Abstract
The nuclear pore complex (NPC) is a multicomponent structure containing a subset of proteins that bind nuclear transport factors or karyopherins and mediate their movement across the nuclear envelope. By altering the expression of a single nucleoporin gene, NUP53, we showed that the overproduction of Nup53p altered nuclear transport and had a profound effect on the structure of the nuclear membrane. Strikingly, conventional and immunoelectron microscopy analysis revealed that excess Nup53p entered the nucleus and associated with the nuclear membrane. Here, Nup53p induced the formation of intranuclear, tubular membranes that later formed flattened, double membrane lamellae structurally similar to the nuclear envelope. Like the nuclear envelope, the intranuclear double membrane lamellae enclosed a defined cisterna that was interrupted by pores but, unlike the nuclear envelope pores, they lacked NPCs. Consistent with this observation, we detected only two NPC proteins, the pore membrane proteins Pom152p and Ndc1p, in association with these membrane structures. Thus, these pores likely represent an intermediate in NPC assembly. We also demonstrated that the targeting of excess Nup53p to the NPC and its specific association with intranuclear membranes were dependent on the karyopherin Kap121p and the nucleoporin Nup170p. At the nuclear envelope, the abilities of Nup53p to associate with the membrane and drive membrane proliferation were dependent on a COOH-terminal segment containing a potential amphipathic α-helix. The implications of these results with regards to the biogenesis of the nuclear envelope are discussed.
Keywords: nuclear pore complex, nucleoporins, nuclear membrane, nuclear transport, karyopherin
Introduction
The nuclear envelope (NE) membrane is composed of two parallel, lamellar membranes, termed the outer and inner nuclear membranes, that surround a flattened cisternal space continuous with the lumen of the ER (Maul 1977; Worman and Courvalin 2000). These inner and outer membranes are connected at multiple locations by cylindrically shaped membranes, ∼90 nm in diameter, that form pores across the NE cisternae, creating a channel connecting the cytoplasm and the nucleoplasm. Each of these “pore membrane” regions wraps around the waist of a nuclear pore complex (NPC), a massive structure that occupies the channel. The NPC is composed of repetitive substructures that form an eightfold (perpendicular to the NE) by twofold (parallel to the NE) symmetrical core attached to the pore membrane, at least in part, by integral membrane proteins that reside there (for reviews, see Ryan and Wente 2000; Wente 2000). Built on the core are two sets of structurally distinct filaments, one set extending into the cytoplasm and the other into the nucleoplasm. It is believed that all transport across the NE occurs through the NPC. In addition to regulating nuclear transport, the NPCs appear to be functionally linked to chromatin (Galy et al. 2000) and to play a role in the maintenance of NE structure (see below).
In yeast, the NPC is composed of ∼30 proteins (termed nucleoporins or nups) present in multiple copies (for reviews, see Ryan and Wente 2000; Rout et al. 2000), which together contribute to the 50–60 MD mass of the NPC (Rout and Blobel 1993; Yang et al. 1998; Rout et al. 2000). In a broad context, these proteins can be divided into two categories, those with repetitive peptide repeats containing Phe-Gly (FG) residues, generally in the context of GLFG or FXFG repeats, and those that lack these repeats. As suggested by the structure of the NPC core, most of its components are symmetrically positioned on both its cytoplasmic and nucleoplasmic faces (Nehrbass et al. 1996; Fahrenkrog et al. 1998; Marelli et al. 1998; Rout et al. 2000). However, several nups are not, and they likely contribute to the asymmetric filamentous structures (Kraemer et al. 1995; Fahrenkrog et al. 1998; Hurwitz et al. 1998; Rout et al. 2000).
The FG-containing repeat nups play a direct role in nuclear transport, acting as binding sites for soluble transport factors, termed karyopherins or kaps (also referred to as importins or exportins), and their cargo (for reviews, see Wozniak et al. 1998; Ryan and Wente 2000). The presence of the repeat nups at sites throughout the NPC likely establishes an interface for the multiple docking and release steps that are believed to occur as the kap and its cargo traverse the NPC (Feldherr et al. 1984; Richardson et al. 1988). However, the mechanism by which the kap/cargo complex moves unidirectionally from nup to nup is not yet resolved. It is clear that the small GTPase Ran, which is believed to exist primarily as Ran-GTP in the nucleus and Ran-GDP in the cytoplasm, plays a role in the terminal release of kaps from nups (Rexach and Blobel 1995; Görlich et al. 1996; Delphin et al. 1997; Floer et al. 1997; Floer and Blobel 1999; Kehlenbach et al. 1999). However, Ran's effects on nup/kap interactions within the NPC are unclear.
As many of the repeat nups share the ability to bind several of the 14 different kaps in yeast, the repeat nups likely form a common pathway through the NPC. However, it is also clear that certain kaps have a higher affinity for particular nups (Aitchison et al. 1996; Pemberton et al. 1997; Rosenblum et al. 1997; Rout et al. 1997; Marelli et al. 1998; Damelin and Silver 2000). A clear example of this is the interaction between Nup53p and Kap121p (Marelli et al. 1998). Although Kap121p binds to many of the same nups as other kaps, it is the only kap that appears to interact with Nup53p. On the basis of these observations, it was proposed that particular kaps may follow distinct routes though the NPC and that this could provide a mechanism for regulating a specific import pathway (Marelli et al. 1998).
The function of the nonrepeat nups in transport through the NPC is less clear. Mutations in several of these nups do affect the active transport of various cargoes including imported proteins and exported mRNA (for a review, see Fabre and Hurt 1997). However, the functional basis for this inhibition remains to be determined, as none of the nups lacking FG repeats have been shown to directly interact with kaps. Their effects on transport may be indirect, as several of the nonrepeat nups have been shown to directly interact with the FG-containing nups suggesting that they may provide a foundation on which the repeat-containing nups are organized (see Fabre and Hurt 1997). In addition, the nonrepeat-containing nups, including Nup170p and Nup188p, may also function in establishing a diffusion channel through the NPC that allows small molecules to cross the NE (Shulga et al. 2000).
Although the role of nups in nuclear transport is well established, several observations have suggested that specific nups are also necessary for the maintenance of normal NE structure. Mutations in several nup genes alter the normal distribution of NPCs along the NE leading to their clustering (summarized in Fabre and Hurt 1997) or a change in the total number of NPCs (Mutvei et al. 1992; Zabel et al. 1996). Other nup gene mutations appear to have more profound effects on the NE membrane. For example, mutations in NUP116 lead to local alterations of the outer nuclear membrane at the NPC that forms a double membrane seal over the NPC (Wente and Blobel 1993). Similar abnormalities have also been documented in strains containing mutations in NUP145, NUP188, and GLE2 (Wente and Blobel 1994; Nehrbass et al. 1996; Zabel et al. 1996; Teixeira et al. 1997; Ho et al. 1998). Other nup gene mutants, including alleles of NUP1, NUP170, NUP188, NUP84, and NUP85, induce massive changes in the shape of the nucleus causing finger-like projections and invaginations of the NE that appear to increase the total surface area of the nucleus (Bogerd et al. 1994; Aitchison et al. 1995; Goldstein et al. 1996; Nehrbass et al. 1996; Siniossoglou et al. 1996). Cumulatively, these observations support the idea that an additional function of NPCs is to regulate the biosynthesis of the NE membrane.
Here we present data from studies examining the role of Nup53p in nuclear transport and NE biogenesis. By overproducing Nup53p, we show that Nup53p is targeted to the NPC and enters the nucleus through a process that is dependent on Kap121p and Nup170p. During or shortly after transport through the NPC, Nup53p associates with membranes and stimulates the formation of intranuclear membranes that develop into double membrane lamellae beneath the NE. These membranes appear to contain pores and pore membrane proteins but not intact NPCs. The implications of these results as they relate to the mechanisms of NPC biogenesis are discussed.
Materials and Methods
Yeast Strains and Media
All strains used in this study are listed in Table or described below. All strains were grown in YPD (1% yeast extract, 2% bactopeptone, and 2% glucose), YPG (1% yeast extract, 2% bactopeptone, and 2% galactose), or in synthetic media (SM) supplemented with the necessary nutrients and containing either 2% glucose or 2% galactose as described in Sherman et al. 1986. All strains were grown at 30°C unless otherwise stated. Yeast manipulations were conducted as described in Sherman et al. 1986 and transformations as described in Delorme 1989.
Table 1.
Yeast Strains
| Strains | Genotype | Derivation |
|---|---|---|
| W303 | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 | |
| DF5 | Mata ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 | |
| NP53-A2 | Mata ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 nup53::HIS3 | Marelli et al. 1998 |
| CNP53 | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 pCUP1- NUP53 (URA3 leu2-d) | W303 transformed with pCUP1-NUP53 |
| CNP53D | Mata ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 pCUP1- NUP53 (URA3 leu2-d) | DF5 transformed with pCUP1-NUP53 |
| GNP53 | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 pBJ244-NUP53 (TRP1) | W303 transformed with pBJ244-NUP53 |
| GNP59 | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 pBJ244-NUP59 (TRP1) | W303 transformed with pBJ244-NUP59 |
| CNP53ΔC | Mata ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 nup53::HIS3 pCUP1- NUP53ΔC (URA3 leu2-d) | NP53-A2 transformed with pCUP1- NUP53ΔC |
| NP170PA-p53 | Mata/Mata ura3-52/ura3-52 his3-Δ200/his3-Δ200 trp1-1/trp1-1 leu2-3,112/leu2- 3,112 lys2-801/lys2-801 nup170-pA/+ (URA3-HIS3) pRCUP1-NUP53 (LEU2) | NP170pA (Aitchison et al. 1995) transformed with pRCUP1-NUP53 |
| NDCPA-p53 | Mata ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys3,112 ndc1-pA (URA3-HIS3) pCUP1-NUP53 (LEU2) | ProA5-4d (Chial et al. 1998) transformed with pRCUP1-NUP53 |
| NP170-p53 | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 nup170-1::HIS3 pCUP1-NUP53 (URA3 leu2-d) | NP170-11.1 (Aitchison et al. 1995) transformed with pCUP1-NUP53 |
| PMY17-p53 | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 pom152-2::HIS3 pCUP1-NUP53 (URA3 leu2-d) | PMY17 (Wozniak et al. 1994) transformed with pCUP1-NUP53 |
| KP123-p53 | Mata ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 kap123:ura3::HIS3 pCUP1-NUP53 (URA3 leu2-d) | Integrative transformation of 123Δ-14-1 (Rout et al. 1997) with ura3::HIS3 switcher, containing pCUP1-NUP53 |
| KP121-41-p53 | ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 kap121::HIS3 pRS314-kap121-41 (TRP1) pCUP1-NUP53 (URA3 leu2-d) | KP121-41 (Leslie et al., manuscript in preparation) transformed with pCUP1-NUP53 |
| KP121-41-pNLS | ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 kap121::HIS3 pRS314-kap121-41 (TRP1) pRS315-p4GFP (LEU2) | KP121-41 transformed with pRS315- p4GFP |
| KP121-41-cNLS | ura3-52 his3-Δ200 trp1-1 leu2-3,112 lys2-801 kap121::HIS3 pRS314-kap121-41 (TRP1) pGADGFP (LEU2) | KP121-41 transformed with pGADGFP (cNLS-GFP; Shulga et al. 1996) |
| CNP53-cNLS | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 pCUP1-NUP53 (URA3 leu2-d) p12-GFP2-NLS (HIS3) | W303 transformed with pCUP1-NUP53 and p12-GFP2-NLS (Lee and Aitchison 1999) |
| CNP53-pNLS | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 pCUP1-NUP53 (URA3 leu2-d) pRS314-p4GFP (TRP1) | W303 transformed with pCUP1-NUP53 and pRS314-p4GFP |
| W303-cNLS | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 pGADGFP (LEU2) | W303 transformed with pGADGFP (cNLS-GFP; Shulga et al. 1996) |
| W303-pNLS | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100) pRS314-p4GFP (TRP1) | W303 transformed with pRS314-p4GFP (pNLS-GFP) |
| CNP53-121 | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 pCUP1-NUP53 (URA3 leu2-d) pK121-GFP (TRP1) | W303 transformed with pCUP1-NUP53 and pK121-GFP |
| W303-121 | Mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 pK121-GFP (TRP1) pYEX-BX (URA3 leu2-d) | W303 transformed with pYEX-BX and pK121-GFP |
Plasmids
The plasmids described in this paper are as follows: pRS314, CEN/TRP1; pRS315, CEN/LEU2; pBJ244, 2μ/TRP1 GAL1/GAL10; pBJ244-NUP53, the NUP53 ORF extending from nucleotide +1 to +1,756 (where +1 is the A of the initiation codon) inserted into a BamHI site in pBJ244; pBJ244-NUP59, the NUP59 ORF, extending from nucleotide +1 to +2,930, inserted into a BamHI site of pBJ244; pYEX-BX, 2μ/URA3/leu2d containing the CUP1 promoter (CLONTECH Laboratories, Inc.); pCUP1-NUP53, a BamHI/BamHI fragment containing the NUP53 ORF in pYEX-BX (CLONTECH Laboratories, Inc.); pCUP1-NUP53ΔC, a fragment of the NUP53 ORF extending form nucleotide +1 to +1,344 followed by a stop codon in pYEX-BX; pRCUP1-NUP53, a 2,035-bp HindIII/PstI fragment containing the CUP1 promoter and the NUP53 ORF in pRS315; pRCUP1, pRCUP1-NUP53 with the NUP53 ORF removed; pK121-green fluorescent protein (GFP), the KAP121–GFP gene fusion was amplified from pPS1069 (Seedorf and Silver 1997; provided by M. Seedorf and P. Silver, Dana Farber Cancer Institute, Boston, MA) inserted into pRS314; pRS314-p4GFP and pRS315-p4GFP (pNLS-GFP), a fragment encoding Pho4140–166-GFP3 from pPho4(140–166) (Kaffman et al. 1998; provided by E. O'Shea, University of California, San Francisco, CA) in pRS314 and pRS315; pGFP-C-FUS (Niedenthal et al. 1996), pNUP53-GFP, pNUP53-ΔC-GFP, and pCT-NUP53-GFP, pGFP-C-FUS containing a DNA fragment encoding the complete NUP53 ORF, amino acid residues 1–450 of Nup53p, and amino acid residues 375–475 of Nup53p, respectively.
Induction of NUP53 Overexpression
For the induction of GAL1 controlled expression of NUP53 and NUP59, GNP53 or GNP59 cells were grown to mid-logarithmic phase in SM-Trp media containing 2% glucose, washed extensively with water, transferred to SM-Trp media containing 2% galactose, and incubated for the indicated times. For the induction of CUP1::NUP53 in strains containing the pCUP1-NUP53 plasmid, cells were grown to mid-logarithmic phase in selection medium and then induced for the indicated times by the addition of copper sulfate to a final concentration of 0.5 mM. Induction of the synthesis of Nup53-GFP, Nup53-ΔC-GFP, and CT-Nup53-GFP was achieved by growing DF5 cells, containing the appropriate plasmid, in SM media lacking URA and methionine for 12–15 h.
Fluorescence Microscopy
Immunofluorescence microscopy was performed as described by Kilmartin and Adams 1984 with modifications described in Wente et al. 1992 and Aitchison et al. 1995. Primary antibody incubations were performed in PBS containing 0.1% Tween 20 (PBS-T) and 5% skim milk. Nup53p was detected using affinity-purified, rabbit anti-Nup53p antibodies and Cy3-conjugated, donkey anti–rabbit antibodies (Jackson ImmunoResearch Laboratories). The monoclonal antibodies mAb118C3 (Strambio-de-Castillia et al. 1995; provided by C. Strambio-de-Castillia and M. Rout, The Rockefeller University, New York, NY) and mAb414 (BAbCo; Berkeley Antibody Co.) were used to detect Pom152p and a subset of FXFG repeat-containing nups, respectively. Monoclonal binding was visualized with goat anti–mouse antibodies conjugated to rhodamine (Cappel Laboratories/Organon Teknika Corp.). Staphylococcus aureus protein A chimeras were detected using rabbit IgG (Cappel Laboratories) followed by Cy3-conjugated, donkey anti–rabbit antibodies (Jackson ImmunoResearch Laboratories). Images were captured using an Olympus BX-50 fluorescence microscope and a SPOT digital camera (Diagnostics Instruments). Confocal images were captured as 0.2-μm thick optical sections on a ZEISS LSM510 confocal microscope.
The distribution of the described GFP fusion proteins was visualized directly by fluorescence microscopy of logarithmically growing cells cultured in the indicated medium.
Electron Microscopy and Immunoelectron Microscopy
The ultrastructural analysis of cells overexpressing NUP53 was performed using two separate techniques. To visualize membrane structures, induced GNP53 cells were washed with water and treated with 1.5% KMnO4 for 20 min at room temperature (Nuttley et al. 1994). Fixed cells were then sequentially incubated at room temperature with 1% sodium periodate for 20 min, 1% NH4Cl for 10 min, and contrasted with 2% aqueous uranyl acetate overnight at 4°C. Samples were then dehydrated with ethanol, embedded in Epon (Electron Microscopy Sciences), and sectioned. A second technique was used to contrast protein structures of the NE. For these experiments, induced GNP53 cells were fixed with 2% glutaraldehyde, converted to spheroplasts, stained with osmium tetroxide and uranyl acetate, and embedded in Epon essentially as described (Byers and Goetsch 1991; Wente et al. 1992).
Immunoelectron microscopy was performed using modifications of a previously described procedure (Wente et al. 1992). GNP53 or W303 cells containing the plasmid pBJ244 were grown for 9.5 h in selection medium containing 2% galactose followed by an additional 2-h incubation in YPG. Cells were then converted to spheroplasts and fixed with 0.075% glutaraldehyde, 4% formaldehyde for 1 h at 4°C. Samples were then dehydrated using a graded ethanol series and resuspended in LR-White resin (Electron Microscopy Sciences) containing 20% (vol/vol) ethanol for 1 h at room temperature. The resin was allowed to infiltrate the sample by incubating overnight in 100% LR-White resin at 4°C. Sections were treated for 5 min in PBS-T containing 0.02 M glycine and blocked for 10 min in PBS-T with 2% BSA (PBS-T/BSA) at room temperature. The grids were probed overnight at 4°C with affinity-purified, anti-Nup53p antibodies in PBS-T with 2% BSA. Antibody binding was detected with goat anti–rabbit antibodies conjugated to 10-nm gold particles (Sigma-Aldrich). After washing, samples were refixed for 5 min in PBS with 2% glutaraldehyde, washed with water, and contrasted by staining with 2% aqueous uranyl acetate for 10 min at room temperature.
The subcellular localization of Pom152p in cells overproducing Nup53p was analyzed by cryo-immunoelectron microscopy. For these experiments, a diploid yeast strain synthesizing a Pom152–protein A fusion (PM152PA; Rout et al. 2000) containing pRCUP1-NUP53 PM152PA-53 were induced for 10 h as described above. Induced cells were washed, converted into spheroplasts, and fixed in 4% paraformaldehyde for 45 min at room temperature. The fixed cells were cryosectioned after infusion in 0.1 M phosphate buffer containing 1.8 M sucrose and 20% polyvinylpyrrolidone for 30 min at room temperature. Sections were blocked in PBS containing 2% FCS for 1 h at room temperature. To detect the Pom152-pA chimera, a rabbit antibody directed against protein A was used. This antibody exhibits a much higher degree of reactivity to protein A than that exhibited by nonspecific rabbit antibodies. The anti-protein A antibodies were derived from antisera of rabbits immunized with a recombinant protein A-Nup59 fusion protein. The reactivity to Nup59p was eliminated by diluting the serum 1:10 in PBS and incubating overnight at 4°C with 250 mg of a yeast whole cell acetone powder per ml of diluted sera. After binding of the anti-protein A antibodies to Pom152-pA, binding was visualized with a secondary antibody conjugated to 10-nm gold particles (Sigma-Aldrich).
All thin section samples were examined using a Philips 410 electron microscope and images were recorded on Eastman Kodak Co. film.
Sodium Carbonate Extractions of Yeast Nuclei and NEs
NEs were isolated as described previously (Strambio-de-Castillia et al. 1995) from CNP53 and W303 cells bearing the pYEX-BX plasmid after 8 h of induction with 0.5 mM copper sulfate. A nuclear fraction from CNP53ΔC and W303 cells bearing the pYEX-BX plasmid was isolated as described (Tcheperegine et al. 1999) following an identical induction procedure. Extractions of both fractions with 0.1 M sodium carbonate, pH 11.4, were performed as described previously (Wozniak et al. 1994).
Preparation of Whole Cell Lysates and Western Blotting
Samples of whole cell lysates for use in Western blot analysis were prepared as follows. After the indicated incubations, cells were collected by centrifugation, washed once in water, and then resuspended in 7.4% β-mercaptoethanol, 1.85 N NaOH for 10 min at 4°C. Proteins were precipitated with TCA at a final concentration of 12%. Protein pellets were then washed with 90% methanol and solubilized in SDS sample buffer. Polypeptides in the extracts were separated by SDS-PAGE, and electrophoretically transferred to a nitrocellulose membrane.
For Western blot analyses, nitrocellulose membranes were blocked in PBS-T containing 5% skim milk. The same buffer was used for antibody incubations. Membranes were probed with the mAb118C3 (α-Pom152p), or rabbit polyclonal antibodies directed against Nup53p, Nup59p (Marelli et al. 1998), or Kar2p (provided by R. Rachubinski, University of Alberta, Edmonton, Canada). Antibody binding was subsequently detected by using the appropriate secondary antibody coupled to HRP and an ECL detection system (Amersham Pharmacia Biotech).
Results
Overproduced Nup53p Accumulates at the NE and Inhibits Nuclear Import
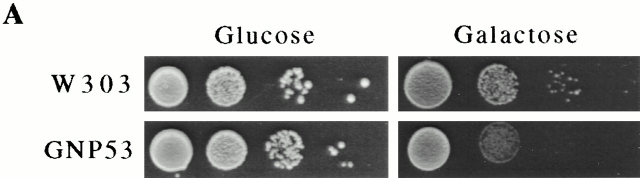
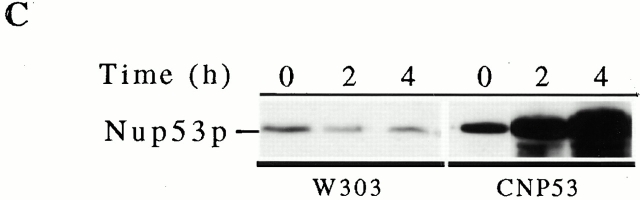
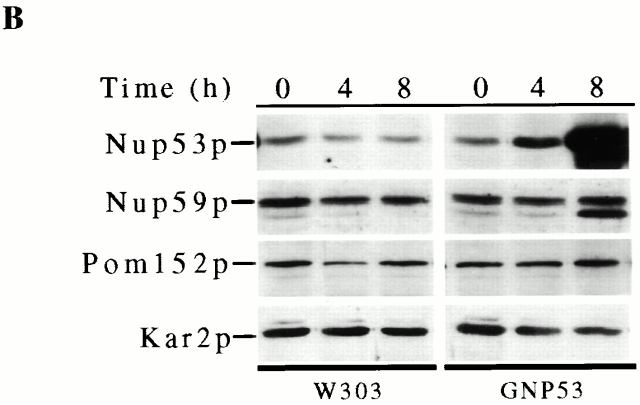
We have investigated the function of the yeast nup Nup53p by altering its expression levels and evaluating the physiological consequences of increased cellular levels of Nup53p. For these experiments, plasmid-born copies of the NUP53 ORF under the control of the inducible GAL1 or CUP1 promoter was introduced into wild-type W303 or DF5 cells. Using both expression systems, we observed that the overexpression of NUP53 retarded cell growth compared with controls (Fig. 1 A, and data not shown). As shown in Fig. 1 B, Western blot analysis of whole cell lysates prepared from galactose-induced GNP53 cells (GAL1::NUP53) showed elevated levels of Nup53p by 4 h after induction, reaching a maximum by 8 h. By comparison, CNP53 cells (CUP1::NUP53) induced with copper sulfate responded more rapidly to the induction conditions with detectable increases in Nup53p within 2 h (Fig. 1 C). CNP53 cells also displayed somewhat higher basal levels of expression. No detectable changes in the levels of Nup59p, Pom152p, or the ER protein Kar2p were observed upon induction of NUP53 using either expression system (Fig. 1 B, and data not shown).
Figure 1.
Overexpression of NUP53 slowed cell growth and increased Nup53p production. (A) Serial dilutions of GNP53 (GAL1::NUP53) and wild-type W303 cells containing the plasmid pBJ244 were spotted onto CM-tryptophan plates containing 2% glucose or 2% galactose and grown at 30°C for 2 d. (B) GNP53 and W303 cells containing pJB244 were shifted to galactose-containing medium for the indicated times. Whole cell lysates were analyzed by Western blotting using rabbit polyclonal antibodies directed against Nup53p, Nup59p, and the ER protein Kar2p or a monoclonal antibody (mAb118C3) that binds Pom152p. Binding was detected with HRP-conjugated antibodies and ECL. The anti-Nup59p antibody reacts weakly with Nup53p leading to the appearance of a 53-kD species after 8 h of induction. (C) CNP53 (CUP1::NUP53) cells and W303 cells containing the pYEX-BX plasmid were induced with 0.5 mM copper sulfate for the indicated times and analyzed as described above using the anti-Nup53p antibodies.
The subcellular localization of overproduced Nup53p was examined by indirect immunofluorescence microscopy using anti-Nup53p antibodies (Fig. 2A and Fig. B). Unlike Nup53p expressed at wild-type levels, which exhibits a distinct punctate perinuclear signal characteristic of nups, induced GNP53 and CNP53 cells showed an intense, continuous perinuclear staining pattern suggesting a uniform distribution along the NE (Fig. 2 A). Approximately 50% of GNP53 and 75% of CNP53 cells exhibited this pattern (data not shown). In this group, cells displaying the strongest signals (∼20% of the population) contained regions of the NE that appeared brighter or contained intensely stained structures adjacent to the NE (Fig. 2A and Fig. B). Serial optical sections obtained by confocal microscopy further documented the continuous NE staining pattern as well as the juxtaposition of a structure rich in Nup53p (Fig. 2 B). In contrast, overproduction of Nup59p, a protein structurally similar to Nup53p, did not associate with the NE but instead was detected in intensely staining cytoplasmic structures (data not shown).
Figure 2.
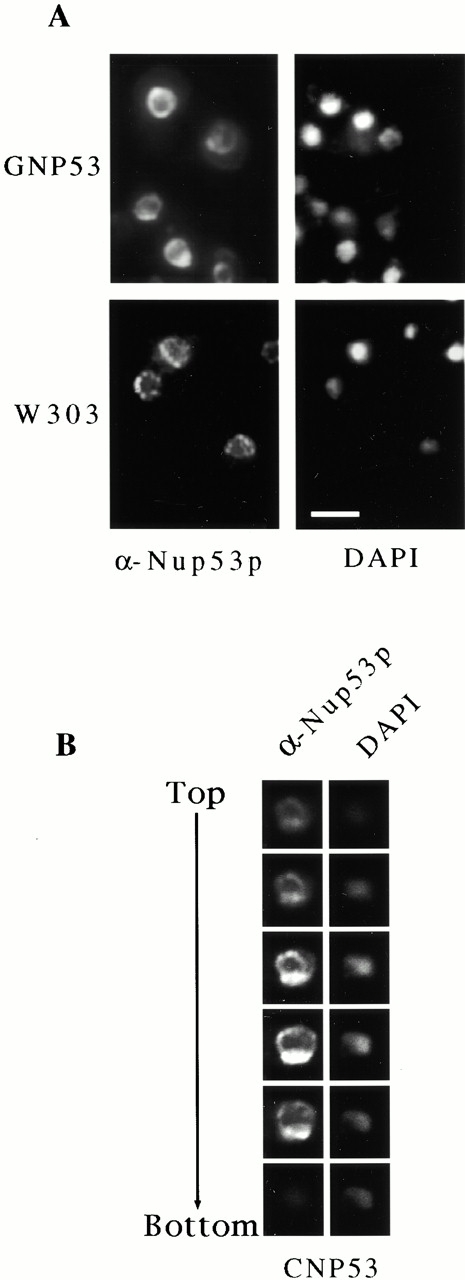
Immunofluorescence localization of overproduced Nup53p. (A) GNP53 and W303 (containing the plasmid pBJ224) cells were grown in galactose-containing media for 8 h and then processed for immunofluorescence microscopy. Samples were probed with affinity-purified, anti-Nup53p antibodies and binding was detected with Cy3-conjugated anti–rabbit antibodies (α-Nup53p). A continuous perinuclear staining pattern was observed in the GNP53 cells that is distinct from the punctate perinuclear staining seen in W303 cells. Note, the exposure time of the photograph of the W303 cells was ∼5 times that of the GNP53 cells. The position of the nuclear DNA was visualized by DAPI staining. (B) Samples of CNP53 cells induced with 0.5 mM copper sulfate and processed as described in panel A were examined by confocal microscopy. A series of 0.2-μm thick serial sections through the nucleus from the top to the bottom show the location of Nup53p and nuclear DNA. Bar, 5 μm.
Previous results have shown that NUP53 mutants exhibit defects in Kap121p-mediated import (Marelli et al. 1998), and more recently a role for Nup53p in Kap95p-mediated transport has also been suggested (Fahrenkrog et al. 2000). These results prompted us to examine the effects of Nup53p overproduction on nuclear import. For these experiments, CNP53 cells synthesizing a reporter cargo for Kap121p (pNLS-GFP, the nuclear localization signal [NLS] of the transcription factor Pho4p fused to GFP; Kaffman et al. 1998) or Kap95p (cNLS-GFP; Shulga et al. 1996) were grown in the presence or absence of copper sulfate (Fig. 3), and the location of the reporter protein was evaluated by fluorescence microscopy. As shown in Fig. 3 A, CNP53 cells grown in medium lacking copper ions accumulated the pNLS-GFP reporter in the nucleus. However, upon induction of CUP1::NUP53, the pNLS-GFP reporter displayed a dramatic mislocalization to the cytoplasm. The effect was specific for Kap121p-mediated import as the Kap95p substrate, cNLS-GFP, was efficiently imported in cells overproducing Nup53p (Fig. 3 A).
Figure 3.
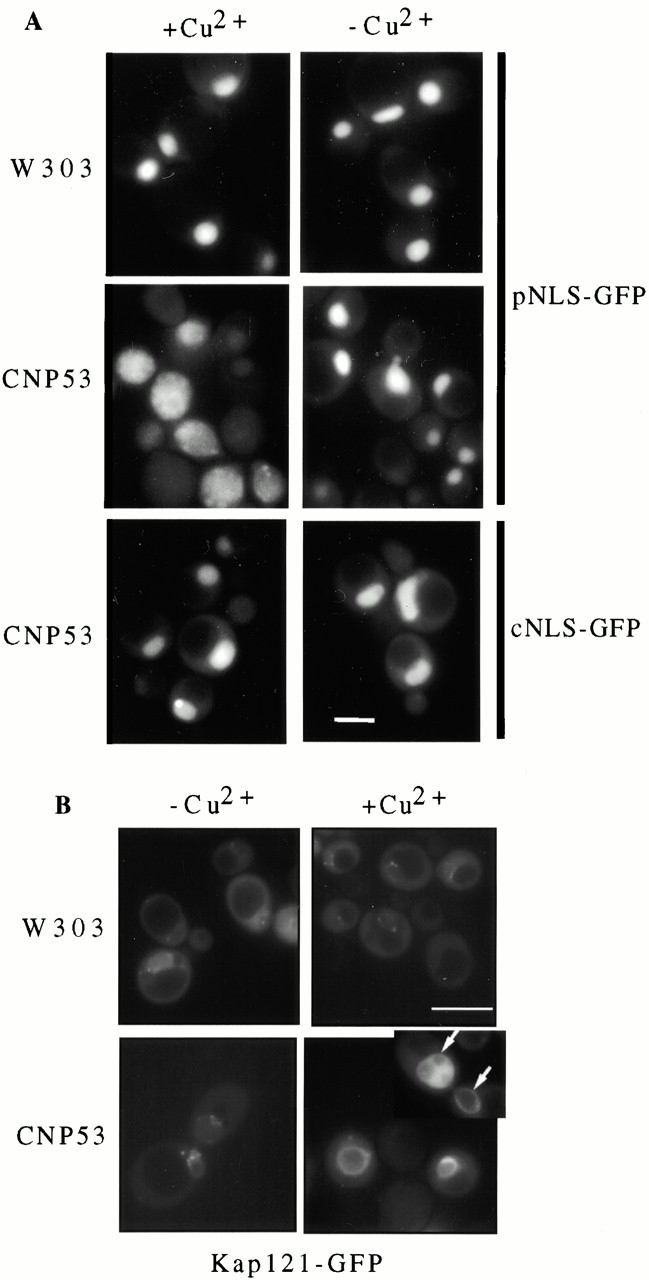
Kap121p-mediated import was specifically inhibited by the overexpression of NUP53. (A) CNP53 (CUP1::NUP53) and W303 (containing the plasmid pYEX-BX) cells expressing either the Kap121p substrate, pNLS-GFP (CNP53-pNLS, W303-pNLS), or the Kap95p substrate, cNLS-GFP (CNP53-cNLS), were grown in the presence (+Cu2+) or absence (−Cu2+) of 0.5 mM copper sulfate for 4 h. The GFP reporters were localized by fluorescence microscopy. (B) CNP53 (CUP1::NUP53) and W303 cells producing the Kap121-GFP reporter (CNP53-121, W303-121) were grown in the presence and absence of copper sulfate for 4 h. The subcellular distribution of the Kap121-GFP reporter was documented by fluorescence microscopy. The inset in the bottom right panel shows the nucleus of a dividing cell with arrows pointing to the NE. The intranuclear Kap121-GFP pattern in one of the dividing nuclei is consistent with its association with the Nup53p-induced intranuclear membranes (see Fig. 4 D). Bars, 5 μm.
As Nup53p acts as a docking site for Kap121p (Marelli et al. 1998), we also examined the effects of Nup53p overproduction on the association of a Kap121-GFP reporter with the NE. For these experiments, a KAP121-GFP chimeric gene was introduced into CNP53 cells (CNP53-121) and the subcellular distribution of the Kap121-GFP fusion was examined by fluorescence microscopy after induction of NUP53. As shown in Fig. 3 B, the overexpression of NUP53 caused a dramatic increase in the amount of perinuclear Kap121-GFP compared with that observed in wild-type cells. The fluorescent pattern of Kap121-GFP mimicked the NE accumulation of excess Nup53p, including an association with structures that appear to reside in the nucleus (Fig. 3 B, see inset). Moreover, the apparent recruitment of Kap121p to the NE by excess Nup53p was specific for this kap, as the distribution of Kap95-GFP was unaffected by NUP53 overexpression (data not shown).
Overproduction of Nup53p Induces the Formation of Intranuclear Membranes
The striking distribution pattern seen with excess Nup53p led us to examine the ultrastructure of the NE using thin-section electron microscopy techniques. For these experiments, GNP53 cells were grown for the indicated times in galactose-containing medium and then processed for electron microscopy with potassium permanganate and uranyl acetate. This methodology intensely stained the NE membrane, and nuclear pores appeared as discontinuities in the NE but the proteinaceous structures of the NPC were not visible (Fig. 4). In uninduced GNP53 cells, the structure of the NE appeared normal (Fig. 4 A). However, after 6 h of induction, an extensive intranuclear network of membranous structures began to appear in ∼50% of the cells. These membranes first appeared as circular or tubular shaped membranes with a regular diameter of 50–100 nm, being generally visible near the periphery of the nucleus (Fig. 4B and Fig. C). Also starting at 6 h, and expanding in numbers by later time points, we observed the appearance of multiple layers (from 2 to 10) of often stacked double membrane lamellae separated from one another by a thin layer of nucleoplasm (Fig. 4 D). In all the sections examined and in serial sections (data not shown), this mass of accumulating membrane structures was surrounded by a single double membrane that appeared to be the NE. The identity of the outer double membrane as the NE was confirmed by the staining of similar samples with osmium tetroxide and uranyl acetate. Using this method, proteinaceous structures such as the NPCs (Fig. 5 A) and spindle pole bodies (Fig. 5 B) were clearly visible in the outer double membrane but not in the underlying double membrane stacks. These results indicated that the Nup53p-induced membranes were contained within the nucleus.
Figure 4.
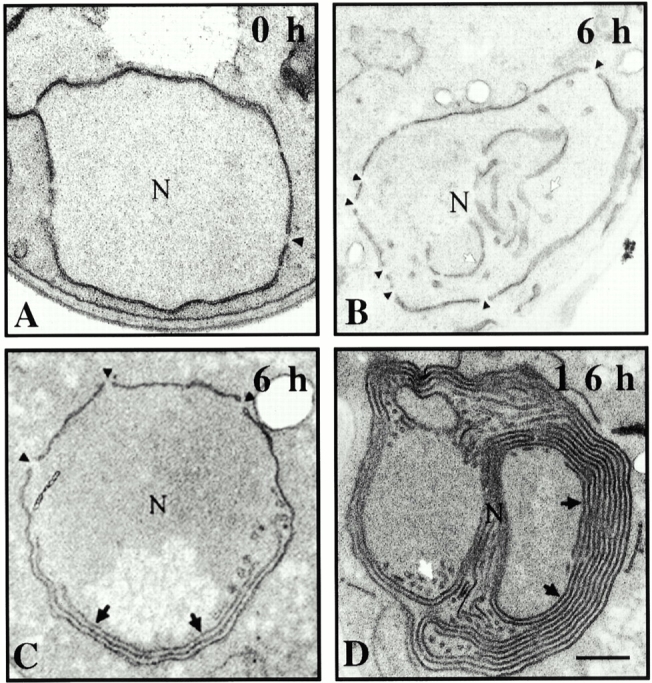
Overproduction of Nup53p induced the formation of intranuclear membranes. GNP53 cells were induced in galactose-containing medium for the indicated times (A–D). Cells were fixed and stained in potassium permanganate and uranyl acetate and processed for electron microscopy. This procedure highlights membrane structures but not proteinaceous structures such as the NPC. Discontinuities in the NE, which reflect the position of NPCs, are highlighted by black arrowheads. In addition to the double membrane NE seen before induction (0 h), induced cells contained tubular and circular shaped membranes (white arrows) as well as flattened, double membrane lamellae (black arrows) that increase in number upon longer induction. N, nucleoplasm. Bar, 0.5 μm.
Figure 5.
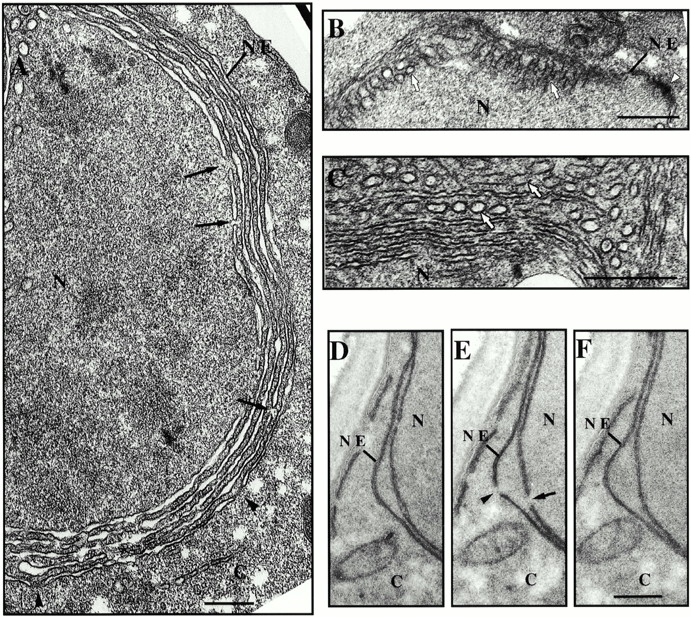
(A–C) GNP53 cells were grown in galactose-containing media for 16 h, fixed, and stained in osmium tetroxide and uranyl acetate. This procedure more clearly stains proteinaceous structures such as the NPC (black arrowheads) and spindle pole bodies (white arrowheads). The position of the NE-containing NPCs is indicated (NE). Intranuclear tubular membranes (white arrows) are visible in cross section (A and C) and in sections parallel to their central axis (B). These structures can be seen radiating from the vicinity of the inner nuclear membrane (B). Double membrane lamellae, having defined cisternae, are visible within the nucleoplasm in stacks often adjacent to the NE (A). The continuity of intranuclear lamellae is occasionally interrupted by pores that lacked darkly staining NPCs (black arrows). (D–F) Induced CNP53 cells were processed for electron microscopy using potassium permanganate and uranyl acetate as described in the legend to Fig. 4. Shown are three images from a set of serial sections where pores of similar size are visible in a single section across the NE (arrowhead) and the intranuclear double membrane lamellae (arrow). N, nucleoplasm; C, cytoplasm. Bars, 0.5 μm.
As vesicular traffic across an intact NE has never been described, nor have we observed this phenomenon upon overexpression of NUP53, the most likely scenario was that the Nup53p-induced membranes arose from the inner nuclear membrane. Consistent with this idea, we observed, in rare tangential sections, the tubular structures radiating from the vicinity of the inner nuclear membrane (Fig. 5 B). However, we were unable to see direct continuity between these membranes and the inner nuclear membrane, as the morphology was generally not well preserved in these regions. The tubular membranes are likely the precursors of the stacked double membrane lamellae, which are more abundant at later time points. Interestingly, multiple tubular membranes were occasionally visible in rows in cross section (Fig. 5 C). A likely scenario is that they fuse, giving rise to the double membrane lamellae.
As shown in Fig. 5 A, like the NE, each intranuclear double membrane lamellae surrounds a clearly defined cisterna. Occasionally we observed in cross sections that the continuity of the cisterna was interrupted by adjacent connecting membranes that appeared similar to the pore membranes seen in the NE (Fig. 5 A). To determine whether the membrane walls of a pore formed these gaps, serial sections were analyzed. As shown in Fig. 5D–F, consecutive sections revealed that the connecting membranes, like those seen in the NE, were visible in a single section and flanked by sections showing a continuous double membrane. Moreover, the distance between the connecting membranes in the induced lamellae was generally similar to the diameter of nuclear pores in the NE (Fig. 5 E). These results suggest that the intranuclear double membranes contain transcisternal pores similar to the NE. However, unlike the NE, the pores across the intranuclear double membranes lacked intensely stained NPC structures (Fig. 5 A), suggesting that they did not contain fully assembled NPCs.
Nup53p, Pom152p, and Ndc1p Are Associated with the Nup53p-induced Intranuclear Membranes
The concentration of overproduced Nup53p at the NE observed by immunofluorescence microscopy suggested that Nup53p might be physically associated with the induced intranuclear membranes. This was directly tested by immunoelectron microscopy. NUP53 expression was induced in GNP53 cells and, after fixation and embedding, sections were labeled using affinity-purified, anti-Nup53p antibodies and binding was detected using secondary antibodies coupled to 10-nm gold particles. As shown in Fig. 6 A, electron micrographs revealed a high concentration of gold particles along the nuclear periphery. The vast majority of gold particles were positioned on the nucleoplasmic side of the NE where many were visible decorating the outer surface of the intranuclear membranes (Fig. 6B and Fig. C). In contrast, this labeling pattern and the intranuclear membrane structures were not present in wild-type cells. These results suggested that Nup53p was physically associated with the membrane structures it induced.
Figure 6.
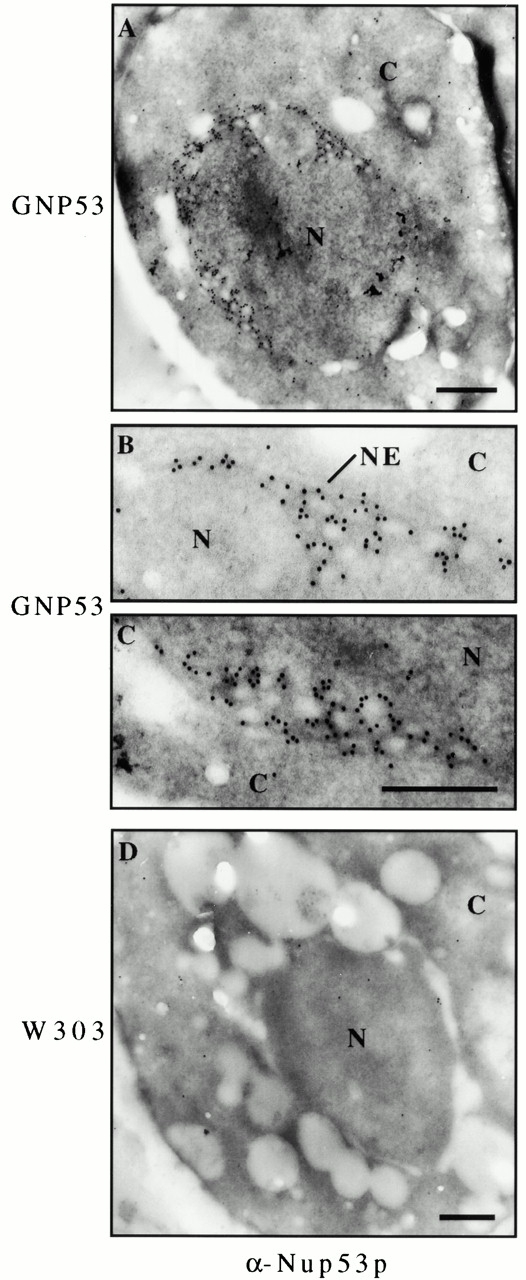
Overproduced Nup53p was associated with the intranuclear lamellar membranes. GNP53 (A–C) and W303 (D) cells were grown in galactose-containing media for 16 h, fixed, embedded, and sectioned. Sections were probed with affinity-purified, anti-Nup53p (α-Nup53p) antibodies and binding was visualized with 10-nm gold-conjugated secondary antibodies. Magnified regions of the image in panel A are shown in panels B and C. Nup53p was detected at the nuclear periphery and associated with lamellar membranes and nucleoplasmic face of the tubular structures. N, nucleoplasm; C, cytoplasm. Bars, 0.5 μm.
We also investigated whether other NPC components were associated with the Nup53p-induced intranuclear membranes. These analyses were provoked by the association of Nup53p with these membranes as well as the presence of pores. The occurrence of pores but the apparent absence of NPCs within the intranuclear lamellar membranes suggested that these membranes might contain a subset of NPC components sufficient for pore formation but not NPC assembly. We tested this hypothesis by immunofluorescence microscopy using a battery of anti-nup antibodies and strains containing epitope-tagged versions of various nups. CNP53D and wild-type cells were induced for 16 h with copper ions and each was probed with the monoclonal antibody mAb414, which binds the FXFG repeat–containing nups Nsp1p, Nup1p, Nup145p, Nup57p, and Nup49p (Rout and Blobel 1993), and mAb118C3, which binds Pom152p (Strambio-de-Castillia et al. 1995). Similarly, strains expressing the protein A–tagged chimera Nup170-pA, Nup59-pA, Nup157-pA, Nic96-pA, Nup2-pA, Gle1-pA, Ndc1-pA, or Pom34-pA (Marelli et al. 1998; Rout et al. 2000), and containing either pRCUP1-NUP53 or the empty pRCUP1 plasmid were also induced and examined by immunofluorescence microscopy. We observed that, for the vast majority of these different NPC proteins, the characteristic punctate NE staining pattern observed in the parent strain was not detectably altered by the overexpression of NUP53. For example, as shown in Fig. 7, the mAb414 reactive nups and the pore membrane protein Pom34-pA exhibited the same uniform punctate distribution along the NE both in wild-type cells and induced CNP53 cells.
Figure 7.
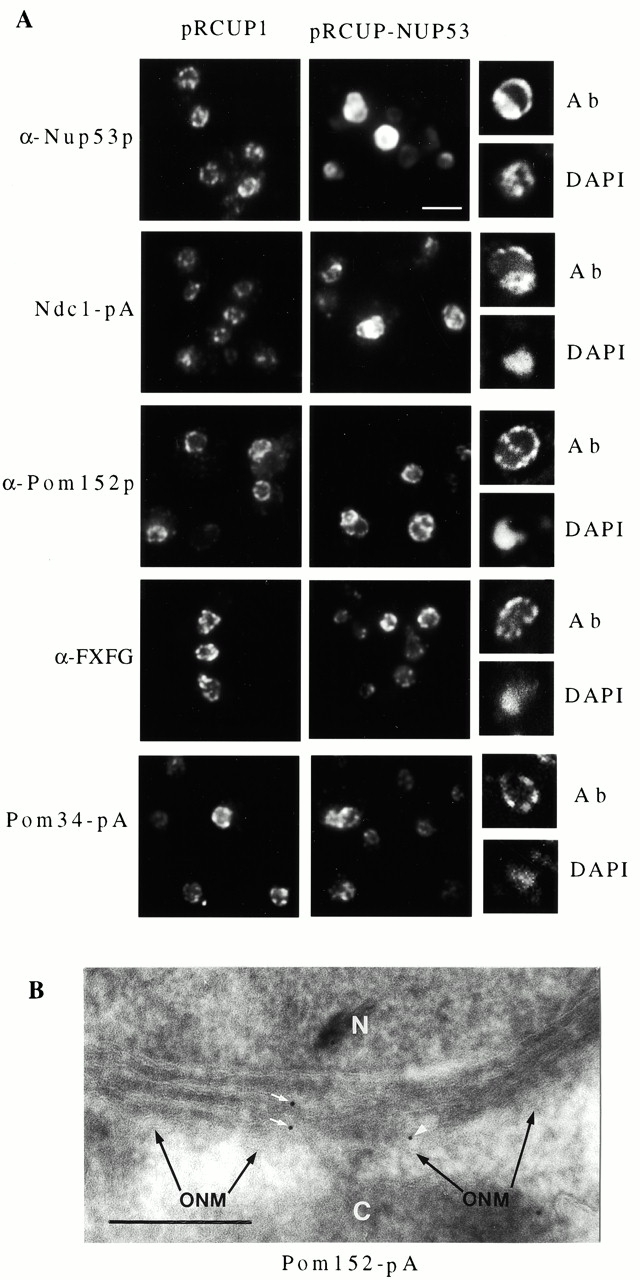
Pom152p and Ndc1p were present in the Nup53p-induced intranuclear membranes. (A) The effect of Nup53p overproduction on the distribution of various NPC proteins was examined by immunofluorescence microscopy. Strains containing the pRCUP1 or the pRCUP1-NUP53 plasmid were grown in media containing 0.5 mM copper sulfate for 16 h and then fixed and permeabilized. Wild-type DF5 cells were probed with either affinity-purified anti-Nup53p antibodies (α-Nup53p) or the monoclonal antibody mAb414 (α-FXFG) or mAb118C3 (α-Pom152p). Binding was detected with Cy3-conjugated secondary antibodies. Strains expressing Ndc1-pA (NDCPA-p53) or Pom34-pA (PM34PA) were probed with rabbit antibodies followed by Cy3-conjugated, donkey anti–rabbit antibodies. Field images were captured with a standard fluorescence microscope. Corresponding single cell confocal microscopy images showing the Cy3 signal (Ab) and DAPI staining of DNA are shown on the right. Note, the region of nucleus containing the nucleolus is not visualized with DAPI. These images were derived from 0.2-μm optical sections through the middle of the nucleus. Bar, 5 μm. (B) Cryosections of PM152PA-53 cells induced to overexpress NUP53 were probed with antibodies directed against protein A to detect Pom152-pA. Binding was visualized with 10-nm gold-labeled secondary antibodies. The location of the outer nuclear membrane (ONM) is indicated. Gold particles associated with two separate intranuclear double membranes (white arrows) and the NE (white arrowhead) are highlighted. N, nucleoplasm; C, cytoplasm. Bar, 0.5 μm.
In contrast, two pore membrane proteins, Pom152p and Ndc1-pA, showed, in addition to a punctate NE pattern, a distinct intranuclear staining pattern in 10–20% of cells overexpressing NUP53 (Fig. 7 A). This pattern was not seen with any of the other NPC proteins examined. This percentage of cells was consistent with that of cells displaying the multiple layers of intranuclear lamellar membranes. When viewed by confocal microscopy, the additional signal appeared bound by the NE, consistent with their localization to the intranuclear lamellar membranes (Fig. 7 A). Most noticeably in the case of Pom152p, the confocal images revealed that the intranuclear pattern often appeared punctate suggesting that it was not uniformly distributed within the intranuclear membranes but instead it might be associated with the pores present in the lamellae. Interestingly, the reactivity of mAb118C3 to Pom152p in these structures was less sensitive to fixation than that observed in wild-type NEs, suggesting that Pom152p was more accessible to the antibodies and probably not as tightly associated with other proteins (data not shown). Conformation of Pom152p's association with the intranuclear membrane was obtained by immunoelectron microscopy. As shown in Fig. 7 B, gold particles bound to Pom152-pA were visible along the double membrane lamellae. However, due to the morphological preservation of the samples, we could not unequivocally determine whether Pom152p was associated with pores in these structures.
Nup53p Import and Membrane Proliferation Are Dependent on Kap121p and Nup170p
On the basis of its intranuclear accumulation, we hypothesized that excess Nup53p was actively imported into the nucleus and that, either at the NPC or after its import into the nucleus, Nup53p associates with the inner nuclear membrane and stimulates membrane proliferation. To investigate the mechanism of these processes, we examined the role of various kaps and nups on the import of Nup53p into the nucleus and its ability to drive membrane proliferation. As Nup53p specifically interacts with Kap121p (Marelli et al. 1998), the possibility that Kap121p may play a role in targeting Nup53p to the nucleus was tested. For these experiments, an allele of KAP121, kap121-41, was isolated that was temperature sensitive for growth. In a strain containing this mutation (KP121-41), the import of the pNLS-GFP reporter was inhibited at both permissive (23°C; Fig. 8 A) and restrictive temperatures (data not shown) whereas the cNLS-GFP reporter was imported normally (Fig. 8 A). The effect of this mutation on the nuclear localization of overproduced Nup53p was tested by introducing the pCUP1-NUP53 plasmid into the KP121-41 strain and inducing NUP53 expression with copper ions. Strikingly, when examined by immunofluorescence microscopy, excess Nup53p was present throughout the cytoplasm and failed to accumulate at the NE (Fig. 8 B). Moreover, when induced cells were examined by thin-section electron microscopy, no intranuclear membranes were observed (data not shown). In contrast, a deletion mutant lacking KAP123 (KP123-p53), a kap known to functionally overlap with Kap121p (Rout et al. 1997), showed no effect on the NE localization of excess Nup53p (Fig. 8 B) or membrane proliferation (data not shown). These data suggest that the nuclear import of Nup53p was dependent on Kap121p in what is likely an early step in the sequence of events that leads to membrane proliferation.
Figure 8.
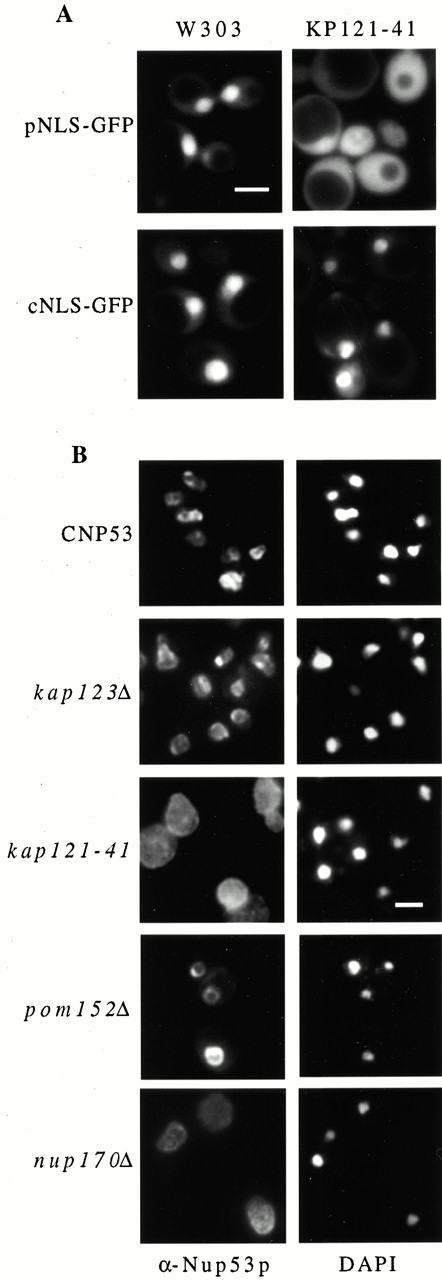
The nuclear localization of overproduced Nup53p was dependent on both Kap121p and Nup170p. (A) Wild-type W303 cells and the strain KP121-41 containing a temperature-sensitive allele of KAP121 (kap121-41) were transformed with plasmids expressing the pNLS-GFP or cNLS-GFP reporter genes. Cells were grown at 23°C and the distribution of the reporter proteins was examined by fluorescence microscopy. (B) The subcellular distribution of overproduced Nup53p was examined in the strains KP121-41-p53 (kap121-41), KP123-p53 (kap123Δ), NP170-p53 (nup170Δ), and PMY17-p53 (pom152Δ). Each of these strains contained the pCUP1-NUP53 plasmid. These strains and CNP53 cells were induced with 0.5 mM copper sulfate for 16 h, fixed, permeabilized, and probed with affinity-purified, anti-Nup53p antibodies (α-Nup53p). Binding was detected with Cy3-conjugated secondary antibodies. Note, in the exposures shown, not all cells are expressing levels of Nup53p that are detectable. The position of the nuclear DNA was visualized by DAPI staining. Bars, 5 μm.
The role that other nups, specifically those that genetically and physically interact with Nup53p, play in the deposition of excess Nup53p on the NE was also examined. For these experiments, null mutants of NUP188, NUP170, NUP157, POM152, or NUP59 were transformed with the pCUP1-NUP53 plasmid and induced with copper sulfate. Immunofluorescence microscopy using anti-Nup53p antibodies revealed that the NE accumulation of excess Nup53p was not affected in strains lacking Pom152p (Fig. 8 B), Nup188p, Nup59p, or Nup157p (data not shown). Moreover, intranuclear membranes were visible in each of these strains (data not shown). In contrast, in a strain lacking Nup170p (nup170Δ; Fig. 8 B), like the KP121-41 strain, excess Nup53p was seen throughout the cytoplasm and did not accumulate at the NE. In this strain, Nup53p overproduction also failed to stimulate the production of intranuclear membranes (data not shown).
A Putative COOH-terminal Amphipathic Helix in Nup53p Is Essential for Membrane Proliferation
To understand the mechanism by which Nup53p associates with the intranuclear membranes, we investigated the biochemical basis for this interaction and defined a region of Nup53p that is required for membrane proliferation. Subcellular fractionation of induced CNP53 cells revealed that excess Nup53p fractionated with nuclei and an enriched fraction of NEs (data not shown). To investigate the physical basis for Nup53p's interaction with the intranuclear membranes, nuclear membrane fractions derived from W303 (wild-type) and induced CNP53 cells were extracted with 0.1 M sodium carbonate, pH 11.4, and then centrifuged to separate peripheral membrane proteins in a soluble supernatant fraction from proteins tightly associated with the membrane pellet, including integral proteins. As shown in Fig. 9 A, essentially all of Nup59p was extracted from the W303 and CNP53 NEs. Similar levels of extraction were observed with Nup170p (data not shown). However, a significant proportion of Nup53p remained associated with the sodium carbonate extracted membrane fraction of W303 NEs. Strikingly, extractions of the CNP53 NEs revealed that ∼50% of the excess Nup53p was resistant to sodium carbonate extraction (Fig. 9A and Fig. B) and remained associated with membranes.
Figure 9.
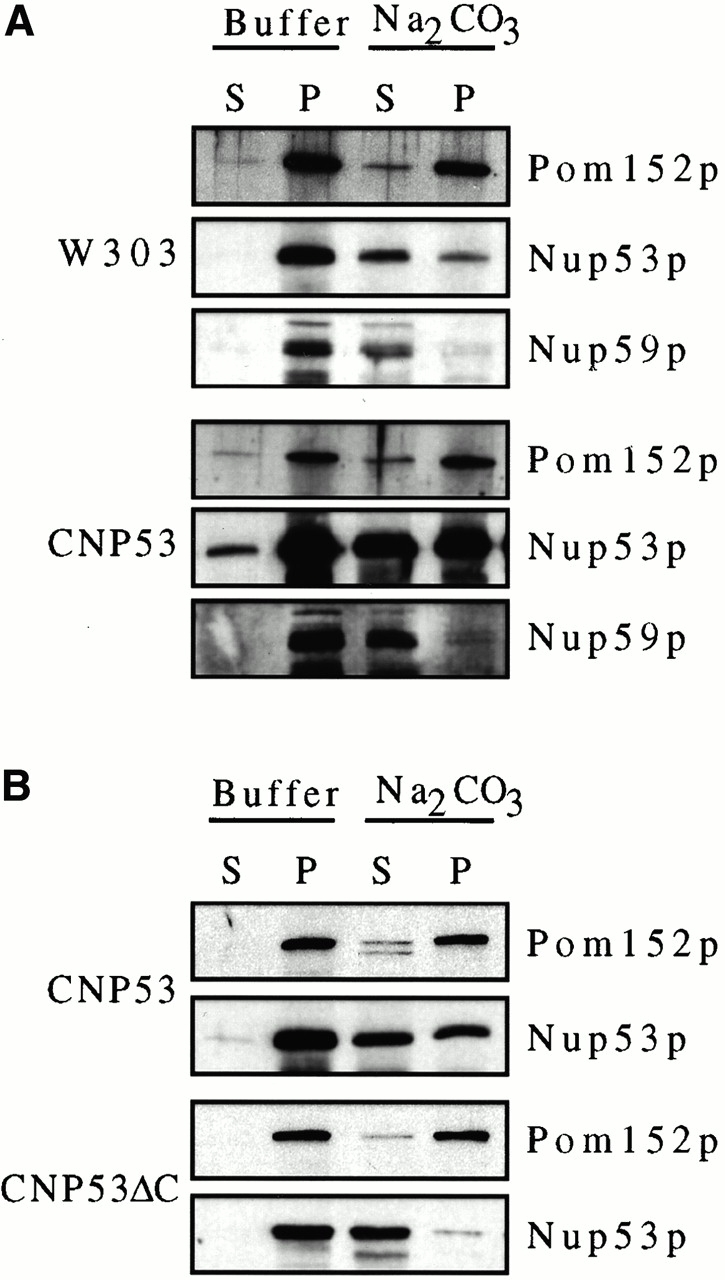
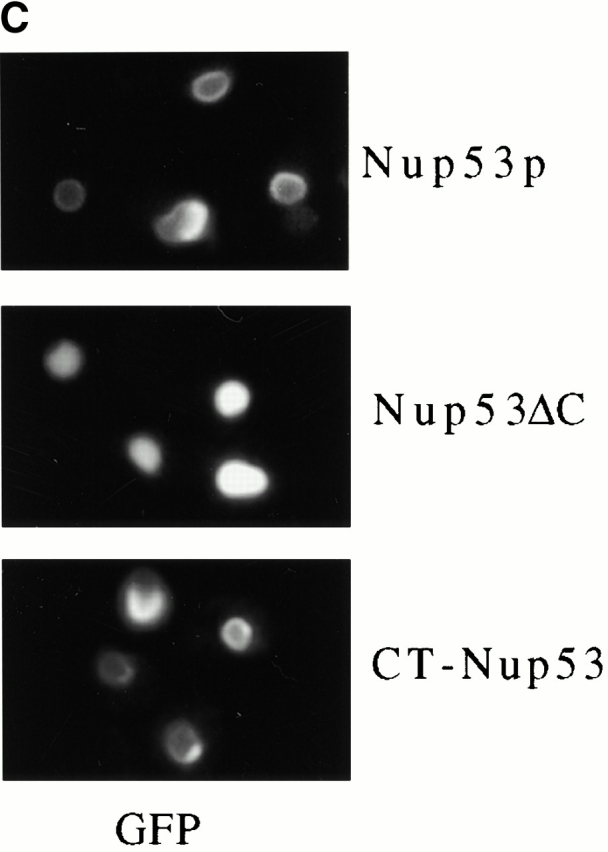
The membrane association of Nup53p was dependent on its COOH-terminal 27 amino acid residues. (A) NEs were isolated from CNP53 cells and W303 cells containing pYEX-BX after 4 h of growth in media containing 0.5 mM copper sulfate. Suspended NEs were treated with buffer alone or extracted with sodium carbonate (Na2CO3), pH 11.4, and centrifuged to produce a supernatant fraction (S) and a membrane-containing pellet (P). Samples were analyzed by Western blotting using affinity-purified, anti-Nup53p antibodies, a rabbit polyclonal antibody directed against Nup59p (note, Nup59p migrates as a doublet at the indicated position), and the monoclonal antibody mAb118C3 (Pom152p). Binding was detected with HRP-conjugated secondary antibodies and ECL. (B) The same analysis was performed on nuclei isolated from induced CNP53 cells and the CNP53ΔC (CUP1::nup53ΔC) strain expressing a truncation of Nup53p lacking its COOH-terminal 27 amino acid residues. (C) DF5 cells producing Nup53-GFP, Nup53-ΔC-GFP (containing amino acid residues 1–450 of Nup53p), or CT-Nup53p-GFP (containing amino acid residues 375–475 of Nup53p) were grown in SM media lacking uracil and methionine to mid-logarithmic phase. The GFP reporters were localized by fluorescence microscopy.
The resistance of endogenous and, to a greater degree, overproduced Nup53p to sodium carbonate extraction suggested that it was more tightly associated with the membrane than other nups with which it physically interacts including Nup59p and Nup170p. However, it is unlikely that Nup53p traverses the membrane, as it lacks a predicted transmembrane segment and it does not show the same level of resistance to extraction that is observed with integral membrane proteins including Pom152p (Wozniak et al. 1994; Fig. 9A and Fig. B). Nup53p's extraction behavior could be explained by a strong association with the periphery of the membrane, either through an interaction with an integral membrane protein or directly with the nucleoplasmic leaflet of the bilayer. An analysis of the secondary structure of Nup53p revealed that its COOH-terminal 17 amino acid residues could form an amphipathic α-helix capable of mediating a direct interaction with the membrane (data not shown). We asked whether this region of Nup53p was necessary for its strong association with the membrane and its ability to drive membrane proliferation by expressing a NUP53 mutation lacking the coding region for its COOH-terminal 27 amino acid residues in wild-type DF5 cells (CNP53ΔC). As shown in Fig. 9 C, the Nup53ΔCp did not concentrate along the NE periphery, but rather it was imported into the nucleus and distributed throughout the nucleoplasm. The apparent inability of the Nup53ΔCp construct to associate with the NE was further supported by sodium carbonate extractions of crude nuclei isolated from induced CNP53ΔC and CNP53 cells. As shown in Fig. 9 B, Nup53ΔCp was efficiently extracted from the nuclear membrane fraction whereas again the full-length Nup53p was only partially extracted. Moreover, thin-section electron microscopy analysis revealed that the overproduction of Nup53ΔCp did not induce the proliferation of intranuclear membranes (data not shown). By comparison, a Nup53p deletion construct (amino acid residues 375–475) containing the COOH terminus and a flanking region required for its nuclear localization (Lusk, C.P., and R.W. Wozniak, unpublished data), localized to the nuclear periphery in a manner similar to Nup53p (Fig. 9 C) and it was capable of driving membrane proliferation (data not shown). These results suggest that the potential COOH-terminal, amphipathic α-helix of Nup53p is required for the formation of the intranuclear membranes.
Discussion
By altering the cellular levels of Nup53p, we have been able to investigate the specific role that this nup plays in nuclear import and we have uncovered an additional Nup53p activity which likely reflects a function for this protein, and probably other nups as well, in NE biogenesis. Nup53p's role in nuclear transport has previously been linked to its function as an important docking site for Kap121p at the NPC. However, it was unclear whether Nup53p had a broader role in nuclear transport. Here we show that increased cellular amounts of Nup53p specifically inhibit Kap121p- but not Kap95p-mediated nuclear import (Fig. 3). Moreover, excess Nup53p is able to specifically sequester Kap121p at the periphery of the nucleus in a pattern that would suggest that Kap121p accompanies Nup53p to the induced intranuclear membranes. The apparent presence of a Kap121p/Nup53p complex in the nucleus is surprising, as we would predict, on the basis of in vitro data (Marelli et al. 1998), that nuclear Ran-GTP would dissociate them. Two possible explanations for these observations are that the Kap121p/Nup53p complex is not accessible to Ran-GTP or that the complex is stabilized in the nucleus by other factors. The observed inhibition of Kap121p-mediated import could therefore occur as a consequence of inhibiting Kap121p's ability to recycle back to the cytoplasm. Taken together, these results support the conclusion that Nup53p plays a specific role in Kap121p-mediated nuclear transport.
The binding of Nup53p to Kap121p does not, however, appear to be restricted to events at the NE. We have shown that the accumulation of excess Nup53p at the nuclear periphery is dependent on Kap121p. A likely scenario is that newly synthesized Nup53p is recognized by Kap121p and targeted to the NPC. At the NPC, the nup Nup170p then plays a critical and specific role in the association of overproduced Nup53p with the nuclear membrane. In strains lacking Nup170p, overproduced Nup53p is largely present in the cytoplasm and fails to accumulate at the NE. This effect is unlikely to be linked to a defect in Kap121p-mediated import, as strains lacking NUP170 import a pNLS-GFP reporter at near wild-type rates (Shulga et al. 2000; Lusk, C.P., M. Marelli, and R.W. Wozniak, unpublished observation). Alternatively, as Nup170p acts as a binding site for Nup53p at the NPC (Marelli et al. 1998), Nup170p could play a direct role in mediating the transfer of Nup53p to the pore membrane where it could subsequently move to the inner nuclear membrane. Presumably, the interaction between excess Nup53p and Nup170p would be transient as the subcellular localization of Nup170p is not altered by the overproduction of Nup53p (see Results). This mechanism would explain the observation that at no time after the overexpression of NUP53 did we see a nucleoplasmic pool of Nup53p.
The steps involving Kap121p and Nup170p that lead to the association of Nup53p with membranes facing the interior of the nucleus are likely a prelude to the events that follow. Here excess Nup53p induces the proliferation of membrane structures within the nucleus. Membrane expansion seems to originate at the inner nuclear membrane with the formation of tubular structures of a defined diameter (∼50 nm) and of varying length (Fig. 5 B). These tubular structures appear to fuse and give rise to flattened, double membrane–enclosed cisternae structurally similar to the NE but often present in multiple layers adjacent to the NE. Interestingly, the series of events that lead to the formation of the intranuclear membranes are similar to those observed when tubular membranes bind to the surface of Xenopus sperm chromatin and later fuse to form a NE (Dreier and Rapoport 2000).
Like the NE, the intranuclear double membrane lamellae induced by Nup53p contain transcisternal pores. These pores are similar in diameter to those in the NE but they did not appear to contain NPCs nor were we able to detect most NPC components in association with these membranes. Notable exceptions are two of three known integral pore membrane proteins, Pom152p and Ndc1p. The selective recruitment of these proteins, the presence of transcisternal pores, and the lack of other NPC markers suggest that the intranuclear membranes contain stalled intermediates of NPC assembly. This incomplete assembly is unlikely to be due to topological constraints, as structures reminiscent of NPCs have been observed previously in the nucleus (Wente and Blobel 1993; Aitchison et al. 1995). An alternative explanation is that these intermediates arise as a result of limiting amounts of other critical nups necessary for the assembly of NPCs. This would not be the case for Pom152p (and possibly Ndc1p), as excess amounts of this protein located in the ER system (Strambio-de-Castilla et al., 1995) could provide a source of material for its recruitment to the intranuclear membranes. By their presence within these membranes, both Pom152p and Ndc1p represent candidates for proteins capable of participating in the formation or maintenance of transcisternal pores present in these structures. The ability to induce pore-containing membranes by NUP53 overexpression should provide a tractable system for examining the role of these and other proteins in pore formation.
Interestingly, both Pom152p and Ndc1p were recently shown to interact with Nup53p by two-hybrid analysis (Uetz et al. 2000), raising the possibility that these proteins may be recruited to the intranuclear membranes by Nup53p. However, as cellular levels of Pom152p and Ndc1p are not markedly altered upon overproduction of Nup53p (Fig. 1, and data not shown), it is likely that only a small percentage of the excess Nup53p is associated with Pom152p and Ndc1p. Thus, the vast majority of excess Nup53p must reside in a separate membrane-bound pool. The association of excess Nup53p with the intranuclear membranes would likely occur either through an unidentified protein or by a direct interaction with the membrane. A clue to this comes from our observation that the COOH-terminal 27 amino acid residues of Nup53p are required for its membrane association (Fig. 9). Interestingly, the last 17 amino acid residues within this segment are predicted to form an amphipathic α-helix. This observation raises the intriguing possibility that this region of Nup53p could directly interact with a lipid bilayer. The specificity for Nup53p's association with the endoplasmic leaflet of the pore membrane and the intranuclear membranes may, as suggested above, be achieved by its interactions with Kap121p and Nup170p. For example, overproduced Nup53p that is mislocalized in the kap121-41 and nup170Δ mutants is distributed throughout the cytoplasm and did not appear to associate with other intracellular membranes. An exception to this was observed in a subpopulation of cells containing the highest concentrations of Nup53p where we also saw Nup53p associated with the plasma membrane (see Fig. 8 B). Indeed, this observation supports the idea that Nup53p can directly interact with membranes, as it is unlikely that receptors for Nup53p are present in the plasma membrane. A notable paradigm for the specific and regulated association of an amphipathic α-helix with a defined membrane system exists in the ER-Golgi network. Here the association of ADP-ribosylation factors (ARFs) with the membrane is mediated by an NH2-terminal, amphipathic α-helix whose association with the membrane is mediated by the ARF's binding to GTP (see Jackson and Casanova 2000). For Nup53p, and possibly other nups as well (see below), the direct binding of an amphipathic region with the pore membrane could provide an alternative mechanism for anchoring NPC substructures to the membrane that is independent of pore membrane proteins.
The intranuclear membrane structures induced by the overproduction of Nup53p represent a phenotype unique to this yeast nup. Several other structural abnormalities in the NE membrane are induced by changes in the stoichiometric balance of nups (see Introduction). However, the structures most closely related to those seen in Nup53p overproducing cells are layers of lamellar membranes, termed karmellae, that were first observed in yeast cells overexpressing the ER membrane protein 3-hydroxy-3-methylglutaryl coenzyme A reductase (Wright et al. 1988). These structures have also been observed in yeast upon overexpression of other integral membrane proteins including the human lamin B receptor (Smith and Blobel 1994) and the canine 180-kD ribosome receptor (Becker et al. 1999). Karmellae appear to arise from the outer nuclear membrane, forming multiple layers of paired, double membrane lamellae that lie adjacent to the NE and surround large portions of the nucleus. Although karmellae resemble the Nup53p-induced membranes, they exhibit several different features that suggest they are formed by separate mechanisms. Karmellae are topologically distinct, being present exclusively in the cytoplasm. In addition, karmellae do not contain transcisternal pores nor have intermediate tubular structures been observed upon their induction (Wright et al. 1988).
Notably, in vertebrate cells, the overproduction of the nup Nup153p also induces the formation of intranuclear membrane structures (Bastos et al. 1996). Like Nup53p, overproduced Nup153p associates with the inner nuclear membrane as well as intranuclear membrane arrays that appear to be composed of tubular structures and membrane lamellae similar to, but much less extensive than, those induced by Nup53p. This phenotype is dependent on the NH2-terminal one-third of Nup153p (Bastos et al. 1996; Enarson et al. 1998). This region contains an M9-like sequence capable of functioning autonomously as an NLS (Nakielny et al. 1999) and a segment between amino acid residues 2–144 that is capable of associating with the inner nuclear membrane (Enarson et al. 1998). Interestingly, the latter segment contains a region (amino acid residues 39–61) capable of forming an amphipathic α-helix similar to the COOH terminus of Nup53p.
In conclusion, we propose that the targeting of excess Nup53p to the NE membrane and the proliferation events that follow may be indicative of a broader range of physiological functions for both kaps and nups. For example, the Nup53p-induced membrane proliferation could reflect a general role for the NPC in regulating the expansion of the inner nuclear membrane; an event that must occur in response to changes in nuclear volume during the cell cycle and nuclear division. The Nup53p overexpression system may provide a means for investigating these membrane proliferation events. In addition, the requirement for Kap121p in directing excess Nup53p to the NPC may reflect a role for kaps in the maintenance of NPC structure or in the formation of new NPCs. Consistent with this idea, we have also shown that in a strain containing the mutant allele kap121-41 or nup170Δ, endogenous Nup53p is inefficiently targeted to the NPC and is partially mislocalized to the cytoplasm (Lusk, C.P., M. Marelli, and R.W. Wozniak, manuscript in preparation). Moreover, other nups, including Nic96p (Grandi et al. 1995) and Nup153p (Nakielny et al. 1999), contain sequences that can function autonomously as NLSs, raising the question of whether kaps play a role in their integration into the NPC. In the case of Nup53p, its targeting to the NPC and its association with the membrane may reflect a more general pathway for localizing a specific subset of nups (which, as Nup53p is not essential for viability, must include other nups) to the inner nuclear membrane. Here the function of such a subcomplex could be to bind specific loci on the underlying chromatin to the inner membrane and, at these sites, seed the initial stages of NPC assembly. Subsequent changes in membrane structure, initiated by these proteins or recruited membrane proteins, could lead to the formation of transcisternal pores and the subsequent assembly of complete NPCs. Such “inside out” pore formation would occur in intact yeast NEs throughout the cell cycle (Winey et al. 1997) and during S-phase in metazoan cells when the number of NPCs doubles (Maul et al. 1972). In both cases, potential NPC assembly intermediates associated with the inner nuclear membrane have been observed (see Maul 1977; Aitchison et al. 1995). Such a mechanism would provide a means of assembling NPCs at specific gene loci as has been proposed previously (Blobel 1985).
Acknowledgments
We thank those individuals listed in the text for providing reagents.
Operating grants and salary support from the Canadian Institutes for Health Research and Alberta Heritage Foundation for Medical Research provided support for this work.
Footnotes
Abbreviations used in this paper: GFP, green fluorescent protein; kap, karyopherin; NE, nuclear envelope; NLS, nuclear localization signal; NPC, nuclear pore complex; nup; nucleoporin; SM, synthetic media.
References
- Aitchison J.D., Rout M.P., Marelli M., Blobel G., Wozniak R.W. Two novel related yeast nucleoporins Nup170p and Nup157pcomplementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J.D., Blobel G., Rout M.P. Kap104pa karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Bastos R., Lin A., Enarson M., Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker F., Block-Alper L., Nakamura G., Harada J., Wittrup K.D., Meyer D.I. Expression of the 180-kD ribosome receptor induces membrane proliferation and increased secretory activity in yeast. J. Cell Biol. 1999;146:273–284. doi: 10.1083/jcb.146.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. Gene gatinga hypothesis. Proc. Natl. Acad. Sci. USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd A.M., Hoffman J.A., Amberg D.C., Fink G.R., Davis L.I. nup1 mutants exhibit pleiotropic defects in nuclear pore complex function. J. Cell Biol. 1994;127:319–332. doi: 10.1083/jcb.127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Preparation of yeast cell for thin-section electron microscopy. Methods Enzymol. 1991;194:602–608. doi: 10.1016/0076-6879(91)94044-d. [DOI] [PubMed] [Google Scholar]
- Chial H.J., Rout M.P., Giddings T.H., Winey M. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 1998;143:1789–1800. doi: 10.1083/jcb.143.7.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damelin M., Silver P.A. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol. Cell. 2000;5:133–140. doi: 10.1016/s1097-2765(00)80409-8. [DOI] [PubMed] [Google Scholar]
- Delorme E. Transformation of Saccharomyces cerevisiae by electroporation. Appl. Environ. Microbiol. 1989;55:2242–2246. doi: 10.1128/aem.55.9.2242-2246.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delphin C., Guan T., Melchior F., Gerace L. RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol. Biol. Cell. 1997;8:2379–2390. doi: 10.1091/mbc.8.12.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier L., Rapoport T.A. In vitro formation of the endoplasmic reticulum occurs independently of microtubules by a controlled fusion reaction. J. Cell Biol. 2000;148:883–898. doi: 10.1083/jcb.148.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enarson P., Enarson M., Bastos R., Burke B. Amino-terminal sequences that direct nucleoporin nup153 to the inner surface of the nuclear envelope. Chromosoma. 1998;107:228–236. doi: 10.1007/s004120050301. [DOI] [PubMed] [Google Scholar]
- Fabre E., Hurt E. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu. Rev. Genet. 1997;31:277–313. doi: 10.1146/annurev.genet.31.1.277. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B., Hurt E.C., Aebi U., Pante N. Molecular architecture of the yeast nuclear pore complexlocalization of Nsp1p subcomplexes. J. Cell Biol. 1998;143:577–588. doi: 10.1083/jcb.143.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B., Hubner W., Mandinova A., Pante N., Keller W., Aebi U. The yeast nucleoporin Nup53p specifically interacts with Nic96p and is directly involved in nuclear protein import. Mol. Biol. Cell. 2000;2000 11:3885–3896. doi: 10.1091/mbc.11.11.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldherr C.M., Kallenbach E., Schultz N. Movement of a karyophilic protein through the nuclear pores of oocytes. J. Cell Biol. 1984;99:2216–2222. doi: 10.1083/jcb.99.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M., Blobel G., Rexach M. Disassembly of RanGTP-karyopherin beta complex, an intermediate in nuclear protein import. J. Biol. Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- Floer M., Blobel G. Putative reaction intermediates in Crm1-mediated nuclear protein export. J. Biol. Chem. 1999;274:16279–16286. doi: 10.1074/jbc.274.23.16279. [DOI] [PubMed] [Google Scholar]
- Galy V., Olivo-Marin J.C., Scherthan H., Doye V., Rascalou N., Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403:108–112. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- Goldstein A.L., Snay C.A., Heath C.V., Cole C.N. Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol. Biol. Cell. 1996;7:917–934. doi: 10.1091/mbc.7.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Pante N., Kutay U., Aebi U., Bischoff F.R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Schlaich N., Tekotte H., Hurt E.C. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A.K., Raczniak G.A., Ives E.B., Wente S.R. The integral membrane protein snl1p is genetically linked to yeast nuclear pore complex function. Mol. Biol. Cell. 1998;9:355–373. doi: 10.1091/mbc.9.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz M.E., Strambio-de-Castillia C., Blobel G. Two yeast nuclear pore complex proteins involved in mRNA export form a cytoplasmically oriented subcomplex. Proc. Natl. Acad. Sci. USA. 1998;95:11241–11245. doi: 10.1073/pnas.95.19.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.L., Casanova J.E. Turning on ARFthe Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- Kaffman A., Rank N.M., O'Shea E.K. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 1998;12:2673–2683. doi: 10.1101/gad.12.17.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehlenbach R.H., Dickmanns A., Kehlenbach A., Guan T., Gerace L. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 1999;145:645–657. doi: 10.1083/jcb.145.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J.V., Adams A.E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces . J. Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer D.M., Strambio-de-Castillia C., Blobel G., Rout M.P. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J. Biol. Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- Lee D.C., Aitchison J.D. Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and RNA. J. Biol. Chem. 1999;274:29031–29037. doi: 10.1074/jbc.274.41.29031. [DOI] [PubMed] [Google Scholar]
- Marelli M., Aitchison J.D., Wozniak R.W. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 1998;143:1813–1830. doi: 10.1083/jcb.143.7.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul G.G., Maul H.M., Scogna J.E., Lieberman M.W., Stein G.S., Hsu B.Y., Borun T.W. Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J. Cell Biol. 1972;55:433–447. doi: 10.1083/jcb.55.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul G.G. The nuclear and cytoplasmic pore complexstructure, dynamics, distribution and evolution. Int. Rev. Cytol. Suppl. 1977;5:75–186. [PubMed] [Google Scholar]
- Mutvei A., Dihlmann S., Herth W., Hurt E.C. NSP1 depletion in yeast affects nuclear pore formation and nuclear accumulation. Eur. J. Cell Biol. 1992;59:280–295. [PubMed] [Google Scholar]
- Nakielny S., Shaikh S., Burke B., Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U., Rout M.P., Maguire S., Blobel G., Wozniak R.W. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J. Cell Biol. 1996;133:1153–1162. doi: 10.1083/jcb.133.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal R.K., Riles L., Johnston M., Hegemann J.H. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Nuttley W.M., Brade A.M., Eitzen G.A., Veenhuis M., Aitchison J.D., Szilard R.K., Glover J.R., Rachubinski R.A. PAY4, a gene required for peroxisome assembly in the yeast Yarrowia lipolytica, encodes a novel member of a family of putative ATPases. J. Biol. Chem. 1994;269:556–566. [PubMed] [Google Scholar]
- Pemberton L.F., Rosenblum J.S., Blobel G. A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J. Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M., Blobel G. Protein import into nucleiassociation and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Richardson W.D., Mills A.D., Dilworth S.M., Laskey R.A., Dingwall C. Nuclear protein migration involves two stepsrapid binding at the nuclear envelope followed by slower translocation through nuclear pores. Cell. 1988;52:655–664. doi: 10.1016/0092-8674(88)90403-5. [DOI] [PubMed] [Google Scholar]
- Rosenblum J.S., Pemberton L.F., Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J. Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Blobel G. Isolation of the nuclear pore complex. J. Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Blobel G., Aitchison J.D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., Chait B.T. The yeast nuclear pore complexcomposition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.J., Wente S.R. The nuclear pore complexa protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 2000;12:361–371. doi: 10.1016/s0955-0674(00)00101-0. [DOI] [PubMed] [Google Scholar]
- Seedorf M., Silver P.A. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc. Natl. Acad. Sci. USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G.R., Hicks J.B. Methods in Yeast Genetics 1986. Cold Spring Harbor Press; Cold Spring Harbor, NY: pp. 186 pp [Google Scholar]
- Shulga N., Roberts P., Gu Z., Spitz L., Tabb M.M., Nomura M., Goldfarb D.S. In vivo nuclear transport kinetics in Saccharomyces cerevisiaea role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N., Mosammaparast N., Wozniak R., Goldfarb D.S. Yeast nucleoporins involved in passive nuclear envelope permeability. J. Cell Biol. 2000;149:1027–1038. doi: 10.1083/jcb.149.5.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer C., Rieger M., Doye V., Tekotte H., Weise C., Emig S., Segref A., Hurt E.C. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homologue, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Smith S., Blobel G. Colocalization of vertebrate lamin B and lamin B receptor (LBR) in nuclear envelopes and in LBR-induced membrane stacks of the yeast Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. USA. 1994;91:10124–10128. doi: 10.1073/pnas.91.21.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel G., Rout M.P. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces . J. Cell Biol. 1995;131:19–31. doi: 10.1083/jcb.131.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcheperegine S.E., Marelli M., Wozniak R.W. Topology and functional domains of the yeast pore membrane protein Pom152p. J. Biol. Chem. 1999;274:5252–5258. doi: 10.1074/jbc.274.8.5252. [DOI] [PubMed] [Google Scholar]
- Teixeira M.T., Siniossoglou S., Podtelejnikov S., Benichou J.C., Mann M., Dujon B., Hurt E., Fabre E. Two functionally distinct domains generated by in vivo cleavage of Nup145pa novel biogenesis pathway for nucleoporins. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:5086–5097. doi: 10.1093/emboj/16.16.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae . Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wente S.R., Rout M.P., Blobel G. A new family of yeast nuclear pore complex proteins. J. Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S.R., Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J. Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S.R., Blobel G. NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J. Cell Biol. 1994;125:955–969. doi: 10.1083/jcb.125.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S.R. Gatekeepers of the nucleus. Science. 2000;288:1374–1377. doi: 10.1126/science.288.5470.1374. [DOI] [PubMed] [Google Scholar]
- Winey M., Yarar D., Giddings T.H., Jr., Mastronarde D.N. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell. 1997;8:2119–2132. doi: 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman H., Courvalin J.-C. The inner nuclear membrane. J. Membr. Biol. 2000;177:1–11. doi: 10.1007/s002320001096. [DOI] [PubMed] [Google Scholar]
- Wozniak R.W., Blobel G., Rout M.P. POM152 is an integral membrane protein of the pore membrane domain of the nuclear envelope. J. Cell Biol. 1994;125:31–42. doi: 10.1083/jcb.125.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R.W., Rout M.P., Aitchison J.D. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- Wright R., Basson M., D'Ari L., Rine J. Increased amounts of HMG-CoA reductase induce “karmellae”a proliferation of stacked membrane pairs surrounding the yeast nucleus. J. Cell Biol. 1988;107:101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Rout M.P., Akey C.W. Three-dimensional architecture of the isolated yeast nuclear pore complexfunctional and evolutionary implications. Mol. Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- Zabel U., Doye V., Tekotte H., Wepf R., Grandi P., Hurt E.C. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J. Cell Biol. 1996;133:1141–1152. doi: 10.1083/jcb.133.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]