Mitochondria are dynamic structures that divide and fuse continually throughout the life of a cell. Mitochondrial division is needed to maintain a full complement of mitochondria when cells divide, but also when they differentiate, increase in size or respond to changes in their environment. Despite a huge body of literature on many aspects of mitochondrial function, very little is known about the mechanisms that control mitochondrial division. This issue contains four papers describing novel proteins that contribute to mitochondrial morphology. Before discussing those new proteins, a short introduction to what was already known about the mechanisms of mitochondrial division is in order.
How Does the Mitochondrial Outer Membrane Divide?
Last year, it was discovered that a dynamin-related protein, called Dnm1 in yeast, and the homologous protein from C. elegans, called DRP-1, contribute to a late stage in the mitochondrial division process (Bleazard et al. 1999; Labrousse et al. 1999; Sesaki and Jensen 1999). Several lines of evidence support this conclusion. Mutations in yeast Dnm1 and C. elegans DRP-1 block mitochondrial division. The defect in mitochondrial division causes a shift in the balance between fission and fusion, resulting in a network of interconnected mitochondria (Bleazard et al. 1999; Sesaki and Jensen 1999). Severing of the mitochondrial inner membrane can still occur in Caenorhabditis elegans, even when severing of the mitochondrial outer membrane is blocked, indicating that DRP-1 acts on the outer membrane (Labrousse et al. 1999). Conversely, the overexpression of wild-type DRP-1 in C. elegans causes an increase in the rate of mitochondrial division (Labrousse et al. 1999). Immunoelectron microscopy shows that yeast Dnm1 colocalizes with constrictions in mitochondria (Bleazard et al. 1999) and time-lapse photography shows that C. elegans DRP-1 is localized in spots on mitochondria where division actually occurs (Labrousse et al. 1999). The appearance of DRP-1 spots before division and their disappearance after division suggests that DRP-1 cycles on and off of mitochondria, similar to the cycling of dynamin between cytosol and the plasma membrane (Labrousse et al. 1999). Therefore, it is likely that Dnm1 and DRP-1 are required to sever the mitochondrial outer membrane in yeast and C. elegans, respectively.
The function of Drp1 in mammalian cells is still not settled. It has been hypothesized that mammalian Drp1, also known as Dlp1, Dvlp1, or Dymple, might mediate vesicle formation, similar to the role of genuine dynamins in the scission of clathrin-coated vesicles (Pitts et al. 1999). However, mutations in mammalian Drp1 specifically affect mitochondrial morphology without affecting the secretory or endocytic pathways (Smirnova et al. 1998). The affected mitochondria form perinuclear clumps, masking a possible defect in mitochondrial division. The latest evidence shows that mitochondria in these clumps are much more interconnected than in wild-type cells, consistent with a defect in mitochondrial division (Smirnova, E., and A.M. van der Bliek, unpublished observations). It is therefore likely that the role of Dnm1/Drp1 is conserved throughout evolution. Analogy to the role of genuine dynamins in the final stages of vesicle formation suggests that Dnm1/Drp1 contributes to mitochondrial division by wrapping around constricted parts of mitochondria where it helps to sever the mitochondrial outer membrane.
Novel Factors Contributing to Mitochondrial Outer Membrane Division
Three of the papers in this issue describe novel factors that may interact with yeast Dnm1 to help mediate scission of the mitochondrial outer membrane (Fekkes et al. 2000; Mozdy et al. 2000; Tieu and Nunnari 2000). These fission genes were isolated as suppressors of defects in mitochondrial fusion. Defects in mitochondrial fusion shift the balance between fission and fusion, causing the mitochondria to become overly fragmented (Bleazard et al. 1999; Sesaki and Jensen 1999). Defective fusion results in loss of mitochondria, because the overly fragmented mitochondria fail to segregate properly. The loss of mitochondria can be prevented by balancing the fusion defect with the block in mitochondrial division caused by mutations in Dnm1. Based on this precedent, Tieu and Nunnari 2000 and Mozdy et al. 2000 looked independently for suppressors of Fzo1 mutants, which affect mitochondrial fusion. Fekkes et al. 2000 isolated their mutants as suppressors of an Mgm1 mutant, which also induces mitochondrial fragmentation. The newly identified second site suppressors completely reverse the excessive fragmentation of mitochondria caused by mutations in Mgm1 or Fzo1, resulting in the formation of closed nets of interconnected mitochondria (Fekkes et al. 2000; Mozdy et al. 2000; Tieu and Nunnari 2000). These closed nets are indicative of a strong defect in mitochondrial division. The three groups identified mutations in overlapping sets of genes. Each group identified additional mutations in Dnm1. In addition, however, several new genes were identified as mutants, and two of those genes were characterized in detail. More components of the division machinery may follow from similar genetic screening strategies.
The first of the two new genes encodes a cytosolic protein with a coiled coil domain and seven WD repeats. The three groups called this protein Mdv1 (for mitochondrial division), Fis2 (for fission), and Gag3 (for glycerol adapted growth) (Fekkes et al. 2000; Mozdy et al. 2000; Tieu and Nunnari 2000). The coiled coil domain of Mdv1/Fis2/Gag3 may be required for dimerization, whereas the WD repeat presumably forms a beta propeller structure, which could bind to other proteins. Mdv1/Fis2/Gag3 can bind to Dnm1 in the yeast two-hybrid system, but binding is too weak to detect by immunoprecipitation. Mdv1/Fis2/Gag3 does, however, colocalize with Dnm1 in spots on mitochondria, suggesting that the binding interactions are physiologically relevant (Fekkes et al. 2000; Tieu and Nunnari 2000). Colocalization of Mdv1/Fis2/Gag3 and Dnm1, their ability to bind to each other, and their similar mutant phenotypes indicate that these two proteins are part of the same mitochondrial division apparatus. Deletion of Mdv1/Fis2/Gag3 had no obvious effect on the localization of Dnm1, but deletion of Dnm1 caused Mdv1/Fis2/Gag3 to redistribute evenly along the mitochondria (Fekkes et al. 2000; Tieu and Nunnari 2000). Thus, it would appear that Dnm1 forms the backbone of the complex. Mdv1/Fis2/Gag3 is still localized to mitochondria without Dnm1, but Dnm1 is needed for assembly into the division complex.
The second gene encodes a transmembrane protein that is inserted in the mitochondrial outer membrane. This protein was identified by two groups and was called Fis1 or Mdv2 (Mozdy et al. 2000; Tieu and Nunnari 2000). A large part of Fis1/Mdv2 is facing the cytosol where it could conceivably interact with cytosolic proteins such as Dnm1 and Mdv1/Fis2/Gag3 (Mozdy et al. 2000). This interaction could be transient, because the Fis1/Mdv2 protein is evenly distributed along the surface of the mitochondria, unlike Dnm1 and Mdv1/Fis2/Gag3, which are concentrated in spots where division is about to occur. Also, there is no evidence that Fis1/Mdv2 can bind to Mdv1/Fis2/Gag3 or Dnm1 (Mozdy et al. 2000). Therefore, it is unlikely that Fis1/Mdv2 is incorporated in a stable complex with Dnm1. Mitochondrial division is, nevertheless, impaired by mutations in Fis1/Mdv2. The effects are similar to the effects of mutations in Dnm1 and Mdv1/Fis2/Gag3. Moreover, mutations in Fis1/Mdv2 cause most of the Dnm1 and Mdv1/Fis2/Gag3 proteins to remain cytosolic (Mozdy et al. 2000). What still goes to mitochondria is localized in a few spots that fail to divide. The persistence of these few spots suggests that Fis1/Mdv2 is not the only determinant for mitochondrial localization, although it does make a major contribution. Thus, Fis1/Mdv2 is required for the assembly of a productive mitochondrial division apparatus, but it is not necessarily incorporated into that apparatus.
How might these two new components of the mitochondrial division apparatus cooperate with Dnm1? The Mdv1/Fis2/Gag3 protein could form a link between Dnm1 and other components of the division apparatus, since it has seven WD repeats, which most likely form a protein interaction domain. This raises the question whether the mitochondrial division apparatus has even more components awaiting discovery. The formation of clathrin coated vesicles could serve as a guide, since the usage of dynamin-related proteins during severing of vesicles at the plasma membrane and during division of the mitochondrial outer membrane suggests a common evolutionary origin. It is not difficult to imagine that the first endosymbiotic bacteria used dynamin of the host cell during entry at the plasma membrane and used it again during division of its outer membrane, before eventually giving rise to modern day mitochondria (Fig. 1). Other components of the endocytic machinery might have also been coopted by the mitochondrial division apparatus. Therefore, it would not be surprising if Mdv1/Fis2/Gag3 and Dnm1/Drp1 were part of a larger mitochondrial division apparatus.
Figure 1.
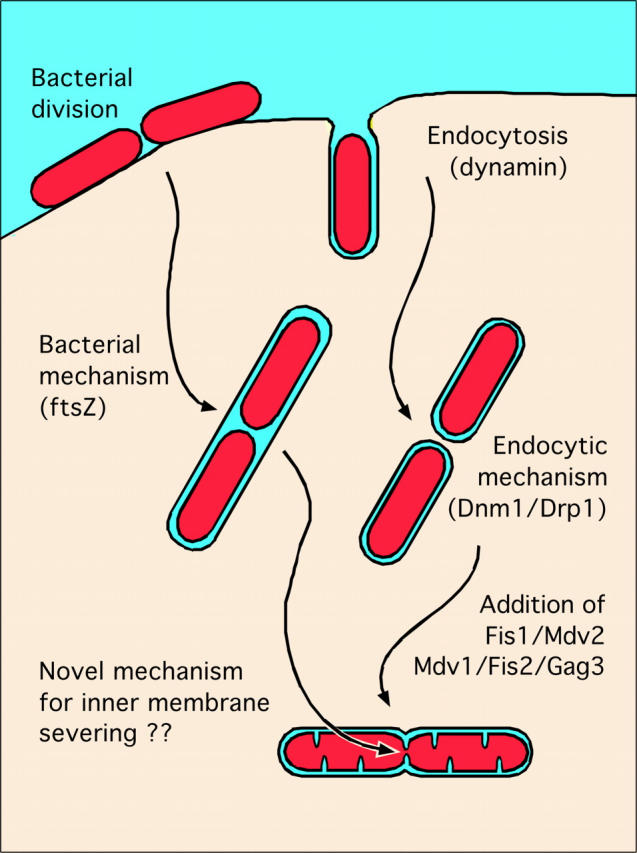
Possible evolutionary origins of the mitochondrial inner and outer membrane division machineries.
The mutant phenotype of Fis1/Mdv2 clearly shows that this protein is required for mitochondrial division, but the distribution along mitochondria and apparent lack of binding to other division proteins suggests that this protein is not a stable part of the mitochondrial division apparatus. One could envision two different functions that are not mutually exclusive (Mozdy et al. 2000). The mislocalization of a large fraction of Dnm1 and Mdv1/Fis2/Gag3 in a Fis1/Mdv2 mutant suggests that Fis1/Mdv2 could serve as a recruitment site for the constituents of the mitochondrial division apparatus (Fig. 2). Once associated with mitochondria, the components of the division apparatus could find the division complex more quickly by lateral diffusion along the surface of the mitochondrial outer membrane. The association with Fis1/Mdv2 would then be transient, as suggested by the lack of in vitro binding. The second possible function of Fis1/Mdv2 is signaling from within mitochondria to the mitochondrial surface, thus triggering the division process. A signaling function would also not necessarily require a concentration of all Fis1/Mdv2 molecules in or near the place of division. Only the activated Fis1/Mdv2 molecules would be near the site of division, whereas inactive molecules could be spread evenly along the surface of mitochondria. Other scenarios are also possible, as time and more experiments will tell.
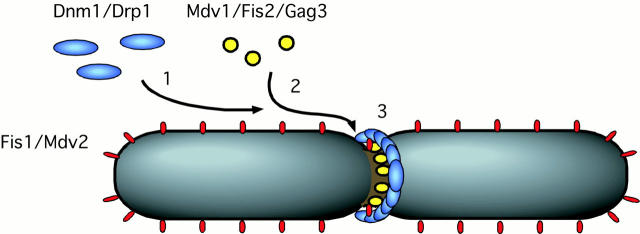
Figure 2.
Model for the functions of Fis1/Mdv2 and Mdv1/Fis2/Gag3 based on their subcellular localizations and mutant phenotypes. In step 1, Fis1/Mdv2 recruits Dnm1 to the surface of the mitochondrion. In step 2, Mdv1/Fis2/Gag3 is added to the complex. In step 3, the mitochondrial division apparatus, consisting of Dnm1, Mdv1/Fis2/Gag3, and possibly other components, help constrict the mitochondrial outer membrane.
The Mitochondrial Inner Membrane and Possible Functions of Mgm1
The mitochondrial inner membrane may well be one of the last frontiers in molecular membrane biology. The morphology of this membrane is difficult to study except by EM. As a consequence, very little is known about the dynamic responses that the mitochondrial inner membrane might have towards changes in the cellular environment. The many different shapes that cristae can attain does suggest that these structures are dynamic (Ghadially 1997). However, the mechanisms controlling cristae morphology are unknown. We also know very little about division of the mitochondrial inner membrane. In most cells, division of the inner membrane appears to be tightly coordinated with division of the mitochondrial outer membrane. There are, however, instances, especially in cells with rapidly dividing mitochondria, where division of the inner membrane occurs without division of the outer membrane (Tandler et al. 1969). The matrix compartment becomes divided by septae, which can only be seen by EM. A difference between inner and outer membrane divisions was also detected in C. elegans muscle cells with mutant DRP-1. In this case, division of the mitochondrial inner membrane can still occur when division of the outer membrane is blocked (Labrousse et al. 1999). These observations indicate that animal mitochondria possess a separate inner membrane division mechanism, which usually, but not always, acts in concert with the external Drp1 complex.
How does the mitochondrial inner membrane divide? Proteins that act on the mitochondrial inner membrane are likely to be of bacterial descent, as this entire compartment originated from a bacterium. The bacterial division apparatus is well characterized. Most bacteria divide using a protein complex built around a ring of ftsZ proteins (Rothfield et al. 1999). The ftsZ ring adheres to the inside of the bacterial membrane and constricts to form a preseptal ingrowth, which then invaginates further to mediate division. Chloroplasts use ftsZ homologues for division (Osteryoung 2000), and it was recently discovered that mitochondria from the algal species Mallomonas splendens also use an ftsZ homologue (Beech et al. 2000). Unfortunately, the complete sequences of C. elegans, Drosophila melanogaster, and yeast do not contain an obvious homologue of ftsZ, suggesting that other proteins have replaced ftsZ in these other eukaryotic species (Erickson 2000).
The possibility existed that another member of the dynamin family, called Mgm1, is responsible for mitochondrial inner membrane division. However, this notion is dispelled by the results of Wong et al. 2000, presented in this issue. They show that mutations in Mgm1 have the opposite effect, inducing excessive fragmentation instead of a block in division. Interestingly, Mgm1 is more closely related to bacterial dynamin-like proteins than to dynamin or Drp1, suggesting that this protein has followed a different evolutionary path (van der Bliek 1999). Mgm1 might have been introduced into eukaryotic cells by the α-proteobacterial progenitor of mitochondria. Unlike other members of the dynamin family, Mgm1 has a mitochondrial targeting sequence. However, the precise localization of Mgm1 is a matter of debate. One group previously suggested that the active form of Mgm1 is an integral protein of the outer membrane (Shepard and Yaffe 1999). A second group suggested that Mgm1 is imported into the mitochondrial matrix (Pelloquin et al. 1999) and in this issue Wong et al. 2000 present compelling evidence that Mgm1 is in the mitochondrial intermembrane space. It will be interesting to see how this issue resolves in the future, because knowing the localization of Mgm1 is essential for a full understanding of the function of Mgm1. Meanwhile, much can be learned from the mutant phenotypes in yeast.
The Mgm1 gene was first discovered in yeast and named Mgm1 to indicate that Mgm1 mutants have a defect in mitochondrial genome maintenance (Jones and Fangman 1992). Mutant Mgm1 also affects mitochondrial morphology (Shepard and Yaffe 1999). The mitochondria form large aggregates in some cells and become excessively fragmented in others (Fekkes et al. 2000; Wong et al. 2000). Conditional mutations showed that loss of mitochondrial DNA occurs at a later stage than changes in mitochondrial morphology (Shepard and Yaffe 1999). Therefore, it would appear that Mgm1 has a morphological function. Subsequent loss of mitochondrial DNA might be a secondary consequence of impaired mitochondrial segregation between daughter cells. The fragmentation phenotype indicates that mitochondrial division is not impaired by mutant Mgm1. However, mitochondrial fusion is also still possible, as Wong et al. 2000 showed elegantly with double mutant analyses. If not fission or fusion, then what is the function of Mgm1? It is possible that Mgm1 affects some other aspect of mitochondrial morphology, such as the shape of cristae, but more experiments will be needed to tell.
Many new questions have been raised and some old questions still remain. What is the precise function of Mgm1? Are there other bacterially derived proteins that control the morphology of the mitochondrial inner membrane? Does Fis1/Mdv2 couple the inner and outer membranes during mitochondrial division? What else does Mdv1/Fis2/Gag3 bind to? Luckily for us mitochondria enthusiasts, there is still plenty of work to do.
Acknowledgments
The author wishes to thank Greg Payne, Patricia Johnson, and members of the lab for helpful suggestions and critical reading of the manuscript.
Work in the authors lab is supported by grants from the National Institutes of Health (GM58166) and the Cancer Research Coordinating Committee (CRCC).
References
- Beech P.L., Nheu T., Schultz T., Herbert S., Lithgow T., Gilson P.R., McFadden G.I. Mitochondrial FtsZ in a chromophyte alga. Science. 2000;287:1276–1279. doi: 10.1126/science.287.5456.1276. [DOI] [PubMed] [Google Scholar]
- Bleazard W., McCaffery J.M., King E.J., Bale S., Mozdy A., Tieu Q., Nunnari J., Shaw J.M. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson H.P. Dynamin and FtsZ. Missing links in mitochondrial and bacterial division. J Cell Biol. 2000;148:1103–1106. doi: 10.1083/jcb.148.6.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekkes P., Shepard K.A., Yaffe M.P. Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J. Cell Biol. 2000;151:333–340. doi: 10.1083/jcb.151.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadially F.N. Ultrastructural Pathology of the Cell and Matrix 1997. Butterworth-Heineman; Boston, MA: pp. 617 [Google Scholar]
- Jones B.A., Fangman W.L. Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 1992;6:380–389. doi: 10.1101/gad.6.3.380. [DOI] [PubMed] [Google Scholar]
- Labrousse A.M., Zapaterra M., Rube D.A., van der Bliek A.M. C. elegans dynamin-related protein drp-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Mozdy A.D., McCaffery J.M., Shaw J.M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000;151:367–379. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W. Organelle fission. Crossing the evolutionary divide. Plant Physiol. 2000;123:1213–1216. doi: 10.1104/pp.123.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloquin L., Belenguer P., Menon Y., Gas N., Ducommun B. Fission yeast msp1 is a mitochondrial dynamin-related protein. J. Cell Sci. 1999;112:4151–4161. doi: 10.1242/jcs.112.22.4151. [DOI] [PubMed] [Google Scholar]
- Pitts K.R., Yoon Y., Krueger E.W., McNiven M.A. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol. Biol. Cell. 1999;10:4403–4417. doi: 10.1091/mbc.10.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L., Justice S., Garcia-Lara J. Bacterial cell division. Annu. Rev. Genet. 1999;33:423–448. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R.E. Division versus fusionDnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard K.A., Yaffe M.P. The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J. Cell Biol. 1999;144:711–720. doi: 10.1083/jcb.144.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E., Shurland D.L., Ryazantsev S.N., van der Bliek A.M. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandler B., Erlandson R.A., Smith A.L., Wynder E.L. Riboflavin and mouse hepatic cell structure and function. II. Division of mitochondria during recovery from simple deficiency. J. Cell Biol. 1969;41:477–493. doi: 10.1083/jcb.41.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q., Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 2000;151:353–365. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek A.M. Functional diversity in the dynamin family. Trends Cell Biol. 1999;9:96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- Wong E.D., Wagner J.A., Gorsich S.W., McCaffery J.M., Shaw J.M., Nunnari J. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 2000;151:341–352. doi: 10.1083/jcb.151.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]