Abstract
Microtubules are linear polymers of α- and β-tubulin heterodimers and are the major constituents of mitotic spindles, which are essential for the separation of chromosomes during mitosis. Here we describe a synthetic compound, 2-fluoro-1-methoxy-4-pentafluorophenylsulfonamidobenzene (T138067), which covalently and selectively modifies the β1, β2, and β4 isotypes of β-tubulin at a conserved cysteine residue, thereby disrupting microtubule polymerization. Cells exposed to T138067 become altered in shape, indicating a collapse of the cytoskeleton, and show an increase in chromosomal ploidy. Subsequently, these cells undergo apoptosis. Furthermore, T138067 exhibits cytotoxicity against tumor cell lines that exhibit substantial resistance to vinblastine, paclitaxel, doxorubicin, and actinomycin D. T138067 is also equally efficacious in inhibiting the growth of sensitive and multidrug-resistant human tumor xenografts in athymic nude mice. These observations suggest that T138067 may be clinically useful for the treatment of multidrug-resistant tumors.
A variety of plant-derived secondary metabolites have been found that bind reversibly to microtubules and inhibit the growth and viability of eukaryotic cells. This group of antimitotic agents includes vincristine, vinblastine, colchicine, maytansin, and paclitaxel (1, 2). Microtubules are dynamic, polymeric structures that sustain cell shape and facilitate the movement and deposition of protein complexes, organelles, and membrane vesicles (3, 4). They are also major constituents of mitotic spindles, which are essential for chromosomal separation during mitosis (5, 6). Microtubules consist of linear polymers of α- and β-tubulin heterodimers (7–9). In humans, several β-tubulin isotypes, all closely related to each other, have been identified (10). Certain microtubule disrupters, including vincristine, vinblastine, and colchicine, bind reversibly to β-tubulin in disassembled tubulin heterodimers, thereby interfering with polymerization (11, 12). In contrast, paclitaxel binds reversibly to β-tubulin subunits in polymerized microtubules and blocks disassembly (12). Several other agents have been described that, by alkylating tubulin sulfhydryl groups, affect microtubule formation (13–16). In general, these compounds lack specificity, because they modify multiple cysteine residues in tubulin and other intracellular proteins, although one such agent displays a slight preference for cysteine residue 239 in β-tubulin (13–15, 17).
Because of the role of microtubules as key components of the mitotic apparatus, microtubule disrupters and stabilizers represent potent inhibitors of mitotic cell growth (5, 12, 18–20). Agents that affect microtubule dynamics have emerged as useful therapeutics for the treatment of human cancer (12, 21, 22). The efficacy of microtubule disrupters and stabilizers for the selective elimination of tumor cells relative to normal cells has been the subject of considerable attention. An attractive hypothesis that may account for the acceptable therapeutic index of such agents evolves from the work of Hartwell and colleagues (23) concerning cell-cycle checkpoints. When normal cells suffer damage in essential macromolecules, protein assemblies, or organelles, checkpoint pathways are activated that lead to cell-cycle arrest. Tumor cells are often defective in these checkpoint pathways. Although checkpoint deficits appear to favor unrestrained mitotic growth, they also attenuate appropriate repair pathways, sensitizing tumor cells to damage caused by radiation and chemical agents (5, 24).
Although several currently used anticancer agents affect microtubule dynamics, tumor cells often develop resistance to these compounds faster than do normal somatic cells. Accordingly, the therapeutic index for antitumor efficacy can be substantially compromised. The molecular basis of drug resistance in human cancers has been studied extensively. A principal and recurrent pathway leading to drug resistance involves the enhanced expression of drug-efflux pumps, including the P-glycoprotein and the multidrug-resistance (MDR) proteins (25–29). These efflux pumps recognize the relatively large, polycyclic microtubule disrupters and stabilizers and actively remove them from tumor cells. Here we describe the identification of an antimitotic compound, 2-fluoro-1-methoxy-4-pentafluorophenylsulfonamidobenzene (T138067), which binds covalently and selectively to a subset of the β-tubulin isotypes, thereby disrupting microtubule polymerization. Covalent modification occurs at a conserved cysteine residue shared by the β1, β2, and β4 tubulin isotypes. T138067 displays cytotoxic effects against MDR human tumor cells in culture and in mouse xenograft models. The covalent interaction of T138067 with β-tubulin may explain its evasion of cellular mechanisms of MDR.
MATERIALS AND METHODS
Cellular Tubulin Modification.
MCF7 cells (2 × 105 cells) were plated in DMEM/F12 (Mediatech, Herndon, VA) containing 10% FCS 24 hr before treatment. After a 3-hr incubation with 400 mM 3H-T138067 (20 Ci/mmol; 1 mCi/ml) at 37°C/5% CO2, the cells were harvested and lysed. Cellular proteins were analyzed by SDS/PAGE. Tubulin was detected with antitubulin antibody (Sigma) and the 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium detection system (Promega). For competition assays, MCF7 or MCF7/ADR cells (2 × 105 cells) were plated as described above and treated for 24 hr with the indicated antimitotic agents before exposure to 3H-T138067. Cells were lysed, and proteins were analyzed by SDS/PAGE. For immunoprecipitation assays, MCF7 cells (5 × 106 cells) were treated with 40 μCi of 3H-T138067 as above. Cell pellets were resuspended and lysed in BRB80 buffer (80 mM Pipes, pH 6.8/0.5 mM MgCl2/1 mM EGTA) supplemented with 10% glycerol, 50 mM NaCl, and 0.05% NP-40. Immunopurification using anti-β2 antibodies (BioGenex Laboratories, San Ramon, CA), 250 μl of extract, and BRB80 buffer was carried out in a manner similar to that described (30). For two-dimensional gel electrophoresis, MCF7/ADR cells (2 × 104 cells) were labeled as described above. Two-dimensional gel electrophoresis (Immobiline DryStrip; pH 7–4 L) was carried out according to the manufacturer’s protocol (Amersham Pharmacia).
Identification of the T138067 Binding Site.
Purified bovine brain tubulin (7.5 μg in BRB80 buffer/10% glycerol; Cytoskeleton, Denver) was incubated at 37°C with 100 μCi of 3H-T138067 in a 10-μl reaction volume for 8 hr. Modified tubulin was partially separated from unincorporated 3H-T138067 on a microspin column equilibrated in BRB80 buffer (P6; Bio-Rad). Recovered protein (20 μl) was treated with cyanogen bromide (20 μg) for 12 hr. Peptides were separated on an Aquapore OD-300/7 mm-C18 reverse-phase column (1 × 220 mm; Applied Biosystems) with a 60-min linear gradient (from 95% [H20 + 0.1% trifluoroacetic acid (TFA) (vol/vol)]/5% [acetonitrile + 0.08% TFA (vol/vol)] to 35% [H20 + 0.1% TFA (vol/vol)]/65% [acetonitrile + 0.08% TFA (vol/vol)]). Radioactive peptides were subjected to Edman degradation.
In Vitro Tubulin Polymerization Reaction.
Ice-cold bovine brain tubulin solution (400 μg in BRB80 buffer/10% glycerol) was mixed with 49 μl of cold BRB80 buffer, 10 μl of cold GTP solution (10 mM), and 1 μl of DMSO or 1 μl of compound solution in DMSO. The mixture was immediately transferred to a quartz cuvette equilibrated at 37°C. Changes in the OD at 340 nm were monitored every 30 sec in a temperature-controlled photospectrometer (Hewlett Packard) at 37°C. For sedimentation experiments, ice-cold bovine brain tubulin solution (20 μg in BRB80 buffer/10% glycerol) was mixed with 5 μl of cold GTP solution (10 mM), 14 μl BRB80 buffer, and 1 μl of DMSO or 1 μl of compound solution in DMSO. The mixture was incubated at 37°C for 10 min, and 20 μl of the mixture was loaded on a 30-μl glycerol cushion (50% glycerol in BRB80 buffer) preequilibrated at 37°C. The mixture was spun for 5 min at 75,000 rpm and 37°C in a TLA100 rotor (Beckman Instruments). Proteins in the 50-μl supernatant (referred to as cushion fraction) and pellet (i.e., proteins that pellet through the cushion) were analyzed by SDS/PAGE.
In Vitro Tubulin Binding Competition Assay.
Ice-cold bovine brain tubulin solution (10 μg in BRB80 buffer/10% glycerol) was mixed with 18 μl of cold BRB80 buffer and 1 μl of DMSO or 1 μl of compound solution in DMSO and incubated at 37°C for 2 hr before addition of 2 μl of 3H-T138067 (20 Ci/mmol; 1 mCi/ml). After a 60-min incubation at 37°C, proteins were analyzed by SDS/PAGE.
Cell-Cycle Analysis.
MCF7 and MCF7/ADR cells (1 × 106 cells per experiment) for FACS analysis were plated, as described above, 24 hr before treatment with T138067 or vehicle control (DMSO). Drug-treated and control cell nuclei were isolated for cell-cycle analysis by using the CycleTEST PLUS DNA Reagent Kit according to the manufacturer’s protocol (Becton Dickinson). Nuclei (n = 20,000) were analyzed with the use of a FACSCalibur and cellquest software (Becton Dickinson).
Cytotoxicity Analysis.
CCRF-CEM and CCRF-CEM/VBL100 cells were obtained from W. T. Beck (University of Illinois, Urbana-Champaign). DC-3F DC-3F/ADII and DC-3F/ADX cells were obtained from J. L. Biedler (Memorial Sloan-Kettering Cancer Center, New York), and P388/0 and P388/ADR cells were obtained from F. A. Schmid (Memorial Sloan-Kettering Cancer Center). MDR sublines were selected against vinblastine (CCRF-CEM/VBL100), actinomycin D (DC-3F/ADX), and doxorubicin (P388/ADR, SK-N-FI/ADR, and MCF7/ADR). The concentration of compound that produced a 50% inhibition of cell growth (IC50 value) was determined by the (2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5carboxanilide) (XTT) microculture tetrazonium method and analyzed as described (31).
Mouse Xenograft Models.
Athymic nude mice (nu/nu) were obtained from Taconic Farms (outbred, Swiss background). Male mice, 6–8 weeks old and weighing from 20 to 25 g, were used. Tumor tissue (6 g) was minced in 15 ml of RPMI medium 1640 (GIBCO/BRL) and implanted (50 μl per mouse) s.c. on day 0. Drugs were dosed in 40 μl of DMSO (i.p.) on days 5, 12, and 19 (weekly schedule). Body weights and tumor node sizes were determined at the intervals indicated. All studies were conducted in accordance with the guidelines of the National Institutes of Health Guide for the Care and Use of Animals and after protocol review by the local Institutional Care and Use Committee. Humane treatment of tumor-bearing mice required their euthanization when tumors reached ≥10% of their total body weight.
RESULTS AND DISCUSSION
T138067 Binds Selectively and Irreversibly to β-Tubulin.
The synthetic compound T138067 (Fig. 1A) represents an optimized analog of a lead identified in a cell-based, high-throughput drug screen (J.C.M., D. Clark, and T.R., unpublished work). Despite the initial appearance of selectivity in this screen, further evaluation of the compound revealed that cells altered their shape and became detached from the culture plate in the presence of the drug, suggesting that the compound might be interfering with their cytoskeleton.
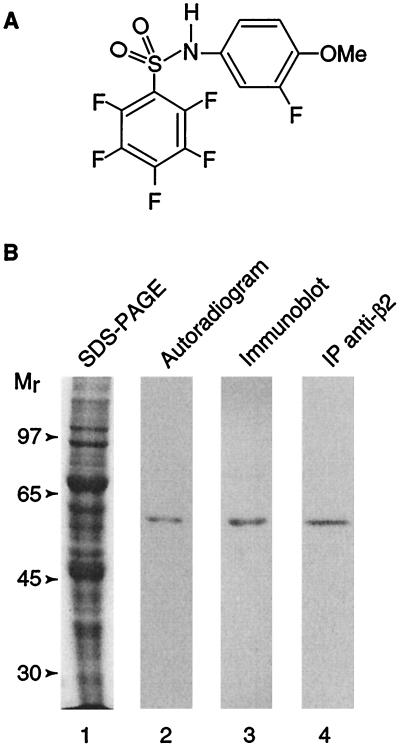
Figure 1.
Covalent and tubulin-specific binding of T138067. (A) Structure of T138067. (B) Binding of 3H-T138067 to cellular β-tubulin. Lane 1, Coomassie-stained proteins derived from MCF7 whole-cell extracts after SDS/PAGE. MCF7 cells were treated with 400 nM 3H-T138067 for 3 hr; lane 2, autoradiogram of membrane with immobilized protein from the gel shown in lane 1; lane 3, immunoblotting of the membrane shown in lane 2 with anti-β-tubulin antibodies; lane 4, autoradiogram of immunopurified cellular 3H-T138067-labeled β-tubulin derived from MCF7 whole-cell extracts (lane 1). Immunopurification was performed with antibodies to β2-tubulin. Molecular size markers are shown on the left.
The chemical structure of T138067 suggests the potential for covalent modification of its target protein(s). To evaluate this possibility, a tritiated derivative of T138067 was incubated with human mammary adenocarcinoma-derived MCF7 cells. After 3 hr, cells were washed, and proteins were analyzed by denaturing SDS/PAGE. Only a single polypeptide, with an apparent molecular size of ≈50 kDa, was labeled under these conditions (Fig. 1B). This labeled polypeptide comigrated with β-tubulin, as determined by Western blot analysis, and was immunoprecipitated with antiserum specific to the β2 tubulin isotype (Fig. 1B). Further cell culture experiments revealed that binding of T138067 to β-tubulin is time and concentration dependent (data not shown).
T138067 Modifies a Subset of the β-Tubulin Isotypes.
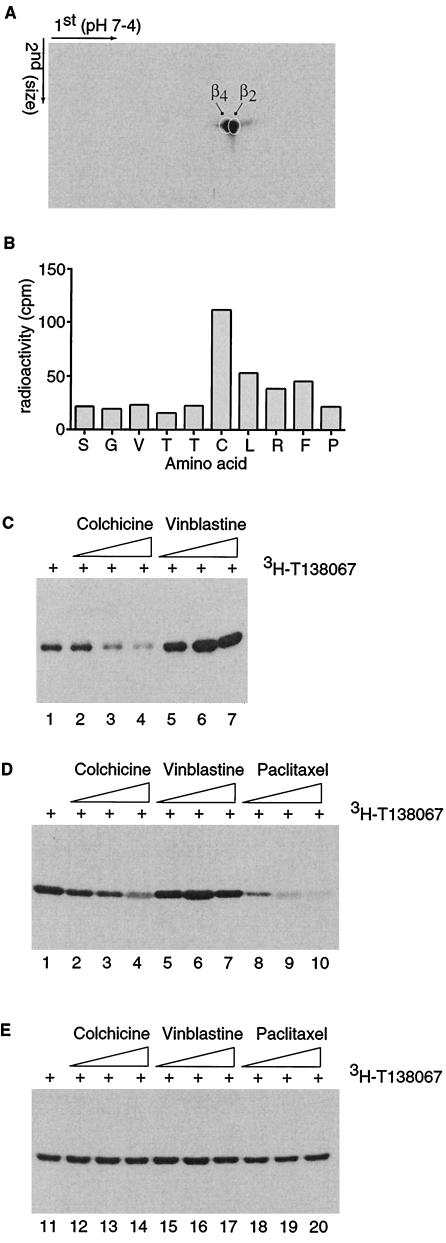
Several β-tubulin isotypes have been identified. To evaluate the β-tubulin isotype specificity of T138067, protein extracts derived from MCF7/ADR cells labeled with 3H-T138067 were further subjected to two-dimensional gel electrophoresis. Separated proteins were transferred onto a poly(vinylidene difluoride) membrane, and the various β-tubulin isotypes were detected with isotype-specific antibodies. The β2 and β4 tubulin isotypes were found to predominate in MCF7/ADR cells, although small amounts of β1 and β3 were detectable (data not shown). Subsequent fluorography and autoradiography revealed that protein spots that colocalized with the β2 and β4 tubulin isotypes were abundantly and specifically labeled with 3H-T138067 (Fig. 2A). A low, but reproducible, level of labeling also was observed at a protein spot that colocalized with the β1 isotype. However, no modification of the β3 isotype was observed (data not shown). In summary, these data indicate that T138067 modifies only a subset of the β-tubulin isotypes.
Figure 2.
Selective binding of T138067 to Cys-239 of β2- and β4-tubulin. (A) Shown is an autoradiogram of a 3H-T138067-labeled cell extract derived from MCF7/ADR cells and separated by two dimensional gel electrophoresis. The first and second dimensions of the gel electrophoresis are shown. The positions of the individual β-tubulin isotypes are indicated. (B) Cys-239 of β2-tubulin bound by 3H-T138067. The 3H-T138067-modified tubulin peptide isolated from a cyanogen bromide digest of tubulin was subjected to Edman degradation. The first 10 aa corresponding to peptide Ser-234 to Met-267 of β2-tubulin and the radioactivity (cpm) coeluting with each of the amino acids (in standard single-letter code) are shown. (C) Colchicine competes for binding of T138067 to Cys-239 in vitro. Purified brain tubulin (5 μM) was incubated in the absence of drug (lane 1) or pretreated for 2 hr with colchicine [lane 2 (0.5 μM), lane 3 (5 μM), and lane 4 (25 μM)] or vinblastine [lane 5 (0.5 μM), lane 6 (5 μM), and lane 7 (25 μM)] before 5 μM of 3H-T138067 was added to the reaction for 60 min. (D and E) Colchicine competes for binding of T138067 to Cys-239 in MCF7 (D), but not in MCF7/ADR (E) cells. Cells were incubated in the absence of drug (lanes 1 and 11) or pretreated for 24 hr with colchicine [lanes 2 and 12 (0.5 μM), lanes 3 and 13 (1 μM), lanes 4 and 14 (1.5 μM)], vinblastine [lanes 5 and 15 (0.5 μM), lanes 6 and 16 (1 μM), and lanes 7 and 17 (1.5 μM)], or paclitaxel [lanes 8 and 18 (0.5 μM), lanes 9 and 19 (1 μM), and lanes 10 and 20 (1.5 μM)] before cells were exposed to 400 nM 3H-T138067 for 3 hr.
T138067 Binds to Cys-239 of β2-and β4-Tubulin.
The sequences of all β-tubulin isotypes are highly conserved (8, 10). However, while the β1, β2, and β4 isotypes possess a cysteine residue at position 239, the β3 isotype has a serine at this position (10). Given the apparently irreversible and subtype-specific nature of the interaction between T138067 and β-tubulin, we reasoned that the compound might act by covalently modifying Cys-239 of the β1, β2, and β4 isotypes. To test this hypothesis, purified bovine brain tubulin, which consists of approximately a 2:1 mixture of β2- and β3-tubulin (32), was exposed to 3H-T138067 for 8 hr, with a 3H-T138067/tubulin ratio of 60:1. Previous studies had shown that β-tubulin labeling did not increase further after 8 hr of incubation (data not shown). Labeled proteins were degraded with cyanogen bromide, and the resulting peptides were separated by reverse-phase HPLC. Among the 40 isolated peptide peaks, only one was found to be radioactive (data not shown). This peptide peak was subjected to Edman degradation sequencing. The peptide sequence started at serine residue 234 of β2-tubulin, and cycle 6 of the sequencing reaction yielded a radiolabeled amino acid, presumably corresponding to Cys-239 (Fig. 2B). Although there are between seven and 11 cysteine residues in the various β-tubulin isotypes, our analysis shows that Cys-239 is the sole target of covalent modification by T138067. Thus, T138067 is rather distinct from other known tubulin alkylating reagents, which, in many cases, modify additional tubulin sulfhydryl groups (14, 15, 17). Because Cys-239 is shared by the β1-, β2-, and β4-tubulin, all of which were labeled by 3H-T138067 in the cell experiments, but not by β3-tubulin, it is likely that Cys-239 represents the common site of covalent modification on all susceptible β-tubulin isotypes. Several lines of evidence indicate that the fluorine atom located at the para-position of the pentafluorophenyl ring of T138067 is displaced by the thiol group of Cys-239, leading to covalent modification. Chemical studies supportive of this reaction mechanism will be presented elsewhere (W.P.F., J.C.M., B.S., D.S., and H.B., unpublished work).
Cys-239 of β-tubulin has been described previously as proximal to or involved in the β-tubulin recognition site for colchicine, but it does not play a role in the paclitaxel or vinblastine sites (16, 33–35). Previous work with other tubulin-alkylating agents has shown that colchicine, but not vinblastine, can block alkylation of Cys-239 (17). To investigate the binding of T138067 further, we performed in vitro and cell culture competition experiments between T138067 and colchicine or vinblastine. Preincubation of purified brain tubulin with colchicine prevents binding of 3H-T138067, in a concentration-dependent manner (Fig. 2C). In contrast, pretreatment of tubulin with increasing concentrations of vinblastine, which like colchicine binds preferentially to nonpolymeric tubulin heterodimers and prevents microtubule formation (11, 12), resulted in an enhanced labeling of β-tubulin by 3H-T138067. Similar results were obtained when MCF7 cells in culture were pretreated with colchicine or vinblastine before addition of 3H-T138067 (Fig. 2D). As before, colchicine, but not vinblastine, blocked the labeling of β-tubulin by 3H-T138067. In contrast, these two agents did not block or enhance tubulin modification by 3H-T138067 in MCF7/ADR cells (Fig. 2E), a MDR subline of MCF7 cells in which colchicine and vinblastine have a strongly reduced efficacy (data not shown, but see below). These results indicate that the binding for T138067 is either in close vicinity or may even overlap with the colchicine binding site. This finding is supported by the observation that T138067, in contrast to other known alkylating agents (14, 17), significantly prevented the binding of colchicine to tubulin (data not shown). Our results also suggest that T138067, like colchicine or vinblastine, binds to heterodimeric tubulin and not to microtubules. This finding is supported by the results of competition assays in MCF7 and MCF7/ADR cells (Fig. 2 D and E). Pretreatment of MCF7 cells with increasing concentrations of paclitaxel, a microtubule stabilizer (11, 12), prevented 3H-T138067 from labeling cellular tubulin (Fig. 2D). Interestingly, in MCF7/ADR cells, which are highly resistant to paclitaxel, paclitaxel pretreatment did not significantly affect β-tubulin labeling by 3H-T138067 (Fig. 2E).
T138067 Prevents Microtubule Formation and Induces Cell-Cycle Arrest.
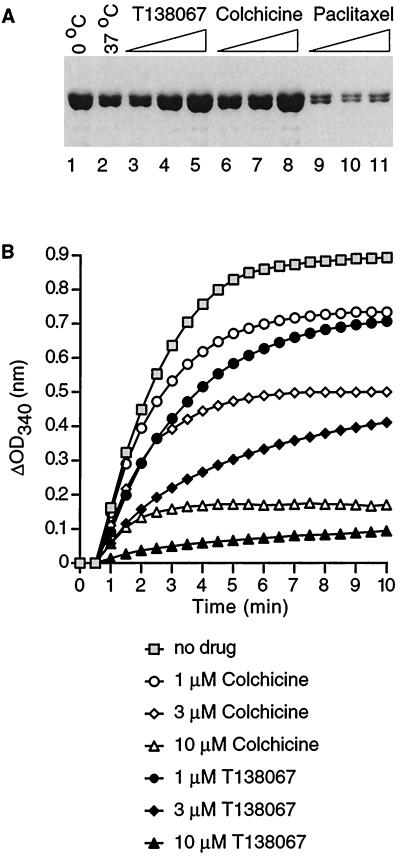
Binding of T138067 to β-tubulin strongly suggests that T138067 also might affect microtubule dynamics. To measure the effects of T138067 on microtubule formation, we used two types of in vitro tubulin polymerization assays. In a sedimentation assay, the polymerization reaction was performed in the absence or presence of T138067, by using colchicine and paclitaxel as comparative agents (Fig. 3A). The reaction products were size-fractionated by spinning the mixture through a glycerol cushion. Heterodimeric tubulin remains primarily in the cushion, whereas larger molecules (microtubules) sediment through the cushion and form a pellet at the bottom of the tube. Tubulin content in the cushion or pellet was analyzed by SDS/PAGE. In the absence of any agent, a substantial reduction in the amount of tubulin in the cushion fraction was observed in a polymerization reaction that was incubated at 37°C, in comparison to one that was kept at 0°C, indicating the formation of microtubules (Fig. 3A, compare lanes 1 and 2). However, increasing concentrations of T138067 at 37°C increased the amount of tubulin in the cushion fraction to levels comparable to those derived from the control reaction at 0°C (Fig. 3A, compare lanes 1 and 3 to 5). Similar results were obtained in reactions that contained colchicine (Fig. 3A). In contrast, polymerization reactions performed in the presence of paclitaxel showed a strong reduction of tubulin in the cushion fraction (Fig. 3A). When the pellet fractions were analyzed, a strong reduction of tubulin was observed in reactions containing either T138067 or colchicine in comparison to reactions that were incubated in the absence of drugs or in the presence of paclitaxel (data not shown). In summary, these results indicate that T138067, like colchicine, prevents microtubule formation.
Figure 3.
Inhibition by T138067 of microtubule formation in vitro. (A) SDS/PAGE of the cushion fraction from a sedimentation assay. Polymerization reactions (40 μM tubulin) were kept on ice (lane 1), 37°C (lane 2), or 37°C in the presence of T138067 [lane 3 (1 μM), lane 4 (3 μM), and lane 5 (10 μM)], colchicine [lane 6 (1 μM), lane 7 (3 μM), and lane 8 (10 μM)], or paclitaxel [lane 9 (1 μM), lane 10 (3 μM), and lane 11 (10 μM)] for 10 min before analyses. (B) Turbidimetric assay. Graphical representation of changes in the OD at 340 nm over time (min) in the absence (no drug) or presence of the indicated compounds.
In a second in vitro assay, microtubule formation was monitored by using the increase in OD of the reaction mixture. At a tubulin/T138067 ratio of 1:40, microtubule formation at 5 min was slightly inhibited relative to a reaction lacking T138067 (Fig. 3B). When tubulin/T138067 ratios were raised to 1:13 or 1:4, microtubule formation after 5 min was inhibited by ≈50% and ≈90%, respectively (Fig. 3B). These results show that T138067 fairly rapidly prevents microtubule formation, with an IC50 value (50% inhibition of microtubule formation) of approximately 2 μM. This finding is in stark contrast to other tubulin-alkylating agents that either require a much longer incubation time, higher drug concentrations, or the presence of premodified tubulin in the polymerization reaction to prevent microtubule formation (14, 20). Interestingly, although the apparent association rate constant (210 ± 90 M−1⋅s−1; data not shown) for T138067 binding to tubulin is approximately 10 times greater than that of colchicine (36), T138067 seems to be more effective in preventing microtubule formation (Fig. 3B). One possible explanation is that T138067-modified tubulin recruits unmodified heterodimeric tubulin into large, amorphous aggregates, and thus quickly depletes the pool of tubulin available for microtubule formation. In fact, T138067 induces the formation of amorphous aggregates under such experimental conditions (data not shown). Similar aggregate formation has been seen with other tubulin-alkylating agents (15).
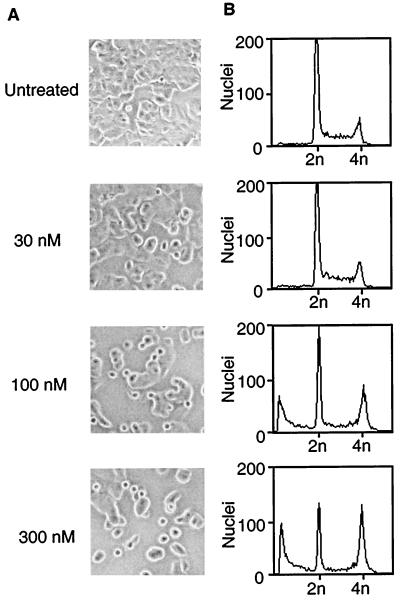
Based on the in vitro polymerization results, we suspected that the mechanism of cytotoxicity of T138067 would involve cytoskeletal collapse. Thus, the effects of T138067 on the cytoskeleton and morphology of MCF7 cells were investigated. In the absence of T138067, cells showed the typical morphology of adherent growing cells (Fig. 4A). However, in the presence of 100 nM T138067 for 9 hr, approximately 40% of the cells adopted a rounded morphology and became detached from the plate surface. At slightly higher drug concentrations (300 nM), almost all cells showed these morphological changes. These morphological changes were accompanied by cytoskeletal disruption, as determined by immunofluorescence microscopy using β-tubulin antibodies (data not shown). Given the pivotal role of microtubules in chromosome segregation during mitosis (5, 6), it was suspected that disruption of the cytoskeleton by T138067 would affect the ploidy of these cells. Indeed, FACS analysis (Fig. 4B) revealed major changes in the ploidy of MCF7 cells after a 24-hr treatment with the compound. In the absence of T138067, almost 90% of the cell population had a diploid (2n) chromosome set. In contrast, at drug concentrations of 100 nM or higher (Fig. 4B), approximately 25–30% of the cells showed a tetraploid (4n) DNA content, indicating an arrest at the G2/M cell-cycle boundary. In addition, 25–30% of cells showed the reduced DNA content (<2n) characteristic of apoptotic cells. After a 48-hr exposure to 100 nM T138067, approximately 50–80% of the cell population was undergoing apoptosis (data not shown). Thus, concentrations of T138067 that affect cell morphology and lead to cell-cycle arrest ultimately induce apoptotic cell death. Interestingly, under these conditions, only about 10% of the cellular β-tubulin pool is modified by T138067 (data not shown).
Figure 4.
T138067-induced collapse of the cytoskeleton and cell-cycle arrest. (A) Phase-contrast images of MCF7 cells after treatment with T138067. MCF7 cells were either incubated in the absence of drug (no drug) or treated for 9 hr with the indicated concentrations of T138067. Representative images were taken at ×100 magnification. (B) FACS analysis of nuclei isolated from MCF7 cells that were untreated (no drug) or treated for 24 hr with T138067 at the concentrations indicated for the corresponding panel in A. DNA content (2n = diploid; 4n = tetraploid) is indicated.
T138067 Is Efficacious Against MDR Tumor Cells in Culture and Mice Xenograft Models.
The development of MDR constitutes a major problem in cancer chemotherapy with most commonly used drugs. Our initial experiments showed that the MDR phenotype of MCF7/ADR breast cancer cells did not affect the ability of 3H-T138067 to modify β-tubulin (Fig. 2A) or its cytotoxic potential. T138067 and four other anticancer compounds (vinblastine, paclitaxel, doxorubicin, and actinomycin D) were tested against drug-sensitive and MDR clonal isolates from MCF7 cells and three additional tumor cell lines (Table 1). To express quantitatively the effect of each MDR phenotype on the cytotoxicity of each drug, the IC50 value (50% inhibition of cell growth) observed with each MDR cell line was divided by that observed with the drug-sensitive, parental cell line. For T138067, this ratio ranged from 1.0 (no difference in the IC50 values) to 2.1, with an average value of 1.55. In contrast, the four other anticancer compounds were considerably less toxic toward the MDR cell lines relative to their drug-sensitive, parental isolates. The average resistance values were 207 for vinblastine, 576 for paclitaxel, 64 for doxorubicin, and 1,193 for actinomycin D. These observations indicate that T138067, by covalently modifying specific β-tubulin isotypes, effectively evades the MDR mechanisms used by the three tumor cell lines tested in this study.
Table 1.
T138067 is efficacious against MDR tumor cells
| Cell lines | T138067 | Vinblastine | Paclitaxel | Doxorubicin | Actinomycin D |
|---|---|---|---|---|---|
| MCF7 | 165 | 0.98 | 3.3 | 57 | 0.68 |
| MCF7/ADR | 165 (1) | 34 (35) | 150 (46) | 216 (3.8) | 1.67 (2.5) |
| CCRF-CEM | 29 | 2.6 | 2.2 | 550 | 0.35 |
| CCRF-CEM/VBL100 | 35 (1.2) | 471 (181) | 3,390 (1,541) | 2,600 (5.1) | 380 (109) |
| DC-3F | 25 | 3 | 51 | 33 | 0.09 |
| DC-3F/ADX | 47 (1.9) | 1,490 (497) | 3,100 (608) | 1,390 (42) | 419 (4,656) |
| P338 | 11 | 3.4 | 2.9 | 7.4 | 0.15 |
| P338/ADR | 23 (2.1) | 39 (115) | 323 (111) | 1,530 (207) | 1.2 (8) |
Comparison of the effects of T138067, paclitaxel, vinblastine, doxorubicin, and actinomycin D on the growth of human mammary adenocarcinoma (MCF7), human lymphoblastic leukemia (CCRF-CEM), hamster lung cell fibroblast (DC-3F), and murine lymphocytic leukemia (P338) cells and their MDR sublines (MCF7/ADR, CCRF-CEM/VBL100, DC-3F/ADX, and P388/ADR, respectively). The IC50 value (nM) for each agent is shown. The fold increase in drug resistance (numbers shown in parentheses) was calculated from the ratio of the IC50 values for each resistant subline relative to that for the parental cell line.
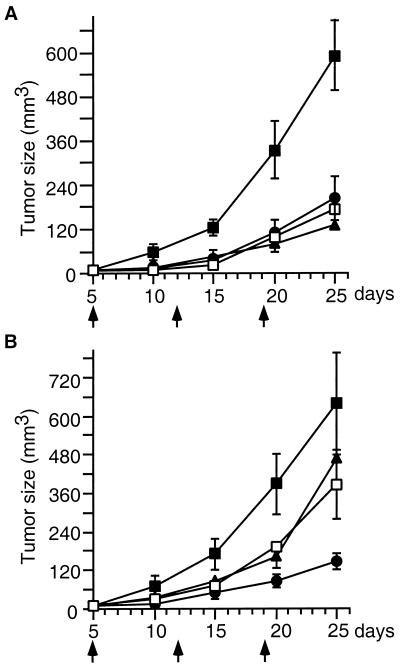
The antitumor efficacies of T138067, paclitaxel, and vinblastine also were evaluated in mouse xenograft models. Drug-sensitive, human T cell leukemia CCRF-CEM cells, and their vinblastine/paclitaxel-resistant subline (CCRF-CEM/VBL100) were grafted separately into athymic nude mice. Animals were dosed i.p. at the maximally tolerated doses of paclitaxel (30 mg/kg), vinblastine (1 mg/kg), and T138067 (40 mg/kg) once per week. After a single i.p. dose, all three drugs impaired the growth of the drug-sensitive CCRF-CEM tumors (Fig. 5A). Two additional weekly doses of each drug further impeded tumor growth, resulting in similar antitumor efficacies for all three drugs. T138067 showed the same degree of efficacy when tested against the MDR subline (Fig. 5B). In contrast, paclitaxel and vinblastine showed approximately 50% reduced efficacy against the MDR tumors (Fig. 5B). Thus, T138067 may be clinically useful for the treatment of human cancers that have developed resistance to standard chemotherapeutic agents.
Figure 5.
Comparison of antitumor effects of T138067 with those of paclitaxel and vinblastine against human tumor xenografts in athymic nude mice. (A) CCRF-CEM lymphoblastic leukemia tumors. (B) CCRF-CEM/VBL100 MDR tumors. Tumor cells were s.c. implanted in athymic nude mice on day 0. T138067 (40 mg/kg; ●); paclitaxel (30 mg/kg; ▴); vinblastine (1 mg/kg; □), or a vehicle control (■) were administered i.p. on days 5, 12, and 19 (indicated by the arrows) after tumor implantation. Tumor size at days 5, 10, 15, 20, and 25 is expressed as mean ± SEM (n = 5).
Acknowledgments
We are thankful to S. L. McKnight for his initial observations that compounds in this series affect cell morphology and have potential anticancer utility. We also thank A. Perlman, C. Case, A. Park, D. Roche, and L. Medin for their support, and L. Belmont and D. Drubin for sharing their outstanding expertise in microtubule function.
ABBREVIATION
- MDR
multidrug resistance
References
- 1.Hamel E. Med Res Rev. 1996;16:207–231. doi: 10.1002/(SICI)1098-1128(199603)16:2<207::AID-MED4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 2.Sackett D L. Pharmacol Ther. 1993;59:163–228. doi: 10.1016/0163-7258(93)90044-e. [DOI] [PubMed] [Google Scholar]
- 3.Gelfand I M, Bershadsky A D. Annu Rev Cell Biol. 1991;7:93–116. doi: 10.1146/annurev.cb.07.110191.000521. [DOI] [PubMed] [Google Scholar]
- 4.Bloom G S. Curr Opin Cell Biol. 1992;4:66–73. doi: 10.1016/0955-0674(92)90060-p. [DOI] [PubMed] [Google Scholar]
- 5.Sorger P K, Dobles M, Tournebize R, Hyman A A. Curr Opin Cell Biol. 1997;9:807–814. doi: 10.1016/s0955-0674(97)80081-6. [DOI] [PubMed] [Google Scholar]
- 6.Mitchinson T J. Annu Rev Cell Biol. 1988;4:527–549. doi: 10.1146/annurev.cb.04.110188.002523. [DOI] [PubMed] [Google Scholar]
- 7.Cleveland D W, Sullivan K F. Annu Rev Biochem. 1985;54:331–365. doi: 10.1146/annurev.bi.54.070185.001555. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan K F. Annu Rev Cell Biol. 1988;4:687–716. doi: 10.1146/annurev.cb.04.110188.003351. [DOI] [PubMed] [Google Scholar]
- 9.Nogales E, Wolf S G, Downing K H. Nature (London) 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 10.Ludueña R F. Mol Biol Cell. 1993;4:445–457. doi: 10.1091/mbc.4.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson L, Jordan M A. Chem Biol. 1995;2:569–573. doi: 10.1016/1074-5521(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 12.Jordan M A, Wilson L. Curr Opin Cell Biol. 1998;10:123–130. doi: 10.1016/s0955-0674(98)80095-1. [DOI] [PubMed] [Google Scholar]
- 13.Kuriyama R, Sakai H. J Biochem. 1974;76:651–654. doi: 10.1093/oxfordjournals.jbchem.a130609. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda Y, Steiner M. Biochemistry. 1978;17:3454–3459. doi: 10.1021/bi00610a005. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y C, Yaple R A, Baldridge R, Kirsch M, Himes R H. Biochem Biophys Acta. 1981;671:71–77. doi: 10.1016/0005-2795(81)90095-7. [DOI] [PubMed] [Google Scholar]
- 16.Ludueña R F, Roach M C. Pharmacol Ther. 1991;49:133–152. doi: 10.1016/0163-7258(91)90027-j. [DOI] [PubMed] [Google Scholar]
- 17.Bai R, Lin C M, Nguyen N Y, Liu T, Hamel E. Biochemistry. 1989;28:5606–5612. doi: 10.1021/bi00439a040. [DOI] [PubMed] [Google Scholar]
- 18.Ramel C. Hereditas. 1967;61:208–230. [PubMed] [Google Scholar]
- 19.Nath J, Rebhun L I. J Cell Biol. 1976;68:440–450. doi: 10.1083/jcb.68.3.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abraham I, Dion R L, Duanmu C, Gottesman M M, Hamel E. Proc Natl Acad Sci USA. 1986;83:6839–6843. doi: 10.1073/pnas.83.18.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowinsky E K, Donehower R C. Pharmacol Ther. 1991;52:35–84. doi: 10.1016/0163-7258(91)90086-2. [DOI] [PubMed] [Google Scholar]
- 22.Rowinsky E K. Annu Rev Med. 1997;48:353–374. doi: 10.1146/annurev.med.48.1.353. [DOI] [PubMed] [Google Scholar]
- 23.Paulovich A G, Toczyski D P, Hartwell L H. Cell. 1987;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 24.Müller H, Eppenberger U. Anticancer Res. 1996;16:3845–3848. [PubMed] [Google Scholar]
- 25.Horwitz S B, Cohen D, Rao S, Ringel I, Shen H J, Yang C P. J Natl Cancer Inst Monogr. 1993;15:55–61. [PubMed] [Google Scholar]
- 26.Zhang Z G, Harstrick A, Rustum Y M. Semin Oncol. 1992;19:4–9. [PubMed] [Google Scholar]
- 27.Gottesman M M, Pastam I. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 28.Gottesman M M, Pastan I, Ambudkar S V. Curr Opin Genet Dev. 1996;6:610–617. doi: 10.1016/s0959-437x(96)80091-8. [DOI] [PubMed] [Google Scholar]
- 29.Deeley R G, Cole S P. Semin Cancer Biol. 1997;8:193–204. doi: 10.1006/scbi.1997.0070. [DOI] [PubMed] [Google Scholar]
- 30.Beckmann H, Chen J-L, O’Brien T, Tjian R. Science. 1995;270:1506–1509. doi: 10.1126/science.270.5241.1506. [DOI] [PubMed] [Google Scholar]
- 31.Chou T-C. In: Synergism and Antagonism in Chemotherapy. Chou T-C, Rideout D C, editors. San Diego: Academic; 1991. pp. 61–102. [Google Scholar]
- 32.Banerjee A, Roach M C, Trcka P, Ludueña R F. J Biol Chem. 1992;267:5625–5630. [PubMed] [Google Scholar]
- 33.Bai R, Pei X, Boyé O, Getahun Z, Grover S, Bekisz J, Nguyen N Y, Brossi A, Hamel E. J Biol Chem. 1996;271:12639–12645. doi: 10.1074/jbc.271.21.12639. [DOI] [PubMed] [Google Scholar]
- 34.Rao S, Orr G A, Chaudhary A G, Kingston D G I, Horwitz S B. J Biol Chem. 1995;270:20235–20238. doi: 10.1074/jbc.270.35.20235. [DOI] [PubMed] [Google Scholar]
- 35.Rai S S, Wolff J. J Biol Chem. 1996;271:14707–14711. doi: 10.1074/jbc.271.25.14707. [DOI] [PubMed] [Google Scholar]
- 36.Bhattacharyya B, Wolff J. Biochemistry. 1976;15:2283–2288. doi: 10.1021/bi00656a006. [DOI] [PubMed] [Google Scholar]