Abstract
It is believed that the hepatitis C virus (HCV)-specific CD8+ cytotoxic T lymphocytes (CTLs) play a role in the development of liver cell injury and in the clearance of the virus. To develop a direct binding assay for HCV-specific CTLs, we generated two peptide-MHC tetramers by using the recombinant HLA A2.1 molecule and A2-restricted T cell epitopes of the HCV NS3 protein. With these reagents we are able to detect specific CD8+ cells in the blood of 15 of 20 HLA-A2+, HCV-infected patients, at a frequency ranging from 0.01% to 1.2% of peripheral CD8+ T cells. Phenotypic analysis of these specific cells indicated that there is a significant variation in the expression of the CD45 isoforms and CD27 in different patients. A 6-hour incubation of one patient’s blood with NS3 peptides resulted in the activation of the epitope-specific CD8+ cells, as indicated by their expression of CD69 and IFN-γ. We also detected NS3-specific CD8+ T cells in the intrahepatic lymphocyte population isolated from liver biopsies of two HCV-infected patients. The frequency of these specific CD8+ cells in the liver was 1–2%, at least 30-fold higher than in the peripheral blood. All of the intrahepatic NS3-specific CD8+ T cells were CD69+, suggesting that they were activated CTLs. Direct quantitation and characterization of HCV-specific CTLs should extend our understanding of the immunopathogenesis and the mechanism of clearance or persistence of HCV.
Hepatitis C virus (HCV) is an enveloped, 9.5-kb positive-strand RNA virus and a member of the Flaviviridae family (1). The virion appears to consist of a nucleocapsid core (C) and two envelope proteins (E1 and E2) embedded in a lipid bilayer envelope. At least six nonstructural proteins (NS2, NS3, NS4 A and B, and NS5 A and B) are involved in replication, transcription, and polyprotein processing. Acute hepatitis infection because of HCV is generally mild and frequently asymptomatic, but the majority of infections result in a chronic carrier state often associated with liver cell injury of varying severity (2).
In HCV-infected individuals, it is believed that T cell responses, including both cytotoxic T lymphocytes (CTLs) and helper T lymphocytes, are critical in determining the outcome (3, 4). In particular, CTLs may play an important role in the development of liver cell injury as well as clearance of the virus. Because the frequency of HCV-specific CTLs in the peripheral blood appears to be quite low, current CTL assays based on 51Cr release require in vitro expansion by peptide stimulation for a period of several weeks to enhance sensitivity (5). The frequency of virus-specific CTLs in HCV-infected liver is believed to be higher than in peripheral blood. However, the limited number of intrahepatic lymphocytes obtained from liver biopsies excludes the possibility of cytolytic function-based assay without in vitro expansion of reactive T cells (6–9). In both cases, the quantitative and qualitative interpretation of current assays is hampered by the need for in vitro expansion.
Recently, a novel method to identify antigen-specific T cells during an immune response has been developed (10). This method involves the engineering of a biotinylation signal sequence onto the C terminus of a recombinant MHC class I or II molecule which, after complexing with a specific peptide, is bound to avidin at a 4:1 ratio. This results in a tetrameric peptide-MHC complex that recognizes T cell receptors (TCRs) on lymphocytes specific for the particular epitope. This has proven to be a powerful tool in enumerating and characterizing antigen-specific T cells, regardless of their function after in vivo or in vitro antigen stimulation, and without the need of in vitro expansion. Such tetramers have been used to study T cell immunity to viruses, including HIV, influenza virus, and Epstein–Barr virus as well as several mouse and monkey viruses (10–17). A similar approach using an Ig Fc backbone to multimerize class I MHC has also been used to stain and characterize CD8+ T cells specific for human T lymphotropic virus type 1 (HTLV-1) in infected patients (18).
In this paper we report the use of peptide-MHC tetramers to analyze CD8+ T cells specific for two epitopes of the HCV NS3 protein in the peripheral blood and liver of HCV-infected patients. Peptide-specific CD8+ T cells at a frequency as low as 0.01% of CD8+ cells were detected from uncultured peripheral blood mononuclear cells (PBMCs). These HCV-specific T cells expressed at least two different phenotypes in the three patients examined. A 6-hour stimulation with specific peptide activated these cells as indicated by their expression of the activated T cell marker CD69 and production of the cytokine IFN-γ. NS3 epitope-specific CD8+ cells also were detected in liver-infiltrating lymphocytes (LILs) isolated from needle biopsy samples. The frequency of such cells in the liver is much higher than in the peripheral blood of the same patient. These cells express CD69, suggesting that they are activated CTLs.
MATERIALS AND METHODS
Patients.
Blood and liver samples of HCV-infected patients were obtained in the outpatient clinics of Veterans Affairs Palo Alto and Veterans Affairs San Francisco Medical Centers, Stanford University Medical Center and the Liver Diseases Section, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. All patients were positive for antibodies to HCV. HCV RNA in the serum was tested by using branched DNA or reverse transcription–PCR assays. Liver tissue was obtained by needle biopsy, which was performed for clinical indications. All specimens were collected with informed consent from the donors. Research protocols were approved by the institutional review boards at Stanford University, University of California at San Francisco, or the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. Blood samples of uninfected donors were obtained from healthy anti-HCV-seronegative volunteers.
Preparation of PBMCs and LILs.
PBMCs were isolated from blood with Ficoll-Paque (Amersham Pharmacia) density gradient or the Vacutainer Cell Preparation Tube (Becton Dickinson). Liver tissues (40–80 mg), obtained as part of a needle biopsy, were washed three times with RPMI 1640 (GIBCO) supplemented with l-glutamine (2 mM) and 10% heat-inactivated fetal calf serum to eliminate contaminating PBMCs and homogenized in 5 ml of the same medium by using a glass dounce tissue homogenizer. The LILs were isolated from the resulting cell suspension by Ficoll-Paque density gradient. Typically, 104–105 mononucleocytes were recovered from each biopsy. The PBMCs or LILs were either studied the same day or frozen in 10% DMSO/90% fetal calf serum at −80°C until used. HLA haplotypes of the donors (A2+ or A2−) were determined by staining PBMCs with the mAbs MA2.1 (19) and BB7.2 (20) and FITC-labeled goat anti-mouse Ig, followed by flow cytometric analysis. Only samples positively stained with both antibodies were considered HLA-A2+.
Peptide-Specific CTL Lines and the Cytotoxicity Assay.
PBMCs were resuspended in RPMI 1640 supplemented with l-glutamine (2 mM), penicillin (100 units/ml), streptomycin (100 μg/ml), and 10% heat-inactivated human AB serum and stimulated in 96-well round-bottom plates at 0.4 × 106 cells per well with 10 μg/ml synthetic peptide, 10 ng/ml rIL-7, and 10 pg/ml rIL-12 (PeproTech, Rocky Hill, NJ). Twenty units/ml rIL-2 (Chiron, Emeryville, CA) was added every 3–4 days. The cultures were restimulated with 10 μg/ml peptide, 20 units/ml rIL-2, and 105 irradiated (3,000 rad) autologous PBMCs in the presence of rIL-7 on day 7 and in the absence of IL-7 on day 14. A standard CTL assay with peptide-pulsed JY-Epstein–Barr virus cells was performed on days 20–24 as described (21). Cells were stained and analyzed fresh or were frozen at −80°C with 10% DMSO/90% fetal calf serum until later analysis.
Synthesis of Peptide-MHC Tetramers.
Peptide-MHC tetramers were prepared as described(10). In brief, recombinant HLA A2.1 with a 15-aa substrate peptide for BirA-dependent biotinylation at its C terminus was expressed in Escherichia coli and isolated from inclusion bodies. It was folded in the presence of β2-microglobulin and a specific peptide to form a peptide-MHC complex, following the procedure of Garboczi et al. (22). After being labeled with biotin by using the enzyme BirA, the complex was mixed with phycoerythrin-labeled strepavidin (PharMingen) at a molar ratio of 4:1 to form the peptide-MHC tetramer.
Cell Staining and Flow Cytometry.
Cells, either freshly prepared or thawed from frozen stocks and washed with medium, were resuspended in 20 μl of fluorescence-activated cell sorter (FACS) buffer (0.5% BSA and 0.05% sodium azide in PBS) supplemented with 1 μg/ml of DNase I. Depending on expected number of tetramer-binding CD8+ cells, 5 × 105–5 × 106 PBMCs, 1 × 105–5 × 105 peptide-stimulated cells, or half of all LILs isolated from a biopsy specimen were used for each staining experiment. For three-color flow cytometric analysis, 23 μl of a staining mixture containing 1.5 μg of tetramer, 0.2 μg of FITC-labeled anti-CD8, and 0.12 μg of each of Tri-color-labeled mAbs to CD4, CD13 or CD19 (Caltag, South San Francisco, CA) was added to the cells. After incubation at room temperature in the dark for 30 min, the cells were washed twice with FACS buffer, fixed with 1% paraformaldehyde, and analyzed on a FACScan flow cytometer. Color compensation settings were made in each round of analysis by using single-stained cells with FITC-, phycoerythrin-, and Tri-color-labeled anti-CD8 (Caltag). In the four-color analysis, the staining mixture was made and used in the same way except that FITC-labeled anti-CD8 was replaced by allophycocyanin-labeled anti-CD8(0.2 μg, Caltag), and a FITC-labeled monoclonal antibody to one of the following markers was also included: CD45RO (0.2 μg), CD45RA (0.2 μg, Caltag), CD27 (10 μl), CD38 (10 μl, PharMingen), CD69 or IFN-γ (20 μl, Becton Dickinson, San Jose, CA). The stained cells were analyzed on a FACSCalibur flow cytometer, whereas color compensation was set with each round of analysis by using single-stained cells with FITC-, phycoerythrin-, Tri-color-, and allophycocyanin-labeled anti-CD8 antibodies. All flow cytometry data were analyzed with cellquest program (Becton Dickinson).
RESULTS
Synthesis and Verification of NS3-Specific Tetramers.
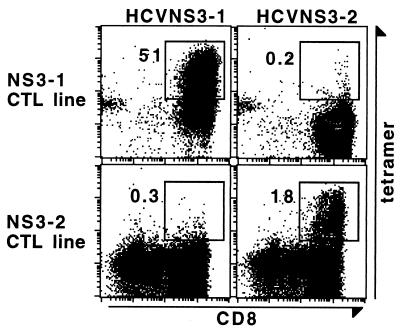
The CTL response against HCV has been studied by in vitro induction of specific CTL lines with peptide stimulation and the 51Cr release-based cytolytic activity assay. These studies suggested that NS3 was one of the HCV proteins that frequently elicits CTL responses in the HCV-infected individuals. Two NS3 peptides, namely NS3-1 (amino acids 1,073–1,081, CVNGVCWTV) (B.R., unpublished result) and NS3-2 (amino acids 1,406–1,415, KLVALGINAV) (23), were identified as CTL epitopes restricted by the MHC class I molecule HLA-A2. We synthesized two peptide-MHC tetramers, HCVNS3-1 and HCVNS3-2, with the recombinant HLA-A2.1 molecule and the peptides NS3-1 or NS3-2, respectively. Their binding specificity was verified by staining CTL lines specific for peptides NS3-1 or NS3-2, generated by in vitro stimulation of PBMCs from HLA-A2-positive (A2+) anti-HCV-seropositive individuals with respective peptides. As shown in Fig. 1, a CD8+ cell population stained by tetramer HCVNS3-1 but not by HCVNS3-2 was detected in the NS3-1 CTL line, whereas a population stained by tetramer HCVNS3-2 but not by HCVNS4-1 was detected in the NS3-2 CTL line. These experiments were repeated for both tetramers with independently generated CTL lines from different individuals. In each case, a population specifically stained by the tetramer was correlated with the peptide-specific cytolytic activity (data not shown), indicating that tetramers HCVNS3-1 and HCVNS3-2 recognize TCRs for the specific epitopes, respectively. In addition, the tetramer HCVNS3-2 was verified by a CTL clone HCV5828.1 which was specific for the peptide NS3-2 (A. Rothman, personal communication). Ninety percent of the cells in this clone stain with the HCVNS3-2 tetramer but not with the HCVNS3-1 tetramer (data not shown).
Figure 1.
Binding specificity of tetramers HCVNS3-1 and HCVNS3-2. NS3 epitope-specific CTL lines were generated by stimulating PBMCs from anti-HCV seropositive individuals with peptides NS3-1 or NS3-2 for 3 weeks. Cytotoxicity of the two lines was determined by 51Cr-release assay, by using peptide-loaded JY-EBV cells as targets. At effector:target = 60:1, the specific lysis by the NS3-1 CTL line was 56%, whereas that by the NS3-2 CTL line was 45%. Spontaneous release as well as nonspecific lysis of target cells was lower than 10%. The CTL lines were stained and analyzed with a three-color flow cytometric assay, by using tetramers HCVNS3-1 or HCVNS3-2 along with mAbs to CD8, CD4, CD13, and CD19. Displayed in the dot plots are lymphocytes which are CD4−CD13−CD19−. The number next to the box is the percentage of tetramer-binding cells of total CD8+ cells.
NS3-Specific CD8+ T Cells in the Peripheral Blood.
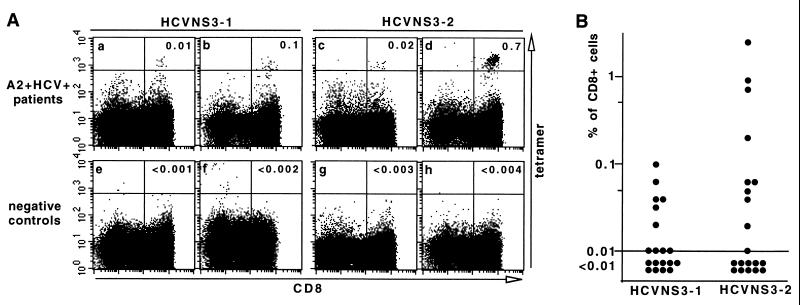
We used the tetramers described above to test PBMCs from 23 A2+ and anti-HCV-seropositive individuals, including 20 patients chronically infected by HCV (A2+HCV+) as indicated by detectable HCV RNA in the serum, 2 previously infected patients who had responded to interferon therapy as indicated by sustained normal serum aminotransferase and undetectable HCV RNA, and 1 healthy anti-HCV-seropositive blood donor (normal serum aminotransferase and undetectable HCV RNA). To assess the background level of tetramer binding, we used two types of individuals as negative controls: HCV-infected HLA A2-negative patients (A2−HCV+) and anti-HCV-seronegative HLA A2-positive individuals (A2+HCV−). The PBMCs from patients and controls were stained with tetramer HCVNS3-1 or HCVNS3-2 along with anti-CD8 and were analyzed by flow cytometry in parallel. A well separated population of tetramer-binding (tetramer+) CD8+ cells with the relative fluorescence intensity of 103 or higher was detected in some of the A2+HCV+ patients (Fig. 2A a–d). The cutoff for tetramer-positive signals was set at a level between this population and the majority of cells in the negative control samples (Fig. 2A e–h). This cutoff was applied to the control samples of both A2+HCV− group (n = 5) and A2−HCV+ group (n = 6) stained with each tetramer. By dividing the total number of tetramer-binding cells with the total number of CD8+ cells in each group (data not shown), we estimated the average background staining to be lower than 0.002% of peripheral CD8+ T cells. The lowest percentage at which five tetramer+CD8+ cells can be detected as a separate population at a relative fluorescence intensity of 103 or higher is 0.01%. Therefore, we consider 0.01% as the limit of detection with our assay. By using 0.01% of CD8+ cell as the cutoff, tetramer+CD8+ cells were detectable in 10 of 19, or 53%, of the A2+HCV+ patients tested with HCVNS3-1, and in 10 of 20, or 50%, of the same group of patients tested with HCVNS3-2 (Fig. 2B). With both tetramers, the overall detection rate of NS3-specific CD8+ T cells was 75%. Interestingly, in the two sustained responders to interferon therapy and in the healthy anti-HCV-seropositive blood donor, no tetramer+ cells could be detected.
Figure 2.
Detection of NS3-specific CD8+ cells in the peripheral blood of chronically infected patients. (A) PBMCs from A2+HCV+ patients (a–d) and control individuals (e and g, A2+HCV−; f and h, A2−HCV+) were stained and analyzed in parallel as described in the legend of Fig. 1, except that a higher cutoff value for fluorescence intensity of the tetramer-binding cells was used to distinguish the tetramer+ cell populations. The number of CD8+ cells acquired from each staining experiment was 2 × 104–1 × 105. The number in the upper-right quadrant is the percentage of tetramer-binding cells of total CD8+ cells. (B) PBMCs from 20 A2+HCV+ individuals were stained and analyzed to determine frequency of tetramer+CD8+ T cells in the peripheral blood of each patient. One of the patients was tested with HCVNS3-2 only. A frequency lower than 0.01% is considered negative.
Surface Phenotype of NS3-Specific CD8+ Cells in the Peripheral Blood.
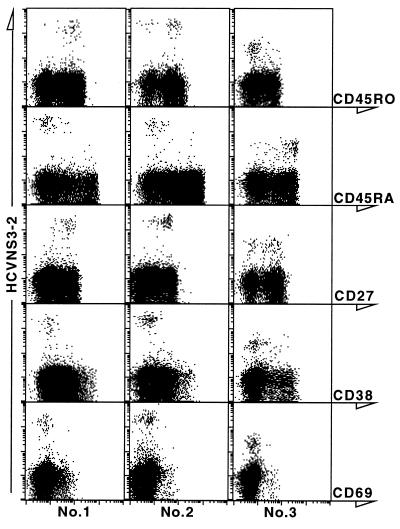
A panel of five surface markers on the NS3-specific CD8+ T cells of three patients was examined by staining with specific antibodies along with the tetramer HCVNS3-2. As shown in Fig. 3, NS3-2-specific CD8+ T cells in the peripheral blood of patients 1 and 2 were predominantly CD45RO+CD45RA−CD27+. In contrast, the NS3-2-specific CD8+ cells from patient 3 were predominantly CD45RO−CD45RA+,and 38% of these cells were CD27−. In all three patients, >90% of the tetramer-binding cells were negative for the activation markers CD38 and CD69. In addition, whereas the percentage of tetramer-binding peripheral CD8+ cells in patient 3 (1.2%) is the highest among the patients we have tested, the relative fluorescence intensity is 102–103, significantly lower than other patients, including patients 1 and 2.
Figure 3.
Surface phenotype of NS3-specific peripheral CD8+ cells. PBMCs from patients 1, 2, and 3 were analyzed with a four-color flow cytometric assay, by using the tetramer HCVNS3-2, mAbs to CD8, CD4, CD13, and CD19, and one of the mAbs against the following markers: CD45RO, CD45RA, CD27, CD38, and CD69. Displayed in the dot plots are CD8+ lymphocytes which are CD4−CD13−CD19−.
Peptide-Induced Activation of the NS3-Specific CD8+ cells.
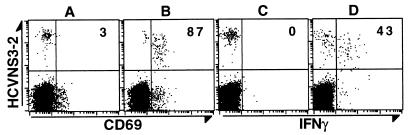
To characterize the response of the NS3-specific CD8+ cells on antigen stimulation, we incubated whole-blood samples from patient 1 with NS3 peptide for 6 hours, in the presence of brefeldin A during the last 4 hours. The cells were then stained with the HCVNS3-2 tetramer in addition to antibodies to CD8 and CD69 or IFN-γ. As shown in Fig. 4, after stimulation with the peptide NS3-2 but not NS3-1, the expression of CD69 was induced in 87% of the tetramer+ cells and that of IFN-γ was induced in 43% of the tetramer+ cells. This result indicates that the NS3-2-specific CD8+ cells in the peripheral blood can be readily activated by specific antigen.
Figure 4.
Antigen-specific activation of NS3-specific peripheral CD8+ cells. Aliquots of heparinized whole blood were incubated with 10 μg/ml HCV peptides NS3–1 (A and C) or NS3-2 (B and D), 1 μg/ml anti-CD28 and 1 μg/ml anti-CD49d (both from Becton Dickinson) at 37°C for 6 hours, with addition of brefeldin A (Sigma) at 10 μg/ml during the last 4 hours, as described (24, 25). The cells were processed and stained with tetramer HCVNS3-2 and antibodies by using a modified procedure (unpublished data; ref. 24). In brief, the cells were stained with tetramer first, followed by lysing red blood cells with FACS Lysing Solution and permeabilyzing white blood cells with FACS Permeabilizing Solution (both from Becton Dickinson). The processed cells were then stained and analyzed as described in the legend of Fig. 3. The number in the upper right quandrant is the percentage of CD69+ or IFN-γ+ cells of the total tetramer+CD8+ cells.
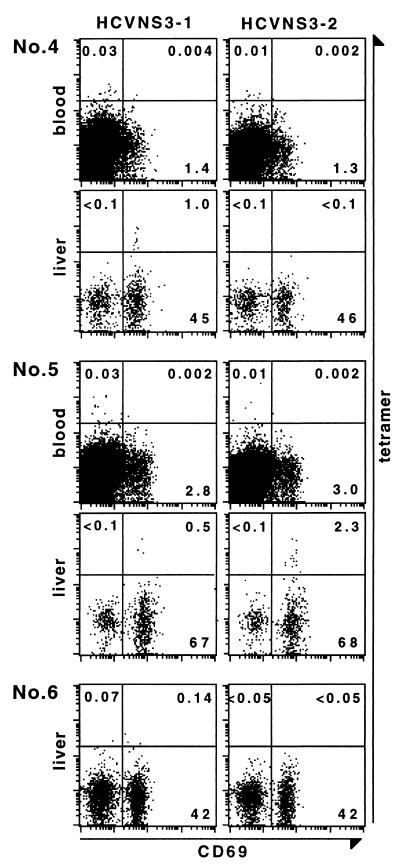
NS3-Specific CD8+ Cells in the HCV-Infected Livers.
We isolated LILs in needle biopsies from livers of A2+HCV+ patients 4 and 5, and A2−HCV+ patient 6. The LILs were stained with tetramers HCVNS3-1 or HCVNS3-2 along with antibodies to CD8 and CD69. PBMCs from patients 4 and 5 obtained on the same day as the liver biopsy also were tested. As shown in Fig. 5, an HCVNS3-1-binding population consisting of 1.0% of CD8+ cells was detected in the liver of patient 4, whereas an HCVNS3-2-binding CD69+ population representing 2.3% of CD8+ cells was detected in the liver of patient 5. When the PBMC samples were analyzed in parallel with the LIL samples by using the same cutoff for tetramer+ signals, the frequency of HCVNS3-1-binding CD8+ cells in the peripheral blood of patient 4 was 0.03%, whereas that of HCVNS3-2-binding CD8+ cells in patient 5 was 0.01%. However, if a more stringent cutoff such as that used in Fig. 2 is applied, both frequencies would be <0.01%, or lower than the limit of detection. These results indicate that the frequency of tetramer+CD8+ cells in the liver is at least 30-fold (patient 4) or 190-fold (patient 5) higher than in the peripheral blood, suggesting that HCV-specific CTLs are greatly enriched in the liver. In contrast to the results obtained from the peripheral blood of patients 1, 2, and 3 where >90% of the NS3-specific CD8+ cells were CD69− (Fig. 3), in the livers of patients 4 and 5, all of the NS3-specific CD8+ T cells were CD69+.
Figure 5.
NS3-specific CD8+ cells in the HCV-infected livers. LILs were isolated from needle biopsies of A2+HCV+ patients 4 and 5 and A2−HCV+ patient 6 (negative control). The biopsies of patients 4 and 6 showed lesions compatible with mild chronic hepatitis, whereas that of patient 5 was compatible with severe chronic hepatitis. Cells were stained and analyzed with a four-color flow cytometric assay, by using tetramers HCVNS3-1 or HCVNS3-2 in addition to antibodies to CD8, CD4, CD13, CD19, and CD69. PBMCs from patients 4 and 5 were analyzed in parallel with the LIL samples. Displayed in the dot plots are CD8+ lymphocytes which are CD4−CD13−CD19−. Numbers in the upper left, upper right, and lower right quadrants indicate percentage of tetramer+CD69−, tetramer+CD69+, and tetramer−CD69+ cells of total CD8+ cells, respectively.
DISCUSSION
With the two peptide-MHC tetramers derived from previously identified CTL epitopes of the HCV NS3 protein, we were able to identify NS3-specific CD8+ T cells in peripheral blood as well as in liver biopsies of HCV-infected patients. Seventy-five percent of chronically infected individuals had circulating CD8+ cells that were recognized by one or both tetramers. The NS3-specific CD8+ cells in the peripheral blood were predominantly negative for the activation markers CD38 and CD69 (26, 27), suggesting that they are memory T cells, whereas those in the liver are all CD69+, suggesting they are activated CTLs.
When the resting HCV NS3-specific CD8+ cells in the peripheral blood were stimulated with specific peptide for 6 hours, 87% expressed CD69, whereas 43% produced IFN-γ, indicating that these cells can be activated and begin to develop effector function when exposed to their specific antigen. Because IFN-γ produced by CTLs may play an important role in the antiviral function of hepatitis virus-specific CTLs (28), this functional assay for the antigen-specific CD8+ T cells should be very useful for studying the CTL response against hepatitis viruses as well as other viruses.
The elucidation of underlying mechanisms for HCV persistence is one of the biggest challenges faced by viral immunologists. Recently Zajac et al. (29) described two mechanisms that may contribute to the chronic infection of lymphocytic choriomeningitis virus in mice: deletion of virus-specific CD8+ T cells and persistence of virus-specific CD8+ T cells lacking antiviral effector functions. Because we have clearly shown that most chronically infected patients (but not the limited number of recovered patients tested) have detectable HCV-specific CD8+ T cells in circulation and that these cells can be activated and can produce IFN-γ when stimulated by their specific antigen (Fig. 4 and data not shown), neither of those two mechanisms seems to apply to the HCV infection, at least in the patients we tested. Other mechanisms for viral immune evasion, such as escape mutations of the virus, are certainly possible.
The surface phenotype of the NS3-specific peripheral CD8+ T cells varied in different patients: in patients 1 and 2, these cells were predominantly CD45RO+CD45RA−CD27+, in agreement with the previously suggested phenotype of memory cells which do not display cytotoxicity without antigen-stimulation (30, 31), whereas in patient 3, the tetramer-binding cells were predominantly CD45RO−CD45RA+ and 38% were CD27−. A similar CD45RA+CD27− phenotype was also observed in CD8+ cells specific for the HCV core131-140 epitope detected with another tetramer (data not shown). Although CD45RA+CD27− has been suggested to characterize the phenotype of effector CTLs (31) and the percentage of the NS3-2 specific CD8+ cells in patient 3 is greater than 1%, a direct 51Cr-release assay for epitope-specific cytotoxicity with this patient’s PBMCs was negative. This is consistent with recent results showing that melanoma antigen-specific T cells with this phenotype are functionally anergic (P. Lee, C. Yee, P. A. Savage, L. Fong, D. Brockstedt, J. S. Weber, D. Johnson, S. Swetter, J. Thompson, P. D. Greenberg, et al., unpublished data). This suggests that HCV may elicit very different immune responses in different individuals. The clinical consequences of this heterogeneity in CTL response are not clear at this time, but given the autoimmune character of this disease these differences in CTL phenotype may be very significant prognostic indicators.
Little is known about the phenotypic and functional state of antigen-specific lymphocytes in HCV-infected livers. Presumably, these are the cells that play an important role in liver cell injury as well as in the clearance of HCV. Nuti et al. (32) report that intrahepatic lymphocytes in HCV-infected individuals display an activated phenotype. These lymphocytes do not proliferate but rather undergo apoptosis in the liver. However, the antigenic specificity of the activated lymphocytes was not defined in their study. The frequency of antigen-specific CTLs has been suggested to be higher in the HCV-infected liver than in the peripheral blood, largely based on the observation that HCV-specific CTLs can be expanded from the liver but not from the peripheral blood by antigen-nonspecific stimulation (4). Direct identification of virus-specific T cells with tetramers should permit a more accurate qualitative and quantitative analysis of the anti-viral cellular immune response. This is especially true in the case of a virus-infected liver, where most of the CTLs are believed to undergo apoptosis and may lack the potential for in vitro expansion. We were able to determine that NS3-specific CD8+ cells in the liver are positive for the activation marker CD69 and that their number accounts for 1.0–2.3% of intrahepatic CD8+ cells but is barely detectable in the blood of the same patients. The degree of enrichment that we see here is far greater than the 2- to 3-fold increase seen in cerebrospinal fluid versus peripheral blood in the HTLV-1-specific T cells (18). Our studies confirm and extend prior indications that anti-HCV lymphocytes are enriched in the liver. Because these cells are apparently short-lived, it is likely that mechanisms exist to recruit specific immune cells into the liver, such as lymphocyte homing. The fact that virus persists in these patients and that the biopsies from which LILs were isolated showed evidence of chronic hepatitis suggests that the presence of HCV-specific CTLs in the liver is sufficient to cause liver cell injury but not sufficient to clear the virus. The number of virus-specific CTLs in these livers may be too small to clear the virus (33). Alternatively, the cytokine environment of the liver, which contributes to liver tolerance, may also suppress the production of antiviral cytokines by the virus-specific CTLs (34). Further quantitative analysis and characterization of HCV-specific T cells in the liver should help to answer these important questions.
The CTL response to HCV is multispecific (5, 6, 8, 23, 35), and the two NS3 epitopes we tested are only a small part of the known CTL epitopes, which do not comprise all potential HLA A2-restricted CTL epitopes of HCV. These findings are in agreement with our observation that approximately half of the CD8+ T cells in the HCV-infected livers carry the activation marker CD69, but only 1–2% of them are specific for the two NS3 epitopes. Presumably, at least some of the remaining CD69+ cells are directed at other HCV peptides, although it is presently unclear whether all are HCV-specific or some are simply activated by nonspecific mechanisms such as bystander effects (36).
Because of the difficulty in obtaining liver samples, most studies on the HCV-specific CTL responses will continue to rely on blood samples that contain far fewer HCV-specific CTLs, as demonstrated in this study. Current estimates are that tetramer staining is 10–500 times more sensitive than the limiting-dilution assay (14). The frequency of HCV peptide-specific CTL precursors in the peripheral blood of three chronically infected individuals was previously determined to be 3.2–12.8 per million PBMCs by limiting-dilution assay (37), approximately equivalent to 0.003–0.01% of circulating CD8+ cells. In contrast, the frequency of NS3-specific peripheral CTLs detected by tetramers appears to range from 0.01 to 1.2% of CD8+ cells, which is significantly higher. However, the number of the HCV NS3-specific CD8+ T cells detected in the peripheral blood of most of the patients we tested is much smaller than the number of the CD8+ cells specific for other pathogenic viruses, including persistent viruses like HIV (10–15). In addition, even though we can reliably detect NS3-specific peripheral CD8+ cells at a frequency as low as 0.01%, we did not detect them in PBMCs from some of the anti-HCV-seropositive individuals, whereas CTL lines with high cytolytic activity can be induced from the same PBMC samples after 3 weeks of in vitro simulation with the same HCV peptides. Induction of peptide-specific cytolytic activity in these lines correlated with appearance of tetramer+ cells which can be readily detected at a percentage as high as 51% (Fig. 1 and data not shown). Thus, HCV-specific CTL precursors do exist in the peripheral blood of some individuals at a frequency of <0.01%. Alternatively, they may be expressing their TCRs at a lower density or with lower affinity to the tetramer, similar to those cells detected from patient 3 (Fig. 3). This latter type of CTL would be identified only if their number was greatly amplified, as in the CTL lines, or if their frequency in the PBMCs was very high, as in patient 3.
The HCV genome is highly variable, as indicated by the existence of different genotypes and virus quasispecies (38). Variant peptide sequences in both of the NS3 epitopes we discuss here have been reported (39) and TCRs specific for such variants may not crossreact with the tetramers used here. However, because PBMCs from the majority (75%) of A2+HCV+ patients appeared to react with at least one of the two tetramers, the use of these reagents should provide significant insight into the overall CTL responses against HCV, whereas use of tetramers made with the variant peptides at these two NS3 epitopes, as well as tetramers made with other HCV epitopes, is certainly warranted. The direct quantitation and characterization of HCV-specific CD8+ T cells in different clinical settings, when correlated with serological, virological, biochemical, and histological analysis of the patients, should extend our understanding of immunopathogenesis as well as the mechanisms of clearance or persistence of HCV.
Acknowledgments
We thank A. Rothman for the CTL clone HCV5828.1, P. Lee for advice on tetramer staining and HLA-A2 typing, V. Maino and Becton Dickinson Immunocytometry Systems for advice and immunological reagents, J. Glenn for collecting clinical samples, Y. Lu for technical assistance and E. Mocarski for critical reading of this manuscript. This work was supported by National Institutes of Health grant AI40034, the Hutchison Program in Translational Medicine at Stanford University and the Howard Hughes Medical Institute. F.X.L. and M.B. were in part supported by grants from the Spanish Association for the Study of the Liver (AEEH). J.B. is supported by National Institutes of Health training Grant GM07276.
ABBREVIATIONS
- CTL
cytotoxic T lymphocyte
- HCV
hepatitis C virus
- LIL
liver-infiltrating lymphocyte
- PBMC
peripheral blood mononuclear cells
- TCR
T cell receptor
References
- 1.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Alter M J. Semin Liver Dis. 1995;15:5–14. doi: 10.1055/s-2007-1007259. [DOI] [PubMed] [Google Scholar]
- 3.Walker C M. Semin Virol. 1996;7:13–21. [Google Scholar]
- 4.Koziel, M. J. (1997) J. Viral. Hepat.4, Suppl. 2, 31–41. [DOI] [PubMed]
- 5.Rehermann B, Chang K M, McHutchinson J, Kokka R, Houghton M, Rice C M, Chisari F V. J Virol. 1996;70:7092–7102. doi: 10.1128/jvi.70.10.7092-7102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koziel M J, Dudley D, Afdhal N, Grakoui A, Rice C M, Choo Q L, Houghton M, Walker B D. J Clin Invest. 1995;96:2311–2321. doi: 10.1172/JCI118287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson D R, Marousis C G, Davis G L, Rice C M, Wong J, Houghton M, Lau J Y. J Immunol. 1997;158:1473–1481. [PubMed] [Google Scholar]
- 8.Wong D K, Dudley D D, Afdhal N H, Dienstag J, Rice C M, Wang L, Houghton M, Walker B D, Koziel M J. J Immunol. 1998;160:1479–1488. [PubMed] [Google Scholar]
- 9.Nelson D R, Marousis C G, Ohno T, Davis G L, Lau J Y. Hepatology. 1998;28:225–230. doi: 10.1002/hep.510280129. [DOI] [PubMed] [Google Scholar]
- 10.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 11.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, et al. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 12.Murali-Krishna K, Altman J D, Suresh M, Sourdive D J, Zajac A J, Miller J D, Slansky J, Ahmed R. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 13.Gallimore A, Glithero A, Godkin A, Tissot A C, Pluckthun A, Elliott T, Hengartner H, Zinkernagel R. J Exp Med. 1998;187:1383–1393. doi: 10.1084/jem.187.9.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuroda M J, Schmitz J E, Barouch D H, Craiu A, Allen T M, Sette A, Watkins D I, Forman M A, Letvin N L. J Exp Med. 1998;187:1373–1381. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callan M F C, Tan L, Annels N, Ogg G S, Wilson J D K, O’Callaghan C A, Steven N, McMichael A J, Rickinson A B. J Exp Med. 1998;187:1395–1402. doi: 10.1084/jem.187.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flynn K J, Belz G T, Altman J D, Ahmed R, Woodland D L, Doherty P C. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- 17.Stevenson P G, Belz G T, Altman J D, Doherty P C. Proc Natl Acad Sci USA. 1998;95:15565–15570. doi: 10.1073/pnas.95.26.15565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greten T F, Slansky J E, Kubota R, Soldan S S, Jaffee E M, Leist T P, Pardoll D M, Jacobson S, Schneck J P. Proc Natl Acad Sci USA. 1998;95:7568–7573. doi: 10.1073/pnas.95.13.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMichael A J, Parham P, Rust N, Brodsky F. Hum Immunol. 1980;1:121–129. doi: 10.1016/0198-8859(80)90099-3. [DOI] [PubMed] [Google Scholar]
- 20.Parham P, Brodsky F M. Hum Immunol. 1981;3:277–299. doi: 10.1016/0198-8859(81)90065-3. [DOI] [PubMed] [Google Scholar]
- 21.Rehermann B, Fowler P, Sidney J, Person J, Redeker A, Brown M, Moss B, Sette A, Chisari F V. J Exp Med. 1995;181:1047–1058. doi: 10.1084/jem.181.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garboczi D N, Hung D T, Wiley D C. Proc Natl Acad Sci USA. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerny A, McHutchison J G, Pasquinelli C, Brown M E, Brothers M A, Grabscheid B, Fowler P, Houghton M, Chisari F V. J Clin Invest. 1995;95:521–530. doi: 10.1172/JCI117694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suni M A, Picker L J, Maino V C. J Immunol Methods. 1998;212:89–98. doi: 10.1016/s0022-1759(98)00004-0. [DOI] [PubMed] [Google Scholar]
- 25.Kern F, Surel I P, Brock C, Freistedt B, Radtke H, Scheffold A, Blasczyk R, Reinke P, Schneider-Mergener J, Radbruch A, et al. Nat Med. 1998;4:975–978. doi: 10.1038/nm0898-975. [DOI] [PubMed] [Google Scholar]
- 26.Holter W, Majdic O, Liszka K, Stockinger H, Knapp W. Cell Immunol. 1985;90:322–330. doi: 10.1016/0008-8749(85)90197-2. [DOI] [PubMed] [Google Scholar]
- 27.Picker L J, Singh M K, Zdraveski Z, Treer J R, Waldrop S L, Bergstresser P R, Maino V C. Blood. 1995;86:1408–1419. [PubMed] [Google Scholar]
- 28.Guidotti L G, Ishikawa T, Hobbs M V, Matzke B, Schreiber R, Chisari F V. Immunity. 1996;4:25–36. doi: 10.1016/s1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- 29.Zajac A J, Blattman J N, Murali-Krishna K, Sourdive D J, Suresh M, Altman J D, Ahmed R. J Exp Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beverley P C L. Semin Immunol. 1992;4:35–41. [PubMed] [Google Scholar]
- 31.Hamann D, Baars P A, Rep M H, Hooibrink B, Kerkhof-Garde S R, Klein M R, van Lier R A. J Exp Med. 1997;186:1407–1418. doi: 10.1084/jem.186.9.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nuti S, Rosa D, Valiante N M, Saletti G, Caratozzolo M, Dellabona P, Barnaba V, Abrignani S. Eur J Immunol. 1998;28:1–10. doi: 10.1002/(SICI)1521-4141(199811)28:11<3448::AID-IMMU3448>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 33.Chisari F V. In: T Lymphocytes in the Liver. Crispe I N, editor. New York: Wiley–Liss; 1999. pp. 117–138. [Google Scholar]
- 34.Crispe I N. In: T Lymphocytes in the Liver. Crispe I N, editor. New York: Wiley–Liss; 1999. pp. 235–249. [Google Scholar]
- 35.Battegay M, Fikes J, Di Bisceglie A M, Wentworth P A, Sette A, Celis E, Ching W M, Grakoui A, Rice C M, Kurokohchi K, et al. J Virol. 1995;69:2462–2470. doi: 10.1128/jvi.69.4.2462-2470.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tough D F, Sprent J. Immunol Rev. 1996;150:129–142. doi: 10.1111/j.1600-065x.1996.tb00699.x. [DOI] [PubMed] [Google Scholar]
- 37.Rehermann B, Lau D, Hoofnagle J H, Chisari F V. J Clin Invest. 1996;97:1655–1665. doi: 10.1172/JCI118592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bukh J, Miller R H, Purcell R H. Semin Liver Dis. 1995;15:41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 39.Chang K M, Rehermann B, McHutchison J G, Pasquinelli C, Southwood S, Sette A, Chisari F V. J Clin Invest. 1997;100:2376–2385. doi: 10.1172/JCI119778. [DOI] [PMC free article] [PubMed] [Google Scholar]