Abstract
p10/NTF2 is a nuclear transport carrier that mediates the uptake of cytoplasmic RanGDP into the nucleus. We constructed a point mutant of p10, D23A, that exhibited unexpected behavior both in digitonin-permeabilized and microinjected mammalian cells. D23A p10 was markedly more efficient than wild-type (wt) p10 at supporting Ran import, but simultaneously acted as a dominant-negative inhibitor of classical nuclear localization sequence (cNLS)-mediated nuclear import supported by karyopherins (Kaps) α and β1. Binding studies indicated that these two nuclear transport carriers of different classes, p10 and Kap-β1, compete for identical and/or overlapping binding sites at the nuclear pore complex (NPC) and that D23A p10 has an increased affinity relative to wt p10 and Kap-β1 for these shared binding sites. Because of this increased affinity, D23A p10 is able to import its own cargo (RanGDP) more efficiently than wt p10, but Kap-β1 can no longer compete efficiently for shared NPC docking sites, thus the import of cNLS cargo is inhibited. The competition of different nuclear carriers for shared NPC docking sites observed here predicts a dynamic equilibrium between multiple nuclear transport pathways inside the cell that could be easily shifted by a transient modification of one of the carriers.
Keywords: nuclear transport, nuclear pore complex, p10, NTF2, karyopherin
Introduction
The p10 protein was first identified as playing a role in nuclear transport by the demonstration that it was a necessary component of the cytosol used to support classical nuclear localization sequence (cNLS)-mediated nuclear import in digitonin-permeabilized cells (Moore and Blobel 1994). This protein was purified from Xenopus cytosol on the basis of its ability to stimulate nuclear import in permeabilized cells (when added with Ran, Kap-α, and Kap-β1) and named p10 because of its migration pattern on SDS-PAGE gels. The predicted molecular mass of human p10 is 14 kD and p10 appears to exist as a homodimer (Moore and Blobel 1994; Bullock et al. 1996; Paschal et al. 1996). p10 binds RanGDP with high affinity, but has almost no affinity for RanGTP (Paschal et al. 1996; Stewart et al. 1998).
The identity of p10 as a nuclear import factor was later verified by the observation that HeLa cytosol passed over a p62 (a nuclear pore complex [NPC] protein) column showed diminished capacity for supporting cNLS cargo nuclear import in permeabilized cells (Paschal and Gerace 1995). The protein that reconstituted this loss of activity was purified from human cytosol, found to be the human homologue of p10, and named NTF (nuclear transport factor)2. In addition to p62, p10/NTF2 has been shown to interact with additional members of the p62 family of NPC proteins from both vertebrates and yeast, the so-called repeat-containing nucleoporins (Nups), or repeat Nups (Clarkson et al. 1996; Hu et al. 1996; Nehrbass and Blobel 1996). Each of these repeat Nups contain the repeat motif FG (PheGly), which is often found as part of a larger repeat motif such as GLFG or FXFG (for a review see Ryan and Wente 2000). Members of the Kap-β superfamily also interact with these repeat Nups, and it has been proposed that this family of NPC proteins provide many or most of the docking sites for the Kap-β class of nuclear transport carriers as they move through the NPC (Radu et al. 1995a,Radu et al. 1995b). One current model for nuclear transport is that nuclear carriers move through the NPC by repeated association–dissociation reactions with NPC proteins, a process that has been called “facilitated diffusion” and that appears not to require an energy source (Kose et al. 1997; Ribbeck et al. 1998; Schwoebel et al. 1998; Englmeier et al. 1999; Talcott and Moore 1999).
One known function of p10 is to serve as a nuclear transport carrier to import RanGDP into the nucleus from the cytoplasm (Ribbeck et al. 1998; Smith et al. 1998). Ran is only 25 kD and thus below the diffusion limit of the NPC, yet at steady-state the cellular distribution of Ran is ∼85% in the nucleus, with the rest in the cytoplasm (Ren et al. 1993). RCC1, the Ran guanine nucleotide exchange factor (GEF), is also found in the nucleus, but the Ran GTPase activating protein (GAP) is found in the cytoplasm, either soluble or bound on the cytoplasmic filaments of the NPC (Ohtsubo et al. 1989; Matunis et al. 1996; Mahajan et al. 1997). This differential localization of Ran's GAP and GEF is thought to keep RanGTP at a low concentration in the cytoplasm, but abundant in the nucleoplasm (Görlich et al. 1996).
All Ran-dependent nuclear transport pathways described thus far utilize a member of the Kap-β/importin β superfamily as a nuclear transport receptor/carrier. These Kap-β family members either bind a nuclear localization sequence (NLS)- or nuclear export sequence (NES)-containing cargo directly, or via an adaptor protein such as Kap-α in the cNLS import pathway (for a review see Nakielny and Dreyfuss 1999). In addition to binding their specific cargo, most (if not all) of the proteins in this family also contain a Ran binding site and preferentially bind RanGTP rather than RanGDP (Rexach and Blobel 1995; Fornerod et al. 1997; Görlich et al. 1997). This Ran binding is key to the assembly and disassembly of Kap-β–containing transport complexes. Export complexes require high RanGTP to assemble, like in the nucleus, and disassemble when RanGTP is low, like in the cytoplasm. Conversely, import complexes will only assemble when RanGTP is low and disassemble when RanGTP is high (Rexach and Blobel 1995; Görlich et al. 1996; Floer et al. 1997; Izaurralde et al. 1997; Kutay et al. 1997a, Kutay et al. 1998; Floer and Blobel 1999).
The p10-mediated accumulation of Ran inside the nucleus requires the GEF activity of RCC1, to generate RanGTP, in whole cells and the addition of GTP to permeabilized cells (Ribbeck et al. 1998; Smith et al. 1998). The evidence suggests that the free GTP is used during the RCC1-stimulated conversion of RanGDP to RanGTP to trigger Ran's release from p10 inside the nucleus, rather than to power movement of the p10–RanGDP complex through the NPC. Also, the addition of Kap-β family members has been shown to stimulate the p10-mediated accumulation of Ran inside the nucleus (Ribbeck et al. 1998). It has been proposed that this is due to binding of nuclear RanGTP by Kap-β family members inside the nucleus, raising Ran's effective size above 25 kD and preventing its diffusion back out of the nucleus. However, the mechanism by which a p10–Ran complex (or any transport complex) moves through the NPC and exactly which NPC proteins it associates with are unknown.
In Saccharomyces cerevisiae, the NTF2 gene is an essential gene, though it was later demonstrated that overexpressing the Ran homologue (GSP1) in these cells would compensate for an NTF2 deletion (Corbett and Silver 1996; Nehrbass and Blobel 1996; Wong et al. 1997). Consistent with this, increasing the concentration of Ran in an in vitro nuclear import assay in permeabilized cells decreases the need for p10 to support the nuclear import of a cNLS cargo (Paschal et al. 1997). Increasing Ran's concentration is thought to increase the amount of Ran that enters the nucleus by p10-independent mechanisms, such as diffusion, thereby decreasing the need for active nuclear import of Ran mediated by p10.
With the goal of examining how a nuclear transport carrier moves through the NPC, we have generated several point mutants of p10. Unexpectedly, we have engineered a mutant p10, D23A, that is more efficient at low concentrations than wild-type (wt) p10 at mediating the nuclear import of its own cargo (RanGDP), but it is a dominant-negative inhibitor of cNLS import. We show that this is due to a change in the relative affinities of p10 and Kap-β1 for the NPC, and we speculate as to how a similar, naturally occurring change in affinity of a nuclear transport carrier for the NPC could alter its function in vivo.
Materials and Methods
Vector Construction and Site-directed Mutagenesis
To make FLAG-tagged p10, p10 was cloned from pTACTL7 (a gift from U. Grundmann, Behringwerke, Marburg, Germany) into pCEP4FLAG(EBNA) using PCR and incorporating HindIII and XhoI sites. Then, standard DNA purification, digestion, and ligation procedures were performed. For His-tag incorporation, FLAG–p10 was subcloned into the pRSET expression vector (Invitrogen), by the method described above, and incorporating KpnI and EcoRI sites. Site directed mutagenesis was performed on pRSET His–FLAG–p10 using a PCR-based strategy using Pfu enzyme from Stratagene, according to their instructions. Both wt and mutated sequences were confirmed by DNA sequencing. The Kap-β1(45–462) fragment was obtained by PCR of the DNA corresponding to amino acids 45–462 of Kap-β1 from pGEX4T-NTF97 (a gift from Steve Adam, Northwestern University Medical School, Chicago, IL) and placed in the pET30 Ek/LIC vector (which will add both NH2-terminal His and S tags) by the method suggested by the manufacturer (Novagen).
Purification of Recombinant Proteins
pRSET–FLAG–p10-expressing wt and mutant p10s were transformed into the BL21 (DE3)-Rep 4 strain. The cells were grown in Luria broth (LB) containing 100 μg/ml ampicillin and 50 μg/ml kanamycin to an optical density at 600 nm (OD600) of 0.6, and then they were induced by the addition of 1 mM isopropyl-β-thioglactoside (IPTG) for 3 h at 37°C. Bacteria were washed and lysed in a French pressure cell in 20 mM Hepes-KOH, pH 7.3, 500 mM potassium acetate, 60 mM imidazole, and 10 mM 2-mercaptoethanol (homogenization buffer) containing 1 mM PMSF and centrifuged at 250,000 g for 90 min at 4°C in a Ti70 rotor (Beckman Coulter). The supernatant was incubated for 2 h at 4°C with 1 ml nickel-agarose (QIAGEN) equilibrated in homogenization buffer. The resin was washed with 60 ml homogenization buffer and eluted with 5 ml homogenization buffer containing 400 mM imidazole. The column eluate was dialyzed against buffer A (20 mM Hepes-KOH, pH 7.3, 100 mM potassium acetate, 1 mM DTT). Ran and Kaps-α2 and -β1 were purified, as described (Schwoebel et al. 1998), from expression vectors obtained from Mark Rush (New York University Medical School, New York, NY), Agnus Lamond (EMBL, Heidelberg, Germany), and Dirk Görlich (Zentrum für Molekulare Biologie der Universität Heidelberg, Heidelberg, Germany), respectively.
Kap-β1(45–462) purification was performed as described for recombinant FLAG p10, except that the homogenization buffer was 50 mM Tris-HCl, pH 7.4, 200 mM NaCl, 15 mM 2-mercaptoethanol. Bound Kap-β1(45–462) was eluted from the nickel resin with sequential elutions of 20, 40, 100, and 200 mM imidazole in homogenization buffer. The eluates were separated on 10% SDS-PAGE gels and those containing Kap-β1 (45–462) were pooled and dialyzed against buffer A.
Permeabilized Cell Assays
Nuclear import assays were performed as described (Moore and Blobel 1992; Schwoebel et al. 1998). In brief, buffalo rat liver (BRL) or HeLa cells were washed once with cold transport buffer (TB, which contains 20 mM Hepes-KOH, pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 2 mM DTT), and permeabilized with 35 μg/ml digitonin (Calbiochem) in TB for 5 min on ice. The permeabilized cells were then washed and incubated with 40 μl of an import mix (in TB) containing the components listed in each figure legend. Permeabilized cells were incubated at room temperature before washing and fixing. Samples were observed and quantitated (Schwoebel et al. 1998) and Xenopus ovarian cytosol was prepared as described (Moore and Blobel 1992).
To observe FLAG-tagged p10 interactions with the nuclear envelope of the permeabilized cells, the permeabilized cells were incubated with 0.25–0.5 μM (dimer) p10 (wt or mutant) in TB containing 2 mg/ml BSA for 20 min at room temperature. Cells were washed twice with 1 ml of TB, fixed, and processed for immunofluorescence microscopy with an anti-FLAG antibody.
Overlay Assays
The Nup98 overlay assay was performed as described (Radu et al. 1995b), except that the nitrocellulose was overlaid with 0.5 μM p10 (dimer) in TB containing 2% gelatin and 0.2% Tween 20, and the p10 binding was detected with an anti-Express tag antibody (Invitrogen).
[α-32P] GTP (3,000 Ci/mmol) (NEN Life Science Products) was converted into [α-32P]GDP by incubation with glucose/hexokinase, and Ran was loaded with hot nucleotide, as described (Schwoebel et al. 1998). Each wt or mutant p10 in 100 μl overlay buffer (20 mM MOPS, pH 7.1, 100 mM potassium acetate, 5 mM magnesium acetate, 1 mM DTT, 0.5% BSA, and 0.05% Tween 20) was dotted onto 0.45-μm nitrocellulose filters using a Minifold II slot-blot apparatus (Schleicher & Schuell, Inc.). Nitrocellulose pieces were incubated 30 min with 7 × 106 cpm [α-32P]RanGTP or [α-32P]RanGDP in 10 ml overlay buffer at room temperature. Blots were washed five times with overlay buffer and exposed to film.
Preparation of Fluorescent Import Substrates
The TRITC–BSA–NLS conjugate was prepared as described (Moore and Blobel 1992). The NLS peptide (CYTPPKKKRKV) contains the NLS of the SV40 T antigen (Lanford et al. 1986). RanGDP was labeled at a 1:1 molar ratio with fluorescein-5-maleimide (Molecular Probes) in 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 2 mM MgCl2 for 2 h on ice, followed by the addition of 2-mercaptoethanol to a final concentration of 50 mM. Uncoupled fluorescein was removed by passing the sample over a Nap-5 column (Amersham Pharmacia Biotech) equilibrated with buffer A containing 2 mM magnesium acetate (Buffer B). FITC–RanGDP was stored at –80°C in single use aliquots.
Microinjection Studies
HeLa cells were injected with proteins at the concentrations indicated in the figure legends. Before injection, all protein injection mixtures were brought to equal volume with Buffer B and centrifuged at 14,000 g for 30 min at 4°C. After injections, cells were incubated for 5 min at room temperature and then fixed in 3% paraformaldehyde in PBS for 15 min on ice.
Results
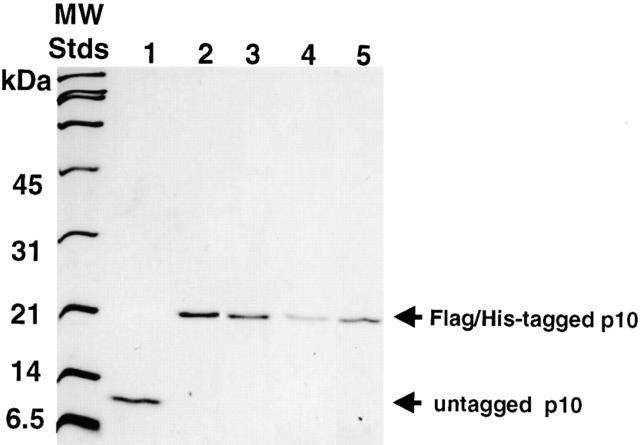
To learn more about the cellular functions of p10, three point mutants of human p10 (E42D, Y19A, and D23A) were created by site-directed mutagenesis. Amino acids in the p10 sequence were selected for mutagenesis based on their position in the crystal structure of p10 (Bullock et al. 1996) and their conservation across species (Katahira et al. 1999). wt and mutant p10s were expressed and purified using an Escherichia coli expression vector that adds both a FLAG- and His- tag to the p10 protein. As shown in Fig. 1, all of the expressed and purified recombinant p10s appeared as single bands by Coomassie blue staining of an SDS-PAGE gel. The addition of the tags raised the predicted monomeric molecular mass of p10 from 14 to 21 kD, but this increase in mass was found to diminish only slightly the ability of wt p10 to stimulate the nuclear import of BSA–NLS in digitonin-permeabilized cells (data not shown).
Figure 1.
Purified recombinant p10 proteins. Purified recombinant p10 proteins were run on a 15% SDS-PAGE gel and stained with Coomassie blue. wt p10 has an apparent molecular mass of 10 kD, but a predicted molecular mass of 14 kD. Tagged p10s have an M r closer to their true predicted mass of 21 kD. Lanes: 1, untagged wt p10; 2, wt p10; 3, E42D p10; 4, Y19A p10; and 5, D23A p10.
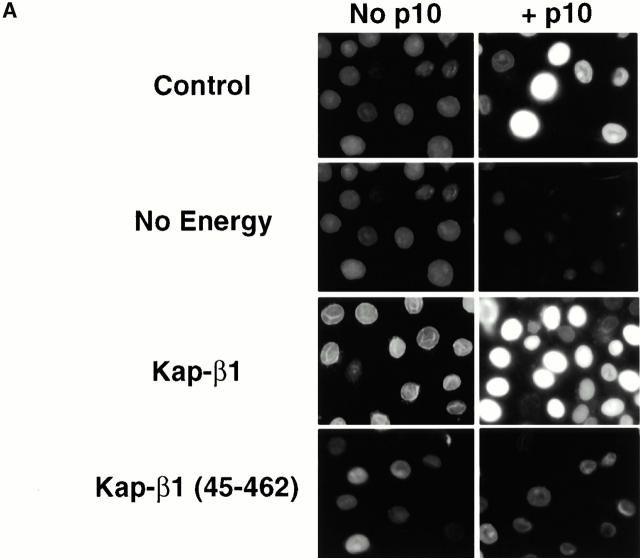
The ability of FLAG–p10 to support nuclear accumulation of Ran in digitonin-permeabilized cells is shown in Fig. 2. In these experiments, FITC–RanGDP was added to the permeabilized cells, either alone or with p10 and various additions. After washing and fixation, the nuclear accumulation of Ran was assessed and quantitated by fluorescence microscopy. In experiments not shown, the ability of FITC–RanGDP to support nuclear import of BSA–NLS in permeabilized cells was found to be unaffected by its labeling by fluorescein-maleimide, indicating that Ran was not damaged by the labeling procedure. Confirming the results of others (Ribbeck et al. 1998; Smith et al. 1998), we found that p10 would support the nuclear accumulation of Ran in permeabilized cells, but only under certain conditions. In the presence of p10, FITC–RanGDP, and an energy mix (consisting of GTP and ATP plus an ATP-regenerating system), nuclear accumulation of the added Ran was observed in ∼30% of the permeabilized cells (Fig. 2 A, top row). If the energy mix was omitted, this accumulation was not observed in any of the cells (Fig. 2 A, second row).
Figure 2.
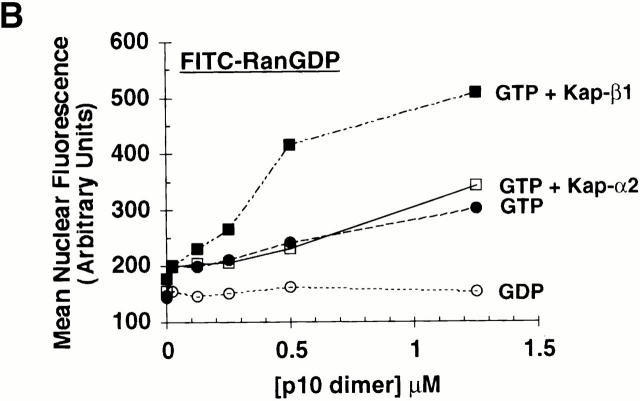
Nuclear accumulation of FITC–RanGDP. (A) Digitonin-permeabilized BRL cells were incubated with 2 μM FITC–RanGDP in TB containing 2 mg/ml BSA for 20 min at room temperature, washed, and fixed. All samples, except those labeled No Energy, also contained 0.5 mM GTP, 1 mM ATP, 5 mM phosphocreatine, and 20 U/ml creatine phosphokinase. Where indicated, the following proteins were also added: 1.0 μM wt p10 (dimer), and 0.25 μM Kap-β1 or Kap-β1(45–462). (B) Quantitation of the nuclear import of FITC–RanGDP in digitonin-permeabilized BRL cells was performed as described in the Materials and Methods. All samples contained 1.5 μM FITC–RanGDP and 2 mg/ml BSA in TB, and the import reaction was incubated for 15 min at room temperature before washing and fixation. Individual samples also contained the indicated concentration of wt p10 dimer and: ○, 0.5 mM GDP; •, 0.5 mM GTP; □, 0.5 mM GTP + 0.25 μM Kap-α2; and ▪, 0.5 mM GTP + 0.25 μM Kap-β1. (C) Quantitation of the nuclear import obtained of FITC–RanGDP in the presence of increasing concentrations of: ○, WT p10; •, D23A p10; and ▪, E42D p10. In addition to 1.5 μM FITC–RanGDP, all the samples contained 0.5 mM GTP, 0.25 μM Kap-β1, and 2 mg/ml BSA in TB. Import was for 7.5 min before washing and fixation.
If Kap-β1 was also added, the percentage of cells showing nuclear accumulation of Ran could be greatly increased, with essentially 100% of the cells showing nuclear accumulation (Fig. 2a and Fig. b). In contrast, substitution of a truncated Kap-β1 lacking its Ran binding domain, Kap-β1 (45–462) (Kutay et al. 1997b), for the full-length Kap-β1 did not result in a stimulation of Ran nuclear accumulation (Fig. 2 A, bottom right) and in fact appeared to block the Ran import obtained when no Kap-β1 was added (Fig. 2 A, top right). Fig. 2 B shows the quantitation of the nuclear import of Ran, obtained under various conditions, and confirms a dependence on added GTP and a further stimulation of nuclear accumulation by Kap-β1. Another nuclear transport factor, Kap-α2, which doesn't bind RanGTP, was found to be unable to stimulate the nuclear accumulation of Ran (Fig. 2 B). As previously proposed (Ribbeck et al. 1998; Smith et al. 1998), we believe it likely that both the GTP and Kap-β1 are being used to stimulate the nuclear accumulation of Ran, rather than being required directly for movement of the p10–Ran complex across the NPC.
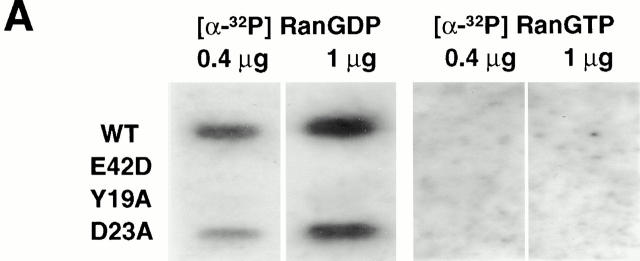
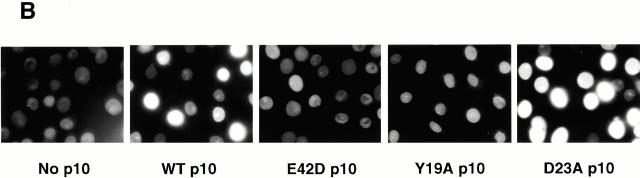
We had anticipated that a mutation in p10 would either have no effect on its biological activity or decrease it. To our surprise, we found that at low concentrations D23A p10 was markedly more efficient at supporting the nuclear accumulation of Ran than wt p10 (Fig. 2 C). In contrast, the mutant E42D p10 was unable to support Ran nuclear import. The reason for the observed inability of E42D p10 to support Ran import can be explained by the results shown in Fig. 3. The binding of wt and mutant p10s to RanGDP or RanGTP was analyzed by Ran overlay assays (Fig. 3 A). Of the p10 mutants tested, only D23A p10 retained its ability to bind RanGDP. Neither E42D nor Y19A p10 were observed to bind RanGDP in this assay. This inability to bind RanGDP explains why E42D and Y19A p10 are also unable to support Ran's nuclear accumulation (Fig. 2 C and 3 B). The crystal structure of p10 bound to RanGDP (Stewart et al. 1998) reveals that p10 amino acid E42 forms a salt bridge between Ran and p10 necessary for the stabilization of RanGDP binding. A different mutation in this amino acid, E42K, was shown previously to abolish Ran binding to p10, without altering the overall structure of p10 (Clarkson et al. 1997). In contrast, amino acids D23 and Y19 are not among the amino acids directly implicated in RanGDP binding by p10, although all of our data suggests that the Y19A mutation results in profound structural changes throughout the p10 molecule (see below).
Figure 3.
p10 mutants that cannot bind RanGDP do not support nuclear accumulation of FITC–RanGDP in digitonin-permeabilized cells. (A) Recombinant p10 proteins were dotted onto nitrocellulose, as described in the Materials and Methods, and the nitrocellulose was incubated with either Ran[α-32P]GDP or Ran[α-32P]GTP, washed, and exposed to film. (B) The ability of different p10 mutants to mediate the import of FITC–RanGDP (2 μM) in permeabilized BRL cells was assayed, as described in the legend to Fig. 2. Samples contained 0.5 mM GTP, 1 mM ATP plus a regenerating system, and 1 μM of the indicated p10 protein (dimer). Import was for 20 min at room temperature before washing and fixation.
Surprisingly, we found that although D23A p10 could support the nuclear import of Ran, this mutant p10 had a marked inhibitory effect on the import of a cNLS cargo (Fig. 4). In Fig. 4 A, the import of BSA–NLS into the nuclei of digitonin-permeabilized BRL cells was supported by Xenopus cytosol as a source of transport factors plus energy. This import substrate contains peptides containing the cNLS of the SV40 T antigen coupled to rhodamine-labeled BSA (Moore and Blobel 1992). The Xenopus cytosol used is known to contain endogenous p10 that is functional in permeabilized mammalian cells (Moore and Blobel 1994). wt p10 had no effect on the nuclear import of BSA–NLS when added to the cytosol at 3 μM (Fig. 4 A). In marked contrast, the addition of D23A p10 at the same concentration severely inhibited the nuclear import of BSA–NLS.
Figure 4.
D23A p10 is a dominant-negative inhibitor of the nuclear import of TRITC–BSA–NLS in vitro. (A) wt p10 or D23A p10 (both at 3.0 μM dimer concentration) were added to an import solution consisting of 10 μg/ml TRITC–BSA–NLS, 10 mg/ml Xenopus ovarian cytosol, 0.5 mM GTP, and 1 mM ATP plus a regenerating system. These mixtures were incubated with permeabilized BRL cells for 20 min at room temperature before washing and fixation. (B) The import assay was done as described in A, except purified transport factors were used to support import, rather than the cytosol, and the reaction was for 15 min. The import reactions contained 10 μg/ml TRITC–BSA–NLS, 0.5 μM Kap-α2, 0.25 μM Kap-β1, 2 μM RanGDP, 0.5 mM GTP, and 2 mg/ml BSA in addition to the indicated concentration of wt or D23A p10 dimer.
Fig. 4 B shows that D23A also inhibits BSA–NLS import in a reaction supported with purified, recombinant transport factors (Kap-β2, Kap-α2, and Ran) rather than cytosol. Because we only wanted to observe inhibitory effects of p10 on BSA–NLS import in this experiment and not stimulatory effects, we added Ran at a sufficiently high concentration such that enough of it can enter the nucleus by diffusion to support cNLS cargo import, thus diminishing an observed stimulatory effect of p10 (Paschal et al. 1997). As can be seen in Fig. 4 B, both wt and D23A p10 slightly stimulated the import of BSA–NLS when added at a low concentration of 1 μM. However, with increasing concentrations, D23A p10 rapidly inhibited the import of BSA–NLS such that at 5 μM D23A p10, the inhibition was almost complete. wt p10 also showed an inhibition of BSA–NLS import at higher concentrations, but the inhibition was not yet complete at the highest concentration of wt p10 tested (20 μM).
We found that D23A p10 has the same inhibitory effect on BSA–NLS nuclear import in vivo as in permeabilized cells (Fig. 5). We had determined previously that FITC–RanGDP injected into the cytoplasm of HeLa cells could be observed to rapidly (within 5 min) accumulate inside the nucleus (data not shown). To simultaneously examine the effects of the p10 mutants on Ran uptake and NLS-mediated import, the following were coinjected into the cytoplasm: FITC–RanGDP, TRITC–BSA–NLS, Cascade blue–labeled BSA (marker), and unlabeled p10 (wt or mutant). Neither the wt nor mutant p10s had any effect on the nuclear accumulation of Ran when coinjected with it into the cytoplasm (Fig. 5, left column); the injected cells would of course contain endogenous wt p10. However, as seen previously in the permeabilized cells, D23A p10 inhibited the nuclear import of coinjected BSA–NLS at the same time that the nuclear accumulation of coinjected Ran was unaffected. As these whole cells at the time of injection obviously contain enough nuclear Ran to support NLS-mediated nuclear import, and also because this mutant appears fully capable of supporting Ran nuclear import in vitro, we concluded that D23A p10 must be inhibiting BSA–NLS import by a mechanism unrelated to the nuclear uptake of Ran.
Figure 5.
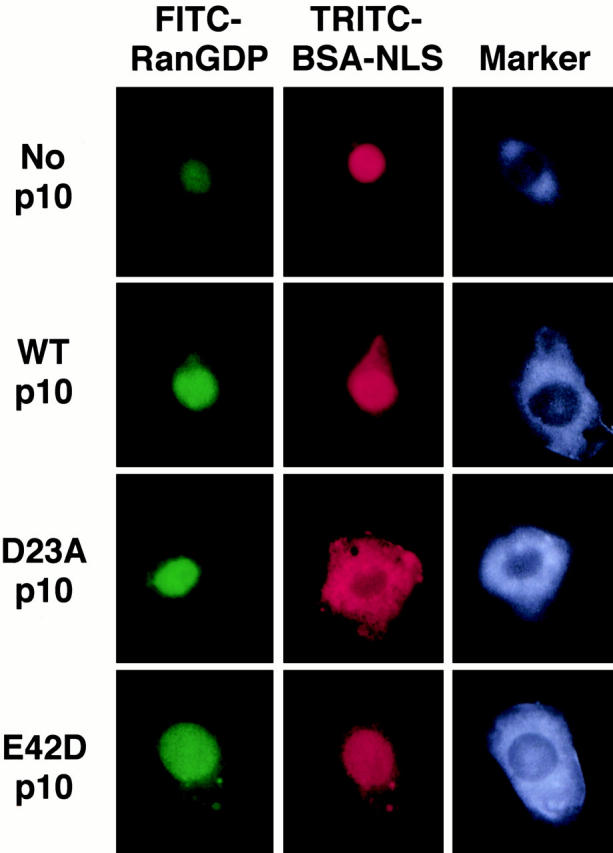
D23A p10 does not inhibit RanGDP nuclear accumulation, but does inhibit BSA–NLS nuclear accumulation in vivo. HeLa cells were coinjected in the cytoplasm with the following: FITC–RanGDP (1 mg/ml), TRITC-BSA–NLS (2 mg/ml), the injection marker Cascade blue–labeled BSA (Molecular Probes) (1 mg/ml), and unlabeled wt or mutant p10 (2.2 mg/ml). The cells were incubated 5 min at room temperature after microinjection, and then fixed for observation.
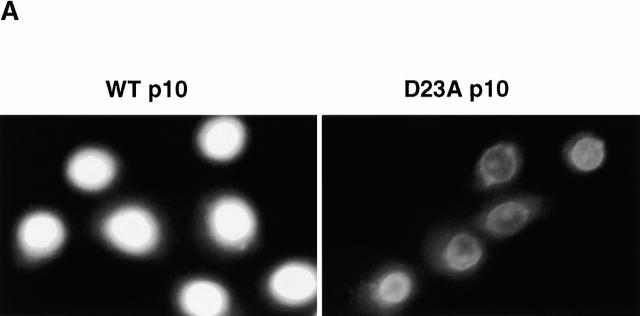
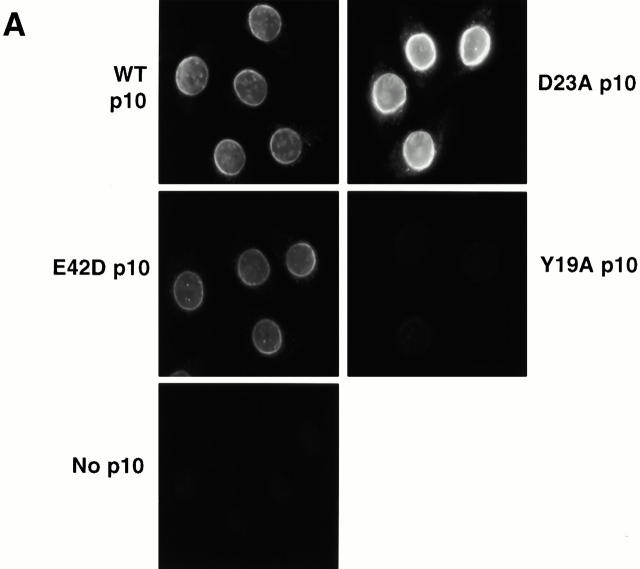
The interaction of wt and mutant p10s with NPC components was assessed using both endogenous, assembled NPCs in permeabilized cells, and fragments of an individual repeat Nup (Nup98). HeLa cells were permeabilized with digitonin and then incubated with purified wt or mutant p10s. After washing and fixation, bound p10 was detected by immunofluorescence microscopy with an anti-FLAG antibody. As shown in Fig. 6 A, wt, E42A, and D23A p10 added in buffer alone bound to the nuclear envelope of the permeabilized cells, however no binding of Y19A p10 was observed. Strikingly, the D23A p10 gave much more intense staining at the nuclear envelope than wt p10. The pattern of staining was identical to that achieved with mAb 414 (data not shown), which binds the FXFG epitope present on a group of NPC proteins (Davis and Blobel 1986; Wente et al. 1992). Thus, D23A p10 showed increased binding to NPCs relative to wt p10.
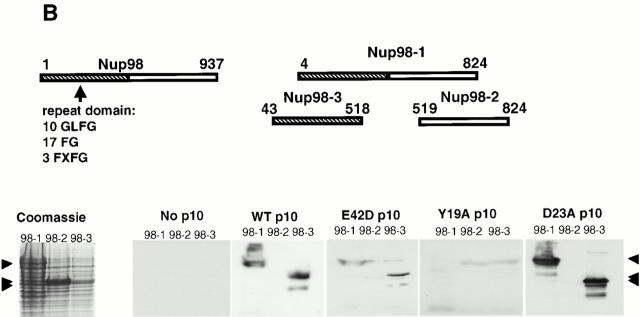
Figure 6.
Interaction of p10 mutants with the NPC. (A) Digitonin-permeabilized HeLa cells were incubated with 0.25 μM (dimer) wt or mutant p10, as described in the Materials and Methods. After washing and fixation, the bound p10 was detected by indirect immunofluorescence microscopy with an anti-FLAG antibody followed by a TRITC-labeled anti–mouse second antibody. (B) Diagram indicates the fragments of Nup98 that were used in the overlay assay. Cell lysates from E. coli expressing different portions of the nucleoporin Nup98 (Nup98-1, Nup98-2, and Nup98-3) were separated on a 10% SDS-PAGE gel. Arrows indicate the migration position of the expressed pieces of Nup98 after staining of a gel with Coomassie blue. Other samples were transferred to nitrocellulose and overlaid with 0.5 μM wt or mutant p10, as described in the Materials and Methods. Bound p10 was detected by immunoblotting with an anti-Express antibody.
p10 is known to bind to various repeat Nups and is also known to be able to bind a repeat Nup and RanGDP at the same time (Paschal and Gerace 1995; Clarkson et al. 1996; Nehrbass and Blobel 1996). To compare the binding of wt and mutant p10s to a specific repeat Nup, an overlay assay was performed against various segments of the vertebrate nucleoporin Nup98 (Fig. 6 B). The Nup98-1 construct contains amino acids 43–824 of rat Nup98, Nup98-2 contains amino acids 519–869, which does not contain repeats, and Nup 98-3 contains amino acids 43–518, which includes 10 GLFG, 17 FG, and 3 FXFG repeats (Radu et al. 1995b). The three Nup98 fragments were expressed in E. coli and the bacterial lysates were separated by SDS-PAGE. The separated proteins were transferred to nitrocellulose, overlaid with either wt or mutant p10, and bound p10 protein was subsequently detected by immunoblotting. wt, E42D, and D23A p10 only bound to the portion of Nup98 that contains repeats (Nup 98-1 and 98-3); they did not bind to the half of Nup98 that does not contain repeats (Nup 98-2) or to any of the endogenous E. coli proteins also present. In contrast, there was no significant binding of Y19A p10 to any of the Nup98 fragments, in agreement with the NPC binding results we observed in permeabilized cells (Fig. 6 A). p10 has been reported to preferentially bind to FXFG-containing nucleoporins rather than nucleoporins that contain only GLFG repeats (Clarkson et al. 1997). Nup98 contains FXFG, GLFG, and FG sequences within the region exhibiting p10 binding. What Nup98 repeat sequences (if any) present in the Nup98-3 construct was responsible for the observed p10 binding was not determined. Kap-β1, another nuclear carrier also known to bind repeat Nups, was shown previously to exhibit the same binding pattern to these Nup98 fragments as wt p10 (Radu et al. 1995b).
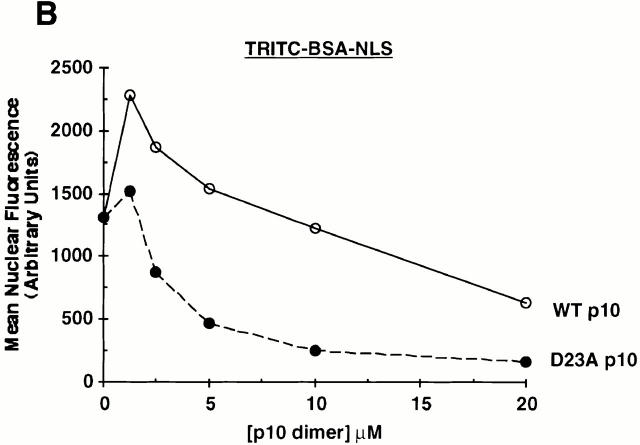
That D23A p10 exhibits increased binding to the NPC relative to wt p10 shown again in Fig. 7. In this experiment, 0.25 μM (dimer) wt or mutant p10 was incubated with the permeabilized cells either alone, or in the presence of 20 μM RanGDP, Kap-β1, or untagged p10. The observed binding of p10 to the nuclear envelope appeared to be specific in that it could be abolished by inclusion of an excess of wt p10 lacking the FLAG epitope (Fig. 7), although the untagged wt p10 appeared somewhat less able to compete away the binding of tagged D23A p10 than tagged wt p10. Interestingly, the binding of wt p10 to NPCs was found to be enhanced by the addition of RanGDP (Fig. 7). The binding of D23A p10 to the NPC in the presence of Ran may also have been marginally higher, but the high signal in the absence of Ran made this determination difficult. This phenomenon of Ran-enhanced p10 binding to the NPC has been reported before (Ribbeck et al. 1998), and is likely due to RanGDP binding sites at the NPC (see Discussion).
Figure 7.
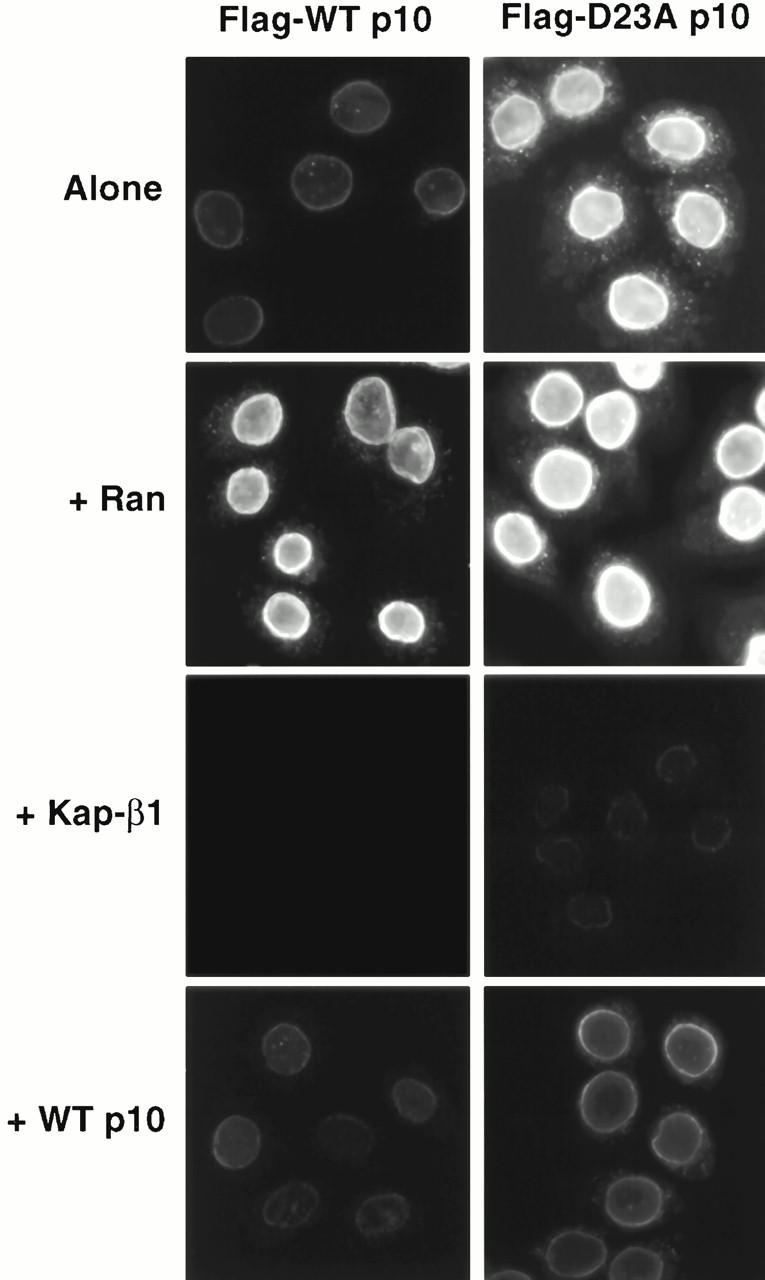
wt p10 and D23A p10 binding to the nuclear envelopes of permeabilized cells in the presence of other nuclear transport factors. Binding of wt and mutant p10 to the nuclear envelope of digitonin-permeabilized HeLa cells was performed as described in the legend to Fig. 6. 0.25 μM (dimer) wt p10 or D23A p10 were added either alone (top), or with 20 μM RanGDP, 20 μM Kap-β1, or 20 μM untagged wt p10.
In contrast to the stimulatory effects of Ran on NPC binding, the inclusion of an excess of Kap-β1 markedly inhibited the binding of both wt and D23A p10 to the NPC (Fig. 7). Others have reported that p10 and Kap-β1 are capable of binding each other directly (Nehrbass and Blobel 1996; Percipalle et al. 1997), but we have been unable to demonstrate such an interaction (data not shown) consistent with the findings of others (Paschal et al. 1996). Instead, we believe that the ability of Kap-β1 to inhibit p10 binding at the NPC indicates that the majority of binding sites of p10 and Kap-β1 at the NPC are either identical, or closely overlapping. We also believe that it is this competition for shared docking sites that explains the ability of D23A p10 to inhibit Kap-β1-mediated cNLS import (see Discussion). In the experiment shown in Fig. 7, 20 μM Kap-β1 was able to greatly diminish any p10 binding when added with 0.25 μM (dimer) p10. In experiments not shown, we found that D23A p10 gave visible NPC binding when Kap-β1 was included in only a tenfold molar excess (rather than the 80-fold molar excess shown in Fig. 7), however wt p10 still did not give visible NPC binding when added with only a twofold molar excess of Kap-β1.
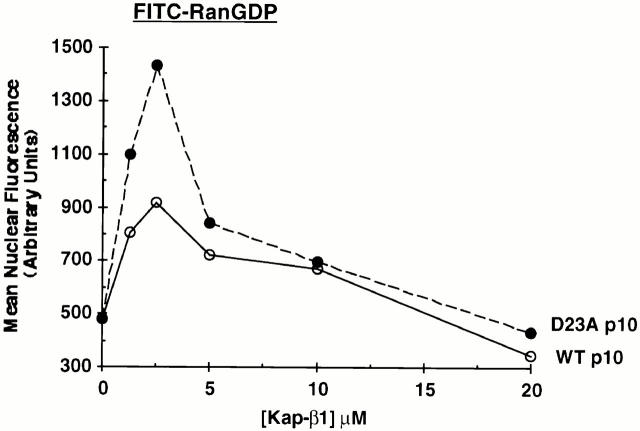
If Kap-β1 and p10 use many of the same docking sites to move their respective cargoes across the NPC, then an excess of one carrier should inhibit the import of the other carrier. As we have already shown in Fig. 2 C, increasing concentrations of D23A or wt p10 do in fact inhibit the import of a cNLS cargo that uses Kap-β1 as a carrier, with D23A p10 showing a greatly increased ability to inhibit relative to wt p10. Fig. 8 shows that the reverse is also true, that increasing amounts of Kap-β1 will inhibit Ran import mediated by p10. In this experiment, increasing amounts of Kap-β1 were added together with wt or D23A p10 and GTP to determine whether adding an excess of Kap-β1 will inhibit the p10-mediated import of FITC–RanGDP. As already discussed, low concentrations of Kap-β1 stimulate p10-mediated Ran nuclear accumulation and this is believed to be due to a trapping of Ran by Kap-β1 inside the nucleus (Ribbeck et al. 1998). As shown in Fig. 8 (see also Fig. 2 C), D23A p10 was measurably more efficient than wt p10 at supporting Ran import when a low level (2.5 μM) of Kap-β1 was present. However, with increasing concentrations of Kap-β1, the nuclear accumulation of Ran mediated by either wt or D23A p10 was progressively inhibited (Fig. 8).
Figure 8.
Inhibition of RanGDP import by high concentrations of Kap-β1. p10-mediated nuclear import of FITC–RanGDP was measured in the presence of increasing concentrations of Kap-β1. All samples contained 1.5 μM FITC–RanGDP, 0.5 mM GTP, 2 mg/ml BSA, and Kap-β1 at the indicated concentrations. In addition, samples contained 1.25 μM (dimer) WT p10 (○) or D23A p10 (•). Import was for 15 min before washing and fixation.
Discussion
We examined the effects of three different point mutations (D23A, E42D, and Y19A) on the activity of the nuclear carrier p10. Surprisingly, we found that one of these mutations, D23A, markedly increased p10's ability to mediate the nuclear import of its cargo RanGDP (Fig. 2 and Fig. 8). Even more surprisingly, we also found that D23A p10, while importing its own cargo more efficiently than wt p10, would inhibit the nuclear import of a cNLS cargo mediated by a different nuclear carrier in a dominant-negative fashion (Fig. 4 and Fig. 5). We further found that D23A p10 shows increased binding to the NPCs of permeabilized cells when compared with wt p10 (Fig. 6 and Fig. 7). We believe that it is this increased NPC binding that is responsible for the ability of low concentrations of D23A p10 to inhibit cNLS import by diminishing the ability of the carrier Kap-β1 to compete effectively for shared NPC docking sites needed for the transport of cNLS cargo. At higher concentrations, wt p10 is also inhibitory to cNLS import (Fig. 4) (Tachibana et al. 1996), thus we believe that the D23A mutation only increases the affinity of p10 for preexisting p10 NPC binding sites rather than causing p10 to bind completely new sites.
Competition studies showed that p10 and Kap-β1 appear to use many of the same, or closely overlapping, NPC docking sites for movement through the NPC. An excess of Kap-β1 will block the binding of p10 to the NPC (Fig. 7), and an excess of either p10 (Fig. 4) or Kap-β1 (Fig. 8) will inhibit both the import of their own cargo as well as cargo transported by the other nuclear transport pathway. Thus, our results support the conclusion that a carrier of one class can compete with a carrier of another class for transport, probably because of competition for shared binding sites at the NPC.
To what extent do different nuclear carriers share docking sites at the NPC? Both biochemical and genetic data have implicated the family of repeat Nups in mediating Kap-β1 transport across the NPC (Iovine et al. 1995; Radu et al. 1995a,Radu et al. 1995b; Rexach and Blobel 1995). Many of the same repeat Nups that can bind Kap-β1 can also bind p10 (though not necessarily at the same site), and other members of the Kap-β family have been shown to bind the same repeat Nups, as well (Aitchison et al. 1996; Nehrbass and Blobel 1996; Bonifaci et al. 1997; Pemberton et al. 1997; Rosenblum et al. 1997; Rout et al. 1997; Nakielny et al. 1999; Ullman et al. 1999; Yaseen and Blobel 1999). Görlich and coworkers engineered a truncated Kap-β1 lacking its Ran binding domain and showed that it accumulates at NPCs and inhibits most nuclear transport pathways, including cNLS and M9-NLS cargo import as well as the export of mRNAs, UsnRNAs, and leucine-rich NES-containing proteins (Kutay et al. 1997b). Here we found that a similar truncated Kap-β1 (45–462) also inhibits the p10-mediated import of RanGDP (Fig. 2).
However, there is good evidence that not all carrier binding sites at the NPC are exactly equivalent. Forbes and coworkers showed that Nup153 binds two different transport carriers (Kap-β1 and transportin/Kap-β2) at different sites of the Nup153 protein (Shah and Forbes 1998). The Gerace laboratory found that the presence of a 100-fold molar excess of p10 did not interfere with the binding of Kap-β1 to three different subunits of the p62 complex (p62, p58, and p54), all of which contain FG repeats (Hu et al. 1996). However, our data shows that in the context of an assembled NPC, p10 is able to compete with Kap-β1 for sites critical for the import of cNLS cargo (Fig. 4 and Fig. 5). Görlich and coworkers also reported that there appear to be at least two separate binding sites for Kap-β1 at the NPC (Kutay et al. 1997a). At least one of these sites affects p10 binding because we observed that not only would Kap-β1 completely prevent p10 binding at the NPC (Fig. 7), but that p10 bound at the NPC could be completely displaced by the subsequent addition of Kap-β1 (data not shown). These results may indicate that p10 and Kap-β1 binding sites at the NPC are overlapping, but not identical, or it might indicate that these two proteins can bind to the same sites but with different affinities. The increased occupancy of these NPC sites by D23A p10 could result in Kap-β1 no longer being able to compete efficiently for shared docking sites, thereby inhibiting cNLS import.
We hypothesize that all transport carriers of the Kap-β or the p10-type are likely to use FG repeats as docking sites at the NPC (heavily, but not necessarily exclusively) and that the amino acids surrounding the FG repeat (such as GLFG or FXFG) can raise or lower a particular carrier's affinity for that docking site. In addition, larger substrates might need more available NPC docking sites than smaller ones during transport. This would explain results that we obtained previously when we examined, by electron microscopy, the effects of microinjected p10 on the ability of Xenopus NPCs to import cNLS–gold conjugates of different sizes (Feldherr et al. 1998). Stage 2 Xenopus oocytes begin to synthesize predominantly 18S and 28S rRNA, rather than the mainly 5S RNA that is synthesized in stage 1 oocytes. This shift is accompanied by an increase in both the rate of nuclear transport and the size of particles able to move through the NPCs (Feldherr et al. 1998). We found that microinjection of p10 into stage 2 oocytes decreases the size of cNLS–gold particles able to be imported through the NPC, such that their import capacity is now identical to that of a stage 1 oocyte and that this effect is dependent on the amount of p10 injected (Feldherr et al. 1998). We concluded that the amount of p10 in the cell may regulate the functional size of the NPC during oogenesis, but at the time we had no idea how this might be accomplished.
Now, we hypothesize that the observed decrease in the size of cNLS–gold that could be imported after microinjection of p10 is directly related to the increased occupation of docking sites at the NPC by p10 that are also needed by Kap-β1 for cNLS import, and that a larger cargo likely needs more empty carrier docking sites to sustain its movement across the NPC than a smaller one. Here we found that microinjected D23A p10 at 2.2 mg/ml into HeLa cells inhibits the import of coinjected BSA–NLS, but that wt p10 microinjected at the same concentration does not (Fig. 5). Because D23A p10 shows increased binding to the NPC relative to wt p10, microinjection of equal concentrations of each would probably result in more D23A p10 bound at the NPC than wt p10. However, in additional experiments not shown, we found that if we increased the microinjected concentration of each to 5 mg/ml, both wt p10 as well as the D23A mutant would inhibit BSA–NLS import. This is in agreement with the findings of Yoneda and coworkers who found that microinjection of p10 (5 mg/ml) into mammalian cells would inhibit both the import of cNLS cargo and the export of leucine-rich NES cargo (Tachibana et al. 1996). If in the Xenopus oocyte study we had microinjected a higher concentration of p10 into the Xenopus oocytes than we did, our prediction now would be that we would have observed a complete shutdown of cNLS–gold import due to an increased occupation of NPC binding sites (Feldherr et al. 1998).
In the crystal structure of p10, amino acid D23 forms an in-line hydrogen bonded relay with another amino acid, H66, which is located at the base of the hydrophobic cavity that is the central feature of the p10 structure (Bullock et al. 1996). Clarkson et al. 1997 showed that D23N p10 (as opposed to the D23A mutant used here) is able to bind both RanGDP and Nsp1p (an FXFG repeat containing yeast nucleoporin), however the ability of D23N p10 to support nuclear import was not determined. With regard to p10's NPC binding ability, an analysis of the p10 crystal structure reveals a hydrophobic patch on the surface of the p10 molecule (centered roughly around Trp7) that was predicted by Stewart and coworkers to possibly interact with the hydrophobic-aromatic side chains of an FXFG repeat (Bayliss et al. 1999). Accordingly, an amino acid constituent of this hydrophobic patch was mutated (W7A) and W7A p10 was shown to have decreased, but not abolished, binding to an FXFG-containing piece of repeat Nup p62. Experiments are in progress in our laboratory to determine the crystal structure of D23A p10 to see what structural changes resulting from this mutation could be responsible for p10's increased binding to the NPC.
Interestingly, a protein (p15/NXT1) that shows significant homology to p10 was identified in HeLa cells and shown to bind to the mRNA export factor TAP (Katahira et al. 1999). p15, like p10, was shown to bind to the nuclear envelope and, perhaps significantly, amino acid D23 (D29 in p15) is conserved between p10 and p15, although H66 is not. The TAP–p15 complex was found to complement a Mex67-Mtr2 double deletion in yeast (both of which have been implicated in mRNA export), even though neither Mex67p nor Mtr2p appear to be the yeast homologue of p15, which does not in fact appear to have a yeast homologue at all. Unlike p10, which preferentially binds RanGDP, p15 strongly and specifically binds RanGTP (Black et al. 1999). Very recently, Dargemont and coworkers reported that p15 strongly stimulates export of U1snRNA, tRNA, and mRNA in an in vitro nuclear export assay in permeabilized HeLa cells, and that the ability of p15 to stimulate export is dependent on its capacity to bind RanGTP (Ossareh-Nazari et al. 2000). These results indicate that p15, like p10, likely uses its NPC binding ability to facilitate the nuclear transport of cargo.
We feel that the results shown here raise the distinct possibility that a cell could increase or decrease the rate of transport of a particular cargo in response to a cellular signal by changing the affinity of a carrier for the NPC by, for example, a posttranslational modification. Slightly changing the affinity of a carrier for its NPC docking sites could substantially increase or decrease the rate of transport of its cargo, both as measured directly and in relation to different cargoes, by altering that carrier's ability to compete with other carriers for shared docking sites. The effect of our D23A mutation on p10 may be mimicking a naturally occurring posttranslational modification that can affect carrier function in vivo.
We have estimated the normal in vivo concentration of p10 to be ∼106 molecules per HeLa cell (data not shown), which would make p10 in HeLa cells ∼1/10th as abundant as Ran (Bischoff and Ponstingl 1991). As p10 is only one of a large number of carriers capable of interacting with the NPC, many of which are probably competing for the same sites, even a slight change in the affinity of one for shared docking sites could shift the equilibrium between different nuclear transport pathways. Whether the rates of nuclear transport inside a cell are in fact modulated by such a change in nuclear carrier affinity for the NPC remains undetermined, but we are examining this possibility.
Acknowledgments
We thank M. Rush, A. Lamond, D. Görlich, U. Grundmann, G. Blobel, and S. Adam for expression plasmids and B. Schwartz for the use of his microinjector. We also thank D. Hoffmaster and Q. Xie for excellent technical assistance and S. Mullican for additional p10 microinjection experiments. We thank E. Schwoebel and B. Talcott for their help preparing some of the figures and also for their comments on the manuscript.
This work was supported by a grant from the National Institutes of Health (GM53678), the Searle Scholar/Chicago Community Trust, and also by Reproductive Training Grant (CML) T32-HD07165.
Footnotes
Abbreviations used in this paper: BRL, buffalo rat liver; cNLS, classical nuclear localization sequence; GEF, guanine nucleotide exchange factor; Kap, karyopherins; NES, nuclear export sequence; NLS, nuclear localization sequence; NPC, nuclear pore complex; NTF, nuclear transport factor; Nups, nucleoporins; TB, transport buffer; wt, wild-type.
References
- Aitchison J.D., Blobel G., Rout M.P. Kap104pa karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Bayliss R., Ribbeck K., Akin D., Kent H.M., Feldherr C.M., Görlich D., Stewart M. Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J. Mol. Biol. 1999;293:579–593. doi: 10.1006/jmbi.1999.3166. [DOI] [PubMed] [Google Scholar]
- Bischoff F.R., Ponstingl H. Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc. Natl. Acad. Sci. USA. 1991;88:10830–10834. doi: 10.1073/pnas.88.23.10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black B.E., Levesque L., Holaska J.M., Wood T.C., Paschal B.M. Identification of an NTF2-related factor that binds Ran-GTP and regulates nuclear protein export. Mol. Cell. Biol. 1999;19:8616–8624. doi: 10.1128/mcb.19.12.8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaci N., Moroianu J., Radu A., Blobel G. Karyopherin β2 mediates nuclear import of an mRNA binding protein. Proc. Natl. Acad. Sci. USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock T.L., Clarkson W.D., Kent H.M., Stewart M. The 1.6 angstroms resolution crystal structure of nuclear transport factor 2 (NTF2) J. Mol. Biol. 1996;260:422–431. doi: 10.1006/jmbi.1996.0411. [DOI] [PubMed] [Google Scholar]
- Clarkson W.D., Corbett A.H., Paschal B.M., Kent H.M., McCoy A.J., Gerace L., Silver P.A., Stewart M. Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran. J. Mol. Biol. 1997;272:716–730. doi: 10.1006/jmbi.1997.1255. [DOI] [PubMed] [Google Scholar]
- Clarkson W.D., Kent H.M., Stewart M. Separate binding sites on nuclear transport factor 2 (NTF2) for GDP-Ran and the phenylalanine-rich repeat regions of nucleoporins p62 and Nsp1p. J. Mol. Biol. 1996;263:517–524. doi: 10.1006/jmbi.1996.0594. [DOI] [PubMed] [Google Scholar]
- Corbett A.H., Silver P.A. The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J. Biol. Chem. 1996;271:18477–18484. doi: 10.1074/jbc.271.31.18477. [DOI] [PubMed] [Google Scholar]
- Davis L.I., Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- Englmeier L., Olivo J.C., Mattaj I.W. Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr. Biol. 1999;9:30–41. doi: 10.1016/s0960-9822(99)80044-x. [DOI] [PubMed] [Google Scholar]
- Feldherr C., Akin D., Moore M.S. The nuclear import factor p10 regulates the functional size of the nuclear pore complex during oogenesis. J. Cell Sci. 1998;111:1889–1896. doi: 10.1242/jcs.111.13.1889. [DOI] [PubMed] [Google Scholar]
- Floer M., Blobel G. Putative reaction intermediates in Crm1-mediated nuclear protein import. J. Biol. Chem. 1999;274:16279–16286. doi: 10.1074/jbc.274.23.16279. [DOI] [PubMed] [Google Scholar]
- Floer M., Blobel G., Rexach M. Disassembly of RanGTP-karyopherin β complex, an intermediate in nuclear protein import. J. Biol. Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- Fornerod M., van Deursen J., van Baal S., Reynolds A., Davis D., Murti K.G., Fransen J., Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Dabrowski M., Bischoff F.R., Kutay U., Bork P., Hartmann E., Prehn S., Izaurralde E. A novel class of RanGTP binding proteins. J. Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Pante N., Kutay U., Aebi U., Bischoff F.R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Hu T., Guan T., Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J. Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine M.K., Watkins J.L., Wente S.R. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J. Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Kutay U., Von Kobbe C., Mattaj I.W., Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J., Strasser K., Podtelejnikov A., Mann M., Jung J.U., Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose S., Imamoto N., Tachibana T., Shimamoto T., Yoneda Y. Ran-unassisted nuclear migration of a 97-kD component of nuclear pore-targeting complex. J. Cell Biol. 1997;139:841–849. doi: 10.1083/jcb.139.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Bischoff F.R., Kostka S., Kraft R., Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor Cell 90 1997. 1061 1071a [DOI] [PubMed] [Google Scholar]
- Kutay U., Izaurralde E., Bischoff F.R., Mattaj I.W., Görlich D. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex EMBO (Eur. Mol. Biol. Organ.) J 16 1997. 1153 1163b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Lipowsky G., Izaurralde E., Bischoff F.R., Schwarzmaier P., Hartmann E., Görlich D. Identification of a tRNA-specific nuclear export receptor. Mol. Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Lanford R.E., Kanda P., Kennedy R.C. Induction of nuclear transport with a synthetic peptide homologous to the SV40 T antigen transport signal. Cell. 1986;46:575–582. doi: 10.1016/0092-8674(86)90883-4. [DOI] [PubMed] [Google Scholar]
- Mahajan R., Delphin C., Guan T., Gerace L., Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Matunis M.J., Coutavas E., Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.S., Blobel G. The two steps of nuclear import, targeting to the nuclear envelope and translocation through the nuclear pore, require different cytosolic factors. Cell. 1992;69:939–950. doi: 10.1016/0092-8674(92)90613-h. [DOI] [PubMed] [Google Scholar]
- Moore M.S., Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc. Natl. Acad. Sci. USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S., Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Nakielny S., Shaikh S., Burke B., Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U., Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M., Okazaki H., Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation, locates in the nucleus and binds to DNA. J. Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B., Maison C., Black B.E., Levesque L., Paschal B.M., Dargemont C. RanGTP-binding protein NXT1 facilitates nuclear export of different classes of RNA in vitro. Mol. Cell Biol. 2000;20:4562–4571. doi: 10.1128/mcb.20.13.4562-4571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal B.M., Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J. Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal B.M., Delphin C., Gerace L. Nucleotide-specific interaction of Ran/TC4 with nuclear transport factors NTF2 and p97. Proc. Natl. Acad. Sci. USA. 1996;93:7679–7683. doi: 10.1073/pnas.93.15.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal B.M., Fritze C., Guan T., Gerace L. High levels of the GTPase Ran/TC4 relieve the requirement for nuclear protein transport factor 2. J. Biol. Chem. 1997;272:21534–21539. doi: 10.1074/jbc.272.34.21534. [DOI] [PubMed] [Google Scholar]
- Pemberton L.F., Rosenblum J.S., Blobel G. A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J. Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P., Clarkson W.D., Kent H.M., Rhodes D., Stewart M. Molecular interactions between the importin α/β heterodimer and proteins involved in vertebrate nuclear protein import. J. Mol. Biol. 1997;266:722–732. doi: 10.1006/jmbi.1996.0801. [DOI] [PubMed] [Google Scholar]
- Radu A., Blobel G., Moore M.S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins Proc. Natl. Acad. Sci. USA. 92 1995. 1769 1773a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A., Moore M.S., Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex Cell 81 1995. 215 222b [DOI] [PubMed] [Google Scholar]
- Ren M., Drivas G., D'Eustachio P., Rush M.G. Ran/TC4a small nuclear GTP-binding protein that regulates DNA synthesis. J. Cell Biol. 1993;120:313–323. doi: 10.1083/jcb.120.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M., Blobel G. Protein import into nucleiassociation and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Ribbeck K., Lipowsky G., Kent H.M., Stewart M., Görlich D. NTF2 mediates nuclear import of Ran. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:6587–6598. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum J.S., Pemberton L.F., Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J. Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Blobel G., Aitchison J.D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Ryan K.J., Wente S.R. The nuclear pore complexa protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 2000;12:361–371. doi: 10.1016/s0955-0674(00)00101-0. [DOI] [PubMed] [Google Scholar]
- Schwoebel E.D., Talcott B., Cushman I., Moore M.S. Ran-dependent signal-mediated nuclear import does not require GTP hydrolysis by Ran. J. Biol. Chem. 1998;273:35170–35175. doi: 10.1074/jbc.273.52.35170. [DOI] [PubMed] [Google Scholar]
- Shah S., Forbes D.J. Separate nuclear import pathways converge on the nucleoporin nup153 and can be dissected with dominant-negative inhibitors. Curr. Biol. 1998;8:1376–1386. doi: 10.1016/s0960-9822(98)00018-9. [DOI] [PubMed] [Google Scholar]
- Smith A., Brownawell A., Macara I.G. Nuclear import of Ran is mediated by the transport factor NTF2. Curr. Biol. 1998;8:1403–1406. doi: 10.1016/s0960-9822(98)00023-2. [DOI] [PubMed] [Google Scholar]
- Stewart M., Kent H.M., McCoy A.J. Structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J. Mol. Biol. 1998;277:635–646. doi: 10.1006/jmbi.1997.1602. [DOI] [PubMed] [Google Scholar]
- Tachibana T., Hieda M., Sekimoto T., Yoneda Y. Exogenously injected nuclear import factor p10/NTF2 inhibits signal- mediated nuclear import and export of proteins in living cells. FEBS Lett. 1996;397:177–182. doi: 10.1016/s0014-5793(96)01180-5. [DOI] [PubMed] [Google Scholar]
- Talcott B., Moore M.S. Getting across the nuclear pore complex. Trends Cell Biol. 1999;9:312–318. doi: 10.1016/s0962-8924(99)01608-6. [DOI] [PubMed] [Google Scholar]
- Ullman K.S., Shah S., Powers M.A., Forbes D.J. The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell. 1999;10:649–664. doi: 10.1091/mbc.10.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S.R., Rout M.P., Blobel G. A new family of yeast nuclear pore complex proteins. J. Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D.H., Corbett A.H., Kent H.M., Stewart M., Silver P.A. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol. Cell. Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaseen N.R., Blobel G. Two distinct classes of Ran-binding sites on the nucleoporin Nup-358. Proc. Natl. Acad. Sci. USA. 1999;96:5516–5521. doi: 10.1073/pnas.96.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]