Abstract
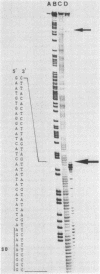
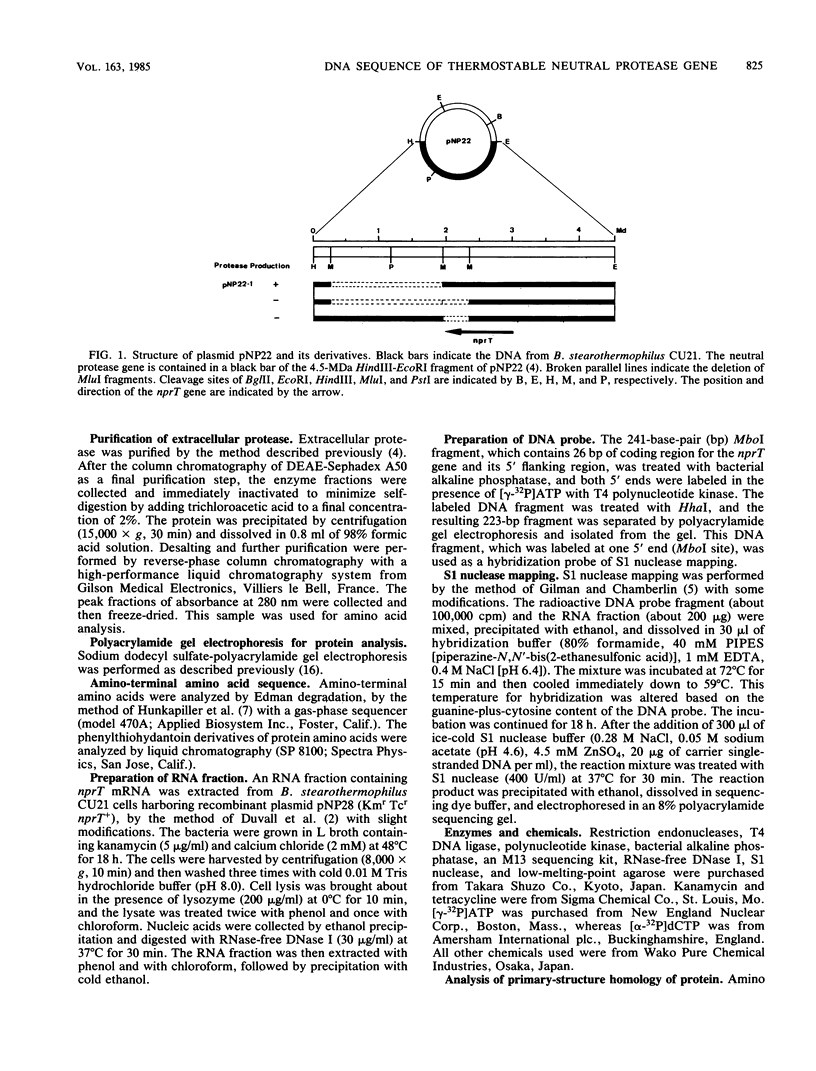
The thermostable neutral protease gene nprT of Bacillus stearothermophilus was sequenced. The DNA sequence revealed only one large open reading frame, composed of 1,644 bases and 548 amino acid residues. A Shine-Dalgarno sequence was found 9 bases upstream from the translation start site (ATG), and the deduced amino acid sequence contained a signal sequence in its amino-terminal region. The sequence of the first 14 amino acids of purified extracellular protease completely matched that deduced from the DNA sequence starting at GTC (Val), 687 bases (229 amino acids) downstream from ATG. This suggests that the protease is translated as a longer polypeptide. The amino acid sequence of the extracellular form of this protease (319 amino acids) was highly homologous to that of the thermostable neutral protease from Bacillus thermoproteolyticus but less homologous to the thermolabile neutral protease from Bacillus subtilis. A promoter region determined by S1 nuclease mapping (TTTTCC for the -35 region and TATTTT for the -10 region) was different from the conserved promoter sequences recognized by the known or factors in bacilli. However, it was very homologous to the promoter sequence of the spo0B gene from B. subtilis. The guanine-plus-cytosine content of the coding region of the nprT gene was 58 mol%, while that of the third letter of the codons was much higher (72 mol%).
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bouvier J., Stragier P., Bonamy C., Szulmajster J. Nucleotide sequence of the spo0B gene of Bacillus subtilis and regulation of its expression. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7012–7016. doi: 10.1073/pnas.81.22.7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Mongkolsuk S., Lovett P. S. Regulatory regions that control expression of two chloramphenicol-inducible cat genes cloned in Bacillus subtilis. J Bacteriol. 1984 Jun;158(3):784–790. doi: 10.1128/jb.158.3.784-790.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliss E. R., Setlow P. Complete nucleotide sequence and start sites for transcription and translation of the Bacillus megaterium protein C gene. J Bacteriol. 1984 Jun;158(3):809–813. doi: 10.1128/jb.158.3.809-813.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M., Takagi M., Imanaka T., Aiba S. Molecular cloning of a thermostable neutral protease gene from Bacillus stearothermophilus in a vector plasmid and its expression in Bacillus stearothermophilus and Bacillus subtilis. J Bacteriol. 1983 May;154(2):831–837. doi: 10.1128/jb.154.2.831-837.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishima A., Honda K. Electrochemical photolysis of water at a semiconductor electrode. Nature. 1972 Jul 7;238(5358):37–38. doi: 10.1038/238037a0. [DOI] [PubMed] [Google Scholar]
- Gilman M. Z., Chamberlin M. J. Developmental and genetic regulation of Bacillus subtilis genes transcribed by sigma 28-RNA polymerase. Cell. 1983 Nov;35(1):285–293. doi: 10.1016/0092-8674(83)90231-3. [DOI] [PubMed] [Google Scholar]
- Gilman M. Z., Wiggs J. L., Chamberlin M. J. Nucleotide sequences of two Bacillus subtilis promoters used by Bacillus subtilis sigma-28 RNA polymerase. Nucleic Acids Res. 1981 Nov 25;9(22):5991–6000. doi: 10.1093/nar/9.22.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller M. W., Hewick R. M., Dreyer W. J., Hood L. E. High-sensitivity sequencing with a gas-phase sequenator. Methods Enzymol. 1983;91:399–413. doi: 10.1016/s0076-6879(83)91038-8. [DOI] [PubMed] [Google Scholar]
- Imanaka T., Fujii M., Aiba S. Isolation and characterization of antibiotic resistance plasmids from thermophilic bacilli and construction of deletion plasmids. J Bacteriol. 1981 Jun;146(3):1091–1097. doi: 10.1128/jb.146.3.1091-1097.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka T., Fujii M., Aramori I., Aiba S. Transformation of Bacillus stearothermophilus with plasmid DNA and characterization of shuttle vector plasmids between Bacillus stearothermophilus and Bacillus subtilis. J Bacteriol. 1982 Mar;149(3):824–830. doi: 10.1128/jb.149.3.824-830.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. C., Moran C. P., Jr, Losick R. Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene. Nature. 1983 Apr 28;302(5911):800–804. doi: 10.1038/302800a0. [DOI] [PubMed] [Google Scholar]
- Kagawa Y., Nojima H., Nukiwa N., Ishizuka M., Nakajima T., Yasuhara T., Tanaka T., Oshima T. High guanine plus cytosine content in the third letter of codons of an extreme thermophile. DNA sequence of the isopropylmalate dehydrogenase of Thermus thermophilus. J Biol Chem. 1984 Mar 10;259(5):2956–2960. [PubMed] [Google Scholar]
- Kester W. R., Matthews B. W. Comparison of the structures of carboxypeptidase A and thermolysin. J Biol Chem. 1977 Nov 10;252(21):7704–7710. [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Pero J. Conserved nucleotide sequences in temporally controlled bacteriophage promoters. J Mol Biol. 1981 Oct 25;152(2):247–265. doi: 10.1016/0022-2836(81)90242-4. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Katakura Y., Imanaka T., Aiba S. Enzymatic and nucleotide sequence studies of a kanamycin-inactivating enzyme encoded by a plasmid from thermophilic bacilli in comparison with that encoded by plasmid pUB110. J Bacteriol. 1984 Oct;160(1):413–420. doi: 10.1128/jb.160.1.413-420.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Moran C. P., Jr, Johnson W. C., Losick R. Close contacts between sigma 37-RNA polymerase and a Bacillus subtilis chromosomal promoter. J Mol Biol. 1982 Dec 15;162(3):709–713. doi: 10.1016/0022-2836(82)90399-0. [DOI] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., Losick R. Nucleotide sequence of a Bacillus subtilis promoter recognized by Bacillus subtilis RNA polymerase containing sigma 37. Nucleic Acids Res. 1981 Nov 25;9(22):5979–5990. doi: 10.1093/nar/9.22.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny J. Matrix program to analyze primary structure homology. Nucleic Acids Res. 1982 Jan 11;10(1):127–131. doi: 10.1093/nar/10.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest F. G. Extracellular enzyme synthesis in the genus Bacillus. Bacteriol Rev. 1977 Sep;41(3):711–753. doi: 10.1128/br.41.3.711-753.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M. L., Ferrari E. Replacement of the Bacillus subtilis subtilisin structural gene with an In vitro-derived deletion mutation. J Bacteriol. 1984 May;158(2):411–418. doi: 10.1128/jb.158.2.411-418.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasantha N., Thompson L. D., Rhodes C., Banner C., Nagle J., Filpula D. Genes for alkaline protease and neutral protease from Bacillus amyloliquefaciens contain a large open reading frame between the regions coding for signal sequence and mature protein. J Bacteriol. 1984 Sep;159(3):811–819. doi: 10.1128/jb.159.3.811-819.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. Z., Doi R. H. Overlapping promoters transcribed by bacillus subtilis sigma 55 and sigma 37 RNA polymerase holoenzymes during growth and stationary phases. J Biol Chem. 1984 Jul 10;259(13):8619–8625. [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. A., Ferrari E., Henner D. J., Estell D. A., Chen E. Y. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 1983 Nov 25;11(22):7911–7925. doi: 10.1093/nar/11.22.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G., Koch G. L., Hartley B. S., Barker D. G. The amino acid sequence of the tyrosyl-tRNA synthetase from Bacillus stearothermophilus. Eur J Biochem. 1983 May 2;132(2):383–387. doi: 10.1111/j.1432-1033.1983.tb07374.x. [DOI] [PubMed] [Google Scholar]
- Wong H. C., Schnepf H. E., Whiteley H. R. Transcriptional and translational start sites for the Bacillus thuringiensis crystal protein gene. J Biol Chem. 1983 Feb 10;258(3):1960–1967. [PubMed] [Google Scholar]
- Wong S. L., Price C. W., Goldfarb D. S., Doi R. H. The subtilisin E gene of Bacillus subtilis is transcribed from a sigma 37 promoter in vivo. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1184–1188. doi: 10.1073/pnas.81.4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M. Y., Ferrari E., Henner D. J. Cloning of the neutral protease gene of Bacillus subtilis and the use of the cloned gene to create an in vitro-derived deletion mutation. J Bacteriol. 1984 Oct;160(1):15–21. doi: 10.1128/jb.160.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]