The AIDS pandemic is spreading unchecked in many parts of the world. Currently available anti-viral regimens are plagued by their high cost, by serious side effects in a significant minority of recipients, and by the increasing problems of drug-resistant HIV variants. Fortunately, rapid advances in understanding how HIV-1 enters cells have lead to the identification of promising new drug targets and have also suggested new strategies for the generation of vaccine candidates. The cellular proteins required for HIV-1 entry have been identified, and structural studies using fragments of the viral envelope protein (Env) that mediates entry have provided views of Env in two conformations. However, placing this information in the appropriate order and kinetic context has not been so straightforward. A paper published in this issue identifies the rate limiting step in Env-mediated membrane fusion and the proximal cause of the membrane fusion reaction (Melikyan et al. 2000). This study provides an important insight into why antibodies to several immunodominant HIV-1 Env epitopes fail to neutralize the virus, information that may help in the development of triggered Env immunogens as vaccine candidates, as well as in the identification of new inhibitors of virus entry. Two additional papers provide new knowledge on the structural properties needed to induce lipid mixing in both viral and cellular membrane fusion systems, together highlighting the essential similarities between the viral and cellular systems (Armstrong et al. 2000; Grote et al. 2000).
The general steps of HIV-1 entry have been elucidated through direct studies of this virus, as well as of better characterized viruses like influenza that elicit membrane fusion using a similar mechanism. HIV-1, like many other viruses, is surrounded by a lipid membrane from which protrudes a virally encoded type I membrane protein (Env). The membrane of the virus and that of the cell present a formidable physical and energetic barrier between the viral genome and the cytoplasm of the host cell. To gain entry, all enveloped viruses mediate a membrane fusion reaction such that their lipid bilayers become contiguous with that of a cellular membrane (Hernandez et al. 1996). This process is invariably mediated by a viral fusion protein, such as HIV-1 Env. This homotrimeric protein is initially synthesized as a single polypeptide precursor that is posttranslationally cleaved into a surface subunit (gp120) that mediates receptor binding and that remains noncovalently attached to a transmembrane domain subunit (gp41; Wyatt and Sodroski 1998). Cleavage liberates the NH2-terminal domain of gp41, a region that constitutes the protein's fusion peptide, a stretch of conserved hydrophobic residues that inserts into the membrane of the host cell during the course of membrane fusion. As such, the cleavage event is a prerequisite for viral infectivity.
For Env to mediate membrane fusion, it must receive a signal that causes it to undergo dramatic conformational rearrangements. For many enveloped viruses, the trigger that results in the fusion-inducing conformational changes is acid pH. As first described in 1980 (Helenius et al. 1980), virus bound to the cell surface is internalized and delivered to endosomes. There, the acidic environment results in protonation of acidic residues in the fusion protein, making the necessary conformational changes possible. Acid-activated viruses include influenza virus and Semliki Forest virus, which have long served as model systems for studying virus-membrane fusion. Other viruses, such as HIV-1, are pH-independent. Here, the information needed to trigger conformational changes results directly from receptor binding. The primary receptor for HIV-1 is the CD4 antigen, to which it binds via the gp120 subunit of Env. This causes structural alterations in gp120, enabling it to subsequently bind to a second receptor, termed a coreceptor. Coreceptor binding is thought to be the final trigger that leads to membrane fusion. All HIV-1 strains use one or both of the seven transmembrane domain chemokine receptors, CCR5 and CXCR4, as coreceptors in conjunction with CD4 for virus entry (Doms et al. 1998). The differential use of these receptors, coupled with their patterns of expression, largely dictate the cell types that are susceptible to virus infection in vivo. Individuals who lack CCR5 due to a deletion in the CCR5 open reading frame are highly resistant to virus infection, but are immunologically normal and healthy (Liu et al. 1996; Samson et al. 1996), indicating that CCR5 antagonists may provide significant protection from virus infection. Indeed, small molecule antagonists of both CCR5 and CXCR4 have been developed, and some have entered clinical trials.
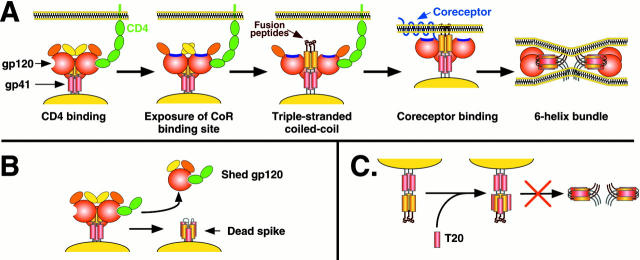
The most widely accepted model describing HIV-1 Env-mediated membrane fusion posits that either CD4 or coreceptor binding results in the formation of a coiled-coil in gp41. This is composed of three NH2-terminal leucine/isoleucine zipper regions, one contributed by each subunit of the Env trimer (Chan et al. 1997; Weissenhorn et al. 1997). The NH2-terminal fusion peptide is thereby displaced in the direction of the target membrane into which it can insert (Fig. 1). As a result, Env transiently becomes an integral component of two membranes: the viral membrane in which it is anchored, and the cellular membrane that it has gaffed. The exterior surface of the coiled-coil contains grooves into which pack a second, more COOH-terminally oriented heptad repeat region of gp41. In other words, the gp41 subunit folds back on itself, forming an exceptionally stable six-helix bundle (first shown for influenza HA; Bullough et al. 1994) in which the fusion peptide and transmembrane domain of gp41 are now oriented at the same end of the molecule (Chan et al. 1997; Weissenhorn et al. 1997). Given the stability of this structure, it is likely that the six-helix bundle represents the terminal conformation of a fusogenic Env. Despite considerable differences in primary sequence, many triggered viral fusion proteins share a common core structure involving a six-helix bundle that has the membrane-associated domains at the same end. This indicates that many viral, and perhaps cellular, proteins induce membrane fusion by essentially the same mechanism (Chan and Kim 1998; Skehel and Wiley 1998).
Figure 1.
A, Model for HIV-1 Env membrane fusion. Binding of CD4 to the gp120 subunit of Env induces exposure of a conserved region in gp120 implicated in coreceptor binding (purple; Rizzuto et al. 1998). In addition, CD4 binding appears to trigger exposure of the triple-stranded coiled-coil, and presumably exposure of the fusion peptide, although coreceptor binding could increase the efficiency and kinetics of this process. It is not known if the more COOH-terminal helical region in each gp41 subunit (red) interact with each other as drawn, but it is known that the extodomain of gp41 in general plays an important role in mediating Env oligomerization. Binding to coreceptor could bring Env in closer proximity to the target membrane, enabling the fusion peptide to insert in the bilayer, or it could impact formation of the six-helix bundle, the transition to which leads to membrane fusion. Note that in the six-helix bundle, the NH2-terminal helices form the core of the helix, with the COOH-terminal helices packing in the grooves on the outside of the structure. It is not known if gp120 remains associated with gp41 throughout the fusion process. B, Formation of dead spikes. Binding of soluble CD4 to Env can induce shedding of gp120 from gp41, and can even induce formation of the six-helix bundle. A similar process is likely to occur at the cell surface. Such modified Env proteins are not fusogenic, but may serve as immunologic decoys. C, Inhibition of fusion by T20. T20 is a small peptide based on the COOH-terminal helical region in gp41. It binds to the grooves on the outside of the triple-stranded coiled-coil formed by the NH2-terminal helices. Therefore, it prevents transition to the six-helix bundle and membrane fusion. Only gp41 is depicted for clarity.
Despite all of this knowledge, it has not been clear what the rate-limiting step of fusion is, whether gp120 remains associated to gp41 during fusion, what are the exact roles of the fusion peptide and transmembrane domains, and whether the stable six-helix bundle is the cause or the result of membrane fusion. In addition, it is likely that several Env trimers are needed for a fusion pore to form, and multiple receptor binding events are needed per trimer to activate their fusion potential, resulting in an additional layer of complexity that is poorly understood.
Melikyan et al. 2000 have used a clever series of interventional strategies to address several of these lacunae. Their approach has been to block fusion at discrete stages, to identify the rate limiting steps in membrane fusion and the order in which they occur. When cells expressing HIV-1 Env are mixed with cells bearing CD4 and an appropriate coreceptor, cell–cell fusion commences, but only after a lag phase of 15-20 min. This makes sense in light of how HIV-1 Env is triggered: unlike acid-activated viruses in which triggering occurs rapidly and synchronously, so that all viral envelope proteins are activated within a short period, HIV-1 Env triggering is apt to be a much slower and stochastic process since it is dependent on receptor binding. Fusion is a highly cooperative process requiring multiple Env proteins and multiple receptor binding events are needed to activate individual Env trimers, and these probably occur over a period of time. Hence, we can predict that the rate of Env triggering will be dependent, in part, on receptor density, and will perhaps also be influenced by the Env-receptor affinity. This asynchrony represents a target of opportunity, because highly conserved regions of Env that are actual or potential targets for antiviral agents are transiently exposed during the course of fusion. For example, a peptide based on the second helical region in gp41 is a potent inhibitor of HIV-1 membrane fusion, both in vitro and in vivo (Kilby et al. 1998). This peptide, termed T20, binds to the exposed grooves on the surface of the triple stranded coiled-coil, preventing formation of the six-helix bundle (Fig. 1). Thus, T20 does not target native Env, but rather a structural intermediate of the fusion process.
An earlier study, confirmed by Melikyan et al. 2000, showed that HIV-1 Env-mediated fusion is temperature dependent (Frey et al. 1995). When cells expressing Env are mixed with target cells at 23°C, fusion does not occur, even after a very long time. Upon warming to 37°C, fusion commences, but without the lag phase that is normally seen with HIV-1 Env-mediated membrane fusion. Clearly, the steps responsible for the lag-phase must be able to occur at 23°C. Therefore, the incubation with receptor-positive cells at 23°C has already allowed Env to achieve an intermediate state. The key finding provided by Melikyan et al. 2000 is that the intermediate state is sensitive to T20, indicating that the triple-stranded coiled coil is accessible under these conditions. This study also supports the idea that this activated form of Env can be achieved by binding to CD4 alone: coreceptor binding is not needed (Furuta et al. 1998). Indeed, it has long been known that the addition of sCD4 to virions, in the absence of coreceptors, is sufficient to cause extensive conformational changes in Env, even leading to the outright dissociation of the gp120 subunits and exposure of the six-helix bundle form of gp41. This latter event, however, does not cause membrane fusion to occur. Indeed, quite the opposite, because fusion is inhibited by sCD4 under these conditions. What role, then, does coreceptor-binding play in the fusion process? One possibility is that the coreceptor association of gp120 accelerates the transition in Env to its six-helix bundle form, and thereby ensures that this event occurs at the correct point in time and space to allow membrane fusion. If the transition were to take place immediately upon CD4 binding, the fusion peptide may be physically too far from the target cell membrane to permit its proper insertion. A fusion peptide left waving in the breeze would be of little use to the virus. However, the coreceptors are flush with the cell membrane, so a rapid triggering of the Env transition when gp120 binds to the coreceptor may ensure that the fusion peptide is close enough to penetrate the target cell when the gaffing process is initiated. In fishing terms, CD4-binding may be analogous to the cocking of the harpoon gun, but coreceptor-binding is the targeting radar that enables the shooter to pull the trigger only when a fish is actually in sight. Eventually, the CD4-Env complex will undergo conformational transitions in the absence of a coreceptor, but this will usually not lead to a successful insertion of the fusion peptide into the target cell membrane: the harpoon will miss the fish. In any event, it appears that the rate-limiting steps in Env-mediated membrane fusion are related to the conformational changes induced by receptor binding that result in the formation of the triple-stranded coiled-coil and/or lateral aggregation of Env-receptor complexes, with the transition to the six-helix bundle occurring the most quickly.
What role does the six-helix bundle play in membrane fusion? By modifying membrane curvature through the addition of exchangeable lipids, Melikyan et al. 2000 show that formation of the six-helix bundle is coincident with membrane fusion. Thus, formation of the six-helix bundle can be considered the proximal cause of the membrane fusion event, and the energy transition involved in the formation of the six-helix bundle must be sufficient for a fusion pore to form. As a result, the binding of CD4 and a coreceptor to HIV-1 Env releases sufficient free energy, such that membrane fusion can occur. But this energy must be released and used at the right time for it to drive membrane fusion; it is not the presence of the six-helix bundle that allows fusion to occur, it is the formation of the six-helix bundle that allows fusion to occur. To return to the gaffing analogy: if one fires a harpoon at a fish, it must be fired at the right time and place. Merely having a harpoon dangling in the waves is unlikely to work, since even fish possess the intelligence not to voluntarily impale themselves on a static object.
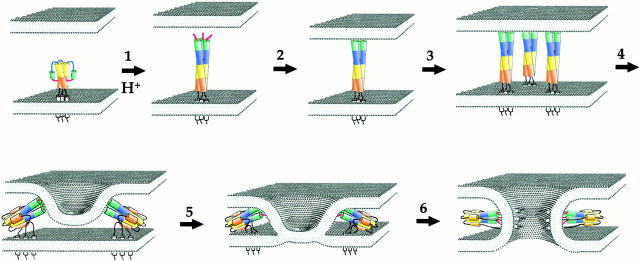
The papers by Armstrong et al. 2000 and Grote et al. 2000 do not directly address the mechanisms of HIV-1 mediated membrane fusion, but instead focus on the influenza hemagglutinin (HA) protein and the soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) fusion system, respectively, the best understood viral and cellular fusion machines. Like HIV-1 Env, HA is a homotrimeric protein in which each monomer consists of a surface (HA1) and transmembrane domain subunit (HA2). At acid pH, the fusion peptide at the NH2 terminus of the membrane-spanning HA2 subunit is first exposed. Then, through formation of a triple-stranded coiled coil, it becomes displaced in the direction of the target membrane into which it inserts (Fig. 2). Several studies have shown that three to six HA trimers are needed to form a fusion pore, indicating that lateral aggregation of HA trimers must occur (Hernandez et al. 1996). Structural studies have shown that the base of the triple stranded coiled-coil folds back on itself, forming the now familiar six-helix bundle (Chen et al. 1999). In doing so, the fusion peptide and transmembrane domains are brought in close proximity, which necessarily brings the target and viral membranes close to each other as well. Fusion mediated by HA may proceed through a hemifusion intermediate, in which lipids in only the outer leaflets of each membrane mix (Kemble et al. 1994). As the six-helix bundle continues to form, bringing the fusion peptide and transmembrane domains into close proximity, this may result in disruption of the hemifusion diaphragm and allow a fusion pore to be made (Fig. 2).
Figure 2.
Model for HA-induced membrane fusion. The HA trimer consists of both HA1 (not shown for clarity) and HA2 subunits. The fusion peptides are hidden in the native conformation, being tucked into the trimer interface. Upon acid activation, the fusion peptides are exposed and directed towards the target membrane by a loop to helix transition (blue), thus extending the triple-stranded coiled-coil (1). Insertion of the fusion peptide into the target membrane (2) links the viral and target membranes. Lateral aggregation of several HA trimers (3) is likely needed for a fusion pore to form. Tilting of the HA trimers may be accomplished by folding of the base of triple stranded coiled-coil upon itself, forming a six-helix bundle, as has been seen for other viral fusion proteins (4). This may bring the membranes in close enough proximity such that hemifusion occurs (5). If the HA trimer is GPI-anchored or if the transmembrane domain is not of sufficient length to span the bilayer, the fusion process does not extend past this point. However, if the TM domain is well anchored in the viral membrane, fusion occurs, and, judging by the work by Melikyan et al. 2000(this issue) may be coincident with the completion of the six-helix bundle that brings the TM domain and fusion peptides into close proximity (6).
Whereas most studies to date have focused on how the ectodomains of viral fusion proteins elicit membrane fusion, it has been clear that the transmembrane domain also plays an important, though poorly defined, role. Substitution of HA's TM domain with a GPI-anchor, for example, does not obviously affect the structure or function of the influenza HA ectodomain. However, GPI-linked HA elicits only hemifusion (Kemble et al. 1994). Under these conditions, fusion pores either do not form, or form slowly and inefficiently. What, then, is the role of the TM domain? Through a thorough and careful series of experiments, Armstrong et al. 2000 show that the TM domain must completely span the lipid bilayer for fusion to occur. If it does not, only hemifusion takes place. Whereas the reason for this is not clear, it is possible that the TM domain must be firmly anchored in the membrane to apply enough tension to the hemifusion diaphragm to promote pore formation. Since the transition to the six-helix bundle is coincident with membrane fusion, the TM domain, and perhaps also the fusion peptide, must be tightly membrane-associated to bring the viral and cellular membranes into close proximity. Thus, the TM domain plays an important role in the fusion process. It will be interesting to determine if the presence of a GPI-anchor or a short TM domain prevents complete formation of the six-helix bundle, or if formation of the six-helix bundle results in extraction of membrane domains that do not span the lipid bilayer, thereby preventing lipid mixing. In this regard, it is notable that some mutations in the fusion peptide allow only hemifusion to occur (Qiao et al. 1999).
While wonderful creatures to study, it must be admitted that viruses are frequently guilty of plagiarism, stealing good ideas from unsuspecting cells for their own nefarious purposes. It is perhaps not surprising then that cellular and viral fusion machines share some features in common. To cause fusion, v-SNAREs that are anchored in a vesicle membrane and t-SNAREs that are anchored in the target membrane associate and form a four-helix bundle that is thought to force the two membranes together. The paper by Grote et al. 2000 shows that substitution of the t-SNARE membrane domains with a gernylgernylation signal inhibits exocytosis after vesicle docking. In fact, this lipid modification proved to be a dominant inhibitor of membrane fusion, consistent with a need for cooperative interactions between multiple SNARE complexes. Fusion could be rescued by addition of lysophosphatidylcholine, a cone-shaped lipid that induces membrane curvature. Altering membrane curvature has been shown to induce full membrane fusion by GPI-HA, so perhaps the activity of the lipid-anchored t-SNARE is blocked at the hemifusion stage. Thus, much like GPI-anchored HA, the presence of a lipid anchor that fails to span the membrane is not compatible with membrane fusion. The papers by Armstrong et al. 2000, Grote et al. 2000, and others (Qiao et al. 1999; McNew et al. 2000) indicate that for membrane fusion to occur, fusion proteins must have both feet firmly planted in, and not just on, the ground.
As the details of the virus entry process have been revealed, it is apparent that structural intermediates of the fusion process represent attractive drug targets. The HIV-1 Env protein is well armored to resist humoral immunity: nearly half of its mass is contributed by N-linked carbohydrate chains that shield antibody epitopes, and surface accessible regions of the protein tend to consist of variable regions. This makes the development of broadly neutralizing antibodies a challenge. The Env protein does contain highly conserved, functionally important domains since all Env proteins bind CD4 and a coreceptor, then cause membrane fusion. Unfortunately, these structures either are not, or may not be, accessible to antibodies until the receptors have been engaged. Structural intermediates of the fusion process may result in the exposure of conserved domains that could be potential drug or vaccine targets. However, even here, there may be little time for an antibody to intervene or only a limited physical space in which it can do so, given the steric constraints of a forming fusion complex. The work by Melikyan et al. 2000 indicates that binding of CD4 to trimeric Env at room temperature is sufficient to not only expose the conserved coreceptor binding site in gp120, but also to render Env susceptible to fusion-inhibiting agents such as T20. The use of coreceptor antagonists may further slow the fusion process by reducing the density of functional coreceptors. This could prolong the exposure of conserved, critically important domains such as those on the surface of the triple stranded coiled-coil. Combination chemotherapy with different classes of entry inhibitors may therefore result in synergistic, and not just additive, inhibition of virus infection.
The study by Melikyan et al. 2000 also helps explain why antibodies to the six-helix bundle are nonneutralizing, yet highly prevalent. One terminology point that needs clarifying as a result of this study is that the six-helix bundle form of gp41 is often referred to as the fusogenic or fusion-competent form of gp41, and nonneutralizing antibodies to this fusion-competent structure have been described (Gorny and Zolla-Pazner 2000). We now know this nomenclature to be incorrect; as outlined above, it is the transition to the six-helix form that drives fusion, not the six-helix form itself. Thus, antibodies to the six-helix form are nonneutralizing because their epitopes are formed coincident with membrane fusion, not before it, so they cannot intervene to stop fusion occurring (i.e., to neutralize the infectivity of the virus). These antibodies are prevalent because the six-helix structure is an antigen that is both stable and accessible on the surface of virions and virus-infected cells. A more accurate terminology for the six-helix form of gp41 is the postfusion form or, colloquially, dead spikes.
Antibodies to such dead spikes are common in the sera of HIV-1-infected humans; the postfusion form of gp41 is immunodominant, but unfortunately irrelevant to an effective virus-neutralizing antibody response (Parren et al. 1999). Thus, some stable Env structures, including monomeric gp120 released from the cell surface, function in part as immunological decoys. These abundant Env-specific antibodies fail to react with the native, fusion-competent trimeric protein, or do so poorly. However, one human anti-gp41 antibody, 2F5, is known to be able to prevent membrane fusion subsequently to virus–cell attachment (Ugolini et al. 1997). It will be important to use the experimental systems outlined by Melikyan et al. 2000 to try to understand at what stage of the fusion process this antibody successfully intervenes. Overall, a goal of vaccine development is to minimize the generation of antibodies to the immunological decoy forms of Env while maximizing the presentation of Env antigens in which the structures important for driving, or actively involved in, the fusion process are exposed. Greater appreciation of the conditions under which Env undergoes its conformational gymnastics, and the requirements for membrane fusion, can only help in identifying targets of opportunity for the use of small molecule inhibitors and in the generation of neutralizing antibodies.
Acknowledgments
The drawing of 2 was kindly provided by Jennifer Gruenke and Judy M. White. More information on the model can be found on Judy White's laboratory web site.
Both authors are supported by Elizabeth Glaser Scientist Awards from the Pediatric AIDS Foundation. R.W. Doms also holds a Burroughs Wellcome Fund Award for Translational Research, and J.P.M. is a Stavros A. Niarchos Scholar. The authors' work in this area is also funded by National Institutes of Health grants AI 40880 and 35383 to R.W. Doms and AI 36082, 41420, and 45463 to J.P. Moore.
Footnotes
Abbreviations used in this paper: Env, viral envelope protein; HA, hemagglutinin; SNARE, soluble N-ethylmaleimide–sensitive factor attachment protein receptor.
References
- Armstrong R.T., Kushnir A.S., White J.M. The transmembrane domain of the influenza virus hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 2000;151:425–437. doi: 10.1083/jcb.151.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullough P.A., Hughson F.M., Skehel J.J., Wiley D.C. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- Chan D.C., Fass D., Berger J.M., Kim P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Chan D.C., Kim P.S. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- Chen J., Skehel J.J., Wiley D.C. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms, R., A. Edinger, and J. Moore. 1998. Coreceptor use by primate lentiviruses. In Human Retroviruses and AIDS. B. Korber, B. Foley, T. Leitner, G. Myers, B. Hahn, F. McCutchan, J. Mellors, and C. Kuiken, editors. Los Alamos National Laboratory. Theoretical Biology and Biophysics, Los Alamos, NM. pp. III-1–III-21.
- Frey S., Marsh M., Gunther S., Pelchen-Matthews A., Stephens P., Ortlepp S., Stegmann T. Temperature dependence of cell–cell fusion induced by the envelope glycoprotein of human immunodeficiency virus type I. J. Virol. 1995;69:1462–1472. doi: 10.1128/jvi.69.3.1462-1472.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta R.A., Wild C.T., Weng Y., Weiss C.D. Capture of an early fusion-active conformation of gp41. Nat. Struct. Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- Gorny M.K., Zolla-Pazner S. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J. Virol. 2000;74:6186–6192. doi: 10.1128/jvi.74.13.6186-6192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote E., Baba M., Ohsumi Y., Novick P.J. Geranylgeranylated SNAREs are dominant inhibitors of membrane fusion. J. Cell Biol. 2000;151:453–465. doi: 10.1083/jcb.151.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A., Kartenbeck J., Simons K., Fries E. On the entry of semliki forest virus into BHK-21 cells. J. Cell Biol. 1980;84:404–420. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez L.D., Hoffman L.R., Wolfsberg T.G., White J.M. Virus–cell and cell–cell fusion. Annu. Rev. Dev. Cell. Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- Kemble G.W., Danieli T., White J.M. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- Kilby J.M., Hopkins S., Venetta T.M., DiMassimo B., Cloud G.A., Lee J.Y., Alldredge L., Hunter E., Lambert D., Bolognesi D. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- Liu R., Paxton W.A., Choe S., Ceradini D., Martin S.R., Horuk R., MacDonald M.E., Stuhlmann H., Koup R.A., Landau N.R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- McNew J.A., Weber T., Parlati F., Johnson R.J., Melia T.J., Sollner T.H., Rothman J.E. Close is not enoughSNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J. Cell Biol. 2000;150:105–117. doi: 10.1083/jcb.150.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan G.B., Markosyan R.M., Hemmati H., Delmedico M.K., Lambert D.M., Cohen F.S. The transition of HIV-1 gp41 into a six-helix bundle configuration induces membrane fusion. J. Cell Biol. 2000;151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren P.W., Moore J.P., Burton D.R., Sattentau Q.J. The neutralizing antibody response to HIV-1viral evasion and escape from humoral immunity. AIDS. 1999;13:S137–S162. [PubMed] [Google Scholar]
- Qiao H., Armstrong R.T., Melikyan G.B., Cohen F.S., White J.M. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol. Biol. Cell. 1999;10:2759–2769. doi: 10.1091/mbc.10.8.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto C.D., Wyatt R., Hernandez-Ramos N., Sun Y., Kwong P.D., Hendrickson W.A., Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- Samson M., Libert F., Doranz B.J., Rucker J., Liesnard C., Farber C.-M., Saragosti S., Lapoumèroulie C., Cogniaux J., Forceille C. Resistance to HIV-1 infection of caucasian individuals bearing mutant alleles of the CCR5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- Skehel J.J., Wiley D.C. Coiled coils in both intracellular and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- Ugolini S., Mondor I., Parren P., Burton D., Tilley S., Klasse P.J., Sattentau Q.J. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J. Exp. Med. 1997;186:1287–1298. doi: 10.1084/jem.186.8.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W., Dessen A., Harrison S.C., Skehel J.J., Wiley D.C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- Wyatt R., Sodroski J. The HIV-1 envelope glycoproteinsfusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]