Abstract
Glycosylphosphatidylinositol-anchored influenza hemagglutinin (GPI-HA) mediates hemifusion, whereas chimeras with foreign transmembrane (TM) domains mediate full fusion. A possible explanation for these observations is that the TM domain must be a critical length in order for HA to promote full fusion. To test this hypothesis, we analyzed biochemical properties and fusion phenotypes of HA with alterations in its 27–amino acid TM domain. Our mutants included sequential 2–amino acid (Δ2–Δ14) and an 11–amino acid deletion from the COOH-terminal end, deletions of 6 or 8 amino acids from the NH2-terminal and middle regions, and a deletion of 12 amino acids from the NH2-terminal end of the TM domain. We also made several point mutations in the TM domain. All of the mutants except Δ14 were expressed at the cell surface and displayed biochemical properties virtually identical to wild-type HA. All the mutants that were expressed at the cell surface promoted full fusion, with the notable exception of deletions of >10 amino acids. A mutant in which 11 amino acids were deleted was severely impaired in promoting full fusion. Mutants in which 12 amino acids were deleted (from either end) mediated only hemifusion. Hence, a TM domain of 17 amino acids is needed to efficiently promote full fusion. Addition of either the hydrophilic HA cytoplasmic tail sequence or a single arginine to Δ12 HA, the hemifusion mutant that terminates with 15 (hydrophobic) amino acids of the HA TM domain, restored full fusion activity. Our data support a model in which the TM domain must span the bilayer to promote full fusion.
Keywords: hemagglutinin, hemifusion, transmembrane domain, glycosylphosphatidylinositol anchor, SNARE proteins
Introduction
Influenza virus fusion is mediated by the hemagglutinin (HA) trimer (for reviews see Stegmann 1994; Gaudin et al. 1995; Hughson 1995; Hernandez et al. 1996). Each HA monomer is composed of two subunits: HA1, which contains the receptor binding and major antigenic sites, and HA2, which is primarily responsible for fusion. HA2 contains an NH2-terminal fusion peptide, a region of high α-helical propensity, a 27–amino acid transmembrane (TM) domain, and a 10–amino acid cytoplasmic tail. Studies have demonstrated the importance of the fusion peptide as well as structural changes within the trimeric coiled coil for fusion (Carr and Kim 1993; Bullough et al. 1994; Steinhauer et al. 1995; Qiao et al. 1998, Qiao et al. 1999).
In previous work, we demonstrated that replacing the TM and cytoplasmic tail domains of HA with a glycosylphosphatidylinositol (GPI) anchor generated an HA trimer that could promote only hemifusion (Kemble et al. 1994). This suggested that the TM domain plays an important role during the hemifusion to fusion transition (Kemble et al. 1994; Melikyan et al. 1995b, Melikyan et al. 1997a; Blumenthal et al. 1996; Nussler et al. 1997; Chernomordik et al. 1998). Other studies have demonstrated that replacing the HA TM domain (and/or cytoplasmic tail) with those from foreign proteins, both viral and nonviral, has no effect on fusion (Roth et al. 1986; Dong et al. 1992; Schroth-Diez et al. 1998; Melikyan et al. 1999). Although the aforementioned conclusions have been based on studies using different assays, the collective findings suggest that there may not be any specific sequence requirements for the HA TM domain to support full fusion (Roth et al. 1986; Dong et al. 1992; Schroth-Diez et al. 1998; Melikyan et al. 1999). However, the HA TM domain may require a minimal length to promote the hemifusion to fusion transition. The major goal of this study was to test the latter hypothesis.
Materials and Methods
Mutagenesis
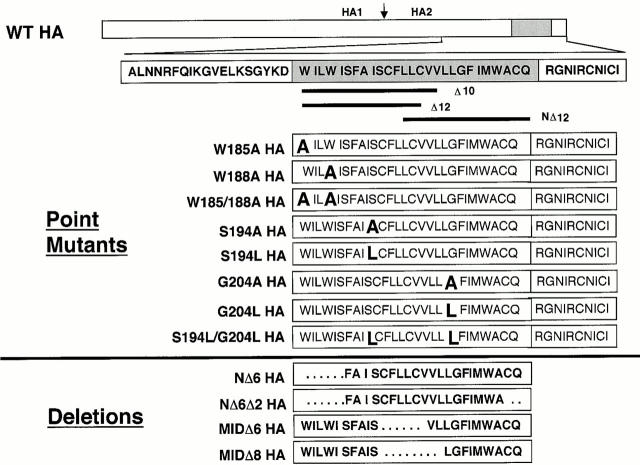
HA mutants were generated in HA cDNA (X:31 strain) present in the pTM1 vector using the Quik-Change Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer's instructions. Oligonucleotide primers with stop codons in the TM domain were used to generate cytoplasmic tail− HA (Tail− HA) and then, sequentially, two–amino acid deletions from the COOH-terminal end of the TM domain (Δ2–Δ14; see Fig. 1). Oligonucleotide primers were also used to create the following additional HA mutants (in the Tail− HA construct): a deletion of 6 amino acids from the NH2-terminal end of the TM domain (Δ185–190; NΔ6); a deletion of 6 amino acids from the NH2-terminal end and 2 amino acids from the COOH-terminal end of TM (Δ185–190/Δ210–211; NΔ6Δ2); a deletion of 12 amino acids from the NH2-terminal end of TM (Δ185–196; NΔ12); deletions of 6 or 8 amino acids from the central region of TM (Δ195–200, MidΔ6; and 195–202, MidΔ8); a deletion of 11 amino acids from the COOH-terminal end of TM (Δ101–111; Δ11); 6 single point mutants (S194L, S194A, G204A, G204L, W185A, and W188A); and two double point mutants (W185A/W188A and S194L/G204L) in the HA TM domain. The point mutants were made in the context of the full-length HA construct (i.e., containing the cytoplasmic tail). We also engineered two HA mutants that contained the TM domain of Δ12 HA (amino acids 185–199), followed by either the entire cytoplasmic tail sequence of HA (Δ12Tail HA) or a single arginine (Δ12Arg HA). In this paper we use the term GPI-HA to refer to the construct BHA-PI (K/S) described in Kemble et al. 1994. When GPI-HA is expressed, a nine–amino acid sequence from the decay-accelerating factor GPI anchor addition signal, containing a lysine to serine substitution, remains with the HA ectodomain (Kemble et al. 1994). GPI-HA was subcloned into the pTM1 vector. Mutant HA cDNAs were sequenced to confirm that the desired mutations had, but that second site mutations had not, been introduced.
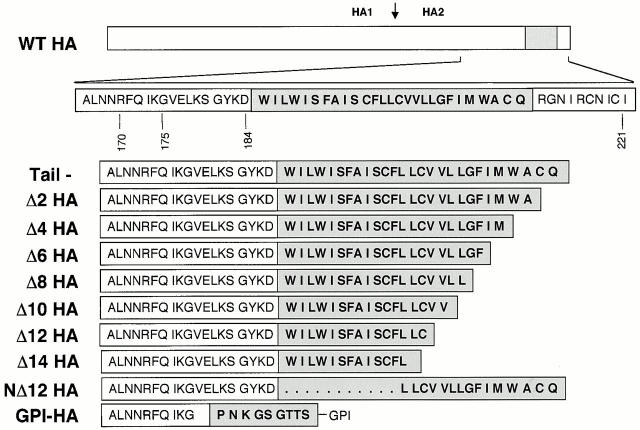
Figure 1.
HA TM domain truncation mutants. Line diagram of the HA gene. The region encompassing the TM domain (gray box) is expanded below. Deletion mutations are shown as sequential removal of two amino acids from the COOH-terminal end of the TM domain (mutants Δ2–14) starting with the tail− construct. NΔ12 HA lacks the NH2-terminal 12 amino acids of the TM domain as well as the cytoplasmic tail. GPI-HA has been aligned with the correct ectodomain sequences of the TM truncation mutants.
Expression of Wild-Type HA and Mutant HAs
CV-1 cells (CCL 70; American Type Culture Collection) were maintained in Iscove's modified Dulbecco's medium (IMDM; GIBCO BRL) containing 10% supplemented calf serum (SCS; Hyclone Laboratories, Inc.), 50,000 U penicillin, 50,000 μg streptomycin (GIBCO BRL), and an additional 146 mg glutamine (GIBCO BRL) per 0.5 liter. Wild-type (WT) and mutant HAs were expressed using the vaccinia virus T7 RNA polymerase transient transfection system (Fuerst et al. 1986). Confluent monolayers of CV-1 cells were infected with modified vaccinia ankara (MVA; a gift of Bernard Moss, National Institutes of Health, Bethesda, MD) at a multiplicity of infection of 10 PFU per cell and incubated at 37°C for 1 h with intermittent rocking. After removing the virus inoculum, the cells were washed once with Dulbecco's PBS (Cellgro; Fisher Scientific) and then transfected with cDNA using 12.0 μl Mirus Transit (Panvera) per 6-cm dish according to the manufacturer's instructions. Unless otherwise stated, we used 5.0 μg cDNA per 6-cm dish except for Δ10, Δ12, and NΔ12, for which we used 7.5 μg cDNA per 6-cm dish. After a 5-h incubation (at 37°C in a 5% CO2 incubator), the DNA/Transit mixture was replaced with IMDM, and the cells were incubated at 31°C for 15–20 h.
Metabolic Labeling
CV-1 cells expressing WT and mutant HAs were metabolically labeled with 35S-Translabel (ICN Biomedicals) essentially as described previously (Kemble et al. 1993). After transfection, the cells were incubated for 2 h in Cys−/Met− DME (GIBCO BRL) containing 2% SCS at 37°C. The medium was then replaced with 1.25 ml Cys−/Met− DME containing 50–75 μCi 35S-Translabel and 2% SCS, and the cells were incubated at 31°C for 14–18 h.
Cell Surface Biotinylation, Immunoprecipitations, and Western Blot Analyses
Biotinylation of cell surface proteins was performed as described previously (Qiao et al. 1999). Trypsin cleavage of HA0 was performed as described previously (Qiao et al. 1998) except that 10 μg/ml trypsin in PBS was used and incubation was for 10 min at room temperature (RT). Cells were then treated with 50 μg/ml soybean trypsin inhibitor (STI; Sigma-Aldrich) for 10 min at RT. CV-1 cells expressing WT and mutant HAs were washed with PBS, lysed in a cell lysis buffer containing 1% NP-40 and protease inhibitors as described previously (Delos et al. 2000), and then immunoprecipitated as described (Kemble et al. 1993). Immune complexes were suspended in SDS gel loading buffer containing 0.14 M β-mercaptoethanol, boiled for 5 min, and separated by 10% SDS-PAGE. The proteins were transferred to nitrocellulose. The membrane was blocked using superblotto (0.5% Tween 20, 3% [wt/vol] BSA, 18% [wt/vol] glucose, 1% [wt/vol] milk, and 10% glycerol in PBS) and probed with antibodies to HA (C-HA1) or streptavidin-HRP (Pierce Chemical Co.) as described previously (Qiao et al. 1999).
Sucrose Gradient Analysis
CV-1 cells expressing WT and mutant HAs were treated with trypsin, then STI, and lysed as described above. Cell lysates were layered on continuous 3–30% sucrose (wt/vol) gradients. After centrifugation, 12 395-μl fractions were collected and prepared as described previously (Qiao et al. 1998).
C-HA1 Conformational Change Assay
Transfected CV-1 cells were metabolically labeled overnight as described above. After treatment with trypsin and STI (see above), the cells were incubated at 37°C for 10 min in fusion buffer (100 mM NaCl, 10 mM Hepes, 10 mM MES, 10 mM succinate, and 2 mg/ml glucose) adjusted to the indicated pH. After reneutralization with pH 7.0 fusion buffer, the cells were lysed and immunoprecipitated with the C-HA1 antibody as described previously (Kemble et al. 1993). Samples were analyzed by SDS-PAGE and PhosphorImager analysis (Molecular Dynamics).
RBC Labeling, Binding, and Lipid and Content Mixing Assays
Freshly collected human RBCs were either colabeled with octadecylrhodamine B chloride (R18) and carboxyfluorescein (CF; Molecular Probes, Inc.) or labeled with CF only (Melikyan et al. 1999). WT and mutant HA-expressing cells were treated with trypsin and STI as described above, washed once with PBS+ (PBS containing 0.1 g/liter CaCl2 and 0.1 g/liter MgCl2), and incubated with a solution containing 0.05% labeled RBCs for 15 min at RT. Unbound RBCs were removed by three washes with PBS+ and fusion was triggered by incubating the HA-expressing cells with pH 5.0 fusion buffer at 37°C for either 2 or 5 min (times indicated in the figure legends). The pH 5.0 solution was replaced with pH 7.0 fusion buffer, and the cells were examined with a fluorescent microscope. Where indicated, cells were treated for 1 min at RT with either 0.1 or 0.5 mM chlorpromazine (CPZ) in fusion buffer, pH 7.0, and then returned to PBS+. Images were collected using an Axioplan 2 microscope (Carl Zeiss, Inc.) equipped with a C4742-95 CCD camera (Hamamatsu), and Openlab (Improvision) software, and were prepared using Adobe Photoshop®.
Preparation of Microsomal Membranes
Transfected CV-1 cells were biotinylated and treated with trypsin and STI as described above. The cells were then released from their dishes by incubation for 10 min at RT in 1.0 ml PEEG (PBS containing 0.5 mM EDTA, 0.5 mM EGTA, and 10 mM glucose), transferred to a 1.5-ml Eppendorf tube, pelleted at 325 g for 2 min at 4°C, resuspended in 0.8 ml DHB buffer (10 mM Tris-HCl, pH 7.5, 1 mM MgCl2), and incubated for 5 min on ice (to induce cell swelling). The cells were then passed 10 times through a 25-gauge needle. Sucrose was added to bring the solution to a final 1.18 M (wt/wt) concentration by the addition of 2.0 M sucrose (in DHB), and the suspension was overlaid with 3.0 ml of 0.25 M sucrose (in DHB). The nuclei were pelleted by centrifugation in an SW55 rotor at 192,000 g for 90 min at 4°C. The interface containing the microsomal membrane fraction was collected, transferred to a tube containing 4.0 ml of 0.25 M sucrose (in DHB), and centrifuged as before. The pellet containing the microsomal membranes was collected.
Carbonate Extraction
The pellet containing the microsomal membranes was resuspended in 0.3 ml 50 mM TEA, pH 7.5 (triethanolamine, pH adjusted with acetic acid). The pH was adjusted to 11.0 by the addition of 0.1 volume 1 M Na2CO3 (pH 11.0), and the suspension was incubated on ice for 20 min. Membranes were layered on top of a 0.68-ml sucrose cushion (0.2 M sucrose, 20 mM Hepes-NaOH (pH 11.0), 150 mM potassium acetate, 2.5 mM magnesium acetate) and centrifuged at 135,000 g in a TLS-55 rotor for 20 min at 4°C. The supernatant was collected and reneutralized by the addition of 30 mM HCl and designated the “supernatant fraction.” The pellet was lysed in 0.5 ml lysis buffer containing protease inhibitors, incubated on ice for 20 min, centrifuged to clear debris at 16,000 g for 10 min at 4°C, and transferred to a fresh 1.5-ml Eppendorf tube and designated “pellet fraction.” HA from the supernatant and pellet fractions was immunoprecipitated using the Site A mAb as described above, resolved by SDS-PAGE on a reducing 10% gel, transferred to nitrocellulose, and probed with streptavidin-HRP as described above.
FACS® Analysis
Transfected CV-1 cells (6-cm dishes) were released from the dish with PEEG (as described above) and transferred to a 1.5-ml Eppendorf tube. The cells were then washed twice with cold PBS+ containing 0.02% azide (PBSA) and centrifuged at 325 g for 2 min at 4°C. The cells were resuspended in 0.2 ml cold PBSA containing 2% SCS and incubated for 30 min on ice with 1.7 μl of 1.0 mg/ml Site A mAb. The cells were then washed twice with PBSA as described above, resuspended in PBSA containing 2% SCS, and incubated with 1.0 μl FITC-conjugated goat anti–mouse IgG for 30 min on ice. The cells were then washed twice with cold PBSA, resuspended in PBS containing 2% paraformaldehyde, and analyzed by FACS® at the University of Virginia Core Facility, using a FACScan™ flow cytometer (Becton Dickinson).
Endo F Treatment
Transfected CV-1 cells (6-cm dishes) were metabolically labeled and treated with trypsin and STI as described above. The cells were released from the dish by a brief treatment with PEEG (as described above) and transferred to a 1.5-ml Eppendorf tube. Lysates were prepared and HA was immunoprecipitated with the Site A mAb as described above. After the immunoprecipitation, 50 μl N-Glycosidase F buffer (1% octylglucoside, 0.2% SDS, 40 mM Tris, pH 8.0, 5 mM EDTA, and 1% β-mercaptoethanol) was added to the protein A–Agarose (PAA) beads. The beads were then treated at 95°C for 3 min, followed by centrifugation for 2 min at 16,000 g. The supernatant was then transferred to a new 1.5-ml Eppendorf tube and treated with 1.0 U N-Glycosidase F (Roche) for 2 h at 37°C. SDS gel loading buffer containing 0.14 M β-mercaptoethanol was added, the samples were reboiled, and the proteins were resolved by SDS-PAGE on a 15% gel. The gel was then fixed in a solution of 40% methanol/10% acetic acid for 20 min at RT, followed by incubation in 1 M salicylic acid for 20 min at RT. The gel was then dried and exposed to film.
Cholesterol Depletion and Triton X-100 Extraction
For fusion experiments, transfected CV-1 cells were treated with trypsin and STI as described above, and depleted of cholesterol by a 30-min treatment with 20 mM methyl β-cyclodextrin (MβCD; Sigma-Aldrich) in PBS+ at 37°C before RBC binding and fusion (see above). For Triton X-100 extraction experiments, transfected CV-1 cells were biotinylated, trypsin treated, cholesterol depleted with MβCD as described above, and lifted off the dish by treatment with PEEG for 10 min at 4°C. The cells were pelleted in a refrigerated microfuge chilled to –2°C for 2 min at 325 g. After centrifugation, the cells were placed on ice in a 4°C coldroom, resuspended in 500 μl cold lysis buffer (50 mM Tris, pH 8.0, 1% Triton X-100, and protease inhibitors), and incubated on ice for 20 min. The insoluble fraction was removed by centrifugation for 15 min at 16,000 g and 4°C, again in a microfuge chilled to –2°C. The supernatant was designated the “soluble fraction.” The insoluble (pellet) fraction was resuspended in 500 μl cold lysis buffer containing 0.1% SDS and protease inhibitors, incubated for 1 h at RT with occasional vortexing, centrifuged to clear debris at 16,000 g for 10 min at 4°C, transferred to a fresh 1.5-ml Eppendorf tube, and designated “pellet fraction.” HA from the soluble fraction and the pellet fraction was immunoprecipitated with the Site A mAb, detected as described above with streptavidin-HRP, and quantitated by PhosphorImager® analysis.
Videomicroscopy Lipid Mixing Assay
Transfected CV-1 cells expressing either WT HA or Δ10 HA were processed for fusion as described above. RBCs (0.05%) labeled with R18 as described above were bound to the CV-1 cells. Fusion was triggered at 37°C with fusion buffer adjusted to the indicated pH. The cells were maintained at 37°C on a warm stage and monitored by videomicroscopy for 5 min using the software package Openlab (Improvision). Fusion was quantified using Scion Image (National Institutes of Health, Bethesda, MD). Care was taken such that one to three RBCs were bound per cell. Each field contained 40 RBCs, and the amount of R18 fluorescence per field was quantified. The values from three to four fields per time point were averaged, and the data were plotted as a function of time. From these plots, values for the lag time, initial rate, and final extent of lipid mixing were calculated.
Results
Biochemical Properties of HA TM Truncation Mutants D2–D14
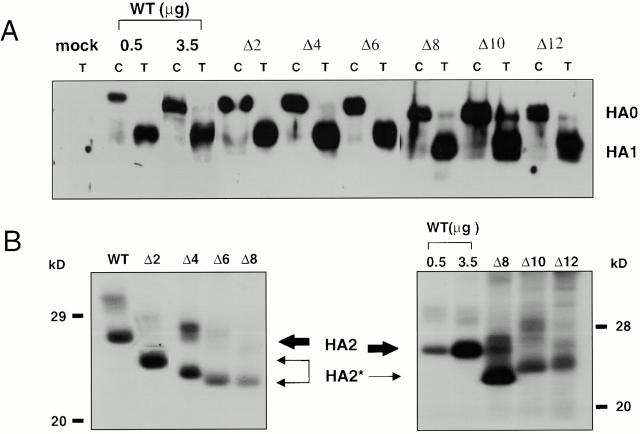
We initially made a set of mutants in which we sequentially deleted 2, 4, 6, 8, 10, 12, and 14 amino acids from the COOH-terminal end of the HA TM domain in the context of a Tail− HA construct (Fig. 1). We first asked whether these mutant HAs could be expressed at the cell surface in a fusion-permissive form (i.e., cleaved from HA0 to HA1-S-S-HA2). We also examined them for a shift in the migration of their HA2 subunits. With the exception of Δ14 (data not shown), all of the mutants were expressed at the cell surface as HA0 and were efficiently cleaved to HA1 and HA2 by the addition of trypsin (Fig. 2 A). Δ2–Δ8 HA exhibited the expected shift in the mobility of the HA2 subunits (Fig. 2 B, left). Δ10 and Δ12 HA exhibited an increased mobility compared with WT HA (and Tail− HA; data not shown), but these mutants migrated more slowly than Δ8 HA (Fig. 2 B, right). The fact that Δ12 was, but that Δ14 was not, expressed at the cell surface is consistent with previous observations that a mutant HA with a 13–amino acid truncation of the TM domain was not transported beyond the cis-Golgi compartment (Doyle et al. 1986).
Figure 2.
Processing of HA truncation mutants. (A) Chymotrypsin (C) or trypsin (T) treated CV-1 cells were biotinylated, lysed, immunoprecipitated with the Site A antibody, resolved by SDS-PAGE, transferred to nitrocellulose, and visualized with streptavidin-HRP. Like WT HA, all mutant HAs exhibit efficient processing. (B) CV-1 cells were metabolically labeled, treated with trypsin, lysed, immunoprecipitated with an HA-specific mAb, treated with 1 U of N-Glycosidase F for 2 h at 37°C, and resolved on 15% SDS-PAGE. The gels were dried and exposed to film. Δ2–12 HA exhibit a mobility shift indicative of a truncated HA2 subunit (HA2*).
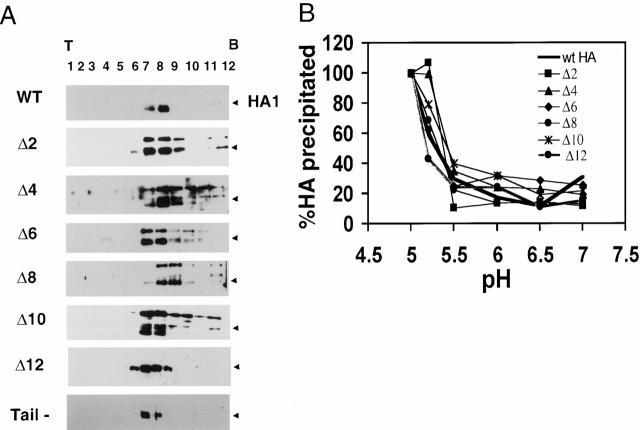
We next asked if the mutant HAs form trimers. Processed forms of Δ2–Δ12 HA migrated to a similar position on sucrose gradients as WT HA (Fig. 3 A, arrows). The higher molecular weight band seen in some of the gradients corresponds to intracellular HA0.
Figure 3.
Biochemical analyses. (A) Sucrose gradient analysis. Transfected CV-1 cells were trypsin treated, lysed, run on 3–30% continuous sucrose gradients, and fractionated as described in Materials and Methods. The samples were precipitated with Con A–agarose, resolved by 10% SDS-PAGE, and analyzed for HA protein by Western blotting. Processed forms of Δ2–12 HA migrate to a similar position on sucrose gradients as the WT HA trimer (black arrowhead). (B) Conformational change assay. Transfected CV-1 cells were metabolically labeled, trypsin treated, incubated at indicated pH values for 10 min at 37°C, reneutralized, and lysed. Cell lysates were then immunoprecipitated with the C-HA1 antibody, which recognizes only the low pH conformation of HA, resolved by 10% SDS-PAGE, and quantitated by PhosphorImager® analysis. The amount of HA precipitated when the total HA precipitated at pH 5 is considered as 100%. Δ2–12 HA change conformation with a pH dependence similar to WT HA. The results presented for WT HA and Δ2–Δ10 are from a typical experiment. The values given for Δ12 HA are the average from three independent experiments.
To address whether the mutant HAs change conformation at the same pH as WT HA, HA-expressing cells were briefly incubated at the indicated pH, lysates were prepared, and HA was immunoprecipitated using C-HA1, a conformation-specific antibody (White and Wilson 1987). As seen in Fig. 3 B, Δ2–Δ12 HA changed conformation with a pH dependence similar to that of WT HA.
Fusion Activity of Δ2–Δ12
We evaluated the fusion activity of Δ2, Δ4, Δ6, Δ8, Δ10, and Δ12 HA using a dye transfer assay. RBCs colabeled with a lipid dye (R18) and a soluble content dye (CF) were bound to HA-expressing cells, and fusion was induced as described in Materials and Methods. After 5 min at 37°C and pH 5, the cells were returned to neutral pH medium and examined with a fluorescence microscope. As seen in Fig. 4, both dyes transferred efficiently to cells expressing Δ2, Δ4, Δ6, Δ8, and Δ10 HA. A different phenotype was seen for cells expressing Δ12 HA: whereas we observed efficient lipid dye transfer (97%), content dye transfer was severely restricted (<10 vs. 97% for WT HA). As expected, we did not observe transfer of R18 or CF by WT or mutant HA-expressing cells at neutral pH or at low pH if the cells had not been pretreated with trypsin to process HA0 (data not shown).
Figure 4.
Quantitation of lipid mixing and content mixing. CV-1 cells expressing WT or mutant HAs were prepared for fusion as indicated in Materials and Methods, except that fusion was triggered at pH 5.0 for 2 min at 37°C and reneutralized. The amount of cDNA used per transfection is indicated. Lipid and content mixing events were averaged from 4–12 random fields (mean ± SEM). Percent lipid dye transfer (hatched bars) was determined by dividing the number of cells receiving lipid dye by the number of cells with bound RBCs in each field. Percent content dye transfer (black bars) was determined by dividing the number of cells receiving content dye by the number of cells with bound RBCs in each field. Relative surface expression of HA in fluorescence units (FU), was obtained by FACS® analyses, and is presented as the mean fluorescence intensity per cell normalized to that of 3.5 μg WT HA cDNA.
Given the striking observation that Δ10 HA mediated robust lipid and content mixing whereas Δ12 HA mediated robust lipid mixing with minimal content mixing, we also tested a mutant lacking the 11 COOH-terminal residues of the TM domain (Δ11 HA). Δ11 HA exhibited biochemical properties similar to WT HA (Table ). Whereas Δ11 HA mediated efficient lipid mixing (Fig. 4), it was significantly impaired in its ability to mediate content dye transfer (Fig. 4).
Table 1.
Summary of Results: Effects of Truncation Mutations in the HA TM
| HA | Trimer | Trypsin cleavage | RBC binding | Lipid mixing | Content mixing | ConfΔ |
|---|---|---|---|---|---|---|
| pH | ||||||
| WT (3.5 μg) | + | + | ++ | ++ | ++ | 5.0 |
| WT (0.5 μg) | + | + | + | ++ | ++ | 5.0 |
| Tail− | + | + | ++ | ++ | ++ | 5.0 |
| Δ2 | + | + | ++ | ++ | ++ | 5.0 |
| Δ4 | + | + | ++ | ++ | ++ | 5.0 |
| Δ6 | + | + | ++ | ++ | ++ | 5.0 |
| Δ8 | + | + | ++ | ++ | ++ | 5.0 |
| Δ10 | + | + | ++ | ++ | ++ | 5.0 |
| Δ11 | ND | + | + | ++ | − | ND |
| Δ12 | + | + | + | ++ | − | 5.0 |
| NΔ12 | ND | + | +/− | + | − | ND |
| Δ12Tail | ND | + | ++ | ++ | ++ | 5.0 |
| GPI-HA | + | + | ++ | + | − | 5.0 |
The transfection efficiency of the HA TM domain mutants ranged from 61 to 78% (SEM was <5%). Expression at the cell surface is indicated in Fig. 4. ND, not done. 3.5 μg WT refers to the higher amounts of cDNA used in transfections and with which comparison with Tail− HA and Δ2–Δ10 should be made. 0.5 μg WT refers to the lower amount of cDNA used in transfections and with which comparison with Δ11, Δ12, NΔ12, and Δ12Tail should be made.
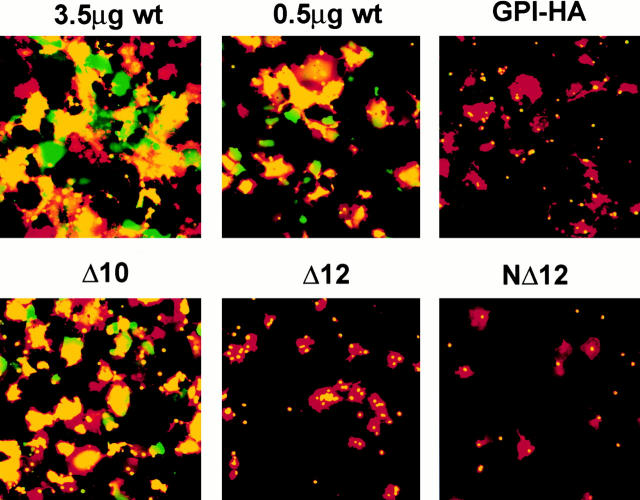
The density of HA at the cell surface can influence the fusion phenotype (Ellens et al. 1990; Melikyan et al. 1995a; Blumenthal et al. 1996; Danieli et al. 1996). Therefore, we took care to analyze the fusion phenotype of mutant HAs expressed at comparable levels to a known amount of WT HA. To do this, we first used FACS® analysis to determine the amount of WT and mutant HA expressed at the cell surface when different amounts of cDNA were used for transfection. We then used an amount of WT HA cDNA such that the level of WT HA expression at the cell surface was equivalent to that of the mutant HA to which it was being compared (see results of FACS® analysis, bottom of Fig. 4). We can therefore say that under conditions of equivalent cell surface expression, whereas WT HA (0.5 μg cDNA) mediates efficient lipid and content mixing, Δ12 HA (7.5 μg cDNA) mediates efficient lipid transfer but very poor content dye transfer. The fusion phenotype of cells expressing Δ12 HA was thus similar to that seen for cells expressing GPI-HA (Fig. 4). Representative micrographs showing the fusion patterns of WT HA, GPI-HA, Δ10 HA, and Δ12 HA are shown in Fig. 5.
Figure 5.
Fusion activity: lipid and content mixing. CV-1 cells were transfected with the indicated amounts of WT HA cDNA, 5.0 μg GPI-HA cDNA, or 7.5 μg mutant HA cDNA. The cells were prepared as described in the legend to Fig. 4 and observed (within 15–30 min) by fluorescence microscopy. Δ10 HA (17–amino acid TM domain) mediates full fusion, whereas Δ12 HA and NΔ12 HA (15–amino acid TM domain) are arrested at hemifusion, as is GPI-HA.
The fusion data presented in Fig. 4 and Fig. 5 suggested that there is a stringent length requirement for the HA TM domain to be able to mediate both lipid and content mixing. To further test this possibility, we generated a second 12–amino acid truncation in the HA TM domain, but in this case we deleted 12 amino acids from the NH2-terminal end of the TM domain (Fig. 1). This mutant, referred to as NΔ12 HA, was then examined for biochemical properties. Like all of the other truncation mutants, NΔ12 HA was expressed at the cell surface, was processed by trypsin into HA1 and HA2, and exhibited a faster migrating HA2 subunit than tail_HA (Table , and data not shown). By all of these criteria, NΔ12 HA resembled Δ12 HA. However, it was not as well expressed at the cell surface as Δ12 HA, as determined by FACS® analysis and RBC binding (∼80% compared with Δ12 HA; Fig. 4 and Table ). In terms of fusion with RBCs, NΔ12 HA mediated significant lipid mixing, albeit less than seen with Δ12 HA (63 vs. 97%). With respect to content mixing, NΔ12 HA mediated <5% dye transfer similar to the behavior of Δ12 HA and GPI-HA (Fig. 4 and Fig. 5). Neither Δ12 HA nor NΔ12 HA promoted significant content mixing (>10%) even after 60 min of incubation at 37°C at either pH 4.8 or 5.0 (data not shown).
Comparison of the Lipid and Content Mixing Ability of D10 HA and WT HA
Given the dramatic decrease in content mixing ability between Δ10 HA and Δ12 HA (and Δ11 HA), we examined the fusion activity of Δ10 HA in more detail. For this purpose, we compared the lag times, initial rate, and final extent of lipid mixing with WT HA and Δ10 HA at different pH values. At all pH values tested, the lag time before the onset of dye transfer was equivalent for Δ10 HA and WT HA (Table ). There was no difference in either the initial rate or the final extent of lipid mixing for Δ10 HA and WT HA at pH 5.0 and 5.25. At pH 5.5, the latter parameter was somewhat lower for Δ10 HA. Hence, the lipid mixing properties of Δ10 HA were very similar to those of WT HA at all pH values tested. In addition, when incubated at the suboptimal pH of 5.25 for 2 min at 37°C, Δ10 HA meditated content mixing to the same extent as WT HA (data not shown).
Table 2.
Lipid Mixing of WT versus Δ10 HA
| pH | HA | Lag time | Initial rate of fusion | Final extent |
|---|---|---|---|---|
| s | ||||
| 5.0 | WT | 45 | 1.0 | 100 ± 4 |
| Δ10 | 50 | 1.13 | 104 ± 1.3 | |
| 5.25 | WT | 65 | 1.01 | 83 ± 6 |
| Δ10 | 70 | 0.96 | 83 ± 6 | |
| 5.5 | WT | 80 | 0.74 | 71 ± 4 |
| Δ10 | 80 | 0.67 | 58 ± 6 |
Lag time indicates the amount of time before initial spread of R18. Initial rate of fusion is the change in intensity over time (ΔI/Δt) for the initial rise in fluorescence relative to WT HA at pH 5.0. Final extent is the percent intensity values (± SEM) relative to WT HA at pH 5.0 at the highest extent of fusion. The data were calculated from two independent experiments. See Materials and Methods for details.
Effect of CPZ on Content Mixing
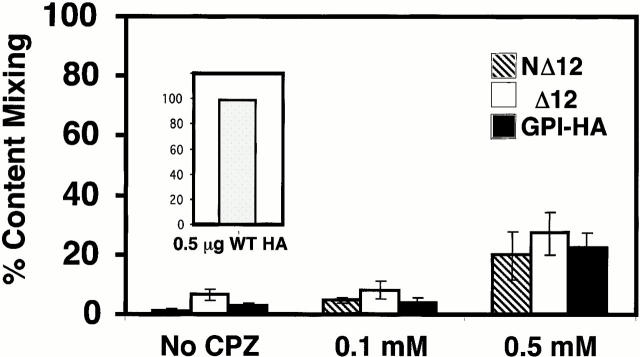
Previous work has shown that treatment with 0.1 mM CPZ, a membrane-permeable amphipathic reagent that partitions preferentially into the inner leaflet of the plasma membrane, efficiently induces full fusion in cases of “stunted fusion” caused by performing fusion experiments under suboptimal conditions (Melikyan et al. 1997a). Stunted fusion is operationally defined as lipid mixing without substantial content mixing due to the formation of small or transient fusion pores. It is thought to occur after hemifusion. In contrast, higher concentrations of CPZ (0.4–0.5 mM) are needed to induce GPI-HA to promote content mixing, and the extent of content mixing seen with GPI-HA in the presence of 0.4–0.5 mM CPZ never reaches that seen with WT HA (Melikyan et al. 1997a). Therefore, we assessed the effects of 0.1 and 0.5 mM CPZ on the ability of Δ12 HA, NΔ12 HA, and GPI-HA to promote content transfer. After binding double-labeled RBCs (R18 and CF) to HA-expressing cells, fusion was triggered by lowering the pH to 5.0 for 5 min at 37°C. The medium was reneutralized, and CPZ was added to the cells at neutral pH. After a 1-min incubation at RT, the CPZ solution was replaced with PBS+. The percentage of R18-stained HA-expressing cells that became labeled with CF was then determined. In the absence of CPZ, ∼1, 7, and 3% of cells expressing NΔ12 HA, Δ12 HA, and GPI-HA, respectively, received aqueous dye (Fig. 6). The addition of 0.1 mM CPZ increased content dye transfer to ∼5, 8, and 4%, respectively. Addition of 0.5 mM CPZ induced a greater extent of CF transfer: ∼20, 27, and 22%, respectively. Representative images of aqueous dye transfer before and after the addition of either 0.1 or 0.5 mM CPZ are shown in Fig. 7. Hence, cells expressing NΔ12 HA and Δ12 HA respond similarly to CPZ as do cells expressing GPI-HA in terms of their ability to promote aqueous dye transfer. The presence of R18 in the RBC membrane augments the transfer of aqueous contents to GPI-HA–expressing cells (Markosyan et al. 2000). Therefore, we assessed content dye transfer from RBCs that were not labeled with R18. As expected, content dye transfer under these conditions was less than with double-labeled RBCs (data not shown). Most importantly, Δ12 HA and NΔ12 HA still responded similarly to CPZ as did GPI-HA: a brief treatment with 0.5 but not 0.1 mM CPZ increased content dye transfer (data not shown).
Figure 6.
CPZ induces transfer of aqueous dye (CF) from RBCs to hemifused NΔ12, Δ12, and GPI-HA–expressing cells. CV-1 cells expressing WT and mutant HAs were prepared as described in the legend to Fig. 4, except that fusion was triggered for 5 min at pH 5.0 and 37°C before reneutralization. After triggering fusion, these cells were exposed to either 0.1 or 0.5 mM CPZ for 1 min at room temperature. The CPZ solution was replaced with PBS+ and the cells were observed as above. In control experiments, virtually all WT HA–expressing cells (0.5 μg WT HA, inset graph) were stained with CF in the absence of CPZ. Percent content mixing was determined by dividing the number of cells receiving CF by the number of cells with bound RBCs. Error bars show the SEM for four to five independent experiments (mean ± SEM). Only 0.5 mM CPZ promoted significant content dye transfer between labeled RBCs and cells expressing NΔ12 HA, Δ12 HA, and GPI-HA.
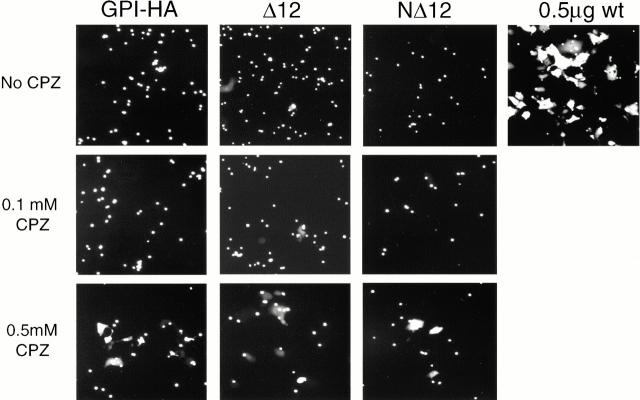
Figure 7.
Effect of CPZ on content mixing. CV-1 cells expressing GPI-HA, NΔ12, and Δ12 HA cDNA were processed for fusion with CF-labeled RBCs in the absence or presence of the indicated amount of CPZ, as described in the legend to Fig. 6. Only 0.5 mM CPZ is able to promote significant content dye transfer (see Fig. 6).
Membrane Association of Δ12 HA
Given the striking phenotype of HA lacking 12 amino acids in the TM domain (lipid, but not content, mixing), we explored how Δ12 HA is anchored in the membrane. Like WT HA, Δ12 HA (as well as GPI-HA) was resistant to carbonate extraction (Fig. 8 A). Given that some GPI-anchored proteins associate with cholesterol and sphingomyelin-rich detergent-insoluble membrane fractions (DIGs; Simons and Ikonen 1997; Friedrichson and Kurzchalia 1998; Varma and Mayor 1998), we examined the solubility of Δ12 HA and NΔ12 HA in Triton X-100 at 4°C before and after treating cells with methyl β-cyclodextrin to remove cholesterol (Scheiffele et al. 1997). Proteins that associate with lipid raft microdomains are often relatively insoluble in 1% Triton X-100 in the cold (Simons and Ikonen 1997). Cholesterol depletion can increase the solubility of these proteins in Triton X-100, presumably by disrupting the raft microdomains (Scheiffele et al. 1997). Both Δ12 HA and NΔ12 HA (data not shown) were readily solubilized by Triton X-100 at 4°C, suggesting that they do not associate with DIGs. In contrast, both WT HA and GPI-HA were partially insoluble in Triton X-100 at 4°C, and depletion of cholesterol appeared to increase their solubility (Fig. 8 B).
Figure 8.
Effect of carbonate extraction and cholesterol depletion on HA TM mutants. (A) Carbonate extraction. Microsomal membranes were prepared, adjusted to pH 11.0, and HA in the supernatant and pellet fractions was prepared, immunoprecipitated with an anti-HA mAb, and detected as described in Materials and Methods. Processed HA (WT and mutant) was only found in the pellet fraction, indicating that it is not extracted by high pH. (B) Triton X-100 insolubility. CV-1 cells expressing WT or mutant HAs were incubated in the absence (−) or the presence (+) of the cholesterol-depleting reagent MβCD (20 mM) for 30 min at 37°C. HA was prepared and divided into insoluble and soluble fractions and detected as described in Materials and Methods. The percentage of HA found in the insoluble fraction in the absence or presence of MβCD was determined. Cholesterol depletion by treatment with MβCD increases the Triton X-100 solubility of WT HA and GPI-HA, but not of Δ12 HA (n = 4). (C) Effect of cholesterol depletion on fusion. CV-1 cells expressing WT or mutant HAs were prepared as described in the legend to Fig. 4, depleted of cholesterol as described in B, bound to labeled RBCs, and triggered for fusion. Cholesterol depletion by treatment with MβCD does not affect the fusion phenotype of WT HA, GPI-HA, Δ12 HA, or NΔ12 HA (see Fig. 5).
Effect of MβCD on Fusion by Δ12 HA and NΔ12 HA
Because of the general interest in glycoprotein localization to plasma membrane microdomains (Scheiffele et al. 1997; Hooper 1999), we asked whether treatment of cells expressing WT HA, GPI-HA, Δ12 HA, or NΔ12 HA with 20 mM MβCD influenced their fusion activity. Treatment of WT HA, GPI-HA, Δ12 HA, and NΔ12 HA-expressing cells with MβCD did not affect the fusion phenotype. WT HA still mediated efficient lipid and content dye transfer, whereas the mutant HAs still demonstrated significant lipid mixing but little or no content mixing (Fig. 8). Similar results were obtained using WT HA of the Japan strain (H2N2; Melikyan et al. 1999).
Why Δ12 HA May Cause Lipid, but Not Content, Mixing
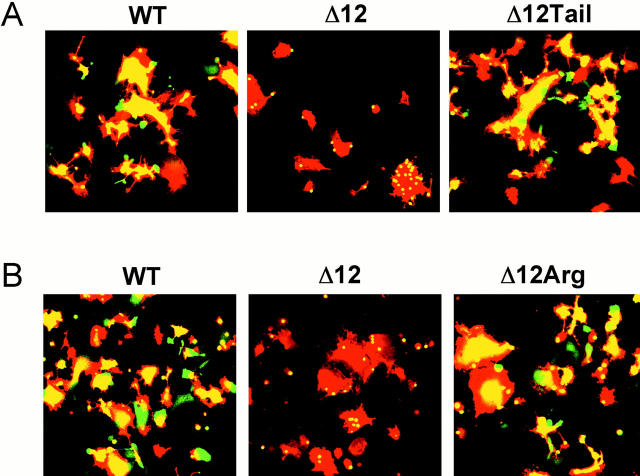
We have considered two general models for why Δ12 HA mediates only lipid mixing whereas Δ10 HA mediates content mixing as well. In the first model (Fig. 9 A, 1), we consider that Δ12 HA has recruited specific (e.g., shorter fatty acyl chain) lipids around it such that it spans a thinned bilayer. The lipids in such a thinned bilayer may not be competent to promote the hemifusion to fusion transition. In the second model, we consider that the TM domain of Δ12 HA is simply too short to span a bilayer; it may be anchored either perpendicularly (Fig. 9 A, 2a), obliquely (Fig. 9 A, 2b), or parallel (Fig. 9 A, 2c) to the membrane normal. To test between these models, we analyzed a mutant HA in which we added back the hydrophilic cytoplasmic tail (10 amino acids) to the end of Δ12 HA. We reasoned that if Δ12 HA spans a thinned bilayer (model 1), addition of the cytoplasmic tail should not affect its fusion phenotype. If, however, Δ12 HA does not span a bilayer (model 2), then addition of the cytoplasmic tail may force Δ12 HA to span a bilayer and it may therefore be able to support full fusion. As seen in Fig. 10 A, the mutant Δ12Tail HA clearly promotes full fusion. Next, we tested whether the first residue of the cytoplasmic tail, an arginine, added to the end of the Δ12 HA TM domain, was sufficient to restore full fusion activity. As seen in Fig. 10 B, Δ12Arg HA clearly promotes full fusion.
Figure 9.
(A) Model of possible interactions between the Δ12 (and NΔ12) TM domains and membranes: (1) The truncated TM domains of Δ12 and NΔ12 may span a thinned bilayer; (2a) the TM domain may project into, but not span, the bilayer in a perpendicular orientation relative to the membrane surface; (2b) the TM domain may project obliquely into the bilayer; (2c) the TM domain may be anchored at the surface parallel to the lipid bilayer. HA is presented as a monomer for clarification. The TM domain is depicted as an α-helix, but it may adopt other structures. (B) Models of HA and SNARE-mediated hemifusion to fusion transition. Multimers of HA trimers promote hemifusion (mixing of the outer, but not inner leaflets of the lipid bilayer). Subsequent interactions between the fusion peptides (white) and the TM domains (dark gray), either alone or in concert, with the hemifusion diaphragm may promote full fusion. In the case of the SNAREs, the TM domains of the t- (gray) and v-SNARE (black) may perform analogous functions.
Figure 10.
Addition of the cytoplasmic tail or a single arginine to Δ12 HA restores fusion. CV-1 cells transfected with 0.5 μg WT, 7.5 μg Δ12, 5.0 μg Δ12Tail, and 5.0 μg Δ12Arg were prepared for fusion as described in Materials and Methods. Fusion was triggered at pH 5.0 for 2 min at 37°C. Images presented in A and B are from separate experiments. Δ12Tail and Δ12Arg were expressed at the cell surface at levels comparable to that using 0.5 μg WT HA cDNA. The COOH-terminal sequences of Δ10, Δ11, Δ12, and Δ12Arg HA mutants are: Δ10WILWISFAISCFLLCVV Δ11WILWISFAISCFLLCV Δ12WILWISFAISCFLLC Δ12Arg WILWISFAISCFLLCR.
Additional HA TM Domain Mutants
To ascertain whether we could detect any specific TM domain sequences needed for HA to promote full fusion, we constructed additional point and deletion mutations within the HA TM domain (Fig. 11). We mutated the tryptophans (to alanines) within the highly conserved WILW sequence at the beginning of the HA TM domain. We mutated a serine at position 194, since this residue is the analogue of a glycine implicated as being important for the fusion activity of Japan HA (Melikyan et al. 1999). We also mutated a glycine at position 204 (singly and in combination with Ser 194), since glycines near the middle of the TM domain have been reported to be important for fusion mediated by the vesicular stomatitis virus envelope glycoprotein (VSV G; Cleverley and Lenard 1998). We also deleted six or eight amino acids at different locations within the TM domain. All mutants were examined for expression at the cell surface, for their ability to be cleaved by trypsin into HA1 and HA2, for RBC binding, and for fusion (both lipid and content mixing). Most of the mutants were also examined for trimer formation and the pH dependence of the conformation change (Table ). As seen in Table , by all of the criteria examined, these additional mutants in the HA TM domain behaved virtually the same as WT HA. Most importantly, all of the 12 additional HA TM domain mutants exhibited efficient lipid and efficient content mixing.
Figure 11.
Additional TM domain deletion and point mutants. Line diagram of the HA gene. In detail is the region surrounding the HA TM domain (gray box). Point mutations were made in the context of full-length HA and are indicated in large font. Deletion mutations were made in the context of tail− HA and are indicated as spaces. Δ10, Δ12, and NΔ12 HA are included as a reference.
Table 3.
Summary of Results: Effects of Point Mutations and Additional Deletions in the HA TM Domain
| HA mutants | Percentage of cells | FU | Trimer | Trypsin cleavage | RBC binding | Lipid mixing | Content mixing | ConfΔ |
|---|---|---|---|---|---|---|---|---|
| % | pH | |||||||
| W185A | 76 | 299 | ND | + | ++ | ++ | ++ | ND |
| W188A | 50 | 270 | ND | + | ++ | ++ | ++ | ND |
| W185A/ W188A | 62 | 137 | + | + | ++ | ++ | ++ | 5.0 |
| S194A | 68 | 204 | ND | + | ++ | ++ | ++ | ND |
| S194L | 66 | 213 | + | + | ++ | ++ | ++ | 5.0 |
| S194L/ G204L | 49 | 130 | ND | + | ++ | ++ | ++ | ND |
| G204A | 22 | 224 | ND | + | ++ | ++ | ++ | ND |
| G204L | 79 | 337 | + | + | ++ | ++ | ++ | 5.0 |
| NΔ6 | 38 | 152 | + | + | ++ | ++ | ++ | 5.0 |
| NΔ6Δ2 | 58 | 87 | + | + | ++ | ++ | ++ | 5.0 |
| MidΔ6 | 30 | 145 | + | + | ++ | ++ | ++ | 5.0 |
| MidΔ8 | 46 | 179 | + | + | ++ | ++ | ++ | 5.0 |
Percentage of cells indicates the expression efficiency (SEM ≤ ±9). Fluorescence units (FU) indicates the mean fluorescence as detected by FACS® analysis (SEM ≤ ±24). ND, not done.
Discussion
Cells expressing the ectodomain of HA linked to the membrane via a GPI anchor (GPI-HA) promote lipid, but not content, mixing (Kemble et al. 1994). The behavior of GPI-HA indicates that the HA TM domain, which is predicted to be 27 amino acids in length, plays an important role in the hemifusion to fusion transition. In this study, we tested whether there is a length or sequence requirement for the TM domain to facilitate this important step in the fusion cascade. To do this, we first engineered stop codons into the TM domain, generating mutant HAs with sequential 2–amino acid deletions from the COOH-terminal end (up to 14 amino acids). We also engineered mutant HAs lacking 6 or 12 amino acids from the NH2-terminal end, mutants lacking 6 or 8 amino acids from the central region, and a mutant lacking 11 amino acids from the COOH-terminal end. These HA mutants lacked the cytoplasmic tail, which has been shown to be dispensable for fusion (Jin et al. 1994; Melikyan et al. 1997b). We also made eight site-specific point mutations within the TM domain of full-length HA. With the exception of Δ14, which did not reach the cell surface (data not shown), all of the mutant HAs possessed biochemical properties similar to WT HA (Table and Table ). In terms of membrane fusion, all HAs studied with ≥17 amino acids in their TM domains were able to efficiently mediate full fusion (both lipid and content mixing). In contrast, HAs with 15 or 16–amino acid TM domains (and no cytoplasmic tail) mediated robust lipid mixing, but were either severely impaired (16–amino acid TM domain) or virtually unable (15–amino acid TM domain) to mediate content mixing. All of the point mutants examined efficiently promoted full fusion. Our findings suggest that there is a stringent length requirement, 17 amino acids, for the HA TM domain to be able to support the hemifusion to fusion transition. Additional experiments (see below) suggested that the HA TM domain must span its bilayer to properly execute the fusion reaction.
Δ12 HA and NΔ12 HA Mediate Hemifusion
Hemifusion is functionally defined as the merger of the outer, but not the inner, leaflets of the fusing bilayers, such that aqueous continuity is not established. Many investigators have proposed that biological fusion events proceed through a hemifusion intermediate (Palade 1975; Pinto da Silva and Nogueira 1977; Kalderon and Gilula 1979; Lucy and Ahkong 1986; Chernomordik et al. 1987; Nanavati et al. 1992).
Studies with GPI-HA indicate that progression to a fusion pore, as monitored by the transfer of small content dyes, does not occur when GPI-HA–expressing cells are induced to fuse with RBCs (Kemble et al. 1994; Melikyan et al. 1995b; Nussler et al. 1997). Combinations of dye transfer and electrophysiological assays also indicated that GPI-HA does not induce fusion pores in planar membranes (Melikyan et al. 1995b; Razinkov et al. 1999). However, more recent electrophysiological studies indicate that under certain conditions (e.g., pH 4.8 with membrane-labeled RBCs), GPI-HA can induce fusion pores during fusion with RBCs, but that these pores occur less frequently than with WT HA, never enlarge, and are strongly influenced by the presence of lipid dyes in the target membrane (Markosyan et al. 2000). These observations indicate that the TM domain is required for efficient fusion pore initiation and for fusion pore enlargement (Markosyan et al. 2000). In the present study, we found that two mutant HAs, Δ12 HA and NΔ12 HA, are severely restricted in their ability to mediate mixing of a 376–mol wt content dye, CF. Since the level of CF mixing seen with Δ12 HA and NΔ12 HA is similar to that seen with GPI-HA (Fig. 6 B), it is likely that Δ12 HA and NΔ12 HA are unable to efficiently promote the hemifusion to fusion transition and are unable to support pore enlargement. Additional evidence that Δ12 HA and NΔ12 HA are blocked at the stage of hemifusion, and not at stunted fusion, is that 0.5 mM CPZ is required to induce appreciable content dye transfer (Fig. 6), as has been seen with GPI-HA (Melikyan et al. 1997a). We propose that Δ12 HA and NΔ12 HA are protein mimetics of GPI-HA.
Length Requirement of the HA TM Domain
We have uncovered a surprisingly stringent length requirement for the HA TM domain to be able to (efficiently) promote the hemifusion to fusion transition. HAs harboring a 17–amino acid (predicted) TM domain promote full fusion, whereas an HA with a 16–amino acid (predicted) TM domain is severely impaired in promoting full fusion and HAs with 15–amino acid (predicted) TM domains appear to arrest at hemifusion.
The finding that there is a stringent length requirement of 17 amino acids for the HA TM domain to efficiently promote the hemifusion to fusion transition suggests that HAs with TM domains ≥17 amino acids are anchored differently in the bilayer than fusion-impaired HAs that have shorter TM domains (≤16 amino acids). Using a synthetic peptide representing the transmembrane segment of X:31 HA, Tatulian and Tamm 1999 have recently shown that the WT HA TM domain (27–amino acid predicted) spans DMPC/DMPG bilayers as an α-helix that aligns roughly perpendicular to the bilayer normal. As discussed in Results (with reference to Fig. 9 A), we have considered two general models for how the fusion-defective TM domain mutants (with TM domains ≤16 amino acids) are anchored in the bilayer. The first (Fig. 9 A, 1) envisions that the short (≤16 amino acids) TM domains are aligned like the WT HA TM domain (as a perpendicular α-helix), but to span the bilayer, they have had to recruit specific lipids (e.g., with short fatty acyl tails) around them. Such lipids may not be able to adopt the necessary curvature to allow fusion to progress beyond hemifusion (Melikyan et al. 1997a; Chernomordik et al. 1998). The second general model proposes that the fusion-incompetent TM domains (≤16 amino acids) cannot span the bilayer (Fig. 9 A, 2a, 2b, and 2c). Our finding that addition of the hydrophilic 10–amino acid cytoplasmic tail sequence to a TM domain of 15 hydrophobic amino acids generated an HA that efficiently promotes full fusion (Fig. 10 A) supports the latter model (Fig. 9 A, 2), as it suggests that the addition of these hydrophilic residues has forced the 15–amino acid TM domain to span the bilayer. Even more striking is the finding that addition of a single arginine to a 15 amino acid TM domain generated an HA that promotes full fusion (Fig. 10 B). We propose that addition of one or more hydrophilic residues forced the 15 amino acid TM domain to span the bilayer. The need for a hydrophilic residue following a 15 amino acid TM domain is underscored by comparing Δ11 HA and Δ12Arg HA (see Fig. 4 and Fig. 10, legend). Collectively, these findings support a model in which the HA TM domain must span a bilayer to efficiently promote full fusion.
Why might it be necessary for the HA TM domain to span its bilayer to efficiently promote fusion? One possibility is based on the concept that the TM domain of HA plays an important role in disrupting the lipid bilayer during fusion (Melikyan et al. 1995b; Tatulian and Tamm 1999). Since bilayer disruption by transmembrane peptides critically depends on the extent of matching between the peptide length and the bilayer thickness (Killian et al. 1996; van Der Wel et al. 2000), aberrant insertion of the short TM domains (Fig. 9 A, 2a, 2b, and 2c) may cause them to be incapable of disrupting the bilayer of the hemifusion diaphragm (Fig. 9 B, top). Another possibility is that the TM domain of HA must apply tension to the hemifusion diaphragm to efficiently promote pore formation. If the short fusion-incompetent TM domains do not span the bilayer, they may be unable to provide the needed tension. And finally, the short TM domains may not be able to adopt α-helical structures that may be required for specific protein–lipid or protein–protein (e.g., TM domain–TM domain or TM domain–fusion peptide) interactions that are required for progression to full fusion. Future biophysical experiments will address how the short fusion-incompetent TM domains are oriented in a bilayer.
Sequence Requirements of the HA TM Domain
We also asked whether we could identify any specific residues within the HA TM domain that are required to promote the hemifusion to fusion transition. During analysis of four additional truncation mutants (of six or eight amino acids) and eight point mutants at different locations in the TM domain (Table ), we were unable to uncover any specific sequence requirement for fusion. In particular, two highly conserved tryptophan residues within the WILW motif at the NH2-terminal end of the TM domain, which appear to be important for targeting HA to the apical surface of epithelial cells (Scheiffele et al. 1997; Lin et al. 1998), do not appear to be required for fusion. We also examined the requirement for fusion of particular residues within the interior of the HA TM domain based on two reports concerning the role of glycine residues within the TM domain of viral fusion proteins. Cleverley and Lenard 1998 suggested the importance of glycine residues within the TM domain of VSV G. Substitution of both glycine residues within the TM domain of VSV G was reported to result in a hemifusion phenotype (Cleverley and Lenard 1998). Furthermore, a study using Japan HA, which contains two glycine residues within the TM domain, demonstrated that mutation of the more NH2-terminal glycine to a leucine (G520L HA) caused a restricted hemifusion phenotype (no pore formation and no transfer of lipid or content dye; Melikyan et al. 1999). We have made the equivalent mutations within the TM domain of X:31 HA (Table ). Our results suggest that neither a serine residue at position 194 (the analogue of G520 in Japan HA) nor a glycine residue at position 204, either individually or jointly, is necessary for fusion (Table ).
If one models the WT HA TM domain as an α-helix, there is a short face of four polar residues (two cysteines and two serines). However, several of our mutant HAs disrupt this motif (i.e., leave only two polar residues), but do not impair fusion. Hence, although we cannot exclude the possibility that there may be a sequence motif that is important for the X:31 HA TM domain to promote the hemifusion to fusion transition, we have not found such a motif. The differences we observe in the sequence requirements for fusion with X:31 HA (H3N2 subtype) and that reported for Japan HA (H2N2 subtype) may be due to differences in the subtype of HA or the techniques used.
Possible Parallel Roles for the Fusion Peptide and the TM Domain in the Hemifusion to Fusion Transition
Four indirect lines of evidence suggest that although they have different net hydrophobicities, the HA fusion peptide and the HA TM domain may play parallel roles in the hemifusion to fusion transition. First, we have recently demonstrated that replacement of the glycine at the first position of the HA fusion peptide with a serine (Ser HA) arrests HA fusion at the hemifusion stage (Qiao et al. 1999). Second, recent work using synthetic peptides corresponding to the fusion peptide and the TM domain indicates that these two peptides have similar effects on synthetic bilayers (Han et al. 1999; Tatulian and Tamm 1999). Both domains appear to order their respective lipid environments, thus decreasing the amount of water bound at the water–bilayer interface as well as increasing the surface hydrophobicity. A third line of evidence that the fusion peptide and the TM domain may be playing parallel roles in breaking the hemifusion diaphragm is the observation that in the lowest energy state of the protein, the fusion peptide and the TM domain are predicted to be very closely opposed (Chen et al. 1999). A fourth line of indirect evidence is that studies on the topology of synthetic versions of the HA fusion peptide as well as studies with HAs containing mutations in the fusion peptide are consistent with the notion that the first 18 residues of the fusion peptide embed in the target bilayer and are important for fusion (Gray et al. 1996; Macosko et al. 1997; Danieli, T., and J.M. White, unpublished data). Hence, the two membrane-interactive domains of HA may have similar length requirements (∼17 amino acids) to efficiently promote full fusion.
Sequence Requirements of the TM Domains of Other Viral Fusion Proteins
We have not observed any specific sequence requirements within the X:31 HA TM domain for full fusion. However, other investigators have suggested that there are specific sequence requirements for fusion within the TM domain of other viral fusion proteins. As discussed above, specific TM domain sequence requirements have been suggested for the VSV G glycoprotein (Cleverley and Lenard 1998) and the HA from the Japan strain of influenza (Melikyan, et al. 1999). In addition, mutation of a specific proline residue within the Moloney murine leukemia virus envelope glycoprotein, to either alanine, glycine, or valine, diminished syncytia formation (Taylor and Sanders 1999). Finally, mutation of a conserved arginine residue to leucine within the TM domain of the HIV gp160 eliminated HIV-1 envelope–mediated syncytia formation (Owens et al. 1994). These findings suggest that for some viral fusion proteins, specific TM domain sequences may be important for fusion. Even for cases where there are specific sequence requirements, we propose that the length of the TM domain will be a critical determinant of fusion for all viral fusion proteins.
Possible Relevance to SNARE-mediated Fusion
Recent structural and biochemical data indicate that the SNARE (soluble N-ethylmaleimide–sensitive factor [NSF] attachment protein receptor) complexes, key players in intracellular fusion events, share similarities with many viral fusion proteins (Hanson et al. 1997; Hohl et al. 1998; Poirier et al. 1998; Skehel and Wiley 1998; Sutton et al. 1998). Formation of a “SNAREpin” structure consisting of a four-helix coiled coil domain is thought to force the fusing membranes together (Katz et al. 1998; Weber et al. 1998). The v- and t-SNARE proteins are each anchored into their respective membranes by a TM domain (21–24 residues in length; Fig. 9 B, bottom). It may therefore be the case that the TM domain of one SNARE performs a function similar to that of the viral fusion peptide while the TM domain of the other SNARE acts in an manner equivalent to the TM domain of the viral fusion protein (Fig. 9 B). Therefore the length of SNARE TM domain anchors may be a critical determinant of fusion. Recent evidence suggests that this may indeed be the case. An analysis of the behavior of Caenorhabditis elegans mutants has suggested that the TM domain of the t-SNARE, unc-64, must span its bilayer to function properly (Saifee et al. 1998). And, very recently McNew et al., using lipid anchors of varying length, provided evidence that the TM domain of the v-SNARE must be anchored in both bilayer leaflets to promote fusion (in this case, lipid mixing; McNew et al. 2000). Hence, it seems likely that the length of the TM domains of both viral fusion proteins and SNAREs will be an important determinant of fusion, and in some cases, of the hemifusion to fusion transition (Fig. 9 B).
Acknowledgments
We are grateful to Drs. F. Cohen, G. Melikyan, D. Castle, L. Tamm, and S. Green for thoughtful discussions and critical reading of the manuscript. The authors also thank Jennifer Gruenke for help preparing 9 and for invaluable assistance with videomicroscopy image capture and quantification.
This work was supported by National Institutes of Health grant AI22470 to J.M. White.
Footnotes
Abbreviations used in this paper: CF, carboxyfluorescein; CPZ, chlorpromazine; GPI, glycosylphosphatidylinositol; HA, hemagglutinin; MβCD, methyl β-cyclodextrin; RT, room temperature; SCS, supplemented calf serum; SNARE, soluble N-ethylmaleimide–sensitive factor attachment protein receptor; STI, soybean trypsin inhibitor; TM, transmembrane; VSV G, vesicular stomatitis virus envelope glycoprotein; WT, wild-type.
References
- Blumenthal R., Sarkar D.P., Durell S., Howard D.E., Morris S.J. Dilation of the influenza hemagglutinin fusion pore revealed by kinetics of individual cell–cell fusion events. J. Cell Biol. 1996;135:63–71. doi: 10.1083/jcb.135.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullough P.A., Hughson F.M., Skehel J.J., Wiley D.C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- Carr C.M., Kim P.S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- Chen J., Skehel J.J., Wiley D.C. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L.V., Melikyan G.B., Chizmadzhev Y.A. Biomembrane fusiona new concept derived from model studies using two interacting planar lipid bilayers. Biochim. Biophys. Acta. 1987;906:309–352. doi: 10.1016/0304-4157(87)90016-5. [DOI] [PubMed] [Google Scholar]
- Chernomordik L.V., Frolov V.A., Leikina E., Bronk P., Zimmerberg J. The pathway of membrane fusion catalyzed by influenza hemagglutininrestriction of lipids, hemifusion, and lipidic pore formation. J. Cell Biol. 1998;140:1369–1382. doi: 10.1083/jcb.140.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleverley D.Z., Lenard J. The transmembrane domain in viral fusionessential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc. Natl. Acad. Sci. USA. 1998;95:3425–3430. doi: 10.1073/pnas.95.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danieli T., Pelletier S.L., Henis Y.I., White J.M. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J. Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delos S.E., Gilbert J.M., White J.M. The central proline of an internal viral fusion peptide serves two important roles. J. Virol. 2000;74:1686–1693. doi: 10.1128/jvi.74.4.1686-1693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Roth M.G., Hunter E. A chimeric avian retrovirus containing the influenza virus hemagglutinin gene has an expanded host range. J. Virol. 1992;66:7374–7382. doi: 10.1128/jvi.66.12.7374-7382.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle C., Roth M.G., Sambrook J., Gething M.-J. Analysis of progressive deletions of the transmembrane and cytoplasmic domains of influenza hemagglutinin. J. Cell Biol. 1986;103:1193–1204. doi: 10.1083/jcb.103.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellens H., Bentz J., Mason D., Zhang F., White J.M. Fusion of influenza hemagglutinin-expressing fibroblasts with glycophorin-bearing liposomesrole of hemagglutinin surface density. Biochemistry. 1990;29:9697–9707. doi: 10.1021/bi00493a027. [DOI] [PubMed] [Google Scholar]
- Friedrichson T., Kurzchalia T.V. Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature. 1998;394:802–805. doi: 10.1038/29570. [DOI] [PubMed] [Google Scholar]
- Fuerst T.R., Niles E.G., Studier F.W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin Y., Ruigrok R.W.H., Brunner J. Low-pH induced conformational changes in viral fusion proteinsimplications for the fusion mechanism. J. Gen. Virol. 1995;76:1541–1556. doi: 10.1099/0022-1317-76-7-1541. [DOI] [PubMed] [Google Scholar]
- Gray C., Tatulian S.A., Wharton S.A., Tamm L.K. Effect of the N-terminal glycine on the secondary structure, orientation and interaction of the influenza hemagglutinin fusion peptide with lipid bilayers. Biophys. J. 1996;70:2275–2286. doi: 10.1016/S0006-3495(96)79793-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Steinhauer D.A., Wharton S.A., Tamm L.K. Interaction of mutant influenza virus hemagglutinin fusion peptides with lipid bilayersprobing the role of hydrophobic residue size in the central region of the fusion peptide. Biochemistry. 1999;38:15052–15059. doi: 10.1021/bi991232h. [DOI] [PubMed] [Google Scholar]
- Hanson P.I., Roth R., Morisaki H., Jahn R., Heuser J.E. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- Hernandez L.D., Hoffman L.R., Wolfsberg T.G., White J.M. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- Hohl T.M., Parlati F., Wimmer C., Rothman J.E., Sollner T.H., Engelhardt H. Arrangement of subunits in 20 S particles consisting of NSF, SNAPs, and SNARE complexes. Mol. Cell. 1998;2:539–548. doi: 10.1016/s1097-2765(00)80153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper N.M. Detergent-insoluble glycosphingolipid/cholesterol-rich membrane domains, lipid rafts and caveolae. Mol. Membr. Biol. 1999;16:145–156. doi: 10.1080/096876899294607. [DOI] [PubMed] [Google Scholar]
- Hughson F.M. Structural characterization of viral fusion proteins. Curr. Biol. 1995;5:265–274. doi: 10.1016/s0960-9822(95)00057-1. [DOI] [PubMed] [Google Scholar]
- Jin H., Leser G.P., Lamb R.A. The influenza virus hemagglutinin cytoplasmic tail is not essential for virus assembly or infectivity. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:5504–5515. doi: 10.1002/j.1460-2075.1994.tb06885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon N., Gilula N.B. Membrane events involved in myoblast fusion. J. Cell Biol. 1979;81:411–425. doi: 10.1083/jcb.81.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Hanson P.I., Heuser J.E., Brennwald P. Genetic and morphological analyses reveal a critical interaction between the C-termini of two SNARE proteins and a parallel four helical arrangement for the exocytic SNARE complex. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:6200–6209. doi: 10.1093/emboj/17.21.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble G.W., Henis Y., White J.M. GPI- and transmembrane-anchored influenza hemagglutinin differ in structure and receptor binding activity. J. Cell Biol. 1993;122:1253–1265. doi: 10.1083/jcb.122.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble G.W., Danieli T., White J.M. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- Killian J.A., Salemink I., de Planque M.R., Lindblom G., Koeppe R.E., II, Greathouse D.V. Induction of nonbilayer structures in diacylphosphatidylcholine model membranes by transmembrane alpha-helical peptidesimportance of hydrophobic mismatch and proposed role of tryptophans. Biochemistry. 1996;35:1037–1045. doi: 10.1021/bi9519258. [DOI] [PubMed] [Google Scholar]
- Lin S., Naim H.Y., Rodriguez A.C., Roth M.G. Mutations in the middle of the transmembrane domain reverse the polarity of transport of the influenza virus hemagglutinin in MDCK epithelial cells. J. Cell Biol. 1998;142:51–57. doi: 10.1083/jcb.142.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucy J.A., Ahkong Q.F. An osmotic model for the fusion of biological membranes FEBS Lett. 199 1986. 1 11[published erratum at 201:180] [DOI] [PubMed] [Google Scholar]
- Macosko J.C., Kim C.-H., Shin Y.-K. The membrane topology of the fusion peptide region of influenza hemagglutinin determined by spin-labeling EPR. J. Mol. Biol. 1997;267:1139–1148. doi: 10.1006/jmbi.1997.0931. [DOI] [PubMed] [Google Scholar]
- Markosyan R.M., Cohen F.S., Melikyan G.B. The lipid-anchored ectodomain of influenza virus hemagglutinin (GPI-HA) is capable of inducing nonenlarging fusion pores. Mol. Biol. Cell. 2000;11:1143–1152. doi: 10.1091/mbc.11.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew J.A., Weber T., Parlati F., Johnston R.J., Melia T.J., Sollner T.H., Rothman J.E. Close is not enoughSNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J. Cell Biol. 2000;150:105–117. doi: 10.1083/jcb.150.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan G.B., Niles W.D., Cohen F.S. The fusion kinetics of influenza hemagglutinin expressing cells to planar bilayer membranes is affected by HA density and host cell surface J. Gen. Physiol. 106 1995. 783 802a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan G.B., White J.M., Cohen F.S. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes J. Cell Biol. 131 1995. 679 691b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan G.B., Brener S.A., Ok D.C., Cohen F.S. Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin–induced hemifusion to full fusion J. Cell Biol. 136 1997. 995 1005a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan G.B., Jin H., Lamb R.A., Cohen F.S. The role of the cytoplasmic tail region of influenza virus hemagglutinin in formation and growth of fusion pores Virology. 235 1997. 118 128b [DOI] [PubMed] [Google Scholar]
- Melikyan G.B., Lin S., Roth M.G., Cohen F.S. Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion. Mol. Biol. Cell. 1999;10:1821–1836. doi: 10.1091/mbc.10.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussler F., Clague M.J., Herrmann A. Meta-stability of the hemifusion intermediate induced by glycosylphosphatidylinositol-anchored influenza hemagglutinin. Biophys. J. 1997;73:2280–2291. doi: 10.1016/S0006-3495(97)78260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens R.J., Burke C., Rose J.K. Mutations in the membrane-spanning domain of the human immunodeficiency virus envelope glycoprotein that affect fusion activity. J. Virol. 1994;68:570–574. doi: 10.1128/jvi.68.1.570-574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade G. Intracellular aspects of the process of protein secretion. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- Pinto da Silva P., Nogueira M.L. Membrane fusion during secretion. A hypothesis based on electron microscope observation of Phytophthora palmivora zoospores during encystment. J. Cell Biol. 1977;73:161–181. doi: 10.1083/jcb.73.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier M.A., Xiao W., Macosko J.C., Chan C., Shin Y.K., Bennett M.K. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat. Struct. Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- Qiao H., Pelletier S., Hoffman L., Hacker J., Armstrong R.T., White J.M. Specific single or double proline substitutions in the “spring-loaded” coiled-coil region of the influenza hemagglutinin impair or abolish membrane fusion activity. J. Cell Biol. 1998;141:1335–1347. doi: 10.1083/jcb.141.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H., Armstrong R.T., Melikyan G.B., Cohen F.S., White J.M. A specific point mutation at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol. Biol. Cell. 1999;10:2759–2769. doi: 10.1091/mbc.10.8.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razinkov V.I., Melikyan G.B., Cohen F.S. Hemifusion between cells expressing hemagglutinin of influenza virus and planar membranes can precede the formation of fusion pores that subsequently fully enlarge. Biophys. J. 1999;77:3144–3151. doi: 10.1016/S0006-3495(99)77144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M., Doyle C., Sambrook J., Gething M. Heterologous transmembrane and cytoplasmic domains direct functional chimeric influenza virus hemagglutinins into the endocytic pathway. J. Cell Biol. 1986;102:1271–1283. doi: 10.1083/jcb.102.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saifee O., Wei L., Nonet M.L. The Caenorhabditis elegans unc-64 locus encodes a syntaxin that interacts genetically with synaptobrevin. Mol. Biol. Cell. 1998;9:1235–1252. doi: 10.1091/mbc.9.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P., Roth M.G., Simons K. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:5501–5508. doi: 10.1093/emboj/16.18.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroth-Diez B., Ponimaskin E., Reverey H., Schmidt M.F.G., Herrmann A. Fusion activity of transmembrane and cytoplasmic domain chimeras of the influenza virus glycoprotein hemagglutinin. J. Virol. 1998;72:133–141. doi: 10.1128/jvi.72.1.133-141.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Skehel J.J., Wiley D.C. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- Stegmann T. Anchors aweigh. Curr. Biol. 1994;4:551–554. doi: 10.1016/s0960-9822(00)00123-8. [DOI] [PubMed] [Google Scholar]
- Steinhauer D.A., Wharton S.A., Skehel J.J., Wiley D.C. Studies of membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J. Virol. 1995;69:6643–6651. doi: 10.1128/jvi.69.11.6643-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton R.B., Fasshauer D., Jahn R., Brunger A.T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Tatulian S.A., Tamm L.K. Secondary structure, orientation, oligomerization and lipid interactions of the transmembrane domain of influenza hemagglutinin. Biochemistry. 1999;39:496–507. doi: 10.1021/bi991594p. [DOI] [PubMed] [Google Scholar]
- Taylor G.M., Sanders D.A. The role of the membrane-spanning domain sequence in glycoprotein-mediated membrane fusion. Mol. Biol. Cell. 1999;10:2803–2815. doi: 10.1091/mbc.10.9.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Der Wel P.C., Pott T., Morein S., Greathouse D.V., Koeppe I.R., Killian J.A. Tryptophan-anchored transmembrane peptides promote formation of nonlamellar phases in phosphatidylethanolamine model membranes in a mismatch-dependent manner. Biochemistry. 2000;39:3124–3133. doi: 10.1021/bi9922594. [DOI] [PubMed] [Google Scholar]
- Varma R., Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- Weber T., Zemelman B.V., McNew J.A., Westermann B., Gmachl M., Parlati F., Sollner T.H., Rothman J.E. SNAREpinsminimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- White J.M., Wilson I.A. Anti-peptide antibodies detect steps in a protein conformational changelow-pH activation of the influenza virus hemagglutinin. J. Cell Biol. 1987;105:2887–2896. doi: 10.1083/jcb.105.6.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]