Abstract
Ryanodine receptors (RyRs), intracellular calcium release channels required for cardiac and skeletal muscle contraction, are macromolecular complexes that include kinases and phosphatases. Phosphorylation/dephosphorylation plays a key role in regulating the function of many ion channels, including RyRs. However, the mechanism by which kinases and phosphatases are targeted to ion channels is not well understood. We have identified a novel mechanism involved in the formation of ion channel macromolecular complexes: kinase and phosphatase targeting proteins binding to ion channels via leucine/isoleucine zipper (LZ) motifs. Activation of kinases and phosphatases bound to RyR2 via LZs regulates phosphorylation of the channel, and disruption of kinase binding via LZ motifs prevents phosphorylation of RyR2. Elucidation of this new role for LZs in ion channel macromolecular complexes now permits: (a) rapid mapping of kinase and phosphatase targeting protein binding sites on ion channels; (b) predicting which kinases and phosphatases are likely to regulate a given ion channel; (c) rapid identification of novel kinase and phosphatase targeting proteins; and (d) tools for dissecting the role of kinases and phosphatases as modulators of ion channel function.
Keywords: leucine zipper, ryanodine receptor, calcium channel, phosphorylation, phosphatase
Introduction
Type 2 ryanodine receptor (RyR2)/Ca2+ release channel macromolecular complexes in the heart include protein kinase A (PKA) and two phosphatases (protein phosphatase [PP]1 and PP2A) that regulate channel function (Marx et al. 2000). Among the largest ion channels, RyRs on the sarcoplasmic reticulum (SR) of striated muscles are required for excitation–contraction (EC) coupling. RyRs have enormous cytoplasmic domains that form scaffolds for multiple proteins that modulate channel function (Marx et al. 2000). The skeletal muscle RyR1 and the cardiac muscle RyR2 are tetramers comprised of four 565,000-D RyR polypeptides, each of which binds one 12,000-D FK506 binding protein (FKBP12 and FKBP12.6, respectively; Marks et al. 1989; Jayaraman et al. 1992; Marks 1996) that stabilize RyR channel function (Brillantes et al. 1994) and facilitate coupled gating between neighboring RyR channels (Marx et al. 1998).
PKA phosphorylation of RyR2 dissociates FKBP12.6 from the channel resulting in increased open probability (P o) of the channel (Marx et al. 2000). PKA phosphorylation of RyR2 is an important part of the “fight or flight” response, as increased channel P o enhances EC coupling gain resulting in stronger cardiac muscle contraction (Marks 2000). In failing hearts we have shown that RyR2 is PKA hyperphosphorylated and have proposed that this represents a maladaptive response to stress resulting in defective (“leaky”) channels with markedly increased P o (Marx et al. 2000).
PKA phosphorylation of RyR2 is regulated by kinases and phosphatases that are bound to the channel via targeting proteins (Marx et al. 2000). In failing hearts, reduced amounts of the phosphatases PP1 and PP2A in the RyR2 macromolecular complex is associated with PKA hyperphosphorylation of RyR2 and defective channels (Marx et al. 2000). We previously identified muscle A-kinase anchoring protein (mAKAP) as the targeting protein for PKA in the RyR2 macromolecular complex (Marx et al. 2000). However, the targeting proteins for PP1 and PP2A in the RyR2 macromolecular complex have not been identified. Moreover, the mechanism by which kinase and phosphatase targeting/adaptor proteins bind to ion channels has not been elucidated.
Leucine/isoleucine zipper (LZ) motifs have been identified in ion channels, including the human potassium channel hSK4 (Joiner et al. 1997), Shaker potassium channel (McCormack et al. 1991), and the intracellular calcium release channel, inositol 1,4,5 trisphosphate receptor (IP3R; Galvan et al. 1999), although their function(s) have not been determined. In this study we show that three highly conserved, previously unidentified, LZ motifs on RyR2 form the binding sites for targeting proteins that bind PKA, PP1, and PP2A to the channel. Using this information, we have identified the targeting proteins for PP1 and PP2A in the RyR2 macromolecular complex. Furthermore, we demonstrate that the bound kinase and phosphatases regulate the phosphorylation of the channel and modulate its function. Based on our finding that LZ motifs form binding sites for kinase and phosphatase targeting proteins, we were able to use this information to identify components of the RyR1 macromolecular complex, which has not been reported previously. In the case of RyR1, we were able to predict that PKA and PP1 but not PP2A would be part of the RyR1 macromolecular complex because the LZs that bind targeting proteins for PKA and PP1 (but not for PP2A) were conserved between RyR1 and RyR2.
Materials and Methods
Glutathione S-Transferase Fusion Proteins
Rabbit RyR2 and RyR1, rat spinophilin, human mAKAP (cDNA template, KIAA0311; Kazusa DNA Research Institute), and PR130 cDNA templates were amplified by PCR and subcloned into pGEX-4T1,-4T2, or -4T3 (Amersham Pharmacia Biotech) for expression as glutathione S-transferase (GST) fusion proteins. All cDNA constructs were confirmed by sequencing both strands. GST fusion proteins were expressed in either DH5α or BL21 cells (Stratagene) and affinity purified with glutathione 4B sepharose (Amersham Pharmacia Biotech).
Site-directed Mutagenesis
Mutagenesis was carried out with either a 5′-3′ mutagenesis kit (Eppendorf) or nested primers. RyR2 /L2,3A mutagenesis primer (L564A, L571A), 3′-CACAATGTAAAACTTCATATACCCGAGCAAGCC-TCAGCTCTCTCCAATCTGC-5′; RyR2 1,347–1,663/I,L2,3A mutagenesis primer (I1610A, L1617A), 5′-AAGGTCGATGTTTCTAGAGC-AAGTGAACGACAAGGCTGGGCAGTGCAGTGTTTGGATCC-3′; RyR2 amino acid (aa) 2,838–3,145/L1,2A mutagenesis primer (L3008A, L3015A), 5′-GGAAATGGTGACTAGCGCATTCTGCAAGCTTGG-AGTTGCAGTCAGGCATAGG-3′; Spinophilin aa 300–634 mutagenesis primer (L492A, L499A), 5′-GAGTCCTTCTCCGCCTCCACAGGAAAGAGCTCCGCCCTCTCCACTCG-3′; and mAKAP aa 1,139–1,479 mutagenesis primer (I1224A, L1231A), 5′-GGATGAAATGGA-CGCTAGCAACAAGTTAATTAGTGCGAATGAGGAATC-3′.
GST Pulldown Assays and Immunoprecipitation
SR membranes were prepared from canine ventricular tissue as described (Marx et al. 2000). GST fusion protein concentrations were normalized using Coomassie staining. GST fusion proteins bound to glutathione sepharose beads were incubated for 3 h at 4°C in modified RIPA buffer with SR membranes. Beads were washed extensively in modified RIPA buffer and proteins separated on SDS-PAGE and immunoblotted (Marx et al. 2000) with the following antibodies: anti-RyR (5029, 1:3,000; Marx et al. 2000), anti-FKBP12 (1:1,000; Jayaraman et al. 1992), anti-PP1, anti-PP2A, and anti-PKA catalytic subunit (1:1,000; Transduction Laboratories), anti-RII (1:1,000; Santa Cruz Biotechnology, Inc.), anti-mAKAP (1:1,000; UBI), anti-spinophilin (1:250; Upstate Biotechnology), and anti-GST (Amersham Pharmacia Biotech). Immunoblots containing 10% of the GST fusion protein input were probed with anti-GST antibody to demonstrate that equivalent amounts of GST fusion proteins were used in all pulldown assays. In all cases, data shown are representative of three or more similar experiments.
Competing PKA Off from RyR2 Using Specific RyR2 Leucine/Isoleucine Peptides
Cardiac SR (200 μg) was suspended in 0.5 ml of buffer containing 20 mM imidazole, pH 7.0, 0.9% NaCl, 1.0 mM NaF, and protease inhibitors (complete protease inhibitor cocktail from Roche Diagnostics). Samples were incubated with GST–RyR2 fusion proteins (on glutathione beads) overnight at 4°C. After removal of beads, the supernatant was centrifuged at 95,000 g for 10 min, and the pellet was washed and resuspended in 100 μl of imidazole buffer. Aliquots were fused to planar lipid bilayers for analyses of RyR2 single channel function or immunoprecipitated with anti-RyR2 antibody, followed by immunoblotting for RyR2 and PKA, or analyzed for PKA phosphorylation of RyR2 as described below. In all cases, data shown are representative of three or more similar experiments.
Phosphorylation
SR membranes (200 μg) were incubated with wild-type or mutant RyR2–GST for 2 h. Supernatants were immunoprecipitated with anti-RyR antibody and protein A sepharose beads. Beads were washed with 1× phosphorylation buffer (8 mM MgCl2, 10 mM EGTA, and 50 mM Tris/piperazine-N,N′-bis[2-ethanesulfonic acid], pH 6.8), resuspended in 10 μl of 1.5× phosphorylation buffer containing cAMP (10 μM; Sigma-Aldrich), with or without protein kinase inhibitor (PKI)5–24 (Sigma-Aldrich) as described (Marx et al. 2000). Phosphorylation of immunoprecipitated RyR2 was initiated with MgATP (33 μM) and 10% [γ-32P]ATP (NEN Life Science Products) and terminated after incubation for 5 min at room temperature with 5 μl stop solution (4% SDS, 0.25 M DTT). As indicated, protamine (1 mg/ml) with or without okadaic acid (5 nM) were added to phosphorylation reactions. For microsome phosphorylation experiments, phosphorylation was with MgATP and cAMP (10 μM) with or without PKI for 15 min at room temperature, and pelleted membranes and supernatant were size fractionated on SDS-PAGE (15%) and immunoblotted with anti-FKBP12 antibody. In all cases, data shown are representative of three or more similar experiments.
Single Channel Recordings
Single channel recordings of RyR2 were performed using black lipid membranes and analyzed as described (Brillantes et al. 1994; Marx et al. 2000). SR membranes were fused to the bilayer after addition to the cis chamber using an osmotic gradient (KCl), which was removed by perfusion. The transmembrane voltage was clamped at 0 mV and the charge carrier was Ca2+. The cis chamber was filled with 1 ml of 250 mM H , 125 mM Tris, 0.05–5 mM CaCl2, pH 7.35, and the trans chamber 53 mM CaOH2. The trans chamber was connected to the head-stage input of an Axon 200 amplifier (Axon Instruments, Inc.) using an Ag/AgCl electrode and agar/KCl bridge. The cis chamber was held at ground with a similar electrode. The single channel currents were filtered at 1 kHz with an 8-pole Bessel filter (Warner Instruments) and digitized at 4 kHz. Data were collected on a Pentium computer, using AxoScope1 (Axon Instruments, Inc.) and a Digidata 2000 (Axon Instruments, Inc.) interface. The pClamp6.01 program (Axon Instruments, Inc.) was used for analyzing single channel data. P
o and the lifetimes of open and closed events were identified by 50% threshold analysis using at least 3 min of continuous record. The baseline level was obtained after leak current subtraction. The Student's t test was used for statistical analyses of the dwell time distributions and open probabilities. At the conclusion of each experiment, ryanodine and/or ruthenium red were applied to confirm channel identity. In all cases, data shown are representative of three or more similar experiments.
, 125 mM Tris, 0.05–5 mM CaCl2, pH 7.35, and the trans chamber 53 mM CaOH2. The trans chamber was connected to the head-stage input of an Axon 200 amplifier (Axon Instruments, Inc.) using an Ag/AgCl electrode and agar/KCl bridge. The cis chamber was held at ground with a similar electrode. The single channel currents were filtered at 1 kHz with an 8-pole Bessel filter (Warner Instruments) and digitized at 4 kHz. Data were collected on a Pentium computer, using AxoScope1 (Axon Instruments, Inc.) and a Digidata 2000 (Axon Instruments, Inc.) interface. The pClamp6.01 program (Axon Instruments, Inc.) was used for analyzing single channel data. P
o and the lifetimes of open and closed events were identified by 50% threshold analysis using at least 3 min of continuous record. The baseline level was obtained after leak current subtraction. The Student's t test was used for statistical analyses of the dwell time distributions and open probabilities. At the conclusion of each experiment, ryanodine and/or ruthenium red were applied to confirm channel identity. In all cases, data shown are representative of three or more similar experiments.
Results
Identification of LZ Motifs in Ion Channels
Ion channel structures are comprised of highly conserved motifs that subserve common functions such as pore formation and ion selectivity. We identified three evolutionary conserved LZ motifs in RyR2 with no known function (Fig. 1 A). Novel LZ motifs were also identified in other ion channels, including IP3R, KvLQT1, and α1c subunit of cardiac dihydropyridine receptor (DHPR; data not shown). Currently available software for detecting LZ and coiled-coils fails to detect most of these leucine/isoleucine motifs because of the presence of isoleucine and occasionally valine residues in the “d” position, and because of occasional nonhydrophobic residue in the “a” position. Therefore, all of the previously unrecognized LZ motifs identified in this paper were identified by manual sequence analyses. We sought to determine the functional role of LZ motifs in mediating protein–protein interactions in ion channels.
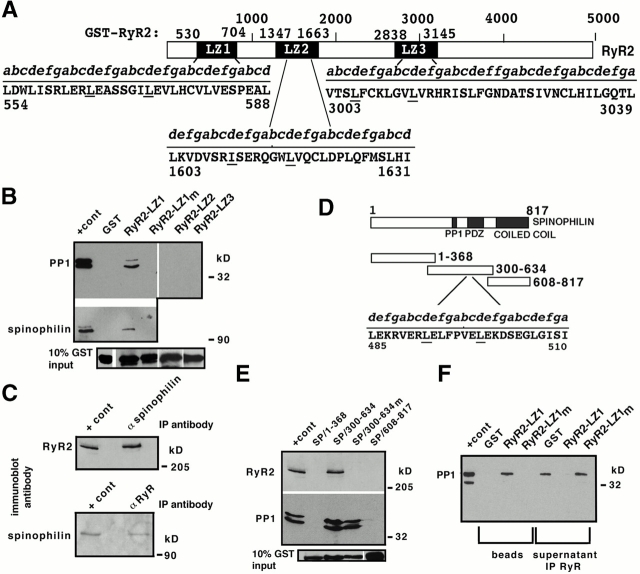
Figure 1.
PP1/spinophilin binds to an LZ motif in RyR2. (A) RyR2–LZ motifs. GST–RyR2-LZ1 (aa 530–704) contains an LZ (aa 555–604); GST-RyR2-LZ2 (aa 1,347–1,663) contains an LZ (aa 1,603–1,631); GST-RyR2-LZ3 (aa 2,838–3,145) contains an LZ (aa 3,003–3,039); underlined residues were substituted with alanines (RyR2–LZXm). (B) Pulldown using cardiac SR (200 μg,) followed by immunoblotting with anti-PP1 or antispinophilin demonstrated specific LZ-dependent binding of PP1 (top panel) and spinophilin (middle panel) to RyR2; “+ cont” represents 5% (spinophilin) or 10% (RyR2) SR input. RyR2–LZ1 but not RyR2–LZ2 or RyR–LZ3 coprecipitated PP1. (C) Coimmunoprecipitation of RyR2 and spinophilin from SR; “+ cont” represents 10% SR input. (D) GST–spinophilin fusion proteins, GST-SP/1–368, GST-SP/300–634, and GST-SP/608–817. GST-SP/300–634 contains the PP1 binding domain (Hsieh-Wilson et al. 1999), a PDZ domain (Allen et al. 1997), and an LZ (aa 485–510). Underlined leucine residues were substituted with alanines (SP/300–634m). (E) Pulldown assays demonstrated a specific interaction between RyR2 and spinophilin; “+ cont” represents either 5% (PP1) or 10% (RyR2) SR input. (F) SR was preincubated with GST alone, RyR2–LZ1, or RyR2–LZ1m before immunoprecipitating supernatants with anti-RyR antibody. Spinophilin/PP1 was competed off RyR2 by GST-RyR2-LZ1, but not by GST alone or RyR2–LZ1m.
LZ Motifs Mediate Targeting of Spinophilin/PP1 to RyR2
GST–RyR2 fusion proteins (Fig. 1 A) were prepared and pulldown assays performed using canine cardiac ventricular muscle SR. The GST fusion protein RyR2–LZ1 coprecipitated with PP1 (Fig. 1 B). This interaction was specific, as GST alone (Fig. 1 B) and other GST–RyR2 fusion proteins that do not contain LZ motifs (data not shown) as well as two other LZ motifs in RyR2 (LZ2 and LZ3) failed to coprecipitate with PP1 (Fig. 1 B). A mutant RyR2–LZ1 (RyR2–LZ1m, containing alanine substitutions for the second and third “d” leucines; Fig. 1 B) failed to coprecipitate with PP1, indicating that the LZ motif is responsible for PP1 binding on RyR2. However, RyR2-LZ1 did not bind recombinant PP1 catalytic subunit (data not shown), indicating that targeting of PP1 to RyR2 was mediated by a third protein and not due to a direct interaction between the PP1 catalytic subunit and RyR2.
PP1 targeting proteins have not been reported to bind to their substrates via LZ motifs. However, we observed that spinophilin, a PP1 targeting protein expressed in cardiac muscle, contains a LZ motif. Spinophilin was originally identified in the neostriatum where it targets PP1 to postsynaptic densities (Allen et al. 1997) and plays a role in the modulation of the ionotropic glutamate receptor, AMPA (Yan et al. 1999). The GST fusion protein RyR2–LZ1, but not GST or GST-RyR2-LZ1m, coprecipitated spinophilin (Fig. 1 B), indicating that spinophilin targets PP1 to RyR2 via binding to a LZ. Spinophilin coimmunoprecipitated with RyR2, indicating the existence of a physical association between these proteins in cardiac SR membranes (Fig. 1 C).
Spinophilin also contains a Src homology 3 binding (SH3) domain, an F-actin binding domain at its NH2 terminus, a predicted coiled coil structure at the COOH terminus, a single PDZ domain (aa 492–583), and a PP1 binding domain (RKIHF aa 447–451; Allen et al. 1997; McAvoy et al. 1999). To identify the binding site for RyR2 on spinophilin, three GST fusion proteins encompassing the NH2 terminus (GST-SP/1–368), mid-region (GST-SP/300–634), and COOH terminus (GST-SP/608–817) of spinophilin were prepared (Fig. 1 D). Only GST-SP/300–634, containing an LZ motif, bound to RyR2 (Fig. 1 E, top panel). Substituting alanines for the second and third “d” leucines (GST-SP/300–634m) eliminated binding to RyR2, indicating that spinophilin interacts directly with RyR2 via LZ motifs. The mutant GST-SP/300–634m bound PP1, indicating that the PP1 binding site on spinophilin is distinct from the LZ motif that binds to RyR2 (Fig. 1 E, middle panel).
To determine whether spinophilin/PP1 is targeted to a single site on RyR2, we incubated SR membranes with GST alone, GST-RyR2-LZ1, or GST-RyR2-LZ1m before immunoprecipitation with anti-RyR antibody. PP1 binding to RyR2 was completely inhibited by preincubation with RyR2–LZ1, but not by preincubation with RyR2–LZ1m or GST alone, indicating that spinophilin targets PP1 to RyR2 at a binding site defined by a unique LZ motif (Fig. 1 F).
LZ Motif Mediates Targeting of PR130/PP2A to RyR2
The LZ2 motif (RyR–LZ2; Fig. 1 A) forms the unique binding site for PP2A on RyR2. Purified GST-RyR2-LZ2 specifically coprecipitated PP2A from cardiac SR membranes (Fig. 2 A), but failed to interact with recombinant catalytic subunit of PP2A (data not shown). This interaction was specific, as GST alone (Fig. 2 A) and other GST–RyR2 fusion proteins that do not contain LZ motifs (data not shown) as well as two other LZ motifs in RyR2 (LZ1 and LZ3) failed to coprecipitate with PP2A (Fig. 2 A). Substituting alanines for the second and third “d” isoleucine/leucines (RyR2–LZ2m) eliminated PP2A coprecipitation (Fig. 2 A), implicating the LZ2 motif on RyR2 as the PP2A binding site. Thus, a targeting protein in cardiac SR mediates PP2A binding to RyR2 via an LZ.
Figure 2.
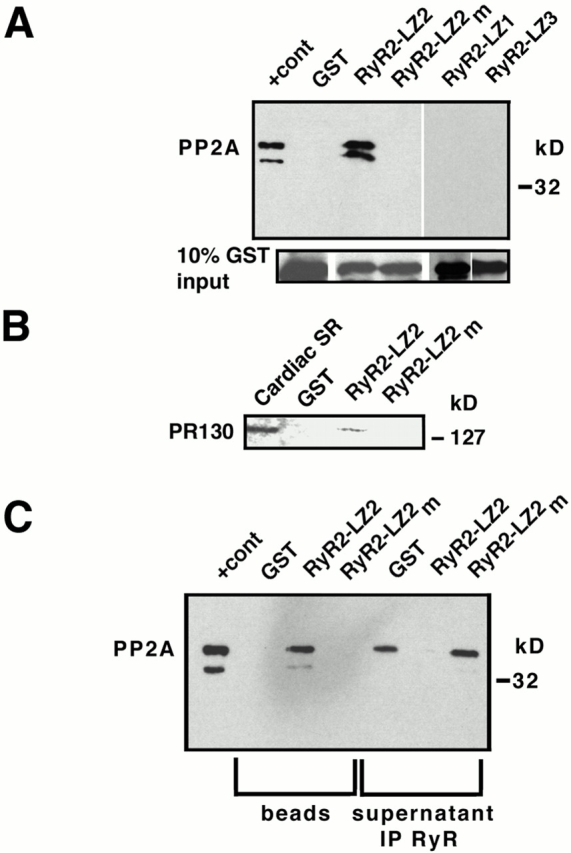
PP2A is targeted to RyR2 via an LZ. (A) PP2A specifically bound GST-RyR2-LZ2 (via PR130), but not RyR2–LZ2m in GST-SR pulldown assays. RyR2–LZ2 but not RyR2–LZ1 or RyR–LZ3 coprecipitated PP2A. “+ cont” represents 5% SR input. (B) PR130 specifically bound GST-RyR2-LZ2, but not RyR2–LZ2m in GST-SR pulldown assays. “Cardiac SR” represents 5% SR input. (C) SR was incubated with GST alone, GST-RyR2-LZ2, or GST-RyR2-LZ2m before immunoprecipitating the supernatant with anti-RyR antibody and immunoblotting with anti-PP2A. PP2A binding to RyR2 was competitively inhibited by incubation with GST-RyR2-LZ2.
PP2A targeting proteins have not been reported to bind to their substrates via LZ motifs. However, we observed that PR130, a PP2A targeting protein expressed in cardiac muscle (Zolnierowicz et al. 1996), contains a LZ motif. To determine whether this LZ in PR130 was responsible for targeting PP2A to RyR2, we showed that a GST fusion protein containing the PR130 LZ motif, but not an alanine substitution mutant of this PR130 LZ, coprecipitated RyR2 in pulldown assays. Moreover, RyR2–LZ2, but not the alanine substitution mutant RyR2–LZ2m, coprecipitated PR130 (Fig. 2 B). To confirm that PR130/PP2A is targeted to RyR2 via LZ-mediated interactions, we incubated cardiac SR membranes with GST alone, GST-RyR2-LZ2, or GST-RyR2-LZ2m before immunoprecipitation with anti-RyR antibody. PP2A binding to RyR2 was completely inhibited by pre-incubation with RyR2–LZ2, but not by preincubation with RyR2–LZ2m or GST alone, indicating that like PP1, PP2A binds to RyR2 via a targeting protein, PR130, via a unique LZ motif (Fig. 2 C).
LZ Motif Mediates Targeting of mAKAP/PKA to RyR2
The LZ3 motif (RyR2–LZ3; Fig. 1 A) specifically coprecipitated PKA, RII, and mAKAP in pulldown assays (Fig. 3 A). This interaction was specific, as GST alone (Fig. 3 A) and other GST-RyR2 fusion proteins that do not contain LZ motifs (data not shown) as well as two other LZ motifs in RyR2 (LZ1 and LZ2) failed to coprecipitate with PKA (Fig. 3 A). LZ3 includes one “skip” residue (eight residues instead of seven between “d” amino acids) as reported previously for some LZ motifs (Brown et al. 1996; Lupas 1996). Substituting alanines for the first and second “d” leucines (RyR2–LZ3m) eliminated coprecipitation of PKA, RII, and mAKAP (Fig. 3 A), indicating that the RyR2 binding site for mAKAP (which anchors PKA and RII) contains an LZ motif. To demonstrate that mAKAP/PKA is targeted to RyR2 via LZs, we incubated cardiac SR membranes with GST alone, GST-RyR2-LZ3, or GST-RyR2-LZ3m before immunoprecipitation with anti-RyR antibody. PKA binding to RyR2 was completely inhibited by preincubation with RyR2–LZ3, but not by preincubation with RyR2–LZ3m or GST alone, indicating that like PP1 and PP2A, PKA binds to RyR2 via a targeting protein, mAKAP, at a unique LZ motif (Fig. 3 B).
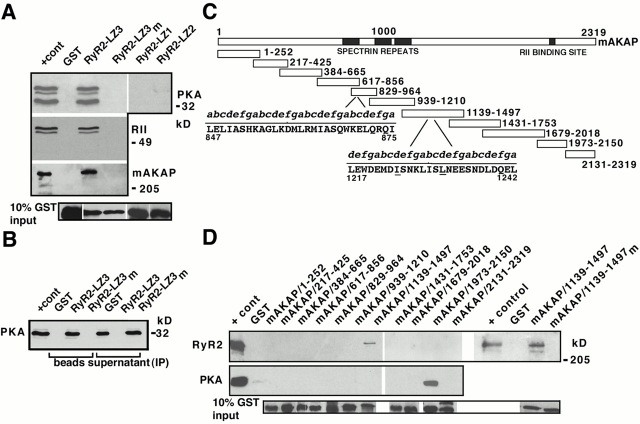
Figure 3.
PKA/RII/mAKAP bind to RyR2 via LZ motifs. (A) GST-SR pulldown demonstrated a specific interaction between mAKAP/RII/PKA and RyR2–LZ3, but not GST or GST-RyR2-LZ3m; immunoblots, anti-PKA (first panel), anti-RII (second panel), and anti-mAKAP (third panel). “+ cont” represents 5% (RII and PKA) or 10% (mAKAP) SR input. (B) SR was incubated with GST, GST-RyR2-LZ3, or GST-RyR2-LZ3m before immunoprecipitation of supernatants with anti-RyR antibody followed by immunoblotting with anti-PKA. RyR2–LZ3 but not RyR2–LZ1 or RyR–LZ2 coprecipitated PKA. Lanes labeled “beads” contain GST-pulldown samples, and lanes labeled “supernatant” are immunoprecipitations. PKA binding to RyR2 via mAKAP was competitively inhibited by incubation with GST-RyR2-LZ3 but not GST or GST-RyR2-LZ3m. “+ cont” represents 5% SR input. (C) mAKAP and GST-mAKAP fusion proteins. GST-mAKAP/1,973–2,150 contains the RII binding domain, GST-mAKAP/829–964 and GST-mAKAP/1,139–1,497 contain LZ motifs. Alanines were substituted for underlined isoleucine/leucines (GST-mAKAP/1,139–1,497m). (D) GST-SR pulldown assays followed by immunoblotting with anti-RyR (top) or anti-PKA (middle). “+ cont” represents 10% (RyR2, top) and 5% (PKA, middle) SR input.
To identify the LZ on mAKAP that binds to RyR2, mAKAP was expressed as 11 GST fusion proteins (Fig. 3 C) and pulldown assays with cardiac SR membranes were performed. GST–mAKAP/1,139–1,497, which contains a LZ motif, specifically bound RyR2 (Fig. 3 D). Substituting alanines for the second and third “d” isoleucine/leucine (mAKAP/1,139–1,497m) eliminated coprecipitation with RyR2 (Fig. 3 D, top panel), indicating that mAKAP binds to RyR2 via LZ motifs. The function of a second LZ motif on mAKAP is currently unknown. In agreement with prior observations (Carr et al. 1991) demonstrating that mAKAP contains an amphipathic helix that binds RII, we found that GST-mAKAP/1,973–2,150 specifically bound PKA (Fig. 3 D, middle panel) and RII (data not shown).
Role of LZ Motifs in the Regulation of RyR Function
PKA phosphorylation of RyR2 dissociates the regulatory protein FKBP12.6, a member of the immunophilin family of peptidyl-prolyl isomerases (Marks 1996) and increases channel P o (Marx et al. 2000). In failing human hearts, RyR2 is PKA hyperphosphorylated, resulting in defective channel function with increased P o that contributes to altered EC coupling (Marx et al. 2000). To establish the functional significance of RyR2-bound PKA, we showed that preincubation with GST-RyR2-LZ3 dissociates PKA/RII/mAKAP from the RyR2 complex (Fig. 3 B and Fig. 4 A, third panel) and prevents cAMP-induced hyperphosphorylation of RyR2 (Fig. 4 A, top panel). A peptide corresponding to RyR2–LZ3 competed PKA off from the channel, whereas a mutated form of the same peptide containing alanine substitutions for two of the leucines in the heptad repeat (RyR2–LZ3m) did not compete PKA off from the channel (Fig. 4 A, third panel). After incubation with GST-RyR2-LZ3, the PKA previously bound to RyR2 was detected by immunoblot bound to GST-RyR-LZ3 beads (Fig. 4 A, bottom panel) and no PKA was detected after incubation with GST-RyR-LZ3 by coimmunoprecipitation with RyR2 (Fig. 4 A, bottom panel). In contrast, after incubation with GST-RyR2-LZ3m, PKA still coimmunoprecipitated with RyR2 and none was detected bound to the GST-RyR-LZ3m beads (Fig. 4 A, bottom panel), indicating the specificity of the LZ-mediated competition.
Figure 4.
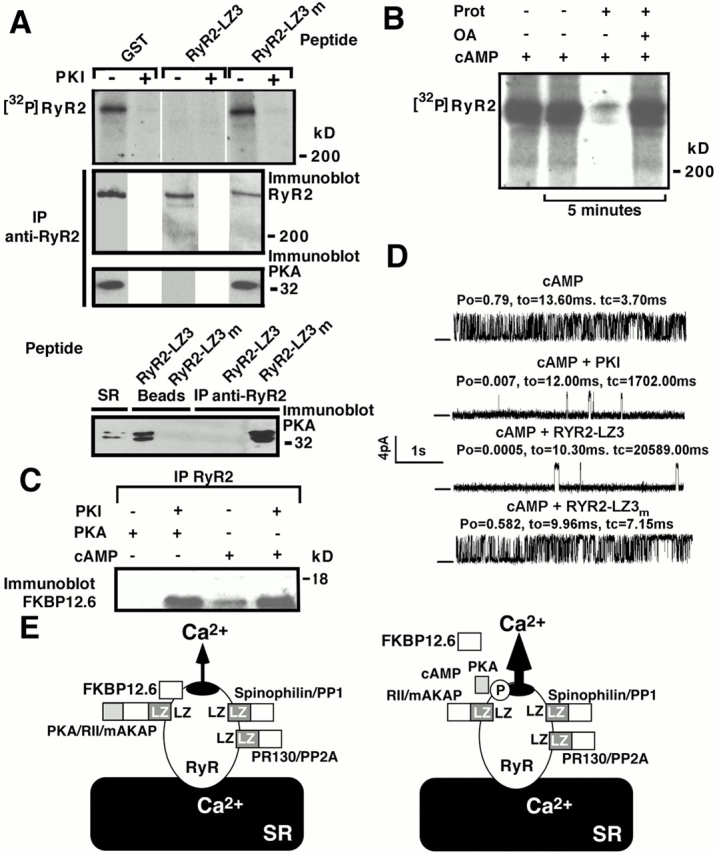
Kinase and phosphatases bound to RyR2 via LZ motifs regulate channel function. (A) Top panel: immunoprecipitated RyR2 was phosphorylated via activation of bound kinase by addition of cAMP (10 μM) and [γ-32P]ATP with or without PKI5–24 (500 nM). RyR2 phosphorylation by bound PKA was inhibited by preincubation with GST-RyR2-LZ3, but not by incubation with either GST alone or GST-RyR2-LZ3m (top panel). Equivalent amounts of RyR2 protein were present in each sample (second panel). Incubation with GST-RyR2-LZ3 but not with GST-RyR2-LZ3m disrupted binding of PKA to RyR2 (via mAKAP) as shown by coimmunoprecipitation (third panel). After incubation with GST-RyR2-LZ3, the PKA previously bound to RyR2 was now bound to GST-RyR-LZ3 beads and none was detected after coimmunoprecipitation with RyR2 (bottom panel). After incubation with RyR2–LZ3m, PKA coimmunoprecipitated with RyR2 and none was detected, by immunoblot, bound to the GST-RyR-LZ3m beads (bottom panel). “SR” denotes a sample of cardiac SR used as a positive control for the anti-PKA antibody. (B) RyR2-bound phosphatases dephosphorylate RyR2. Immunoprecipitated RyR2 was phosphorylated with cAMP for 15 min, and further phosphorylation was inhibited by PKI. RyR2 was dephosphorylated for 5 min by activation of bound phosphatases with protamine (Prot; 1 mg/ml), which was inhibited by okadaic acid (OA; 5 nM). (C) cAMP-induced activation of RyR2-bound PKA caused dissociation of FKBP12.6. (D) Pelleted membranes were introduced into planar lipid bilayers and RyR2 single channel properties determined. cAMP-induced phosphorylation of RyR2 by bound PKA increased P o. This was inhibited by the PKA inhibitor PKI. Preincubation with GST-RyR2-LZ3 (RYR2–LZ3) but not with GST-RyR2-LZ3m (RYR2–LZ3m) disrupted binding of PKA to RyR2 (see A) and inhibited the cAMP-dependent increase in channel P o. (E) Model of RyR2 macromolecular signaling complex, one subunit of the tetrameric RyR channel is shown. Targeting of kinases and phosphatases to RyR2 requires LZ interactions between RyR2 and targeting proteins. cAMP activates bound PKA, phosphorylating RyR2 and increasing calcium release.
Thus, cAMP-induced RyR2 phosphorylation requires LZ anchoring of PKA/RII/mAKAP to RyR2. In failing human hearts, the amounts of RyR2-bound phosphatases (PP1 and PP2A) were diminished compared with controls and were restored after left ventricular assist device (LVAD) implantation (Marx et al. 2000). This suggests that the PKA hyperphosphorylation of RyR2 in heart failure may be due, at least in part, to a reduction in the amount of RyR2-associated phosphatases. To establish the functional role of LZ-mediated association of RyR2 and phosphatases, we showed that protamine-induced activation of RyR2-bound phosphatases dephosphorylated RyR2 (Fig. 4 B). Moreover, RyR2-bound phosphatases were able to reverse the phosphorylation mediated by cAMP activation of RyR2-bound PKA.
Phosphorylation of RyR2 by cAMP-induced activation of RyR2-bound PKA caused dissociation of FKBP12.6 from RyR2 using both immunoprecipitated RyR2 (Fig. 4 C) and cardiac microsomes (data not shown). To establish the functional role of RyR2-bound PKA, we showed that cAMP-induced phosphorylation of RyR2 significantly increased RyR2 P o (Fig. 4 D, top tracing) compared with channels treated with cAMP plus the PKA inhibitor PKI (Fig. 4 D, second tracing) (cAMP-treated RyR2, P o = 0.25 ± 0.23, n = 12; cAMP plus PKI, P o = 0.019 ± 0.02, n = 10, P < 0.015; e.g., Fig. 4 D, first and second tracings). Moreover, RyR2 channels incubated with GST-RyR2-LZ3 (Fig. 4 D, third tracing), but not with GST-RyR2-LZ3m (Fig. 4 D, fourth tracing) were no longer activated by cAMP-induced PKA phosphorylation (GST-RyR2-LZ3-treated RyR2, P o = 0.014 ± 0.011, n = 5; GST-RyR2-LZ3m, P o = 0.38 ± 0.24, n = 4, P < 0.01; e.g., Fig. 4 D, third and fourth tracings). Taken together, these data show that peptides that disrupt binding of PKA (via mAKAP) to RyR2 via the LZs also prevent regulation of RyR2 by the bound PKA. RyR–LZ3 specifically competed mAKAP/RII/PKA off from RyR2 (Fig. 3 B and 4 A). Moreover, competing mAKAP/RII/PKA off from RyR2 with RyR–LZ3 prevented cAMP-induced phosphorylation of RyR2 (Fig. 4 A). Thus, cAMP-induced phosphorylation of RyR2 depends on LZ-mediated targeting of mAKAP/RII/PKA to RyR2 and likely plays a key role in the regulation of SR Ca2+ release via cAMP-induced phosphorylation of RyR2 (Fig. 4 E).
Using LZs to Predict Kinase and Phosphatase Targeting Protein Binding Sites on Ion Channels
We compared LZ motifs in RyR2 and RyR1 (skeletal muscle) and found that the binding sites for PP1 and PKA, but not for PP2A are highly conserved between the two channel isoforms (Fig. 5 A). Thus, we predicted that RyR1 would be associated with PP1 and PKA but not PP2A. Indeed, immunoprecipitation and GST pulldown studies demonstrated that RyR1 is complexed with PKA and PP1, but not PP2A in skeletal muscle SR (Fig. 5 B). A GST–RyR1 fusion protein (aa 497–759) containing a putative leucine/isoleucine/valine zipper (aa 554–603) (Fig. 5 C) specifically coprecipitated PP1 (Fig. 5 D, top panel) in skeletal muscle SR pulldown assays. A GST–RyR1 fusion protein (aa 2,968–3,216) containing a putative leucine/valine zipper with a “skip” (aa 3,039–3,075) (Fig. 5 C) specifically coprecipitated PKA in skeletal muscle SR pulldown assays (Fig. 5 D, middle panel). These data suggest that, like RyR2 (Marx et al. 2000), RyR1 is a macromolecular complex that includes PKA, and PP1 bound to the channel via targeting proteins that contain LZ motifs. By analogy to the cardiac RyR2 channel interaction with PP1 and PKA, we predict that there are targeting proteins (containing LZ motifs) for PP1 and PKA that mediate their binding to RyR1 and are in the process of identifying them using the RyR1 LZ motif binding sites as bait in yeast two-hybrid screens. Identification of conserved LZ motifs in IP3Rs, KvLQT1, and the DHPR α1c subunit has enabled us to identify kinases and phosphatases targeted to these ion channels as well (Marx, S., S. Reiken, Y.-M. Yang, and A.R. Marks, manuscript in preparation).
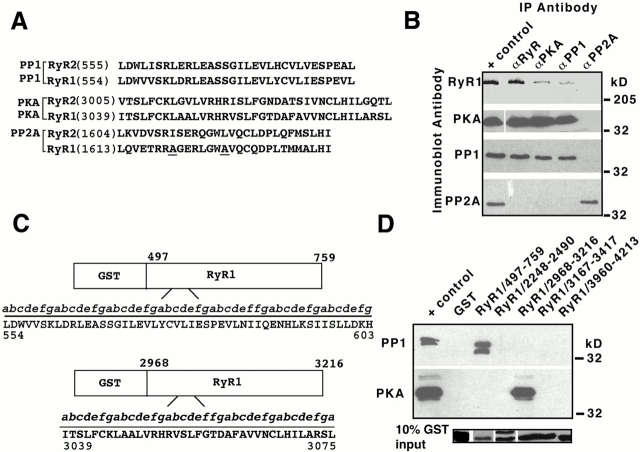
Figure 5.
Components of the RyR1 macromolecular signaling complex bind via LZ motifs. (A) Comparison of aa sequences required for targeting of PP1, PKA, and PP2A to RyR2 and PP1 and PKA to RyR1. The second and third “d” position of RyR2–PP2A targeting motif is not conserved in RyR1 (underlined residues), implying that PP2A is not targeted to RyR1 through this motif. Numbers in parentheses correspond to the first residue in each sequence. (B) RyR1 macromolecular signaling complex contains PKA and PP1, but not PP2A. Components of the RyR1 complex, PKA and PP1, were coimmunoprecipitated from purified skeletal SR (200 μg; Marx et al. 1998). Positive (+) control represents 5% (PKA, PP1, and PP2A) and 10% (RyR) of SR input. Data shown are representative of more than three similar experiments. (C) Schematic diagram of PP1 and PKA binding domain on RyR1. The GST fusion protein corresponding to aa 497–759 of RyR1 contains a leucine/valine heptad repeat between aa 554–603. The GST fusion protein corresponding to aa 2,968–3,216 of RyR1 contains a leucine/valine heptad repeat between aa 3,039–3,075. The motif is comprised of a periodic repeat (labeled “abcdefg”) of a leucine at every seventh or “d” position with a “skip” labeled “ff” between repeats 3 and 4. (D) GST pulldown assays demonstrated a specific interaction between RyR1 and PP1 and RyR1 and PKA through putative LZ targeting motifs. GST fusion proteins were incubated with rabbit skeletal SR membranes (200 μg). Bound proteins were analyzed with SDS-PAGE. The positive (+) control represents 5% of SR input. GST fusion protein, GST-RyR1/497–759 demonstrated specific interaction with PP1 (top panel). GST fusion protein, GST-RyR1/2,968–3,216 demonstrated specific interaction with PKA (middle panel). Separate immunoblots containing 10% of the GST fusion protein input were probed with anti-GST antibody to demonstrate that equivalent amounts of GST fusion proteins were used in pulldown assays (bottom panel). As GST fusion proteins are different molecular weights, bands were cut from immunoblot and realigned for purposes of comparison.
Striking evolutionary conservation of the LZ motifs that bind PP1, PP2A, and PKA in RyRs provides clues regarding signaling pathways that regulate these channels (Fig. 6). For example, the RyR LZ motif responsible for targeting PP1 is remarkably conserved in all three known forms of RyRs (RyR1, RyR2, and RyR3) from Homo sapiens to Caenorhabditis elegans (although there is a two residue shift in the Drosophila sequence that might prevent binding). These data suggest that dephosphorylation of RyRs by PP1 is highly conserved and predicts that spinophilin or a closely related homologue is also highly conserved. The RyR LZ motif responsible for targeting PKA is also completely conserved from human to fish but an alanine replaces the third leucine in the heptad repeat in Drosophila and C. elegans. These data imply that PKA-mediated phosphorylation of RyRs is a generalized regulatory mechanism in vertebrates and that mAKAP homologues containing LZ motifs exist that target PKA to each of the RyR isoforms. In contrast, the PP2A LZ motif is conserved only in the RyR2 isoform in vertebrates. These data suggest that dephosphorylation of RyRs by PP2A is specific to the RyR2 isoform in vertebrates. RyR, PP1, PP2A, PKA, and AKAPs have all been identified in C. elegans (Maryon et al. 1996; Angelo and Rubin 1998; Zeke et al. 1998).
Figure 6.
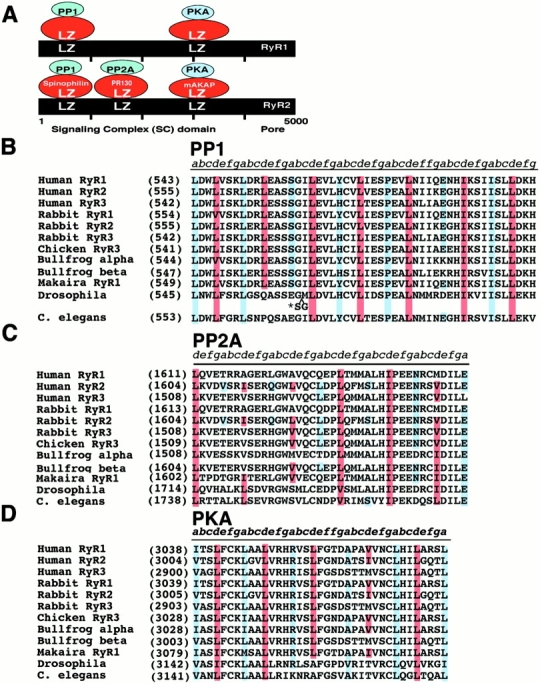
Conservation of LZ motifs in RyRs. (A) Location of the LZ motifs on RyR1 and RyR2 that bind to LZ motifs in adaptor/targeting proteins for PKA, PP1, and PP2A. The numbers below the RyR2 refer to aa residues. RyR sequences corresponding to the LZ motifs that bind (B) PP1, (C) PP2A, and (D) PKA were aligned. Numbers in parentheses correspond to the first residue in each sequence. Blue shaded residues indicate conserved “a” and red shaded residues indicate conserved “d” positions of the leucine/isoleucine heptad repeats. There are three forms of RyR identified to date, RyR1 (Marks et al. 1989; Takeshima et al. 1989; Zorzato et al. 1990), RyR2 (Nakai et al. 1990; Otsu et al. 1990), and RyR3 (Hakamata et al. 1992). The frog alpha corresponds to RyR1 and beta to RyR3 (Oyamada et al. 1994). The fish RyR (blue marlin, Makaira nigricans) corresponds to RyR1 (Franck et al. 1998). The Drosophila RyR is ∼45% homologous to mammalian RyRs (Takeshima et al. 1994; Xu et al. 2000), and C. elegans RyR corresponds to RyR1 (Maryon et al. 1996). In B, the (*) indicates two residues inserted in the Drosophila sequence that displace the alignment of the LZ motif. The proline in the middle of the PP1 LZ motif would introduce a bend in the helix (Hendrickson and Love 1971) which is followed by a phase shift “skip” (eight residues instead of seven between the leucine/isoleucine in the repeat). The phase shift could bring the repeats back into register following the bend in the helix to permit alignment with the corresponding heptad repeats in the LZ motif in spinophilin.
Discussion
A common theme in signal transduction is the close association of signaling molecules in multienzyme complexes, presumably to spatially and temporally regulate signaling in response to localized stimuli (Hunter 1995). However, the mechanism of targeting kinase and phosphatase adaptor proteins to ion channels has remained unclear. These studies demonstrate a novel role for LZ motifs in the targeting of kinases and phosphatases to ion channels. Signaling via protein–protein interactions mediated by LZ motifs in ion channel macromolecular complexes is analogous to targeted signaling involving SH2 and SH3 domains (Koch et al. 1991; Pawson and Gish 1992).
Identification and characterization of the substrate binding sequences of targeting proteins for kinases and phosphatases has significant implications for the study of other ion channels. For instance, we have used this information to identify kinases, phosphatases, and targeting proteins that bind to novel LZ sequences in other cardiac ion channels that are modulated by phosphorylation. Understanding the mechanism by which kinases and phosphatases are targeted to ion channels may lead to novel approaches for the understanding and treatment of human heart diseases, such as heart failure and arrhythmogenesis (Kass and Davies 1996; Marks 2000; Marx et al. 2000).
The LZ domain is an α helical structure that forms coiled coils and was originally identified as a highly conserved motif mediating the binding of transcription factors to DNA (Landschulz et al. 1988). Its role in promoting homo- and heterodimerization of transcription factors has been well documented (Turner and Tjian 1989). LZ motifs have also been shown to mediate protein–protein interactions (e.g., myosin binding subunit/cGKIα; Surks et al. 1999) and intraprotein oligomerization (e.g., phospholamban, the phosphoprotein that regulates Ca2+ uptake across the SR; Simmerman et al. 1996). However, no prior studies have identified a role for LZs in the formation of ion channel macromolecular complexes.
Mutagenesis has been used to study the sequence specificity of interacting helices in the GCN4 DNA binding domain (Harbury et al. 1993), phospholamban (Simmerman et al. 1996), and myosin binding subunit/cGKIα (Surks et al. 1999). Substitution of an alanine for one or more of the “d” position leucines in the LZ motif diminishes the ability of the LZ to mediate protein–protein interaction without disrupting its native α helical structure (Simmerman et al. 1996; Moitra et al. 1997). Using site-directed mutagenesis with alanine substitutions of two “d” position residues, we showed that LZ motifs in RyR and in kinase/phosphatase targeting proteins are responsible for regulation of the phosphorylation and function of the channel. These findings indicate a novel role for conserved LZ motifs in the targeting of phosphorylation modulatory molecules to ion channels.
The specificity of each LZ motif for a unique targeting protein is imparted largely via the amino acids in the “a” position and in other positions in the coiled-coil forming the α helix (Moitra et al. 1997), as the leucines/isoleucines in the “d” positions alone cannot explain the observed degree of specificity.
It is interesting to note that a proline (aa 496) is located at an “a” position within the putative LZ motif on spinophilin. Introduction of a proline into an α helical structure is known to introduce a bend but does not necessarily “break” the helix (Hendrickson and Love 1971; Alber et al. 1988; Barlow and Thornton 1988). Nevertheless, a proline substitution in an “a” position of a heptad repeat has been demonstrated to inhibit α helical folding based on circular dichroism spectroscopy (Wagschal et al. 1999; Tripet et al. 2000). However, substitution of a proline for a leucine or isoleucine in the “a” position of a LZ in the transmembrane protein gp41 of HIV-1 did not disrupt oligomerization of this protein, although it did reduce the stability of the coiled coil (Chang et al. 1999).
The LZ motifs responsible for targeting PKA and PP1 to RyR2 are conserved among RyR isoforms, but only the RyR2 isoform contains the PP2A targeting motif. The finding that PKA and PP1, but not PP2A coimmunoprecipitate with RyR1 support our model for using conserved LZ motifs to predict which kinases and phosphatases regulate members of an ion channel family.
Recently, we reported that PKA hyperphosphorylation of RyR2 obtained from failing hearts was associated with defective channel function (Marx et al. 2000). The amounts of PP1 and PP2A associated with RyR2 obtained from heart failure patients were diminished compared with controls, suggesting that dephosphorylation of RyR2 might be impaired in heart failure patients (Marx et al. 2000). The abnormal phosphorylation and function were reversed in patients receiving an LVAD, implying that the processes mediating the maladaptive responses seen in heart failure may be reversible (Marx et al. 2000). The elucidation of the mechanism(s) underlying the specific phosphorylation and dephosphorylation of ion channels in general and specifically RyR2 may have significant ramifications in terms of treating heart failure and prevention of arrhythmias and sudden cardiac death (Marks 2000). Enhanced dephosphorylation of RyR2 by PP1 and/or PP2A could restore normal phosphorylation and function of RyR2 and normal EC coupling gain. Abnormal regulation of phosphorylation of RyR2 in the intact heart is believed to be one of the principal mechanisms underlying electrical and mechanical alternans caused by impairment of glycolysis (i.e., ischemia; Huser et al. 2000).
This study elucidates a novel mechanism for targeting kinases and/or phosphatases to ion channels. LZ motif–mediated targeting of kinases and phosphatases to ion channels provides specific physiologic and pathophysiologic regulation of the phosphorylation and function of RyR in cardiac and skeletal muscle. The findings that these motifs are evolutionary conserved supports the premise that LZ motifs are critically important for the regulation of intracellular Ca2+ release in the heart and skeletal muscle. Identification of this novel role for LZ motifs provides roadmaps for locating binding sites and for dissecting the function of ion channel macromolecular signaling complexes in the heart and other tissues.
Acknowledgments
We thank W. Hendrickson, L. Kay, and J. Forman-Kay for helpful discussions, and F. Huang and A. Prieto for assistance.
Supported by the National Institutes of Health, American Heart Association, and Whitaker Foundation.
Footnotes
Abbreviations used in this paper: aa, amino acid(s); EC, excitation–contraction; FKBP, FK506 binding protein; GST, glutathione S-transferase; IP3R, inositol 1,4,5 trisphosphate receptor; LZ, leucine/isoleucine zipper; mAKAP, muscle A-kinase anchoring protein; PKA, protein kinase A; PKI, protein kinase inhibitor; PP, protein phosphatase; RyR, ryanodine receptor; SR, sarcoplasmic reticulum.
References
- Alber T., Bell J.A., Sun D.P., Nicholson H., Wozniak J.A., Cook S., Matthews B.W. Replacements of Pro86 in phage T4 lysozyme extend an alpha-helix but do not alter protein stability. Science. 1988;239:631–635. doi: 10.1126/science.3277275. [DOI] [PubMed] [Google Scholar]
- Allen P., Ouimet C., Greengard P. Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc. Natl. Acad. Sci. USA. 1997;94:9956–9961. doi: 10.1073/pnas.94.18.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo R., Rubin C.S. Molecular characterization of an anchor protein (AKAPCE) that binds the RI subunit (RCE) of type I protein kinase A from Caenorhabditis elegans . J. Biol. Chem. 1998;273:14633–14643. doi: 10.1074/jbc.273.23.14633. [DOI] [PubMed] [Google Scholar]
- Barlow D.J., Thornton J.M. Helix geometry in proteins. J. Mol. Biol. 1988;201:601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- Brillantes A.B., Ondrias K., Scott A., Kobrinsky E., Ondriasova E., Moschella M.C., Jayaraman T., Landers M., Ehrlich B.E., Marks A.R. Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell. 1994;77:513–523. doi: 10.1016/0092-8674(94)90214-3. [DOI] [PubMed] [Google Scholar]
- Brown J.H., Cohen C., Parry D.A.D. Heptad breaks in α-helical coiled coilsstutters and stammers. Proteins Struct. Funct. Genet. 1996;26:134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Carr D.W., Stofko-Hahn R.E., Fraser I.D., Bishop S.M., Acott T.S., Brennan R.G., Scott J.D. Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J. Biol. Chem. 1991;266:14188–14192. [PubMed] [Google Scholar]
- Chang D.-K., Cheng S.-F., Trivedi V.D., Lin K.-L. Proline affects oligomerization of a coiled coil by inducing a kink in a long helix. J. Struct. Biol. 1999;128:270–279. doi: 10.1006/jsbi.1999.4182. [DOI] [PubMed] [Google Scholar]
- Franck J.P.C., Morrissette J., Keen J.E., Londraville R.L., Beamsley M., Block B.A. Cloning and characterization of fiber type-specific ryanodine receptor isoforms in skeletal muscles of fish. Am. J. Physiol. 1998;275:C401–C415. doi: 10.1152/ajpcell.1998.275.2.C401. [DOI] [PubMed] [Google Scholar]
- Galvan D.L., Borrego-Diaz E., Perez P.J., Mignery G.A. Subunit oligomerization, and topology of the inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 1999;274:29483–29492. doi: 10.1074/jbc.274.41.29483. [DOI] [PubMed] [Google Scholar]
- Hakamata Y., Nakai J., Takeshima H., Imoto K. Primary structure and distribution of a novel ryanodine receptor/calcium release channel from rabbit brain. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1992;312:229–235. doi: 10.1016/0014-5793(92)80941-9. [DOI] [PubMed] [Google Scholar]
- Harbury P.B., Zhang T., Kim P.S., Alber T. A switch between two-, three- and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Hendrickson W.A., Love W.E. Structure of lamprey haemoglobin. Nat. New Biol. 1971;232:197–203. doi: 10.1038/newbio232197a0. [DOI] [PubMed] [Google Scholar]
- Hsieh-Wilson L.C., Allen P.B., Watanabe T., Nairn A.C., Greengard P. Characterization of the neuronal targeting protein spinophilin and its interaction with protein phosphatase-1. Biochemistry. 1999;38:4365–4373. doi: 10.1021/bi982900m. [DOI] [PubMed] [Google Scholar]
- Hunter T. Protein kinases and phosphatasesthe yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- Huser J., Wang Y.G., Sheehan K.A., Cifuentes F., Lipsius S.L., Blatter L.A. Functional coupling between glycolysis and excitation-contraction coupling underlies alternans in cat heart cells. J. Physiol. 2000;524:795–806. doi: 10.1111/j.1469-7793.2000.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman T., Brillantes A.-M.B., Timerman A.P., Erdjument-Bromage H., Fleischer S., Tempst P., Marks A.R. FK506 binding protein associated with the calcium release channel (ryanodine receptor) J. Biol. Chem. 1992;267:9474–9477. [PubMed] [Google Scholar]
- Joiner W.J., Wang L.-Y., Tang M.D., Kaczmarek L.K. hSK4, a member of a novel subfamily of calcium-activated potassium channels. Proc. Natl. Acad. Sci. USA. 1997;94:11013–11118. doi: 10.1073/pnas.94.20.11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass R.S., Davies M.P. The roles of ion channels in an inherited heart diseasemolecular genetics of the long QT syndrome. Cardiovasc. Res. 1996;32:443–454. [PubMed] [Google Scholar]
- Koch C.A., Anderson D., Moran M.F., Ellis C., Pawson T. SH2 and SH3 domainselements that control interactions of cytoplasmic signaling proteins. Science. 1991;252:668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- Landschulz W.H., Johnson P.F., McKnight S.L. The leucine zippera hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lupas A. Coiled coilsnew structures and new functions. TIBS (Trends Biochem. Sci.). 1996;21:375–382. [PubMed] [Google Scholar]
- Marks A.R. Cellular functions of immunophilins. Physiol. Rev. 1996;76:631–649. doi: 10.1152/physrev.1996.76.3.631. [DOI] [PubMed] [Google Scholar]
- Marks A.R. Cardiac intracellular calcium release channelsrole in heart failure. Circ. Res. 2000;87:8–11. doi: 10.1161/01.res.87.1.8. [DOI] [PubMed] [Google Scholar]
- Marks A.R., Tempst P., Hwang K.S., Taubman M.B., Inui M., Chadwick C., Fleischer S., Nadal-Ginard B. Molecular cloning and characterization of the ryanodine receptor/junctional channel complex cDNA from skeletal muscle sarcoplasmic reticulum. Proc. Natl. Acad. Sci. 1989;86:8683–8687. doi: 10.1073/pnas.86.22.8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx S.O., Ondrias K., Marks A.R. Coupled gating between individual skeletal muscle Ca2+ release channels (ryanodine receptors) Science. 1998;281:818–821. doi: 10.1126/science.281.5378.818. [DOI] [PubMed] [Google Scholar]
- Marx S.O., Reiken S., Hisamatsu Y., Jayaraman T., Burkhoff D., Rosemblit N., Marks A.R. PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor)defective regulation in failing hearts. Cell. 2000;101:365–376. doi: 10.1016/s0092-8674(00)80847-8. [DOI] [PubMed] [Google Scholar]
- Maryon E.B., Coronado R., Anderson P. unc-68 encodes a ryanodine receptor involved in regulating C. elegans body-wall muscle contraction. J. Cell Biol. 1996;134:885–893. doi: 10.1083/jcb.134.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAvoy T., Allen P.B., Obaishi H., Nakanishi H., Takai Y., Greengard P., Nairn A.C., Hemmings H.C., Jr. Regulation of neurabin I interaction with protein phosphatase 1 by phosphorylation. Biochemistry. 1999;38:12943–12949. doi: 10.1021/bi991227d. [DOI] [PubMed] [Google Scholar]
- McCormack K., Tanouye M.A., Iverson L.E., Lin J.W., Ramaswami M., McCormack T., Campanelli J.T., Mathew M.K., Rudy B. A role for hydrophobic residues in the voltage-dependent gating of Shaker K+ channels. Proc. Natl. Acad. Sci. USA. 1991;88:2931–2935. doi: 10.1073/pnas.88.7.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra J., Szilak L., Krylov D., Vinson C. Leucine is the most stabilizing aliphatic amino acid in the d position of a dimeric leucine zipper coiled coil. Biochemistry. 1997;36:22567–22573. doi: 10.1021/bi971424h. [DOI] [PubMed] [Google Scholar]
- Nakai J., Imagawa T., Hakamata Y., Shigekawa M., Takeshima H., Numa S. Primary structure and functional expression from cDNA of the cardiac ryanodine receptor/calcium release channel. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1990;271:169–177. doi: 10.1016/0014-5793(90)80399-4. [DOI] [PubMed] [Google Scholar]
- Otsu K., Willard H.F., Khanna V.K., Zorzato F., Green N.M., MacLennan D.H. Molecular cloning of cDNA encoding the Ca2+ release channel (ryanodine receptor) of rabbit cardiac muscle sarcoplasmic reticulum. J. Biol. Chem. 1990;265:13472–13483. [PubMed] [Google Scholar]
- Oyamada H., Murayama T., Takagi T., Iino M., Iwabe N., Miyata T., Ogawa Y., Endo M. Primary structure and distribution of ryanodine-binding protein isoforms of the bullfrog skeletal muscle. J. Biol. Chem. 1994;269:17206–17214. [PubMed] [Google Scholar]
- Pawson T., Gish G.D. SH2 and SH3 domainsfrom structure to function. Cell. 1992;71:359–362. doi: 10.1016/0092-8674(92)90504-6. [DOI] [PubMed] [Google Scholar]
- Simmerman H.K., Kobayashi Y.M., Autry J.M., Jones L.R. A leucine zipper stabilizes the pentameric membrane domain of phospholamban and forms a coiled-coil pore structure. J. Biol. Chem. 1996;271:5941–5946. doi: 10.1074/jbc.271.10.5941. [DOI] [PubMed] [Google Scholar]
- Surks H.K., Mochizuki N., Kasai Y., Georgescu S.P., Tang K.M., Ito M., Lincoln T.M., Mendelsohn M.E. Regulation of myosin phosphatase by a specific interaction with cGMP- dependent protein kinase I alpha. Science. 1999;286:1583–1587. doi: 10.1126/science.286.5444.1583. [DOI] [PubMed] [Google Scholar]
- Takeshima H., Nishimura S., Matsumoto T., Ishida H., Kangawa K., Minamino N., Matsuo H., Ueda M., Hanaoka M., Hirose T., Numa S. Primary structure and expression from complementary DNA of skeletal muscle ryanodine receptor. Nature. 1989;339:439–445. doi: 10.1038/339439a0. [DOI] [PubMed] [Google Scholar]
- Takeshima H., Nishi M., Iwabe N., Miyata T., Hosoya T., Masai I., Hotta Y. Isolation and characterization of a gene for a ryanodine receptor/calcium release channel in Drosophila melanogaster . FEBS (Fed. Eur. Biochem. Soc.) Lett. 1994;337:81–87. doi: 10.1016/0014-5793(94)80634-9. [DOI] [PubMed] [Google Scholar]
- Tripet B., Wagschal K., Lavigne P., Mant C.T., Hodges R.S. Effects of side-chain characteristics on stability and oligomerization state of a de novo-designed model coiled-coil20 amino acid substitutions in position “d”. J. Mol. Biol. 2000;300:377–402. doi: 10.1006/jmbi.2000.3866. [DOI] [PubMed] [Google Scholar]
- Turner R., Tjian R. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science. 1989;243:1689–1694. doi: 10.1126/science.2494701. [DOI] [PubMed] [Google Scholar]
- Wagschal K., Tripet B., Lavigne P., Mant C., Hodges R.S. The role of position a in determining the stability and oligomerization state of alpha-helical coiled coils20 amino acid stability coefficients in the hydrophobic core of the proteins. Protein Sci. 1999;8:2312–2329. doi: 10.1110/ps.8.11.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Bhat M.B., Nishi M., Takeshima H., Ma J. Molecular cloning of cDNA encoding a drosophila ryanodine receptor and functional studies of the carboxyl-terminal calcium release channel. Biophys. J. 2000;78:1270–1281. doi: 10.1016/S0006-3495(00)76683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Hsieh-Wilson L., Feng J., Tomizawa K., Allen P.B., Fienberg A.A., Nairn A.C., Greengard P. Protein phosphatase 1 modulation of neostriatal AMPA channelsregulation by DARPP-32 and spinophilin. Nat. Neurosci. 1999;2:13–17. doi: 10.1038/4516. [DOI] [PubMed] [Google Scholar]
- Zeke T., Gergely P., Dombradi V. The catalytic subunits of Ser/Thr protein phosphatases from Caenorhabditis elegans . Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1998;119:317–324. doi: 10.1016/s0305-0491(97)00341-6. [DOI] [PubMed] [Google Scholar]
- Zolnierowicz S., Van Hoof C., Andjelkovic N., Cron P., Stevens I., Merlevede W., Goris J., Hemmings B.A. The variable subunit associated with protein phosphatase 2A0 defines a novel multimember family of regulatory subunits. Biochem. J. 1996;317:187–194. doi: 10.1042/bj3170187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorzato F., Fujii J., Otso K., Phillips M., Green N.M., Lai F.A., Meissner G., MacLennan D.H. Molecular cloning of cDNA encoding human and rabbit forms of the Ca2+ release channel (ryanodine receptor) of skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 1990;265:2244–2256. [PubMed] [Google Scholar]