Abstract
Mutations in the dynamin-related GTPase, Mgm1p, have been shown to cause mitochondrial aggregation and mitochondrial DNA loss in Saccharomyces cerevisiae cells, but Mgm1p's exact role in mitochondrial maintenance is unclear. To study the primary function of MGM1, we characterized new temperature sensitive MGM1 alleles. Examination of mitochondrial morphology in mgm1 cells indicates that fragmentation of mitochondrial reticuli is the primary phenotype associated with loss of MGM1 function, with secondary aggregation of mitochondrial fragments. This mgm1 phenotype is identical to that observed in cells with a conditional mutation in FZO1, which encodes a transmembrane GTPase required for mitochondrial fusion, raising the possibility that Mgm1p is also required for fusion. Consistent with this idea, mitochondrial fusion is blocked in mgm1 cells during mating, and deletion of DNM1, which encodes a dynamin-related GTPase required for mitochondrial fission, blocks mitochondrial fragmentation in mgm1 cells. However, in contrast to fzo1 cells, deletion of DNM1 in mgm1 cells restores mitochondrial fusion during mating. This last observation indicates that despite the phenotypic similarities observed between mgm1 and fzo1 cells, MGM1 does not play a direct role in mitochondrial fusion. Although Mgm1p was recently reported to localize to the mitochondrial outer membrane, our studies indicate that Mgm1p is localized to the mitochondrial intermembrane space. Based on our localization data and Mgm1p's structural homology to dynamin, we postulate that it functions in inner membrane remodeling events. In this context, the observed mgm1 phenotypes suggest that inner and outer membrane fission is coupled and that loss of MGM1 function may stimulate Dnm1p-dependent outer membrane fission, resulting in the formation of mitochondrial fragments that are structurally incompetent for fusion.
Keywords: fission, membrane remodeling, inner membrane, organelle, morphology
Introduction
During cell division, copies of both the mitochondrial organelle and its genome must be inherited for progeny to survive and be respiratory competent. The accurate partitioning of these components into daughter cells is dependent on the morphology of the mitochondrial organelle. In S. cerevisiae, mitochondria form a reticulum distributed at the cell cortex (Hoffman and Avers 1973; Stevens and White 1979; Nunnari et al. 1997). The maintenance of this structure is a complex process dependent on cytoskeletal elements and mitochondrial-associated proteins (Hermann and Shaw 1998; Yaffe 1999).
The continuity and reticular nature of yeast mitochondria is also established and maintained by balanced fission and fusion events (Nunnari et al. 1997). The coordinate fusion of the mitochondrial outer and inner membranes requires the evolutionarily conserved Fzo family of mitochondrial outer membrane GTPases (Hales and Fuller 1997; Hermann et al. 1998; Rapaport et al. 1998). In Drosophila, mutations in Fzo block a developmentally regulated mitochondrial fusion event during spermatogenesis (Hales and Fuller 1997). In yeast, a conditional fzo1 mutation causes mitochondrial reticuli to fragment, a phenotype consistent with ongoing mitochondrial fission and a block in mitochondrial fusion, and also causes mitochondrial DNA (mtDNA) loss in cells during vegetative growth (Hermann et al. 1998). In addition, mitochondrial fusion is blocked in fzo1 cells during yeast mating (Hermann et al. 1998).
Mitochondrial fission is mediated by the dynamin-related GTPase, Dnm1p (Otsuga et al. 1998; Bleazard et al. 1999; Sesaki and Jensen 1999). Dnm1p is a cytoplasmic protein that is found concentrated in punctate structures localized to the tips and sides of mitochondrial tubules in cells (Otsuga et al. 1998). These structures are associated with the mitochondrial outer membrane and are found at sites where both the inner and outer membranes are coordinately constricted (Bleazard et al. 1999). In yeast, deletion of DNM1 causes mitochondria to form a net-like structure, resulting from a defect in mitochondrial division, but has no effect on mtDNA inheritance (Bleazard et al. 1999; Sesaki and Jensen 1999). Deletion of DNM1 in fzo1 cells blocks mitochondrial fragmentation, consistent with their respective antagonistic roles in fission and fusion (Bleazard et al. 1999; Sesaki and Jensen 1999). Dnm1p homologues in higher eucaryotes also have been shown to control mitochondrial fission, indicating their role is evolutionarily conserved (Smirnova et al. 1998; Labrousse et al. 1999).
In the case of double-membraned organelles such as mitochondria and chloroplasts, mechanisms likely exist to coordinate fission and fusion events of the outer and inner membranes, and to maintain the separation of these membranes. Data suggest that coordination of chloroplast outer and inner membrane fission is accomplished by the differential localization and action of two distinct homologues of the bacterial cell division GTPase, FtsZ (FtsZ1 and FtsZ2; Osteryoung and Vierling 1995; Osteryoung et al. 1998). Chloroplast FtsZ2, like mitochondrial Dnm1p, is predicted to localize to the cytosolic face of the outer membrane. In contrast, the chloroplast FtsZ1 protein is associated with the inner membrane and localized within the stromal compartment. Thus, the requirement for two similar components and their unique localization to each membrane might serve as a mechanism to coordinate these membranes during the fission process. These observations raise the question of whether, in addition to outer membrane associated Dnm1p, components localized to the mitochondrial inner membrane are required for mitochondrial fission. Thus, it is of interest that a second dynamin-related GTPase, Mgm1p, has been shown to localize to mitochondria and play a role in the maintenance of mitochondrial structure in yeast cells (Jones and Fangman 1992; Guan et al. 1993; Shepard and Yaffe 1999).
Despite the structural similarities between Dnm1p and Mgm1p, the phenotypes associated with dnm1 cells and mgm1 cells are distinct. Unlike dnm1 cells, mutations in MGM1 cause mitochondrial aggregation and mtDNA loss in cells (Jones and Fangman 1992; Guan et al. 1993; Shepard and Yaffe 1999). Structural Mgm1p homologues have been identified in higher eucaryotes, such as Schizosaccharomyces pombe, where the Mgm1p homologue, Msp1p, is required for the maintenance of mtDNA during cell division and its overexpression causes mitochondrial aggregation (Pelloquin et al. 1998, Pelloquin et al. 1999). Thus, the role of this subfamily of dynamin-related GTPases in mitochondrial maintenance is also conserved. However, the reported submitochondrial localization of Mgm1p homologues in these organisms is different. Mgm1p was reported to localize to the mitochondrial outer membrane (Shepard and Yaffe 1999), whereas Msp1p was reported to localize to the matrix, associated with the mitochondrial inner membrane (Pelloquin et al. 1999). Progress towards understanding the function of Mgm1p in mitochondrial maintenance has been limited by both the mutant phenotypes of mgm1 cells and the contrasting localization data in different organisms.
In contrast to published observations, our localization data indicate that Mgm1p is in the intermembrane space compartment of mitochondria, associated with the inner membrane. We also show that loss of MGM1 function results in the rapid fragmentation of mitochondrial reticuli with secondary aggregation of mitochondrial fragments. In addition, we have found that mitochondrial fusion in mgm1 cells is blocked during mating. However, MGM1 does not appear to function directly in fusion, because its requirement can be bypassed by the deletion of DNM1. Instead, our data are consistent with the idea that loss of MGM1 function triggers Dnm1p-dependent outer membrane fission, resulting in the formation of fusion incompetent mitochondrial fragments. Taken together, our data suggest a model where Mgm1p functions in the intermembrane space in coordination with Dnm1p-dependent outer membrane fission to regulate inner membrane division and/or structure.
Materials and Methods
Yeast Genetic Techniques
Media preparations and genetic techniques were performed as described (Guthrie and Fink 1991). Double mutants were constructed by mating, sporulation, and dissection. Yeast strains used in this study are listed in Table .
Table 1.
Yeast Strains Used in This Study
| Strain | Genotype | Source |
|---|---|---|
| W303 | ade2-1, leu2-3, his3-11,15, trp1-1, ura3-1, can1-100, MATa or MATα | R. Rothstein |
| JNY177 | same as W303, except mgm1-5, MATa | This study |
| JNY543 | same as JNY177, MATα | This study |
| JNY537 | same as W303, except Δmgm1::HIS3, MATα | This study |
| JNY539 | same as W303, except fzo1-1, MATa | This study |
| JNY540 | same as JNY539, except MATα | This study |
| JNY524 | Δdnm1::HIS3, mgm1-5, MATa | This study |
| JNY525 | same as JNY524, except MATα | This study |
| JNY500 | same as W303, except Δdnm1::HIS3, fzo1-1 mgm1-5, MATa | This study |
| JNY502 | same as JNY500, except MATα | This study |
| JSY836 | ura3-52, his3Δ200, trp1Δ63, MATa | Bleazard et al. 1999 |
| JSY2519 | same as JSY836, except MGM1:3xHA, MATa | This study |
| JSY3753 | ura3-52, his3Δ200, Δdnm1::HIS3, MATα | Bleazard et al. 1999 |
| JSY3754 | same as JSY3753, MATa | Bleazard et al. 1999 |
| JSY3752 | ura3-52, his3Δ200, Δdnm1 fzo1-1, MATa | Bleazard et al. 1999 |
| JSY3755 | same as JSY3752, except MATα | Bleazard et al. 1999 |
Isolation and Characterization of mgm1 Alleles
Mutant alleles of MGM1 were isolated in a previously described screen for conditional, recessive nuclear mutations that cause loss of mtDNA at the nonpermissive temperature of 37°C (Meeusen et al. 1999). MGM1 alleles isolated in this screen were identified by complementation analysis with a null allele of MGM1. Sporulation and tetrad analysis of diploids confirmed mutations were linked to the MGM1 locus. All mgm1 alleles isolated were recessive for both glycerol growth and mitochondrial morphology defects.
To characterize mgm1 alleles, cultures were grown at the permissive temperature (25°C) to log phase in YPG, cells were plated onto YPD plates, and colony color was assessed and quantified. Because all strains analyzed contained the ade2 mutation, on YPD, white colonies were classified as respiratory deficient and red colonies were classified as respiratory competent. To directly visualize mtDNA, cells were fixed in 70% ethanol containing 10 ng/ml DAPI and washed with PBS. Mitochondrial morphology was visualized and quantified either by incubating cells in YPG with MitoTracker CMXR (Molecular Probes) or by mitochondrial matrix targeted GFP (mito-GFP, kindly provided by B.Westerman and W. Neupert, Ludwig Maximilians Universitaet Muenchen, Muenchen, Germany).
Sequence Analysis of mgm1-5
The mutation at the MGM1 locus in mgm1-5 cells was identified by DNA sequence analysis. Yeast genomic DNA prepared from mgm1-5 and W303 strains as described (Guthrie and Fink 1991) was used as template for PCR with the following sets of primers derived from MGM1 locus: 5′-tcc ccc ggg gga ctc ttt ttg ggt agt aca gat atc a-3′ and 5′-tgc tgg cct ttt gaa aat atc c-3′; 5′-acg ata ggt gtc att acc aaa ctg g -3′ and 5′- gct cta gag caa ggg tat gcc ttt att ttt tca ga -3′ (Custom Primers Inc.). The PCR products from eight independent reactions with a given set of primers were pooled, gel purified, and sequenced using the above listed primers and 5′-ggg atg gct gct gca ggg agt tat-3′ and 5′-aag agt tgg atg ata ctt ctt att-3′ by Davis Sequencing.
In Vivo Mitochondrial Fusion Assay
To assess mitochondrial fusion, haploid strains of opposite mating type were labeled with either mito-GFP or MitoTracker CMXR, mated at 25 or 37°C, and analyzed as described by Nunnari et al. 1997.
Construction of HA-tagged Mgm1p
JSY2519 was generated by integrating a 3X-HA-URA3-3X-HA (hemagglutinin) cassette between codons 173 and 174 of the MGM1 open reading frame in JSY836 (Schneider et al. 1995). Cells that had undergone recombination and loss of the URA3 portion of the cassette were selected by growth on 5-fluro orotic acid. The resulting JSY2519 strain expressed full-length Mgm1 protein containing 3X-HA epitope inserted in frame between codons 173 and 174 of the polypeptide chain as confirmed by DNA sequencing and Western analysis.
Biochemical Analyses
Mitochondria were prepared from JSY2519 and protease protection experiments were conducted on enriched mitochondrial fractions as described (Nunnari et al. 1993). Proteins were analyzed by SDS-PAGE and by Western blotting. For Western analysis, the following antibodies were used at a 1:1,000 dilution: anti-Fzo1p, monoclonal anti-HA (Covance Inc.), anti-Tim23p (R. Jensen, Johns Hopkins University, Baltimore, MD), and anticytochrome b2, anti-KDH, and anti-ADP/ATP carrier (AAC; C. Koehler, UCLA, Los Angeles, CA). Western blots were developed using HRP-conjugated secondary antibodies and enhanced chemiluminescence reagents (Amersham Pharmacia Biotech).
Indirect Immunofluorescence
JSY2519 cells were grown to log phase in YPG and stained with MitoTracker CMXR for 30 min at 25°C as described (Nunnari et al. 1997). Cells were fixed overnight in 3.7% formaldehyde and processed for indirect immunofluorescence (Meeusen et al. 1999). Monoclonal HA antibodies (Covance, Inc.) were preadsorbed with JSY2519 cells as described, and used at a 1:5 dilution (Hermann et al. 1998). Oregon green-conjugated anti-mouse secondary (Molecular Probes) was used at 1:200.
Microscopy
All samples were imaged using a Leica confocal microscope with a 100× 1.4 NA objective (Leica, Inc.). Cryoimmunoelectron microscopy was performed as described (Bleazard et al. 1999). Images were analyzed using Image Space software and figures were prepared using Adobe Photoshop.
Results
The Primary Defect Associated with Loss of MGM1 Function Is Fragmentation of Mitochondrial Reticuli
Previous work showed that MGM1 is required for wild-type mitochondrial morphology and respiratory competence. Specifically, mtDNA loss and mitochondrial aggregation and inheritance defects were observed in temperature sensitive mgm1 and Δmgm1 cells (Guan et al. 1993; Jones and Fangman 1992; Shepard and Yaffe 1999). To gain more insight into the exact role of MGM1 in morphology maintenance, we analyzed new temperature-sensitive alleles of MGM1 isolated in a screen for conditional mutants that are unable to maintain mtDNA (Meeusen et al. 1999).
To determine the primary defect associated with loss of MGM1 function, we focused on one conditional allele of MGM1, mgm1-5. This allele was chosen because analysis of our collection of mgm1 mutants indicated that mgm1-5 cells exhibited the greatest and most rapid phenotypic change upon shifting to nonpermissive temperature. Specifically, under permissive conditions, no difference in the generation of respiratory incompetent colonies was observed between mgm1-5 and wild-type cells, indicating that Mgm1p is functional under these conditions (99% respiratory competent, n = 100). In contrast, under restrictive conditions, 100% of mgm1-5 cells were respiratory incompetent and devoid of mtDNA as compared with 3% of wild-type cells, indicating a complete loss of MGM1 function (n = 100). In heterozygous mgm1-5 MGM1 strains, mgm1-5 phenotypes were recessive, indicating that the mutation causes a loss of MGM1 function at nonpermissive temperature. The MGM1 locus in mgm1-5 cells contains a single point mutation that changes G408 to D408. This residue is contained within the GTPase domain of the protein, consistent with previously published observations indicating its importance for MGM1 function (Shepard and Yaffe 1999).
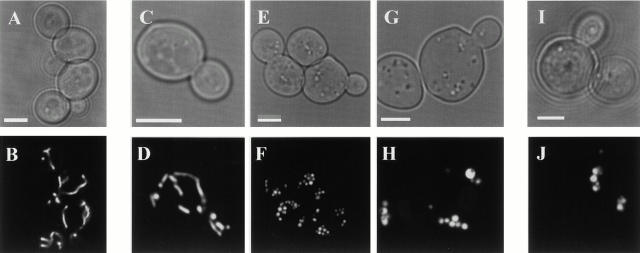
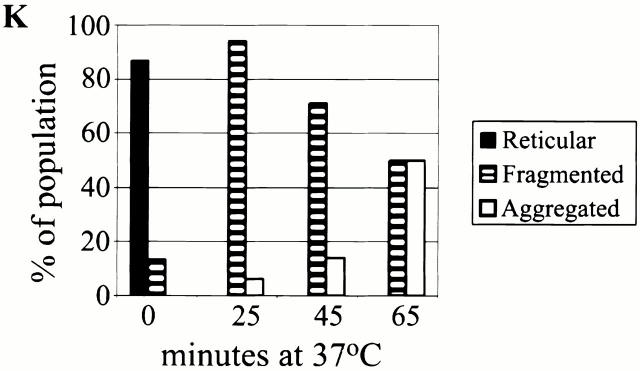
To gain more insight into MGM1 function, we examined mitochondrial morphology in mgm1-5 cells using mito-GFP. Under permissive conditions, the majority of mgm1-5 cells contained reticular mitochondria, similar to that observed in wild-type cells (Fig. 1, compare B to D). Within 20 min after shifting to the nonpermissive temperature, mitochondrial reticuli fragmented into many smaller mitochondria that remained distributed at the cell cortex in mgm1-5 cells (Fig. 1, compare D to F). Extended exposure (60 min) of mgm1-5 cells to nonpermissive temperature resulted in aggregation of mitochondrial fragments (Fig. 1 H). This terminal morphological phenotype resembles the mitochondrial morphology defect observed in Δmgm1 cells (Fig. 1, compare H to J). Thus, the primary phenotype associated with loss of MGM1 function is mitochondrial fragmentation and a secondary phenotype is aggregation of these mitochondrial fragments (see Fig. 1 K for quantification).
Figure 1.
The primary phenotype resulting from loss of MGM1 function is fragmentation of mitochondrial reticuli. Mito-GFP was used to visualize mitochondria in wild-type (A and B), mgm1-5 (C and D), and Δmgm1 (I and J) cells grown overnight at 25°C to log phase, and after shifting to 37°C for 25 min (E and F) or 65 min (G and H). Quantification of mitochondrial morphology phenotypes at 37°C (K). Bars, 2 μm.
A previous study reported a mitochondrial inheritance defect in mgm1 cells and suggested that Mgm1p might be involved in the movement of mitochondria into daughter cells (Shepard and Yaffe 1999). We observed mitochondrial inheritance defects in mgm1-5 cells only after prolonged exposure to nonpermissive temperature. Specifically, 5% (n = 130) of buds lacked mitochondria after a 20-min incubation at 37°C versus 45% (n = 73) at 60 min. However, the majority of cells that lacked mitochondria in the bud contained a single mitochondrial aggregate. In addition, in a small percentage of cells, the mitochondrial aggregate was present in the bud and absent from the mother, indicating that in these cases the aggregate was transported into the yeast bud. Thus, Mgm1p does not function directly to mediate mitochondrial movement into daughter cells.
Mitochondrial Fusion Is Blocked in mgm1-5 Cells during Mating
Both mitochondrial fragmentation and mtDNA loss phenotypes observed in mgm1-5 cells are similar to the phenotypes associated with loss of FZO1 function (Hermann et al. 1998). In fzo1-1 cells, mitochondrial fragmentation results from a block in fusion and unopposed, ongoing mitochondrial fission. To determine if MGM1, like FZO1, has a role in mitochondrial fusion, we examined mitochondrial fusion in mgm1-5 cells during mating. Mitochondrial fusion was assayed as previously described by labeling mitochondria in haploid cells of opposite mating type with either a mito-GFP or a covalent vital probe, MitoTracker (Nunnari et al. 1997). Mitochondrial fusion was assessed by examining the distribution of these probes in large-budded zygotes formed at both permissive and nonpermissive temperature. Consistent with previously published observations, in wild-type zygotes, both haploid-derived mitochondrial probes were colocalized at permissive and nonpermissive temperatures indicating that haploid mitochondria had fused and their contents had mixed (Nunnari et al. 1997; Fig. 2, A–D; Table ). Haploid mitochondrial content mixing also occurred in the majority of the mgm1-5 zygotes formed at the permissive temperature (Fig. 2, E–H; Table ). In contrast, at the nonpermissive temperature, mitochondrial reticuli fragmented in mgm1-5 zygotes and the resulting fragments failed to fuse, even though mitochondria were closely associated with one another (Fig. 2, I–L; Table ). This fusion defect is similar to that observed in fzo1-1 zygotes, supporting the idea that MGM1 function is important for mitochondrial fusion (Hermann et al. 1998; Fig. 2, M–P; Table ).
Figure 2.
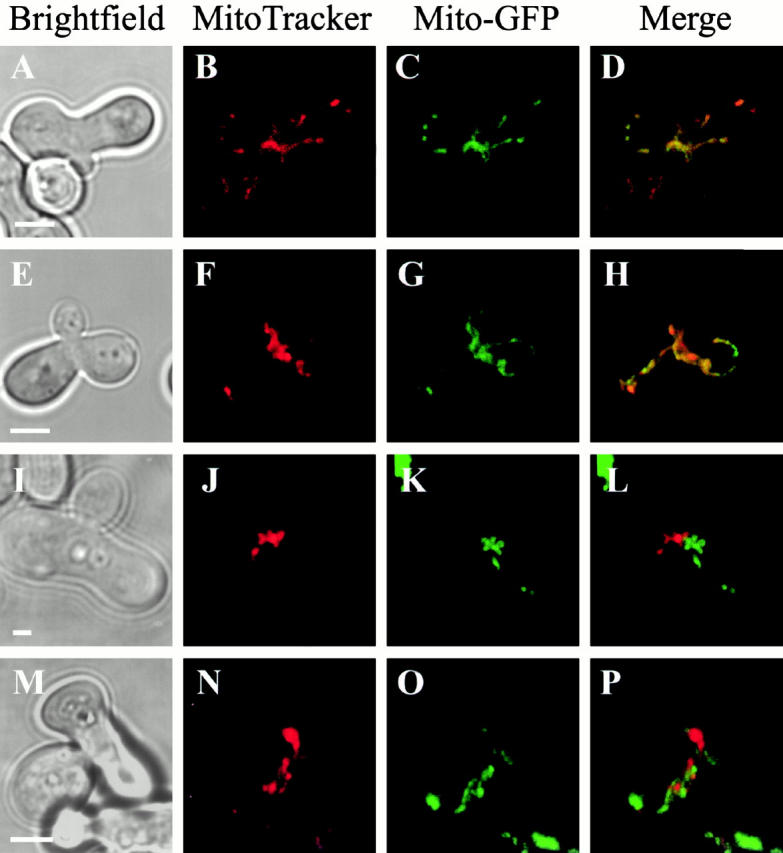
Mitochondrial fusion is blocked in mgm1-5 cells during mating. Cells of opposite mating type were grown to log phase at 25°C, labeled with either mito-GFP or MitoTracker and mated at 25°C (E–H) or 37°C (A–D, I–P). Mitochondrial fusion was assessed by examining in merged mito-GFP and MitoTracker images of large-budded homozygous zygotes formed from wild-type (A–D), mgm1-5 (E–L), and fzo1-1 (M–P) cells. Bars, 2 μm.
Table 2.
Quantification of Fusion in Large-budded Zygotes during Mating
| Homozygous cross | Temperature | Fused | Not fused |
|---|---|---|---|
| (n) | % | % | |
| MGM1 FZO1 DNM1 | 25°C (53) | 98 | 2 |
| 37°C (51) | 88 | 12 | |
| mgm1-5 FZO1 DNM1 | 25°C (52) | 69 | 31 |
| 37°C (54) | 4 | 96 | |
| MGM1 fzo1-1 DNM1 | 25°C (54) | 91 | 9 |
| 37°C (55) | 0 | 100 | |
| MGM1 FZO1 Δdnm1::HIS3 | 25°C (55) | 95 | 5 |
| 37°C (60) | 92 | 8 | |
| mgm1-5 FZO1 Δdnm1::HIS3 | 25°C (59) | 92 | 8 |
| 37°C (66) | 77 | 23 | |
| MGM1 fzo1-1 Δdnm1::HIS3 | 25°C (53) | 89 | 11 |
| 37°C (58) | 2 | 98 | |
| mgm1-5 fzo1-1 Δdnm1::HIS3 | 25°C (55) | 100 | 0 |
| 37°C (53) | 0 | 100 |
Deletion of DNM1 in mgm1-5 Cells Blocks Mitochondrial Fragmentation and Restores Mitochondrial Fusion during Mating
To further test the hypothesis that fragmentation of mitochondrial reticuli in mgm1-5 cells is the result of a block in fusion, we determined whether mitochondrial tubules were restored in mgm1-5 cells under conditions where mitochondrial fission is abolished. To block mitochondrial fission in mgm1-5 cells, we deleted DNM1, which encodes a dynamin-related GTPase required for mitochondrial division. Mitochondrial morphology was examined in mgm1-5 Δdnm1 cells at permissive and nonpermissive conditions using mito-GFP. Under permissive conditions, where Mgm1p is functional, mitochondrial net-like structures are observed in mgm1-5 Δdnm1 cells (Table ). These net-like mitochondrial structures are characteristic of Δdnm1 cells and likely arise because mitochondrial tubules fuse and new tubule ends cannot be generated by fission (Fig. 3E and Fig. F; Table ). At the nonpermissive temperature in mgm1-5 Δdnm1 cells, where mitochondrial fragmentation is observed in mgm1-5 cells, mitochondrial tubules and net structures are maintained (Fig. 3, compare C and D to G and H; Table ). Thus, abolishing Dnm1p-dependent mitochondrial fission blocks mitochondrial fragmentation resulting from loss of MGM1 function. In addition, the mgm1-5 glycerol growth and mtDNA loss phenotypes observed at nonpermissive temperature are suppressed in mgm1-5 Δdnm1 cells (not shown). Deletion of DNM1 in fzo1 cells also has been shown to block mitochondrial fragmentation and mtDNA loss.
Table 3.
Quantification of Mitochondrial Morphology at 37°C
| Cells | |||
|---|---|---|---|
| Strain | Wild-type/Reticular | Fragmented | Tubular/nets |
| % | % | % | |
| MGM1 FZO1 DNM1 | 99 | 1 | 0 |
| mgm1-5 FZO1 DNM1 | 0 | 100 | 0 |
| MGM1 FZO1 Δdnm1::HIS3 | 0 | 0 | 100 |
| MGM1 fzo1-1 DNM1 | 0 | 100 | 0 |
| mgm1-5 FZO1 Δdnm1::HIS3 | 0 | 0 | 100 |
| mgm1-5 fzo1-1 Δdnm1::HIS3 | 0 | 0 | 100 |
Total number of cells equals 100.
Figure 3.
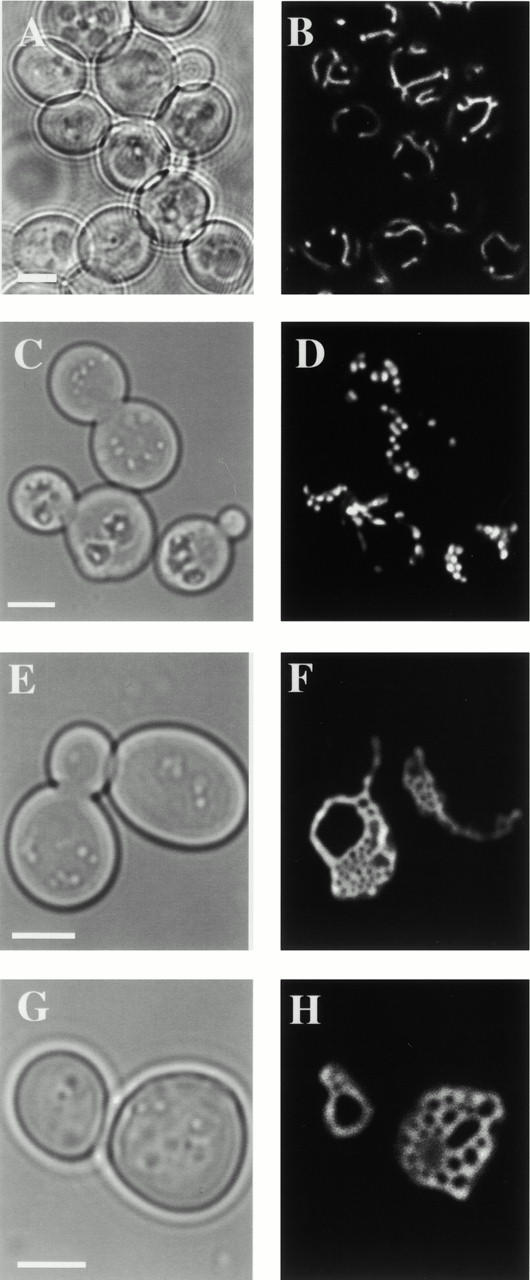
Deletion of DNM1 blocks mitochondrial fragmentation in mgm1-5 cells. Cells were grown to log phase at 25°C and shifted to 37°C for 40 min. Mitochondria were visualized using mito-GFP by fluorescence confocal microscopy in wild-type (A and B), mgm1-5 (C and D), Δdnm1 (E and F), and mgm1-5Δdnm1 (G and H) cells. Bars, 2 μm.
Although these data suggest that MGM1 might play a role in the fusion process, the structural similarity of Mgm1p to dynamin-like proteins suggests that it is involved in membrane remodeling and/or fission events. Thus, we considered the alternative explanation, that mitochondrial fragmentation in mgm1-5 cells results from an increase in Dnm1p-dependent mitochondrial fission. In this scenario, mitochondrial fragments might fail to fuse because of their geometry/structure and not as a primary consequence of loss of MGM1 function. To test this possibility, we assayed mitochondrial fusion during mating in mgm1-5 Δdnm1 cells, where deletion of DNM1 blocks mitochondrial fragmentation and restores mitochondrial tubular structures (Fig. 3G and Fig. H; Table ). As shown previously, deletion of DNM1 has no effect on mitochondrial fusion during mating, consistent with its role in fission (Bleazard et al. 1999; Fig. 4, A–D; Table ). Thus, as expected at the permissive temperature where MGM1 is functional, mitochondrial fusion and content mixing occurred in zygotes formed from mgm1-5 Δdnm1 haploids (Table ). In contrast to mgm1-5 cells at nonpermissive temperature, where both mitochondrial fragmentation and a block in mitochondrial fusion are observed, deletion of DNM1 in mgm1-5 cells restored mitochondrial fusion during mating (Fig. 4, E–H; Table ). Thus, the presence of mitochondrial tubular and net structures in mgm1-5 Δdnm1 cells correlates with the restoration of mitochondrial fusion (Table ). In contrast, FZO1 function is still required for mitochondrial fusion, even when tubules are restored in fzo1-1 Δdnm1 zygotes, consistent with Fzo1p's proposed direct role in the fusion process (Bleazard et al. 1999; Fig. 4, I–L; Table ).
Figure 4.
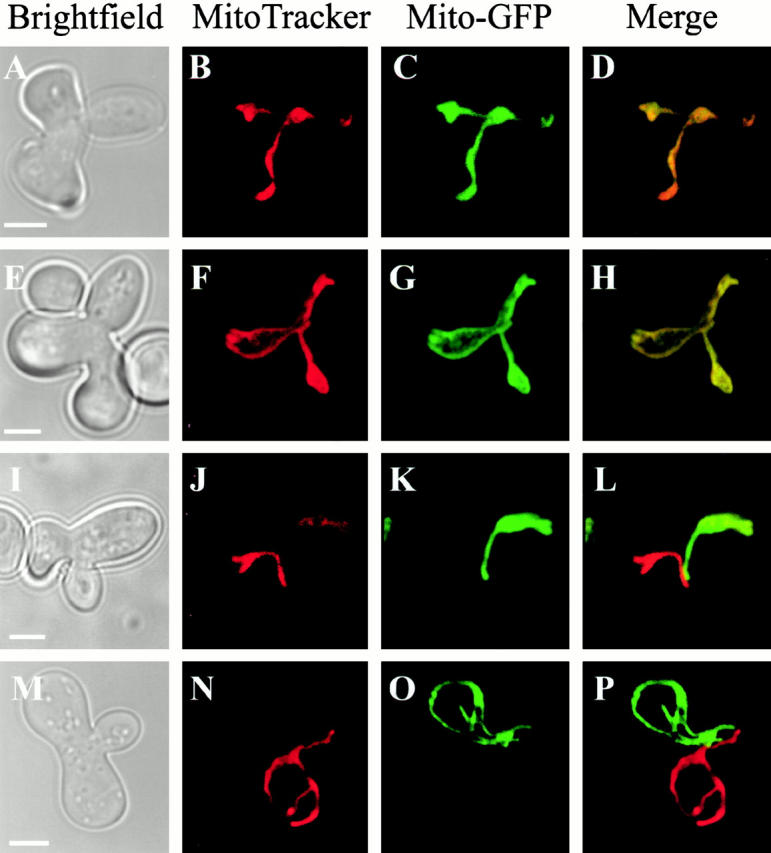
Mitochondrial fusion does not require MGM1 function. Mitochondrial fusion was assessed as described in Fig. 2. Mitochondria in homozygous zygotes formed at 37°C from Δdnm1 (A–D), mgm1-5 Δdnm1 (E–H), fzo1-1 Δdnm1 (I–L), and mgm1-5 fzo1-1 Δdnm1 (M–P) cells were visualized by fluorescence confocal microscopy. Bars, 2 μm.
To test whether restoration of mitochondrial fusion in mgm1-5 Δdnm1 zygotes was FZO1-dependent and not due to activation of an alternate fusion pathway, we examined mitochondrial fusion in the mgm1-5 fzo1-1 Δdnm1 triple mutant. Morphologically, mitochondria in these triple mutant cells were similar to those in both Δdnm1 and mgm1-5 Δdnm1 cells (Fig. 4, M–P). As expected, at permissive temperature, mitochondria fused in zygotes formed from mgm1-5 fzo1-1 Δdnm1 haploids (Table ). However, at nonpermissive temperature, mitochondria failed to fuse in mgm1-5 fzo1-1 Δdnm1 zygotes, confirming previous evidence indicating a direct role of Fzo1p in mitochondrial fusion (Hermann et al. 1998; Fig. 4, M–P; Table ). Taken together these observations indicate that MGM1 does not play a direct role in mitochondrial fusion.
Mgm1p Is Localized to the Mitochondrial Intermembrane Space
Given that MGM1 does not play a direct role in mitochondrial fusion, we suggest three possible explanations for the mitochondrial fragmentation observed in mgm1-5 cells. In wild-type cells, Mgm1p may function indirectly in mitochondrial fusion by remodeling and/or maintaining mitochondrial membranes in fusion competent tip and tubular structures. Alternatively, given that both Mgm1p and Dnm1p are dynamin-related GTPases predicted to assemble into oligomeric structures, Mgm1p may interact directly with Dnm1p to negatively regulate mitochondrial fission. This latter possibility is potentially supported by a recent report that Mgm1p is localized to the mitochondrial outer membrane (Shepard and Yaffe 1999). However, experiments examining the S. pombe Mgm1p homologue, Msp1, suggested that this protein is localized inside mitochondria, specifically in the matrix compartment (Pelloquin et al. 1999). A third possibility is that Mgm1p may function in inner membrane organization and/or fission and that these events may be coordinated with outer membrane fission. In this scenario, loss of MGM1 function might stimulate Dnm1p-dependent outer membrane fission, resulting in mitochondrial fragmentation. To help distinguish between these possibilities, we reexamined the submitochondrial localization of Mgm1p using subcellular fractionation, indirect immunofluorescence, and cryoimmunoelectron microscopy analyses.
To detect and examine its subcellular localization, we epitope-tagged Mgm1p. Neither NH2- or COOH-terminal tagged Mgm1p complemented the mitochondrial morphology and growth defects of mgm1 cells (not shown). A tagged version of Mgm1p that retained function was created by inserting three tandem copies of the HA epitope (3XHA) within the coding sequence between amino acids 216–217, directly proceeding the predicted GTPase domain (Mgm1:3XHAp). In a strain where Mgm1:3XHAp replaced the wild-type MGM1 locus (JSY2519), mitochondrial morphology was indistinguishable from wild-type cells (n = 443).
Western blot analysis of extracts made from JSY2519 with anti-HA antibodies detected a predominant 92-kD species (Fig. 5 A, lane 1). In contrast, no species were detected by Western blotting in the absence of tagged Mgm1p, indicating that anti-HA antibodies specifically recognize Mgm1:3XHAp (Fig. 5 A, lane 2).
Figure 5.
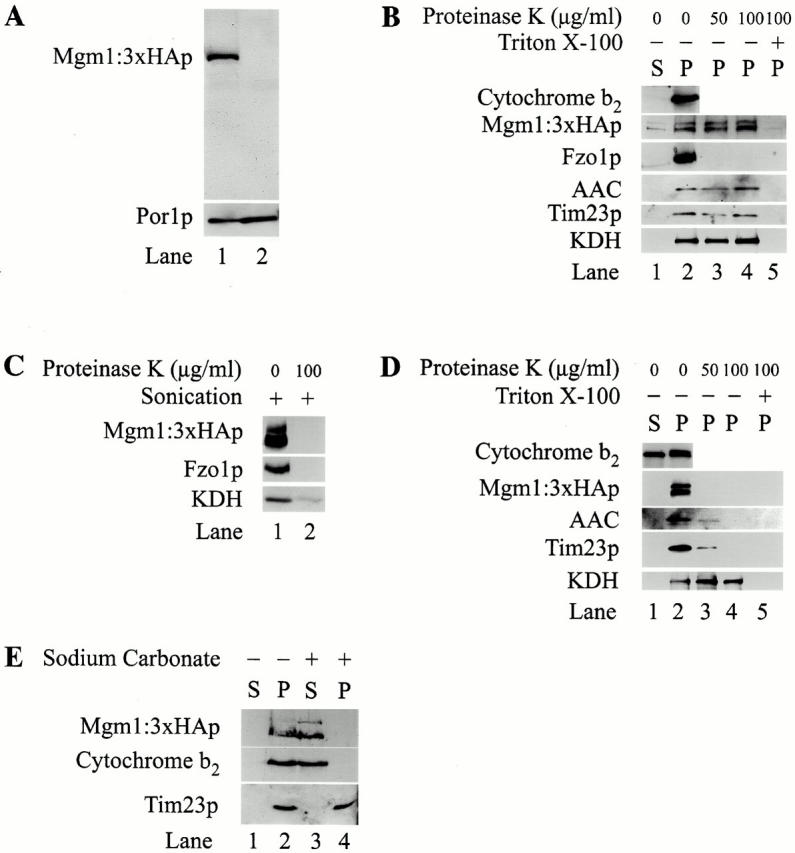
Mgm1p is a mitochondrial intermembrane space protein peripherally associated with the inner membrane. Whole cell extracts of JSY836 and JSY2519 were prepared as described in Materials and Methods and analyzed by SDS-PAGE and Western blotting with indicated antibodies (A). Subcellular fractions of enriched intact mitochondria were isolated, treated with PK after mock-treatment (B), sonication (C), or hypoosmotic shock (D) and analyzed by SDS-PAGE and Western blotting as described. E, Intact mitochondria were treated with 0.1 M sodium carbonate (pH 10.5) as described and fractions were analyzed by SDS-PAGE and Western blotting.
To examine Mgm1p's subcellular localization, JSY2519 extracts were fractionated by differential centrifugation and analyzed by SDS-PAGE and Western blotting. Consistent with the previously reported mitochondrial localization of Mgm1p, Mgm1:3XHAp was highly enriched in mitochondrial pellet (Fig. 5 B, lane 2). In addition to the predominant 92-kD form of Mgm1:3XHAp, we also detected a less abundant, slower migrating 116-kD form of Mgm1:3XHAp in enriched mitochondrial fractions (Fig. 5 B, lane 2). These data are in agreement with a previous study of Mgm1p localization, where two forms of Mgm1p were detected with antibodies raised against a COOH-terminal Mgm1p-derived peptide (Shepard and Yaffe 1999). Both the 92- and 116-kD forms of Mgm1:3XHAp are localized to the same mitochondrial compartment (see below).
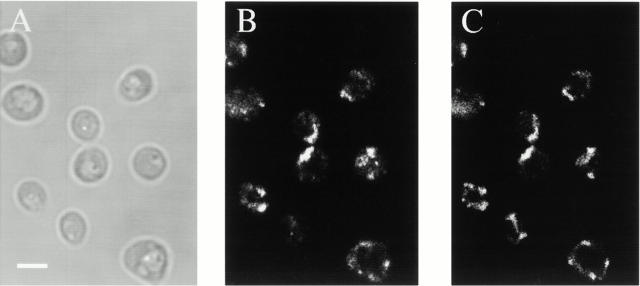
Indirect immunofluorescence of JSY2519 cells using anti-HA antibodies revealed that Mgm1p:3XHAp was localized to reticular structures at the cell cortex (Fig. 6 B). Mgm1:3XHAp staining colocalized with structures labeled with the covalent vital mitochondrial fluorescent probe, MitoTracker (Fig. 6B and Fig. C). Thus, localization of Mgm1p by both biochemical and indirect immunofluorescence indicate that it is a mitochondrial protein, consistent with previous observations (Shepard and Yaffe 1999).
Figure 6.
Mgm1:3xHAp localizes to mitochondria in vivo. JSY2519 cells were grown in YPD to log phase, labeled with MitoTracker (B), processed for indirect immunofluorescence with anti-HA (C), and were imaged using fluorescence confocal microscopy as described. Bar, 2 μm.
We examined the submitochondrial localization and topology of Mgm1:3XHAp by treating isolated mitochondria with exogenous proteases. In intact mitochondria, both the 92- and 116-kD forms of Mgm1:3XHAp were inaccessible to even high concentrations of proteinase K (PK; Fig. 5 B, lanes 2–4) and trypsin (not shown). Under these conditions, however, the outer mitochondrial marker, Fzo1p, was digested completely, indicating that the outer membrane was fully accessible to protease (Fig. 5 B, lanes 2–4). In addition, both intermembrane space (AAC and Tim23p) and matrix (KDH) marker proteins were protected from proteolysis, confirming that the mitochondrial outer and inner membranes were intact (Fig. 5 B, lanes 2–4). When both the inner and outer mitochondrial membranes were disrupted by treatment with detergent or by sonication, complete proteolysis of Mgm1:3XHAp and marker proteins in all mitochondrial compartments was observed (Fig. 5 B, lane 5, and C, lanes 1 and 2). These results indicate that Mgm1p is not a component of the mitochondrial outer membrane and is localized inside mitochondria. Thus, Mgm1p does not directly interact with Dnm1p to regulate mitochondrial outer membrane fission.
To determine whether Mgm1p resides in the intermembrane space or matrix compartment, we converted intact mitochondria to mitoplasts by selectively rupturing the mitochondrial outer membrane by hypoosmotic shock. Upon osmotic shock, a significant fraction of the soluble intermembrane space protein, cytochrome b2, was released into the supernate fraction after centrifugation of treated mitochondria (Fig. 5 D, lanes 1 and 2). In contrast, the inner membrane markers, AAC and Tim23p, and the soluble matrix protein, KDH, was recovered in the pellet fraction, indicating that the inner membrane of the mitoplasts remained intact (Fig. 5 D, lanes 1 and 2). Both forms of Mgm1:3XHAp also were recovered in the pellet fraction, indicating that they are either membrane-associated or soluble in the matrix.
To determine if Mgm1:3XHAp is localized to the matrix compartment or membrane-associated and exposed to the intermembrane space, we treated mitoplasts with PK. In mitoplasts, the matrix marker, KDH, was protected from PK digestion, indicating that the inner membrane was impermeant to protease and intact (Fig. 5 D, lanes 2–4). Although both AAC and Tim23p are inner membrane proteins, they contain epitopes exposed to the intermembrane space. Thus, as predicted, both were accessible to PK in mitoplasts (Fig. 5 D, lanes 2–4). Under these conditions, both forms of Mgm1:3XHAp behaved exactly like these intermembrane space markers and were also sensitive to PK digestion in mitoplasts (Fig. 5 D, lanes 2–4). In addition, identical results were observed for both forms of Mgm1:3XHAp and marker proteins when mitoplasts were treated with trypsin (not shown), indicating that the substrate specificity did not influence the susceptibility to proteolysis. These results indicate that both forms of Mgm1p are not localized to the matrix compartment, but instead, are membrane-associated and exposed to the intermembrane space.
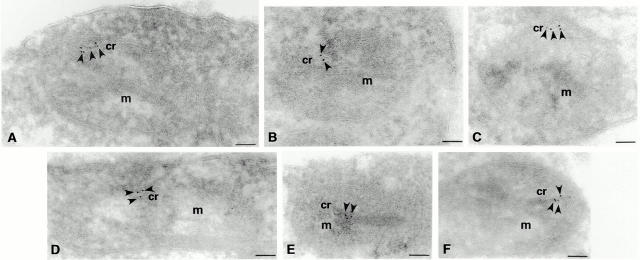
To determine whether Mgm1p is associated with the outer or inner mitochondrial membrane, we examined Mgm1:3XHAp's localization by immunoelectron microscopy using anti-HA antibodies. Immunogold-labeling was performed as described on cryosections of JSY2519 cells (Bleazard et al. 1999). Consistent with data from both indirect immunofluorescence and biochemical analyses, the majority of gold particles were found associated with mitochondrial structures (90%, n = 71; Fig. 7, see m). Further analysis of mitochondrial-associated gold particles indicates that the vast majority were found within the interior of the organelle and, in most cases, clearly associated with the inner membrane (95%, n = 64; Fig. 7, see arrows). This inner membrane association is especially apparent in sections where gold particles are associated with inner membrane folds or cistae (Fig. 7, see cr and arrow). In addition, we did not observe gold particles dispersed throughout mitochondria, unlike the distribution reported for Msp1p, the S. pombe Mgm1p homologue (Pelloquin et al. 1999).
Figure 7.
Mgm1p is associated with the inner membrane. JSY2519 cells were grown in YPD to log phase and processed for cryoimmunoelectron microscopy using anti-HA. Mitochondrial profiles (m) from JSY2519 cells are shown (A–F). Arrows indicate gold particle labeling and critae decorated by gold particles are indicated (cr). Bars, 0.1 μM.
To determine whether Mgm1p is an integral inner membrane protein, we examined Mgm1p's sensitivity to sodium carbonate extraction. Mitochondria containing Mgm1:3XHAp were extracted with 0.1 M Na2CO3 (pH 10.5) or mock-treated and fractionated into supernate and pellet fractions by centrifugation (Fig. 5 E, lanes 1–4). As expected, the inner membrane protein Tim23p was resistant to sodium carbonate extraction and was quantitatively recovered in the pellet fraction (Fig. 5 E, compare lanes 3 and 4). In contrast, both forms of Mgm1:3XHAp and the soluble intermembrane space protein cytochrome b2 were released into the supernatant upon sodium carbonate treatment, but not in control samples treated with buffer (Fig. 5 E, lanes 1–4). Based on this analysis, Mgm1:3XHAp is peripherally associated with the inner membrane, and not an integral membrane protein.
Discussion
S. cerevisiae Mgm1p was previously reported to be an integral mitochondrial outer membrane protein (Shepard and Yaffe 1999). In contrast, Msp1p, the S. pombe Mgm1p homologue, was reported to be an integral inner membrane protein, with its COOH-terminal GTPase domain localized in the matrix (Pelloquin et al. 1999). Our data indicate that Mgm1p is an intermembrane space protein that is tightly, but peripherally, associated with the mitochondrial inner membrane. Consistent with this interpretation, the NH2 terminus of Mgm1p contains a predicted matrix targeting signal, followed by a stretch of hydrophobic amino acids, features indicative of an intermembrane space targeting signal (Glick et al. 1992). Our conclusions are based on the susceptibility of Mgm1p to proteolysis in intact mitochondria and mitoplasts, where the outer membrane is selectively ruptured. Data from these experiments indicate that Mgm1p behaves in a manner identical to inner membrane proteins that contain protease-accessible intermembrane space domains. In contrast, in both previously published reports, the proteolytic susceptibility of intermembrane space marker proteins was not assessed, making it impossible to monitor the integrity of the outer membrane (Pelloquin et al. 1999; Shepard and Yaffe 1999). Moreover, our conclusions from protease protection experiments are substantiated by the pattern of localization observed for Mgm1p on the ultrastructural level, where gold particles are found closely associated with the inner membrane cristae.
The picture that emerges from these data is that the dynamin-related GTPases, Dnm1p and Mgm1p, are regulating the morphology of the outer and inner mitochondrial membranes, respectively (Otsuga et al. 1998). Given that Dnm1p has been shown to act on the outer membrane to regulate mitochondrial fission, we propose that Mgm1p plays a similar role in mediating fission and/or remodeling of the inner membrane. Indeed, in C. elegans, under conditions where the action of the Dnm1p homologue, Drp1, is blocked and outer membrane fission is abolished, inner membrane fission still occurs, suggesting that separate machinery governs the division of each membrane (Labrousse et al. 1999).
Precedence for a role of Mgm1p in inner membrane remodeling also can be found in observed similarities between mitochondria and chloroplast fission. Chloroplast fission requires the coordinate action of two homologues of the bacterial cell division GTPase FtsZ, called FtsZ1 and FtsZ2 (Osteryoung and Vierling 1995; Osteryoung et al. 1998; Osteryoung 2000). Like Dnm1p, FtsZ2 is predicted to localize to the cytosolic face of the outer membrane and, like Mgm1p, FtsZ1 is associated with the inner membrane (Osteryoung et al. 1998). In addition, both pairs of chloroplast and mitochondrial GTPases are predicted to self assemble into higher order ring and spiral structures that mediate membrane remodeling events (Sweitzer and Hinshaw 1998). Thus, in higher eucaryotes the dynamin-related GTPases have replaced the procaryotic-related FtsZ GTPases in the regulation of both mitochondrial outer and inner membrane remodeling events (Erickson 2000). The one characterized exception is a chromophytic alga, which contains a mitochondrial FtsZ (Beech et al. 2000).
Although FtsZ1 and Mgm1p may play related roles in remodeling inner organellar membranes, their topologies with respect to the inner membrane are different. FtsZ1 localizes to the stromal (matrix-equivalent) face of the chloroplast inner membrane, while Mgm1p localizes to the intermembrane space side of the mitochondrial inner membrane (Osteryoung and Vierling 1995; Osteryoung et al. 1998). This difference makes sense when one considers the topologies of FtsZ and dynamin relative to the membranes they remodel. In bacteria, the filamentous FtsZ ring on the inner membrane is thought to mediate division by pulling the membrane inward. In contrast, during endocytosis, dynamin mediates division by pinching the membrane inward (Sweitzer and Hinshaw 1998; Sever et al. 1999, Sever et al. 2000). By analogy, the most likely role of Mgm1p is to remodel inner membranes by a dynamin-like pinching event.
This Mgm1p-mediated pinching activity could affect inner membrane structure in several ways. First, Mgm1p might function in the formation or stabilization of inner membrane cristae by self-assembling into ring-like structures around the inner membrane (Fig. 8A and Fig. B). A second possibility is that Mgm1p facilitates inner membrane fission, similar to dynamin and Dnm1p (Fig. 8 B). Finally, a third possibility is that Mgm1p is required sequentially for cristae formation and inner membrane fission as depicted in Fig. 8 B.
Figure 8.
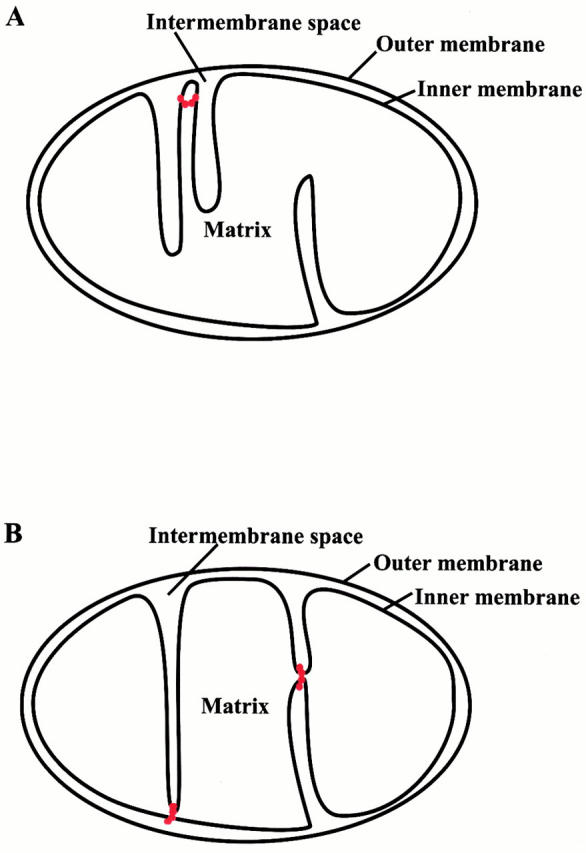
Models for Mgm1p function in mitochondrial inner membrane remodeling events. Mgm1p oligomeric ring structures are depicted in red. Mgm1p may help form and/or stabilize inner membrane cristae (A and B) or regulate inner membrane fission (B). See Discussion for details.
If inner membrane division is mediated by machinery separate from the Dnm1p-dependent outer membrane machinery, as is suggested from observations in C. elegans, then mechanisms that coordinate inner and outer membrane fission must exist. Taking this into account, the mitochondrial fragmentation we observe in mgm1 cells may indicate that the loss of MGM1 function can somehow be sensed by the outer membrane, causing the rate of Dnm1p-dependent outer membrane fission to increase. Consistent with this model, mitochondrial fragmentation is blocked and fusion is restored during mating in mgm1 cells when fission is abolished by deletion of DNM1. Alternatively, it is possible that loss of MGM1 function could either directly or indirectly affect the structure of the mitochondrial membrane tubules, rendering them fusion incompetent, and ultimately resulting in fission-mediated fragmentation observed in mgm1 cells. In this case, an Mgm1p-independent pathway for tubule formation could be operating in cells lacking Dnm1p, explaining the observed restoration of mitochondrial tubules and fusion in the dnm1 mgm1 double mutant.
Independent of the exact mechanism of mitochondrial fragmentation in mgm1 cells, our data are consistent with a role of Mgm1p in inner membrane remodeling events. We recently used genetic approaches to identify several new components of the Dnm1p outer mitochondrial membrane fission machinery (Mozdy et al. 2000; Tieu and Nunnari 2000). These molecules appear to act in a multistep pathway with Dnm1p on the outer membrane to mediate fission. The identification and analysis of components that act together with Mgm1p should help to reveal how this GTPase regulates inner membrane dynamics.
Acknowledgments
We thank Voytek Okreglak and T.J. Holewinske for their technical and intellectual contributions, and the Nunnari lab for discussions and critical reading of the manuscript. We also thank C. Koehler and R. Jensen for antibodies.
This work was supported by the National Science Foundation (MCB-9724143) to J. Nunnari and the National Institutes of Health (NIH; GM-53466) to J. Shaw. E. Wong was supported by an NIH Training Grant (GM-07377).
Footnotes
Edith D. Wong and Jennifer A. Wagner contributed equally to this work.
Abbreviations used in this paper: mito-GFP, mitochondrial matrix-targeted green fluorescent protein; mtDNA, mitochondrial DNA; PK, proteinase K.
References
- Beech P.L., Nheu T., Schultz T., Herbert S., Lithgow T., Gilson P.R., McFadden G.I. Mitochondrial FtsZ in a chromophyte alga. Science. 2000;287:1276–1279. doi: 10.1126/science.287.5456.1276. [DOI] [PubMed] [Google Scholar]
- Bleazard W., McCaffery J.M., King E.J., Bale S., Mozdy A., Tieu Q., Nunnari J., Shaw J.M. The dynamin-related GTPases, Dnm1, regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson H.P. Dynamin and FtsZmissing links in mitochondrial and bacterial division. J. Cell Biol. 2000;148:1103–1105. doi: 10.1083/jcb.148.6.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B.S., Brandt A., Cunningham K., Mueller S., Hallberg R.L., Schatz G. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 1992;69:809–822. doi: 10.1016/0092-8674(92)90292-k. [DOI] [PubMed] [Google Scholar]
- Guan K., Farh L., Marshall T.K., Deschenes R.J. Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr. Genetics. 1993;24:141–148. doi: 10.1007/BF00324678. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. Guide to Yeast Genetics and Molecular Biology. Academic Press; San Diego: 1991. [Google Scholar]
- Hales K.G., Fuller M.T. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Hermann G.J., Shaw J.M. Mitochondrial dynamics in yeast. Annu. Rev. Cell Dev. Biol. 1998;14:265–303. doi: 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., Shaw J.M. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 1998;143:359–374. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman H.-P., Avers C.J. Mitochondrion of yeastultrastructural evidence for one giant, branched organelle per cell. Science. 1973;18:1749–1751. doi: 10.1126/science.181.4101.749. [DOI] [PubMed] [Google Scholar]
- Jones B.A., Fangman W.L. Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 1992;6:380–389. doi: 10.1101/gad.6.3.380. [DOI] [PubMed] [Google Scholar]
- Labrousse A.M., Zappaterra M.D., Rube D.A., van der Bliek A.M. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Meeusen S., Tieu Q., Wong E., Weiss E., Schieltz D., Yates J.R., Nunnari J. Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA. J. Cell Biol. 1999;145:291–304. doi: 10.1083/jcb.145.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A.D., McCaffery J.M., Shaw J.M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000;151:367–379. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., Fox T.D., Walter P. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science. 1993;262:1997–2005. doi: 10.1126/science.8266095. [DOI] [PubMed] [Google Scholar]
- Nunnari J., Marshall W., Straight A., Murray A., Sedat J.W., Walter P. Mitochondrial transmission during mating in S. cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mtDNA. Mol. Biol. Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W. Organelle fissioncrossing the evolutionary divide. Plant Physiology. 2000;123:1213–1216. doi: 10.1104/pp.123.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., Vierling E. Conserved cell and organelle division. Nature. 1995;376:473–474. doi: 10.1038/376473b0. [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Stokes K.D., Rutherford S.M., Percival A.L., Lee W.Y. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell. 1998;10:1991–2004. doi: 10.1105/tpc.10.12.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D., Keegan B.R., Brisch E., Thatcher J.W., Hermann G.J., Bleazard W., Shaw J. The dynamin GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelloquin L., Belenguer P., Menon Y., Ducommun B. Identification of a fission yeast dynamin-related protein involved in mitochondrial DNA maintenance. Biochem. Biophys. Res. Comm. 1998;25:720–726. doi: 10.1006/bbrc.1998.9539. [DOI] [PubMed] [Google Scholar]
- Pelloquin L., Belenguer P., Menon Y., Gas N., Ducommun B. Fission yeast msp1 is a mitochondrial dynamin-related protein. J. Cell Sci. 1999;112:4151–4161. doi: 10.1242/jcs.112.22.4151. [DOI] [PubMed] [Google Scholar]
- Rapaport D., Brunner M., Neupert W., Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae . J. Biol. Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- Schneider B.L., Seufert W., Steiner B., Yang Q.H., Futcher A.B. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae . Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R.E. Division versus fusionDnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever S., Muhlberg A.B., Schmid S.L. Impairment of dynamin's GAP domain stimulates receptor-mediated endocytosis. Nature. 1999;398:481–486. doi: 10.1038/19024. [DOI] [PubMed] [Google Scholar]
- Sever S., Damke H., Schmid S.L. Garrotes, springs, ratchets, and whipsputting dynamin models to the test. Traffic. 2000;In press doi: 10.1034/j.1600-0854.2000.010503.x. [DOI] [PubMed] [Google Scholar]
- Shepard K., Yaffe M. The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J. Cell Biol. 1999;144:711–719. doi: 10.1083/jcb.144.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E., Shurland D., Ryazantsev S., van der Bliek A. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B.J., White J.G. Computer reconstruction of mitochondria from yeast. Methods Enzymol. 1979;56:718–728. doi: 10.1016/0076-6879(79)56064-9. [DOI] [PubMed] [Google Scholar]
- Sweitzer S.M., Hinshaw J.E. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- Tieu Q., Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 2000;151:353–365. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe M.P. The machinery of mitochondrial inheritance and behavior. Science. 1999;283:1493–1497. doi: 10.1126/science.283.5407.1493. [DOI] [PubMed] [Google Scholar]