoint when added at 50–100% of the level of the endogenous protein. (B) Immunoblot of Bub1 for samples used in A.
Discussion
We have isolated a Xenopus Bub1 homologue and studied its role in the spindle checkpoint in Xenopus egg extracts. Four lines of evidence indicate that the cDNA we have isolated encodes a Bub1 homologue. First, this sequence shows a high degree of homology with Bub1 from other species. Second, antibodies made against recombinant Bub1 abolish the spindle checkpoint in egg extract, demonstrating the essential role of Bub1 in the spindle checkpoint (Fig. 2). Third, Xenopus Bub1 coimmunoprecipitates with Bub3, consistent with the interaction found in other species (Fig. 1 A). Fourth, the sequence shows a higher degree of homology to the known Bub1 proteins from other species than to Bub1-related proteins (BubR1). Although the human BubR1 also interacts with Bub3 (Taylor et al. 1998), our antibodies are not expected to recognize a potential Xenopus BubR1 because the antibodies are raised against a region unique to Bub1. This region is <11% homologous to known BubR1 sequences. In addition, a unique Xenopus EST sequence (GenBank/EMBL/DDBJ accession no. BE025630) predicts a polypeptide with similarity to BubR1 and may be derived from Xenopus BUBR1. While our manuscript was being reviewed, another study independently isolated a Xenopus BUB1 cDNA (GenBank/EMBL/DDBJ accession no. AF11978) (Schwab et al. 2001). Our sequence differs from AF11978 by 13 nucleotides scattered in the coding region, which leads to a difference of nine amino acids and one extra amino acid in length. We believe that these two clones represent the same gene, and that the small difference may come from a genetic variation.
The mass of Xenopus Bub1 recognized by our anti-Bub1 antibodies appears to be larger than its predicted molecular mass. This apparent size is due at least in part to phosphorylation. The protein is phosphorylated at both interphase and mitosis, and under spindle checkpoint–active conditions (Fig. 1 B). Studies in human cells have demonstrated that BubR1 is phosphorylated during mitosis and becomes hyperphosphorylated when microtubules are disrupted by nocodazole treatment (Chan et al. 1999). Bub1 in interphase and mitotic egg extracts shows a similar mobility on SDS-PAGE (Fig. 1 C), suggesting a similar extent of phosphorylation under these conditions. Even though the bulk of Bub1 may not change at different stages of the cell cycle, it is conceivable that a fraction of Bub1 may be regulated only at kinetochores. A recent study shows that Xenopus Bub1 becomes phosphorylated during oocyte maturation via the NOS pathway. It has thus been hypothesized that Bub1 may be involved in the CSF-induced metaphase arrest (Schwab et al. 2001). We now demonstrate that immunodepletion of Bub1 has no effect on this arrest (Fig. 7), indicating that Bub1 is not necessary for maintaining the CSF activity.
Both Xenopus Bub1 and Bub3 associate with kinetochores (Fig. 3 and Fig. 5), consistent with studies in other species (Taylor and McKeon 1997; Chan et al. 1998; Jablonski et al. 1998; Basu et al. 1999) and in Xenopus oocytes (Schwab et al. 2001). Bub1 remains associated with kinetochores that have attached microtubules. Disruption of microtubules increases the level of Bub1 at kinetochores (Fig. 3 A). The protein dissociates completely from kinetochores during anaphase (Fig. 3 B). Unlike Bub1, kinetochore binding of Mad1 and Mad2 is controlled by microtubule attachment.
The kinetochore localization of the checkpoint proteins Bub1, Bub3, Mad1, Mad2, and CENP-E suggests a potential interaction among these proteins. Our previous study has demonstrated that kinetochore binding of Mad2 is dependent on Mad1 (Chen et al. 1998). We have now determined the dependency between CENP-E and the Mad1–Mad2 and Bub1–Bub3 complexes. Immunodepletion of Bub1 from egg extracts abolishes kinetochore binding of Bub1, Bub3, Mad1, Mad2, and CENP-E (Fig. 5), and the binding is restored upon adding back wild-type Bub1 protein (Fig. 8). This demonstrates that Bub1 rather than a Bub1-interacting protein is required for these checkpoint proteins to bind kinetochores. Consistent with our finding, Drosophila Bub3 fails to localize to kinetochores when the BUB1 gene is deleted (Basu et al. 1998). Studies in human cells also show that interaction with Bub3 is required for Bub1 to localize to kinetochores (Taylor et al. 1998), indicating a role for Bub3 in targeting Bub1 to kinetochores. All together, these results demonstrate a mutual dependency between Bub1 and Bub3 in kinetochore binding.
One of the possible roles for Bub1 is to physically provide a platform on the kinetochore to which checkpoint proteins, including Mad1–Mad2 complex and CENP-E, can bind (Fig. 9). In the absence of Bub1, other checkpoint proteins are unable to assemble onto the kinetochore. Consistent with this notion, human Bub1 has been found to assemble onto the kinetochore before CENP-F, BubR1, and CENP-E (Jablonski et al. 1998). The order of kinetochore binding between CENP-E and the Mad1–Mad2 complex is not clear. However, a potential interaction among these proteins is indicated by the observation that anti–CENP-E antibodies dissociate Mad1 and Mad2 from kinetochores during checkpoint maintenance (Abrieu et al. 2000). Mad1–Mad2 may interact directly with Bub1 (Fig. 9 A) or indirectly with Bub1 through CENP-E (Fig. 9 B). Furthermore, CENP-E is known to bind BubR1 in human cells (Chan et al. 1998). We did not detect CENP-E in a Bub1 immunoprecipitate prepared from CSF-arrested extract (Fig. 4). It remains a possibility that CENP-E may interact with Bub1 only at the kinetochore, or that our anti-Bub1 antibody may disrupt any weak interaction between CENP-E and Bub1. The loss of CENP-E staining at the kinetochore in Bub1-depleted extract suggests that BubR1 alone is not sufficient to localize CENP-E or that Bub1 may also be required for BubR1 to bind kinetochores.
Figure 9.
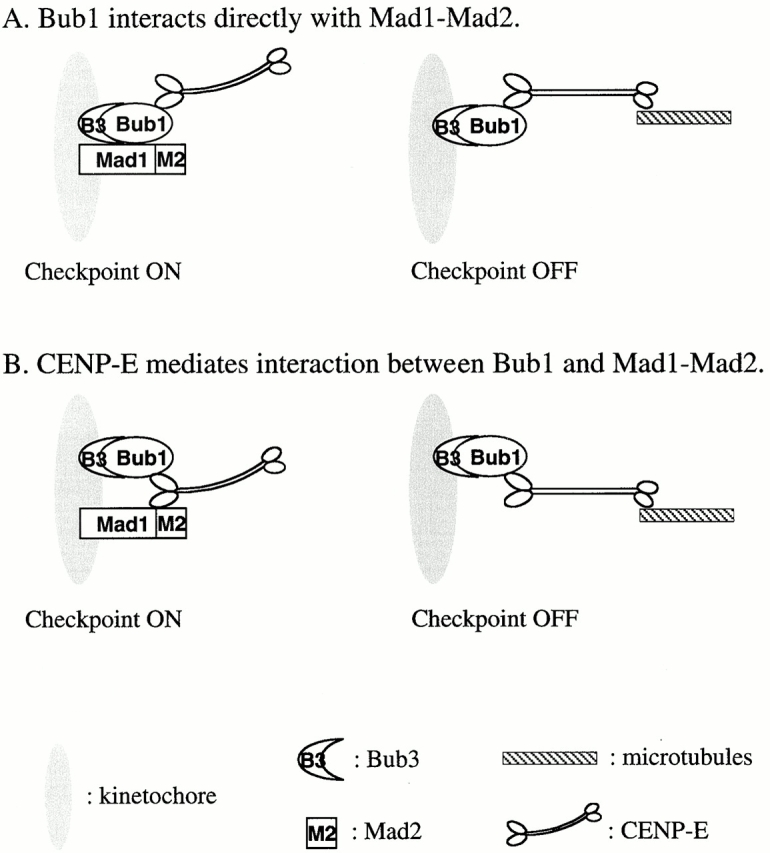
Models for the interaction of spindle checkpoint proteins at the kinetochore. (A) Bub1 interacts directly with CENP-E and Mad1–Mad2 on unattached kinetochores. Binding of CENP-E to microtubules may induce a conformational change in Bub1 that disrupts its interaction with the Mad1–Mad2 complex. The anti–CENP-E antibody may produce a similar effect on Bub1, thus dissociating Mad1–Mad2 from kinetochores (Abrieu et al. 2000). (B) Bub1 interacts with Mad1–Mad2 through CENP-E. Loss of Mad1–Mad2 on kinetochores in Bub1-depleted extracts may be a result of a lack of CENP-E on kinetochores.
The spindle checkpoint is abolished when egg extract is incubated with anti-Bub1 antibodies before the checkpoint establishment (Fig. 2). Under this condition, Bub1 is still at kinetochores, whereas kinetochore staining of Mad1 is diminished (our unpublished result). Yeast Mad1 forms complexes with Bub1 and Bub3 under checkpoint-active conditions, and the complex can be coimmunoprecipitated (Brady and Hardwick 2000). We also detect a low level of interaction between Bub1 and Mad1 by immunoprecipitation (Fig. 4). It remains a possibility that our anti-Bub1 antibody may have been raised against the region of Bub1 that is critical for a stable interaction between Bub1 and Mad1 at kinetochores, so that the antibody blocks the Mad1 kinetochore binding during check-point establishment.
Addition of anti-Bub1 antibodies to egg extracts during checkpoint maintenance also abolishes the checkpoint (Fig. 2). Interestingly, under this condition Bub1 as well as Mad1, Mad2, and CENP-E remain on kinetochores (our unpublished result), indicating that anti-Bub1 antibodies still have access to Bub1 on the kinetochore when added after checkpoint establishment. The result also suggests that once Bub1 and Mad1 have established their interaction with kinetochores or with each other, this interaction cannot be easily disrupted by anti-Bub1 antibodies.
Besides a direct physical interaction, Bub1 may phosphorylate other kinetochore components or checkpoint proteins. Interestingly, we find that a kinase-deficient Bub1 mutant is capable of restoring the spindle checkpoint and the kinetochore binding of other checkpoint proteins in Bub1-depleted extracts (Fig. 7 and Fig. 8), indicating that the kinase activity of Bub1 is not essential for its checkpoint function. In yeast, Bub1 with a similar mutation also partially rescues the sensitivity of bub1-1 to the microtubule poison benomyl (Roberts et al. 1994), suggesting that the kinase-deficient Bub1 in yeast retains a partial function. Even though the kinase activity of Xenopus Bub1 is not essential for the checkpoint, we cannot exclude the possibility that the kinase activity may allow the checkpoint to work in an efficient way that is not detectable in our assays. Alternatively, phosphorylation by Bub1 may regulate mitotic events other than the spindle checkpoint. Identification of the physiological substrates for Bub1 shall address these possibilities. In yeast, hyperphosphorylation of Mad1 during mitosis is dependent on its physical interaction with Mad2 as well as functional Bub1 and Bub3 (Hardwick and Murray 1995; Chen et al. 1999). Together with our study, this demonstrates that Bub1 works upstream of Mad1 and regulates Mad1 function. In yeast, the upstream kinase for Mad1 is thought to be Mps1 rather than Bub1 (Hardwick et al. 1996), whereas human Bub1 translated in vitro is able to phosphorylate Mad1 (Seeley et al. 1999). It remains to be determined whether vertebrate Bub1 is a physiological kinase for Mad1 and whether it modulates Mad1 function through phosphorylation.
Immunodepletion of Mad1 abolishes kinetochore binding of both Mad1 and Mad2 without affecting Bub1 and Bub3 (Fig. 4), indicating that the Mad1–Mad2 complex is not required for binding of Bub1 and Bub3 to the kinetochore. This is consistent with the finding that Bub1 and Bub3 remain on kinetochores that have attached to microtubules and have very little or no Mad1–Mad2 proteins. Interestingly, in Mad1-depleted extract the level of CENP-E at kinetochores is reduced but not abolished, suggesting that Mad1 may help to stabilize CENP-E at kinetochores. A recent study has shown that CENP-E is required for Mad1 and Mad2 to bind kinetochores (Abrieu et al. 2000). Thus, a mutual dependency likely exists between CENP-E and the Mad1–Mad2 complex.
Our study demonstrates that Bub1 plays a central role in generating the spindle checkpoint signal from the kinetochore. The protein sets the stage for binding of other checkpoint components, including Mad1, Mad2, Bub3, and CENP-E, to the kinetochore. Identification of the Xenopus Bub1 homologue allows future investigation of its biochemical and molecular mechanism by the use of Xenopus egg extracts.
Acknowledgments
We thank Dr. T. Huffaker for critically reading the manuscript.
This work was supported by grants from the National Institutes of Health and the David and Lucille Packard Foundation to R.-H. Chen.
Footnotes
Abbreviations used in this paper: APC, anaphase-promoting complex; CSF, cytostatic factor; LPC, leupeptin, pepstatin, and chymostatin; LPP, lambda protein phosphatase; MAP, mitogen-activated protein; XB, extract buffer.
References
- Abrieu A., Kahana J.A., Wood K.W., Cleveland D.W. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell. 2000;102:817–826. doi: 10.1016/s0092-8674(00)00070-2. [DOI] [PubMed] [Google Scholar]
- Amon A. The spindle checkpoint. Curr. Opin. Genet. Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- Basu J., Logarinho E., Herrmann S., Bousbaa H., Li Z., Chan G.K., Yen T.J., Sunkel C.E., Goldberg M.L. Localization of the Drosophila checkpoint control protein Bub3 to the kinetochore requires Bub1 but not Zw10 or Rod. Chromosoma. 1998;107:376–385. doi: 10.1007/s004120050321. [DOI] [PubMed] [Google Scholar]
- Basu J., Bousbaa H., Logarinho E., Li Z., Williams B.C., Lopes C., Sunkel C.E., Goldberg M.L. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila . J. Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady D.M., Hardwick K.G. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 2000;10:675–678. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- Cahill D.P., Lengauer C., Yu J., Riggins G.J., Willson J.K., Markowitz S.D., Kinzler K.W., Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- Chan G.K., Schaar B.T., Yen T.J. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J. Cell Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G.K., Jablonski S.A., Sudakin V., Hittle J.C., Yen T.J. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 1999;146:941–954. doi: 10.1083/jcb.146.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.-H., Waters J.C., Salmon E.D., Murray A.W. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Chen R.H., Shevchenko A., Mann M., Murray A.W. Spindle checkpoint protein xmad1 recruits xmad2 to unattached kinetochores. J. Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Brady D.M., Smith D., Murray A.W., Hardwick K.G. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell. 1999;10:2607–2618. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobles M., Liberal V., Scott M.L., Benezra R., Sorger P.K. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- Fang G., Yu H., Kirschner M.W. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr K.A., Hoyt M.A. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol. Cell. Biol. 1998;18:2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky G.J., Chen R.H., Murray A.W. Microinjection of antibody to mad2 protein into mammalian cells in mitosis induces premature anaphase. J. Cell Biol. 1998;141:1193–1205. doi: 10.1083/jcb.141.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T., Kinoshita T. Maternal transcripts of mitotic checkpoint gene, Xbub3, are accumulated in the animal blastomeres of Xenopus early embryo. DNA Cell Biol. 1999;18:227–234. doi: 10.1089/104454999315448. [DOI] [PubMed] [Google Scholar]
- Hardwick K., Murray A.W. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K.G., Weiss E., Luca F.C., Winey M., Murray A.W. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Hoyt M.A., Totis L., Roberts B.T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hwang L.H., Lau L.F., Smith D.L., Mistrot C.A., Hardwick K.G., Hwang E.S., Amon A., Murray A.W. Budding yeast Cdc20a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Jablonski S.A., Chan G.K., Cooke C.A., Earnshaw W.C., Yen T.J. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 1998;107:386–396. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- Jin D.Y., Spencer F., Jeang K.T. Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- Kalitsis P., Earle E., Fowler K.J., Choo K.H. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000;14:2277–2282. doi: 10.1101/gad.827500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan K.B., Burds A.A., Swedlow J.R., Bekir S.S., Sorger P.K., Nathke I.S. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat. Cell Biol. 2001;3:429–932. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Lin D.P., Matsumoto S., Kitazono A., Matsumoto T. Fission yeast Slp1an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- Li R., Murray A.W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li X., Nicklas R.B. Mitotic forces control a cell cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Li Y., Benezra R. Identification of a human mitotic checkpoint genehsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Minshull J., Sun H., Tonks N.K., Murray A.W. MAP-kinase dependent mitotic feedback arrest in Xenopus egg extracts. Cell. 1994;79:475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- Murray A.W. Cell cycle extracts. Methods Cell Biol. 1991;36:573–597. [PubMed] [Google Scholar]
- Page A.M., Hieter P. The anaphase-promoting complexnew subunits and regulators. Annu. Rev. Biochem. 1999;68:583–609. doi: 10.1146/annurev.biochem.68.1.583. [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Cole R.W., Khodjakov A., Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R.T., Farr K.A., Hoyt M.A. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol. Cell. Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar B.T., Chan G.K., Maddox P., Salmon E.D., Yen T.J. CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 1997;139:1373–1382. doi: 10.1083/jcb.139.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M.S., Roberts B.T., Gross S.D., Tunquist B.J., Taieb F.E., Lewellyn A.L., Maller J.L. Bub1 is activated by the protein kinase p90Rsk during Xenopus oocyte maturation. Curr. Biol. 2001;11:141–150. doi: 10.1016/s0960-9822(01)00045-8. [DOI] [PubMed] [Google Scholar]
- Seeley T.W., Wang L., Zhen J.Y. Phosphorylation of human MAD1 by the BUB1 kinase in vitro. Biochem. Biophys. Res. Commun. 1999;257:589–595. doi: 10.1006/bbrc.1999.0514. [DOI] [PubMed] [Google Scholar]
- Shapiro P.S., Vaisberg E., Hunt A.J., Tolwinski N.S., Whalen A.M., McIntosh J.R., Ahn N.G. Activation of the MKK/ERK pathway during somatic cell mitosisdirect interactions of active ERK with kinetochores and regulation of the mitotic 3F3/2 phosphoantigen. J. Cell Biol. 1998;142:1533–1545. doi: 10.1083/jcb.142.6.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka K., Gotoh Y., Nishida E. MAP kinase is required for the spindle assembly checkpoint but is dispensable for the normal M phase entry and exit in Xenopus egg cell cycle extracts. J. Cell Biol. 1997;136:1091–1097. doi: 10.1083/jcb.136.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S., McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Ha E., McKeon F. The human homologue of bub3 is required for kinetochore localization of bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Prinz S., Amon A. CDC20 and CDH1a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Wang X.M., Zhai Y., Ferrell J.E., Jr. A role for mitogen-activated protein kinase in the spindle assembly checkpoint in XTC cells. J. Cell Biol. 1997;137:433–443. doi: 10.1083/jcb.137.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J.C., Chen R.H., Murray A.W., Salmon E.D. Localization of mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E., Winey M. The S. cerevisiae SPB duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood K.W., Sakowicz R., Goldstein L.S., Cleveland D.W. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- Wu H., Lan Z., Li W., Wu S., Weinstein J., Sakamoto K.M., Dai W. p55CDC/hCDC20 is associated with BUBR1 and may be a downstream target of the spindle checkpoint kinase. Oncogene. 2000;19:4557–4562. doi: 10.1038/sj.onc.1203803. [DOI] [PubMed] [Google Scholar]
- Yao X., Abrieu A., Zheng Y., Sullivan K.F., Cleveland D.W. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- Zecevic M., Catling A.D., Eblen S.T., Renzi L., Hittle J.C., Yen T.J., Gorbsky G.J., Weber M.J. Active MAP kinase in mitosislocalization at kinetochores and association with the motor protein CENP-E. J. Cell Biol. 1998;142:1547–1558. doi: 10.1083/jcb.142.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]