Abstract
Tom20 is a major receptor of the mitochondrial preprotein translocation system and is bound to the outer membrane through the NH2-terminal transmembrane domain (TMD) in an Nin-Ccyt orientation. We analyzed the mitochondria-targeting signal of rat Tom20 (rTom20) in COS-7 cells, using green fluorescent protein (GFP) as the reporter by systematically introducing deletions or mutations into the TMD or the flanking regions. Moderate TMD hydrophobicity and a net positive charge within five residues of the COOH-terminal flanking region were both critical for mitochondria targeting. Constructs without net positive charges within the flanking region, as well as those with high TMD hydrophobicity, were targeted to the ER-Golgi compartments. Intracellular localization of rTom20-GFP fusions, determined by fluorescence microscopy, was further verified by cell fractionation. The signal recognition particle (SRP)–induced translation arrest and photo–cross-linking demonstrated that SRP recognized the TMD of rTom20-GFP, but with reduced affinity, while the positive charge at the COOH-terminal flanking segment inhibited the translation arrest. The mitochondria-targeting signal identified in vivo also functioned in the in vitro system. We conclude that NH2-terminal TMD with a moderate hydrophobicity and a net positive charge in the COOH-terminal flanking region function as the mitochondria-targeting signal of the outer membrane proteins, evading SRP-dependent ER targeting.
Keywords: mitochondria, protein import, sorting signal, signal recognition particle, signal-anchor sequence
Introduction
Most mitochondrial proteins are synthesized in the cytosol as preproteins bearing NH2-terminal extensions (presequences) that direct them to the mitochondria (Schatz and Dobberstein 1996; Neupert 1997; Omura 1998). The proteins are delivered to the import receptors of the mitochondrial outer surface by cytosolic import factors such as mitochondrial import stimulation factor and hsp70 (Mihara and Omura 1996), and are then transported to the intramitochondrial compartments by the preprotein translocation machinery of the outer and inner membranes (the TOM and TIM complexes, respectively; Lill and Neupert 1996; Meijer et al. 1996; Schatz and Dobberstein 1996; Neupert 1997; Voos et al. 1999).
In contrast to the import of preproteins into the interior compartments of the mitochondria, the mechanisms of targeting and membrane insertion of the outer membrane proteins are poorly understood. All the mitochondrial outer membrane proteins are synthesized without a cleavable presequence (Shore et al. 1995; Mihara 2000). Most known outer membrane proteins span the membrane either once or several times by α-helical hydrophobic transmembrane domains (TMD) within the molecule (Shore et al. 1995). On the other hand, Tom40, the core component of the Tom machinery, and mitochondrial porin are predicted to span the outer membrane by 12–14 antiparallel β strands in a β-barrel structure (Stanley et al. 1995; Mannella et al. 1996).
Mitochondrial outer membrane proteins such as Tom5 (Dietmeier et al. 1997), Tom6 (Cao and Douglas 1995), monoamine oxidases (Mitoma and Ito 1992), outer membrane cytochrome b5 (OMb) (Kuroda et al. 1998), metaxin (Armstrong et al. 1997), and VAMP-1B (Isenmann et al. 1998) are tail-anchored proteins, whereas proteins such as Tom70 (Hurt et al. 1985; McBride et al. 1992), and Tom20 (Schneider et al. 1991) bind to the outer membrane through the NH2-terminal TMD. The mitochondria-targeting signal of the outer membrane proteins seems to be contained in both the TMD and in the flanking regions. For example, the COOH-terminal 10 amino acid residues of OMb have sufficient information to direct OMb to the mitochondria. When either of the two basic amino acid residues in the COOH-terminal tail is replaced by alanine, the mutant OMb becomes targeted to the ER (Kuroda et al. 1998), probably by the signal recognition particle (SRP)- and Sec61-independent pathway (Kutay et al. 1995). Tom22 is anchored to the membrane by the TMD located at the middle portion of the molecule in an Ncyt-Cin orientation. The TMD and NH2-terminal flanking region of 13 amino acid residues of Saccharomyces cerevisiae Tom22 carry sufficient mitochondria-targeting information (Egan et al. 1999). Membrane insertion of Tom22 strictly depends upon Tom20 and Tom70 (Keil and Pfanner 1993). The signal for mitochondria targeting and insertion has been most extensively studied in S. cerevisiae Tom70. It contains NH2-terminal targeting information consisting of a short presequence-like segment (residues 1–10), followed by a stretch of uncharged, mostly apolar, amino acids that functions as the stop-transfer sequence (Hurt et al. 1985). Subsequent studies indicate that the TMD is required for both targeting and membrane integration, whereas the NH2-terminal “presequence-like” region cooperates with the TMD to enhance the overall rate of insertion (McBride et al. 1992). Insertion of Tom20 and Tom70 into the outer membrane does not require import receptors, but is directly targeted to Tom40 and is assembled into the TOM complex (Schneider et al. 1991; Schlossmann and Neupert 1995; Neupert 1997).
Despite these advances, the precise molecular mechanism of mitochondria targeting of the outer membrane proteins remains largely unknown. In the present study, we used rat Tom20 (rTom20) to characterize the mitochondria-targeting signal of the outer membrane proteins with the NH2-terminal TMD. Mitochondria targeting of this type of protein is intriguing because TMDs are composed of hydrophobic stretches that resemble the signal-anchor sequence of the ER-targeted membrane proteins (Shore et al. 1995; Sakaguchi et al. 1992). The classical signal peptides are generally composed of a hydrophobic core (h-) region of 6–15 hydrophobic amino acids (Martoglio and Dobberstein 1998). When the signal sequences contain a sufficient hydrophobic h-region and are not cleaved by signal peptidase, they function as the signal-anchor sequence. Features of signal-anchor sequences that determine type I (Nin-Ccyt) or type II (Ncyt-Cin) orientation have been identified (Sakaguchi et al. 1992; Martoglio and Dobberstein 1998; Kida et al. 2000). It is not known how the outer membrane proteins with such a structural feature are targeted from the ribosome to the mitochondria, without involving SRP-dependent ER targeting.
rTom20 consists of the NH2-terminal TMD (residues 7–24), the tetratricopeptide repeat motif (TPR; residues 70–103), a charged amino acid–rich flanking region between the TMD and TPR (residues 25–69), and a short cluster of acidic amino acids at the COOH terminus (142–145; Goping et al. 1995; Seki et al. 1995; Hanson et al. 1996; Iwahashi et al. 1997). The segment 1–69 of rTom20 is sufficient to complement the function of Δtom20 yeast cells (Iwahashi et al. 1997).
To define the mitochondria-targeting signal of rTom20, we constructed a series of rTom20–green fluorescence protein (GFP) fusion proteins carrying deletions, insertions, or point mutations within the TMD or flanking region and examined their intracellular localization in COS-7 cells. The post- and cotranslational integration into rat liver mitochondria and dog pancreas microsomes, respectively, were also examined in vitro. Moderate TMD hydrophobicity and at least one net positive charge in the subsequent segment of five amino acid residues functioned together as the necessary and sufficient mitochondria-targeting signal; when hydrophobicity of the TMD was increased, or a net positive charge in the COOH-terminal flanking region was dissipated, the Tom20-GFP fusions were then targeted to the ER-Golgi compartments, or sometimes from there to the plasma membrane.
Measurement of SRP-induced translation arrest of rTom20-GFP constructs and photo–cross-linking of the nascent rTom20-GFP peptides to SRP indicated that: (a) SRP recognized, although with low affinity, the TMD of the rTom20-GFP construct in which the positive charges in the COOH-terminal flanking region had been deleted, thus inhibiting the translation; and (b) introduction of positive charges to the COOH-terminal flanking region released the SRP-induced translation arrest, presumably without dissociating SRP from the nascent chain. Furthermore, truncated rTom20-GFP in the ribosome nascent chain complexes (RNCs) was not able to be inserted into the mitochondrial outer membrane unless it was released from the ribosomes. Because SRP first scans nascent chains for the presence of a signal sequence as they emerge from the ribosome and targets them to the ER cotranslationally in mammalian cells (Walter and Johnson 1994; Ng et al. 1996), these results suggest that SRP potentially recognizes the TMD of nascent rTom20. Basic amino acid residues in the subsequent segment inhibit the function of SRP and, together with the TMD, enable correct post-translational targeting of rTom20 to mitochondria.
Materials and Methods
Construction of Expression Plasmids
pRc/CMV (Invitrogen), a mammalian expression vector, was digested with HindIII and XbaI, treated with a Klenow fragment, and self-ligated using T4 DNA ligase. cDNA of Aequoria victoria GFP was subjected to PCR using the primers GSX (5′-TATTCTAGAATGAGTAAAGGAGAAGAACTTTTCAC-3′) and GAA (5′-TATGGGCCCTTATTTGTATAGTTCATCCATGC-3′). The PCR products were digested with XbaI and ApaI at the sites underlined, and introduced to the expression vector, which had been opened with XbaI and ApaI. The resulting plasmid was called pRcG. To construct pRc20G, the cDNA of rat Tom20 (Iwahashi et al. 1997) was amplified by PCR with the primers RSX (5′-TATTCTAGAATGGTGGGCCGGAACAGCGCC-3′) and RAX (5′-TATTCTAGATTCCACATCATCCTCACCCAAG-3′). The PCR products were digested with XbaI and inserted into an XbaI site of pRcG. The rTom20-GFP mutants carrying deletions were constructed by PCR with pRc20G as the template and complementary pairs of oligonucleotides as primers. The rTom20-GFP constructs carrying point mutations or insertions were generated using Pfu DNA polymerase as described previously (Edward et al. 1998). In brief, complementary pairs of oligonucleotides carrying the desired mutation were used as primers for the complete synthesis by Pfu Turbo DNA polymerase (Stratagene) of both strands of the template plasmid. The product was treated with DpnI to digest the methylated parental DNA template and then used for transformation of XLI-blue cells. The Tom20-GFP constructs thus generated were all confirmed to carry the desired mutations or deletions by DNA sequencing. Information on the nucleotide sequences of the primers used in this study is available on request.
Transfection of COS-7 Cells and Fluorescence Microscopy
COS-7 cells were cultured on coverslips in 35-mm dishes in 2 ml of DME supplemented with 10% FCS at 37°C overnight under an atmosphere of 10% CO2 in air. Transfection was performed using FuGene 6 reagent (Roche Molecular Biochemicals). The cells were incubated for 24 h. When mitochondria were to be stained, 100 nM MitoTracker (Molecular Probes) was added to the medium and incubated for 20 min before fixation. The cells cultured on coverslips were fixed with 2 ml of 50% acetone/50% methanol for 5 min at room temperature. The coverslips were washed with 2 ml of PBS three times. To stain the Golgi apparatus or the ER, the fixed cells on coverslips were incubated with rabbit anti–rat GM130 antibodies (Nakamura et al. 1995) or rabbit anti–rat Calnexin antibodies (Stress Gen) for 1 h, and then with Texas red–conjugated goat antibodies against rabbit IgG for 1 h. Fluorescent images were taken and analyzed by a confocal laser microscope Radiance 2000 (Bio-Rad Laboratories).
Preparation of Mitochondria and Golgi Membranes from Cultured Cells
COS-7 cells cultured in a 10-cm dish were washed with PBS, and then scraped off with 5 ml of PBS. Collected cells were precipitated by centrifugation at 600 g for 5 min, and washed with HE buffer (10 mM Hepes-KOH, pH 7.5, and 1 mM EDTA) containing 10% (wt/vol) sucrose. The cells were resuspended in 1 ml of the same buffer containing 20 μg/ml α2-macroglobulin, homogenized by five-times aspiration through a 27-gauge needle, and then centrifuged at 600 g for 5 min to obtain a post-nuclear supernatant. The supernatant was layered over a discontinuous gradient of 40 and 60% sucrose in HE buffer (3 and 1 ml, respectively). After centrifugation at 100,000 g for 3 h, 0.5-ml aliquots were collected from the top of the tube. 100 μl of each fraction was precipitated with 10% TCA and subjected to SDS-PAGE and immunoblotting using antibodies against rTom20, rat GM130, or rat Golgin97.
In Vitro Insertion of rTom20-GFP Fusion Proteins into the ER and Mitochondria
mRNAs for rTom20-GFP fusion proteins transcribed in vitro were translated in rabbit reticulocyte lysate in the presence of dog pancreas rough microsomes, as described previously (Ota et al. 1998). Membrane integration was assessed by alkali treatment of the reaction mixture followed by alkali-sucrose gradient centrifugal flotation (alkali flotation; Ota et al. 1998). Insertion of the fusion proteins into mitochondria was performed as follows. Protein import mixture (20 μl final vol) containing 5 μl of unlabeled translation product in reticulocyte lysate, 20 μg of isolated mitochondria, 10 mM Hepes-KOH, pH 7.5, 250 mM sucrose, and 100 μg/ml α2-macroglobulin was incubated at 30°C for 60 min. After the reaction, 400 μl of 0.1 M sodium carbonate, pH 11.5, was added to each reaction mixture and cooled on ice for 30 min. After centrifugation at 100,000 g for 10 min, the supernatant was TCA precipitated. The membrane fraction was dissolved in 100 μl 0.1 M sodium carbonate and TCA precipitated.
SRP-induced Translation Arrest of mRNAs for rTom20-GFP Constructs
mRNA for wild-type rTom20-GFP, H22GFP, 4SGFP, or mouse synaptotagmin II (Syt II; Fukuda et al. 1994; Kida et al. 2000) transcribed in vitro was cotranslated with GFP mRNA in the wheat germ lysate system (25 μl) in the presence or absence of SRP (Sakaguchi et al. 1984). SRP was purified according to the method of Walter and Blobel 1980.
Photo–cross-linking of SRP to the Truncated rTom20-GFP Constructs in RNCs
The truncated mRNA for rTom20-GFP-74 (residues 1–74), 4SGFP-74 (1–74), H22GFP-78 (1–78), or preprolactin-86 (1–86) was translated in the wheat germ lysate system for 20 min at 25°C in the presence of 35S-methionine, 4-(3-trifluoromethyl-diazirino) benzoic acid (TDBA)-Lys-tRNA, and SRP where indicated (Görlich et al. 1991). After termination of the translation with 2 mM cycloheximide, the translation mixtures were transferred to a 96-well plate and UV-irradiated at 0°C for 10 min. The irradiated samples were resolved by SDS-PAGE (16% gel) and analyzed by digital autoradiography.
Results
A Net Positive Charge in the COOH-terminal Flanking Region of the TMD Determines Mitochondrial Localization of rTom20-GFP
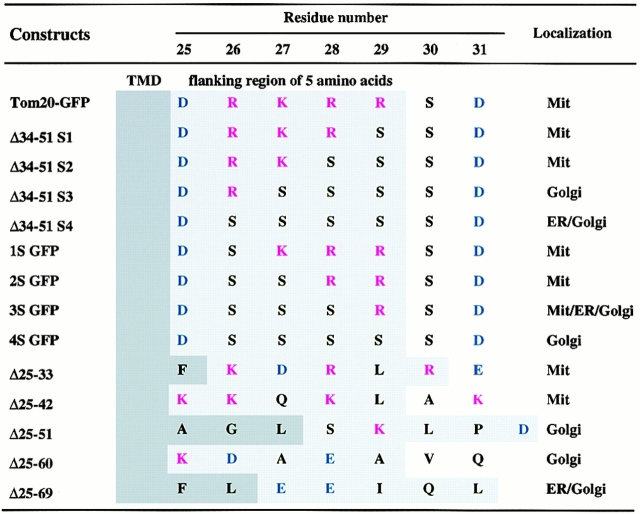
To characterize the mitochondria-targeting signal of rTom20, we constructed rTom20-GFP in which rTom20 (Fig. 1 A) was fused to the NH2 terminus of GFP, and several mutants in which the flanking region of rTom20-GFP was deleted to various extents; the flanking region was divided into five segments, which were deleted in combination, as shown in Fig. 1 B. These constructs were expressed in COS-7 cells and their intracellular localization was examined by a confocal microscope with MitoTracker staining as the reference. rTom20-GFP was colocalized with MitoTracker in the dispersed filamentous structure (Fig. 2), sometimes in the aggregated mitochondria at the perinuclear region, which were dispersed in the presence of nocodazole, which breaks microtubules (data not shown). The merged confocal images of fluorescence of GFP and MitoTracker for the various deletion mutants are summarized in Fig. 3 A. Δ25-33GFP, Δ25-42GFP, Δ43-51GFP, and Δ34-51GFP colocalized with MitoTracker, whereas Δ25-51GFP, Δ25-60GFP, and Δ25-69GFP localized to the ER-Golgi compartments (see below). These results suggest that the number of positive charges proximal to the COOH terminus of the TMD is important for mitochondria targeting (see Table ). The residues 1–33 of rTom20 (N33-GFP) function as an efficient mitochondria-localization signal, as shown in Fig. 3 A.
Figure 1.
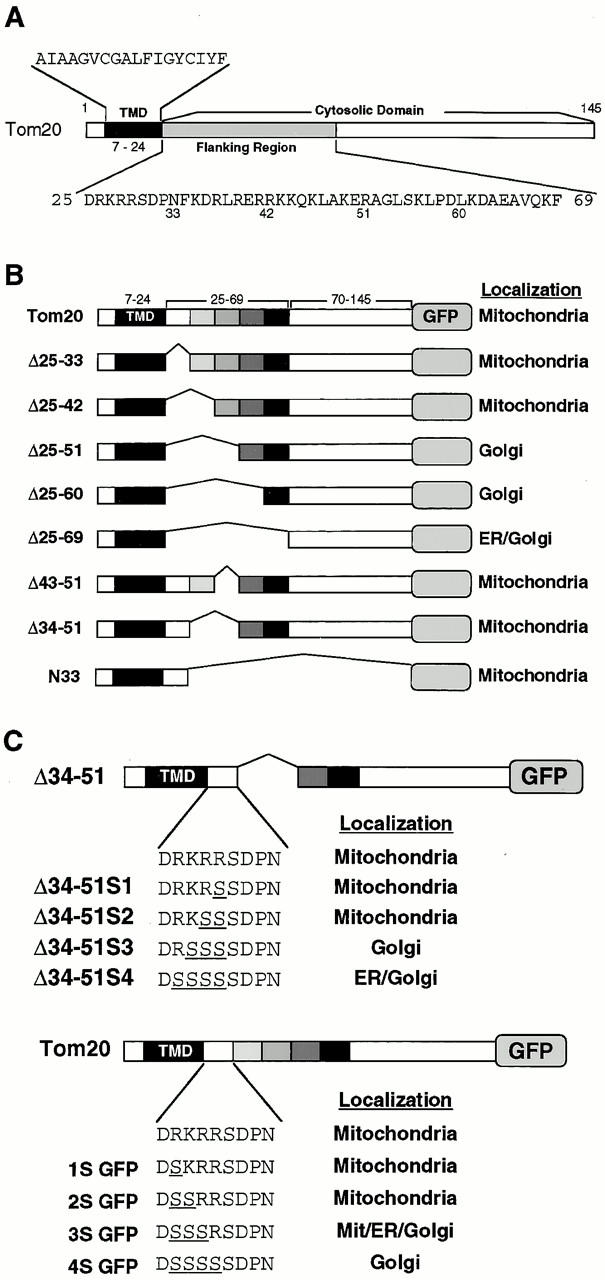
Schematic presentation of rTom20 and the flanking region-deleted mutants of rTom22-GFP fusions. (A) Schematic structure of rTom20. Amino acid sequences of the TMD and the flanking region are shown. (B) Schematic structure of the flanking region–deleted rTom20-GFP fusions. (C) Structure of the constructs carrying mutations in the COOH-terminal flanking region.
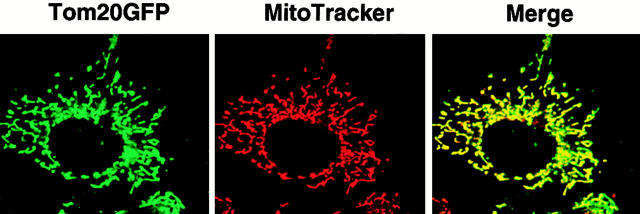
Figure 2.
Expression of wild-type rTom20-GFP in COS-7 cells. COS-7 cells were transfected with the expression vector containing the rTom20-GFP cDNA as described in Materials and Methods. The cells were incubated with MitoTracker, and fluorescent images of GFP (green) and MitoTracker (red) were taken by a confocal microscope.
Figure 3.
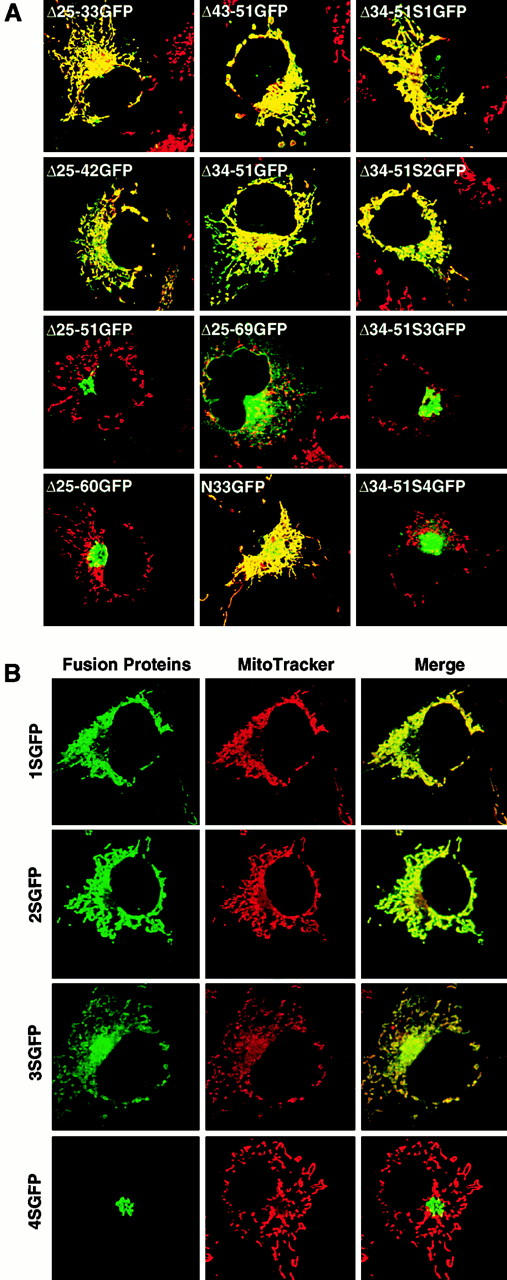
Basic amino acids at the COOH-terminal flanking region of the TMD are critical for targeting rTom20-GFP fusions to mitochondria. COS-7 cells were transfected with the indicated constructs in the expression vectors. The cells were costained with MitoTracker. Other conditions were the same as in Fig. 2. (A) Localization of rTom20-GFP constructs carrying mutations in the COOH-terminal flanking region. Merged fluorescent images of GFP and MitoTracker are shown. (B) Fluorescent images of MitoTracker and rTom20-GFP mutants in which the basic amino acids in the flanking region were replaced by serine residues.
Table 1.
Amino Acid Sequence of the COOH-terminal Flanking Region of the TMD of Tom20–GFP Proteins
To further assess the importance of basic amino acid residues in the COOH-terminal flanking region, arginine or lysine residues in this segment were changed to serine and the obtained constructs (Fig. 1 C) were examined for their intracellular localization. Δ34-51S1GFP and Δ34-51S2GFP localized to the mitochondria, whereas Δ34-51S3GFP and Δ34-51S4GFP were transported to the ER-Golgi compartments (Fig. 3 A, and see below). Similarly, the constructs 1SGFP and 2SGFP localized exclusively to the mitochondria, whereas 3SGFP exhibited mixed localization to the mitochondria, the ER, and the Golgi apparatus (Fig. 3 B; see also subfractionation in Fig. 5 B). On the other hand, 4SGFP exclusively localized to the Golgi compartment (Fig. 3 B). Taken together, these results suggest that at least one net positive charge within the region containing five amino acid residues following the TMD determines the mitochondrial localization of the constructs (summarized in Table ). Localization of some fraction of 3SGFP to the mitochondria might reflect insufficient neutralization of the positive charge by a positional effect between Asp-25 and Arg-29.
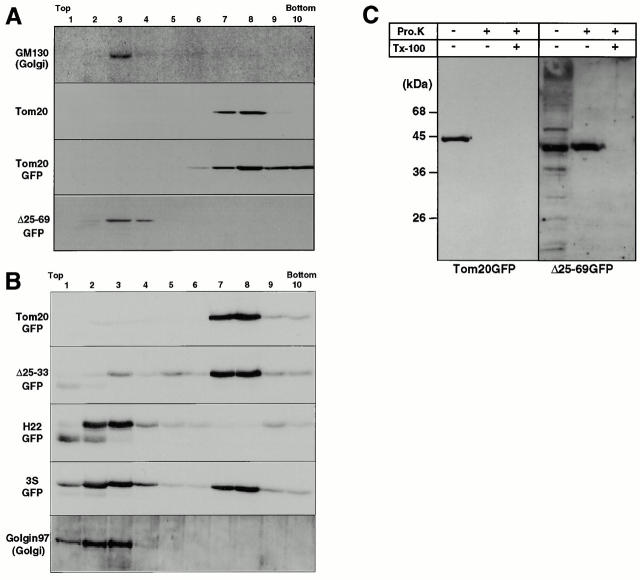
Figure 5.
Subcellular localization of rTom20-GFP constructs as examined by discontinuous sucrose gradient centrifugation. (A) Localization of Δ25-69GFP to the ER-Golgi compartments. rTom20-GFP– or Δ25-69GFP–transfected COS-7 cells were subfractionated by sucrose density gradient as described in Materials and Methods. GM130 (Golgi marker), rTom20, rTom20-GFP, and Δ25-69GFP in each fraction were detected by Western blotting. (B) Subcellular localization of Δ25-33GFP, H22GFP, and 3SGFP. Δ25-33GFP-, H22GFP-, or 3SGFP-transfected COS-7 cells were subfractionated by sucrose density gradient centrifugation. The Golgi-enriched fractions were detected using anti–Golgin97 antibodies. Other conditions were the same as those described in A. (C) Membrane topology of rTom20-GFP and Δ25-69GFP expressed in COS-7 cells. Fractions 3 and 8 in A were treated with 100 μg/ml proteinase K for 30 min at 0°C in the presence or absence of 1% Triton X-100. The reaction mixtures were analyzed by SDS-PAGE and immunoblotting with anti–rTom20 IgG.
Mutant rTom20-GFP Fusions that Failed to be Targeted to Mitochondria Are Directed to the ER-Golgi Compartments
The constructs that failed to be targeted to the mitochondria (Δ25-51GFP, Δ25-60GFP, Δ25-69GFP, Δ34-51S3GFP, Δ34-51S4GFP, and 4SGFP) were transported to the ER-Golgi compartments in COS-7 cells, because they partly colocalized with the immunofluorescence of GM130, a peripheral membrane protein of cis-Golgi stacks (Nakamura et al. 1995) and partly with an ER membrane protein, Calnexin (Wada et al. 1991). A typical example for Δ25-69GFP is shown in Fig. 4 A. In the presence of nocodazole, which breaks microtubules, the perinuclear localized signal of GFP was dispersed and dot-like structures became visible: known behavior of proteins of the Golgi apparatus (Fig. 4 B). In the presence of brefeldin A, which blocks anterograde vesicular transport from the ER to the Golgi compartment (Lippincott-Schwartz et al. 1990), Δ25-69GFP became colocalized with Calnexin, but not with MitoTracker, indicating that the fusion protein was transported back from the Golgi compartment to the ER (Fig. 4 B).
Figure 4.
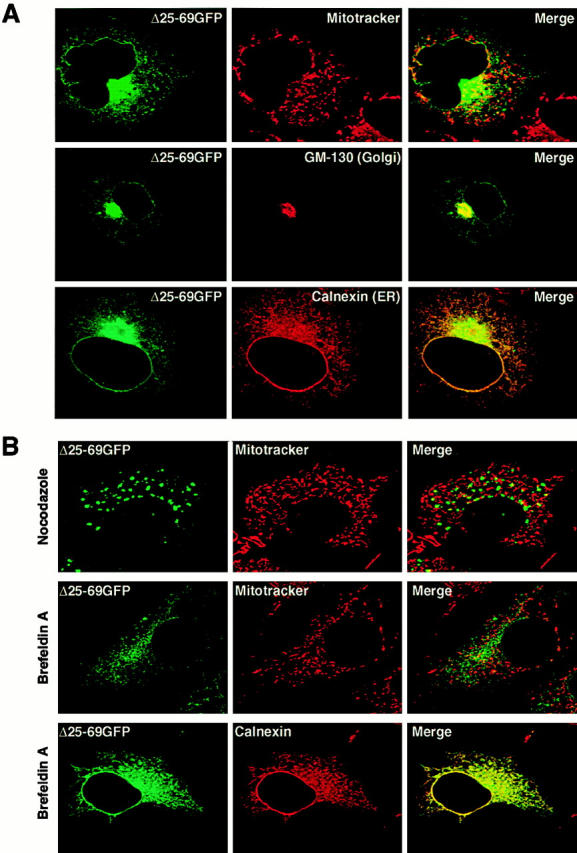
The rTom20-GFP constructs that are incompetent for mitochondria-targeting localized to the ER-Golgi compartments. (A) Localization of Δ25-60GFP to the ER-Golgi compartments. COS-7 cells expressing Δ25-60GFP were immunostained with IgGs against GM130 (Golgi marker) or Calnexin (ER marker). Fluorescent images of GFP (green), GM130 (red), and Calnexin (red) were taken by a confocal microscope. (B) Effect of nocodazole or brefeldin A (BFA) on intracellular localization of Δ25-60GFP. Δ25-69GFP-expressing COS-7 cells were treated with 20 μg/ml nocodazole for 2 h or with 5 μg/ml BFA for 1 h. The cells were then stained with MitoTracker. In a different experiment, Δ25-69GFP-expressing COS-7 cells were treated with BFA as described above, and then immunostained with IgGs against Calnexin (red).
We then performed cell fractionation for several constructs that exhibited distinct subcellular localization images of fluorescence. As shown in Fig. 5A and Fig. B, wild-type rTom20-GFP colocalized with authentic mitochondrial Tom20. Δ25-69GFP colocalized with endogenous GM130, however, and no signal was detected in the fractions where mitochondria were located. Δ25-33GFP, which exhibited a mitochondrial fluorescence pattern, localized mainly to mitochondrial fractions and only a small fraction was detected in the ER-Golgi fraction. On the other hand, 3SGFP, which exhibited mixed fluorescence images of mitochondria and the ER-Golgi compartments, had clear binary localization to the mitochondria and the ER-Golgi fractions (Fig. 5 B). H22GFP, the construct with a longer TMD, was detected almost exclusively in the ER-Golgi–enriched fractions, confirming the results obtained using fluorescence microscopy. Thus, the results obtained by subcellular fractionation for several rTom20-GFP constructs coincided well with those obtained by fluorescence microscopy.
We next examined the topology of the rTom20-GFP fusion proteins in the mitochondria and the ER-Golgi fractions using proteinase K as the probe. As shown in Fig. 5 C, rTom20-GFP localizing to the mitochondria was completely digested by a low concentration of proteinase K, indicating that rTom20-GFP was inserted into the mitochondrial outer membranes in the type I orientation, extruding the bulk of the COOH-terminal domain to the cytosol. On the other hand, Δ25-69GFP localizing in the ER-Golgi compartments was resistant to the externally added proteinase. It was completely digested, however, by proteinase K in the presence of 1% Triton X-100. It was also resistant to sodium carbonate, pH 11.5, extraction (data not shown). Thus, Δ25-69GFP was inserted into the membrane in the type-II orientation with the bulk portion translocated across the membrane to the lumen. Not all the ER-targeted constructs assumed type-II orientation in the ER or Golgi apparatus; so far examined, Δ34-51S4 and the rTom20-GFP constructs with a longer TMD, H20, and H22 (see Fig. 6 A) were inserted into the ER in the type-I orientation (data not shown). The COOH-terminal flanking region of the TMD might influence the topology of rTom20-GFP constructs (Kida et al. 2000).
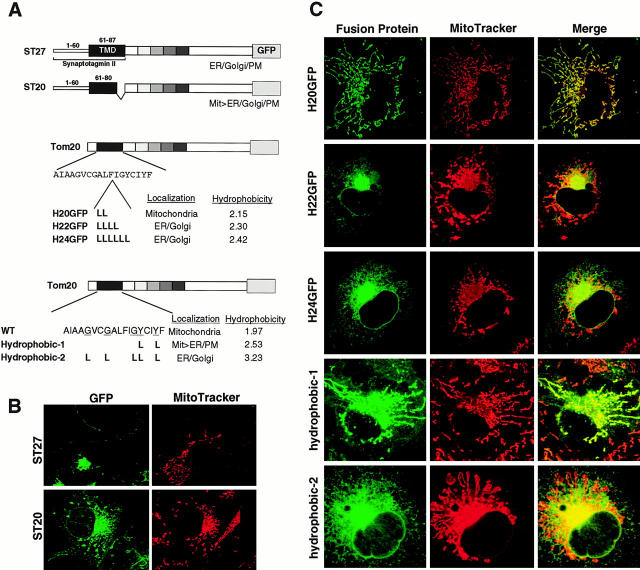
Figure 6.
TMD length or hydrophobicity is a critical determinant for mitochondria localization. (A) Schematic presentation of rTom20-GFP constructs in which the NH2-terminal hydrophilic segment and the TMD were replaced by those of Syt II (ST27 and ST20), leucine repeats were inserted into the TMD (H20, H22 and H24), or glycine or tyrosine residues in the TMD were replaced by leucine residues (Hydrophobic-1 and -2). Hydrophobicity, hydropathic index/residue; Mit, mitochondria; PM, plasma membrane. (B) Localization of ST27 and ST20 in COS-7 cells. Confocal images of GFP (green) and MitoTracker (red) are shown. (C) Mitochondria localization of rTom20-GFP fusions depends on the TMD length or hydrophobicity. The indicated rTom20-GFP constructs were expressed in COS-7 cells. The cells were costained with MitoTracker and fluorescence images were taken by a confocal microscope.
TMD Hydrophobicity Is also Critical for the Function of the Mitochondria-targeting Signal
We addressed whether the signal-anchor sequence of the ER-targeted membrane protein could function as the mitochondria-targeting signal. For this purpose, the segment composed of a hydrophilic leader (residues 1–6) and the following TMD (7–24) of rTom20-GFP was replaced by the NH2-terminal segment of synaptotagmin II (Syt II) containing the hydrophilic lumenal segment (residues 1–60) and the following TMD segment (residues 61–87) to create ST27 (Fig. 6 A). Syt II is a protein of the synaptic vesicles and during synthesis it is inserted first into the ER via the NH2-terminal signal-anchor sequence in type-I orientation (Kida et al. 2000). Upon expression in COS-7 cells, ST27 was localized throughout the secretory organelles, the ER, the Golgi, and the plasma membranes (Fig. 6 B). When the COOH-terminal segment of seven hydrophobic amino acid residues of the TMD in ST27 was deleted, however, the resultant protein (ST20) was localized mainly to the mitochondria with some fraction localized to the ER (Fig. 6 B), suggesting that either lower hydrophobicity or a shorter TMD decreased the ER-targeting efficiency, which in turn activated the mitochondria targeting. To further confirm this, we constructed H20GFP, H22GFP, and H24GFP (Fig. 6 A), in which respective leucine repeats, two-, four-, and six-times Leu, were inserted into the TMD of rTom20-GFP, and examined their localization in COS-7 cells. As shown in Fig. 6 C, H20GFP localized exclusively to dispersed mitochondria. In marked contrast, H22GFP and H24GFP assumed the ER- and Golgi-localization pattern; this was verified by cell fractionation (see Fig. 5 B for H22GFP).
We further examined whether the TMD hydrophobicity is a determinant of the mitochondria-targeting signal. We constructed rTom20-GFP mutants in which glycine or tyrosine residues in the TMD were changed to two and five leucine residues, without changing the length of the TMD, to create Hydrophobic-1 and -2, respectively (Fig. 6 A). Upon expression in COS-7 cells, Hydrophobic-1 exhibited a mixed localization profile to the mitochondria and ER, whereas Hydrophobic-2 exhibited an ER- and Golgi-localization pattern (Fig. 6 C), indicating that TMD hydrophobicity rather than TMD length is the critical factor, although contribution of the latter cannot be ruled out.
To summarize, the degree of TMD hydrophobicity or TMD length and net positive charges in the COOH-terminal proximal region were critical for the function of the mitochondria-targeting signal of rTom20-GFP. When TMD hydrophobicity or TMD length were increased, the TMD then functioned as a dominant ER-targeting signal-anchor sequence.
Effect of Proteasome Inhibitor on the Expression and Localization of rTom20-GFP Constructs in COS-7 Cells
In the present study, intracellular localization of the in vivo–expressed rTom20-GFP fusions at the steady state level was analyzed by fluorescence microscopy, which might ignore the fractions of the constructs that are rapidly turning over. The ubiquitin-proteasome system is mainly responsible for the rapid protein degradation (Bonifacino and Weissman 1998). We examined this and found that a proteasome inhibitor lactacystin did not induce noticeable changes in the expression level of rTom20-GFP and H24GFP in COS-7 cells (Fig. 7 A) or in their intracellular localization (B, and data not shown), under the condition in which rapid degradation of factor XII mutants in the secretory pathway was strongly blocked (data not shown). Thus, the rTom20-GFP constructs that are degraded by the proteasome system in COS-7 cells are few, if any, and the obtained fluorescence images reflected behavior of majority of the expressed constructs in COS-7 cells.
Figure 7.
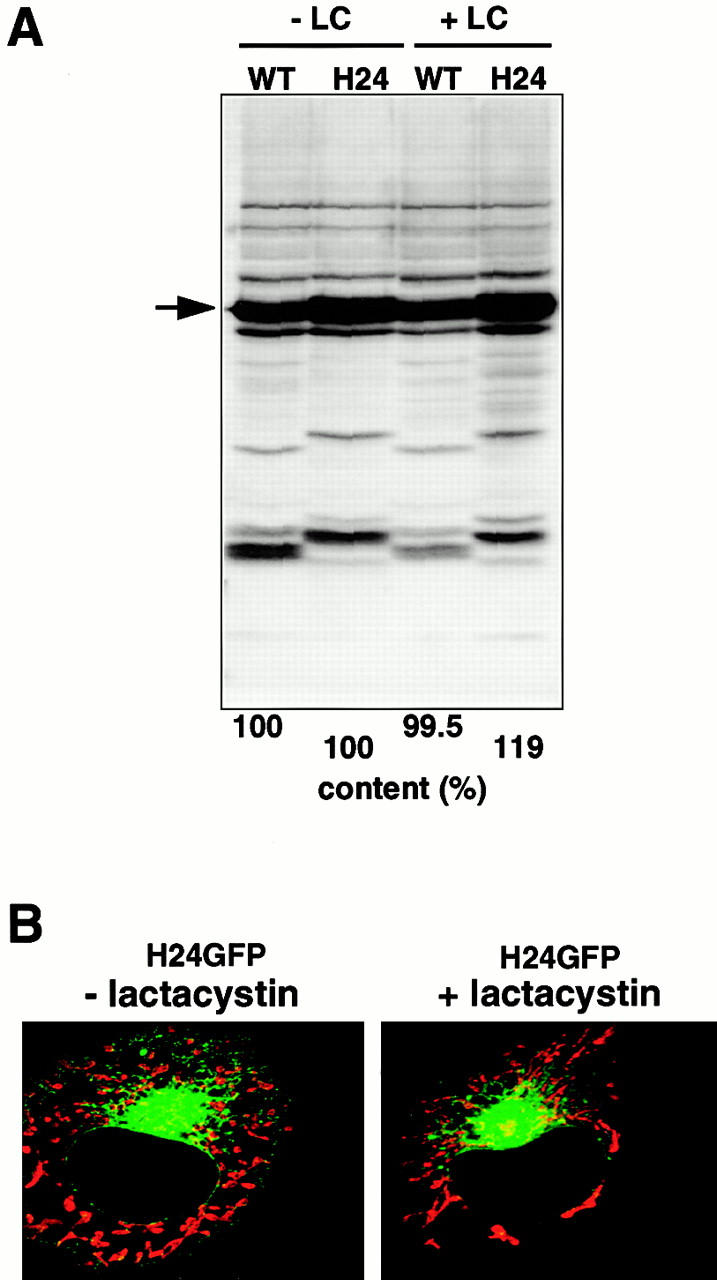
Proteasome inhibitor lactacystin does not affect the expression level and subcellular localization of rTom20-GFP constructs in COS-7 cells. (A) Effect of lactacystin on the expression of rTom20-GFP and H24GFP in COS-7 cells. COS-7 cells transfected with wild-type rTom20-GFP (WT) or H24GFP were grown at 37°C for 24 h, which were then incubated with or without 20 μM lactacystin (LC) for 10 h at 37°C. The cell lysates were subjected to SDS-PAGE followed by Western blotting with anti–rTom20 IgG. WT and H24 proteins are shown by an arrow. (B) Effect of lactacystin on the localization of H24GFP in COS-7 cells. H24GFP-expressed COS-7 cells prepared as in A were stained with MitoTracker. Merged confocal images of GFP (green) and MitoTracker (red) are shown.
Mitochondria-targeting Signal as Assessed by the In Vitro Import System
We then assessed the mitochondria-targeting signal of rTom20-GFP constructs in the in vitro insertion system. Various rTom20-GFP constructs were expressed in the in vitro transcription and translation system using reticulocyte lysate. Their post-translational insertion into mitochondria or cotranslational insertion into dog pancreas microsomes was assessed using sodium carbonate, pH 11.5, extractability as the criterion (Fig. 8).
Figure 8.
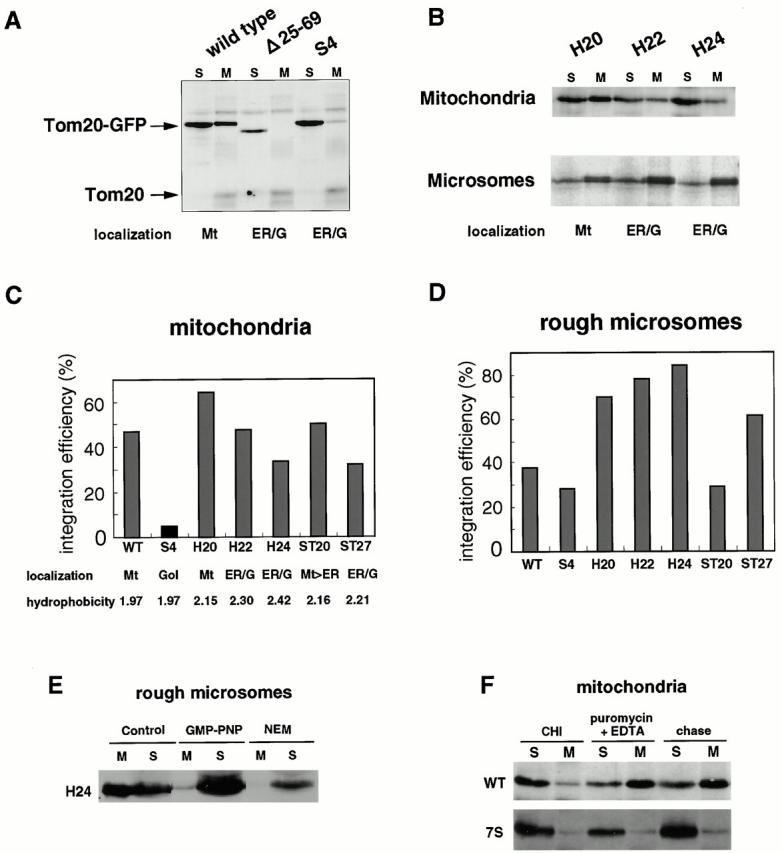
Insertion of rTom-GFP fusion proteins into rat liver mitochondria or dog pancreas microsomes in vitro. (A) Post-translational insertion of rTom20-GFP fusions into mitochondria. Wild-type rTom20-GFP, Δ25-69GFP, and Δ34-51S4GFP were synthesized in vitro and subjected to mitochondrial import. The reaction mixtures were subjected to sodium carbonate, pH 11.5, extraction. The mitochondria (M) and supernatant (S) fractions were analyzed by SDS-PAGE and subsequent Western blotting with anti–rTom20 IgG. Endogenous Tom20 is also shown. Mt, mitochondria; ER/G, ER-Golgi compartments. (B) Post-translational insertion into mitochondria and cotranslational insertion into the ER exhibit distinct dependency upon the TMD length. In vitro–synthesized H20GFP, H22GFP, and H24GFP were subjected to post-translational mitochondrial import and cotranslational ER import. Other conditions were the same as those described in A. The efficiency of post-translational mitochondria-insertion (C) and cotranslational ER-integration (D) of various rTom20-GFP constructs was quantified from the data in A and B. Integration efficiency (%) = [signal in M/ signal in (M + S)] × 100. (E) Insertion of nascent H24GFP into microsomal membrane. Truncated H24GFP (202 residues) was synthesized in the reticulocyte lysate system at 28°C for 25 min. After addition of 2 mM cycloheximide, the reaction mixture was divided into three aliquots. Two aliquots were incubated with microsomes in the absence (Control) or presence of 25 mM GMP-PNP (GMP-PNP) at 28°C for 20 min. The other aliquot was incubated with 10 mM NEM at 0°C for 10 min, followed by incubation with microsomes at 28°C for 20 min. All the reaction mixtures were subjected to the alkali floatation to separate the membrane (M) and unfloated (S) fractions. Both fractions were subjected to SDS-PAGE followed by Western blotting using anti–rTom20 IgG. (F) Insertion of truncated rTom20-GFP or 7SGFP into mitochondria. Truncated wild-type rTom20-GFP (WT) or 7SGFP (202 residues each) was synthesized in the reticulocyte lysate at 28°C for 25 min and divided into three aliquots. Two aliquots were incubated with mitochondria in the presence of 2 mM cycloheximide (CHI) or 2 mM puromycin plus 25 mM EDTA at 28°C for 60 min. The other aliquot was incubated with mitochondria at 28°C for 40 min, and then incubated in the presence of puromycin plus EDTA for 20 min (chase). Other conditions are as described in E.
Wild-type rTom20-GFP was inserted post-translationally into rat liver mitochondria with significant efficiency, whereas Δ25-69GFP and Δ34-51S4, which were transported to the ER-Golgi compartments in vivo, failed to be inserted into the mitochondria, confirming the in vivo results that a positive net charge of the COOH-terminal flanking region is critical for mitochondria-targeting (Fig. 8 A). H20GFP with a TMD of 20 amino acids exhibited maximum insertion efficiency, but the efficiency clearly decreased as the TMD hydrophobicity increased (Fig. 8B and Fig. C). In contrast, the efficiency of cotranslational ER insertion increased depending on the TMD hydrophobicity. The TMD with 24 amino acid residues functioned as the most efficient ER signal thus far examined (Fig. 8 D).
It should be noted that H20GFP and wild-type rTom20-GFP, which were targeted exclusively to the mitochondria in COS-7 cells, were also inserted cotranslationally into the ER with significant efficiency (Fig. 8B and Fig. D). Similarly, H22GFP and H24GFP, which were targeted exclusively to the ER-Golgi compartments in vivo were inserted post-translationally into the mitochondria to a significant extent (Fig. 8B and Fig. C). These results suggest that some factors that interfere with nonspecific ER targeting and assist with correct mitochondria targeting are limiting in the in vitro import system, or vice versa.
The truncated H24GFP (202 residues) in RNCs was inserted into the ER without being released from the ribosomes (Fig. 8 E, control). This reaction was inhibited either in the presence of GMP-PNP, or using NEM-treated RNCs, the conditions that inhibit the function of SRP (Walter and Johnson 1994; Fig. 8 E, GMP-PNP and NEM), indicating that the ER insertion of these constructs occurs cotranslationally and is SRP dependent. In marked contrast, mitochondrial insertion of the truncated rTom 20-GFP in RNCs (202 residues; truncated at the 58th residue of GFP) did not occur unless it was released from the ribosomes by puromycin and EDTA, although the entire portion of rTom20 had already emerged from the ribosomes (Fig. 8 F). In contrast, the Golgi-targeted construct 7SGFP, in which seven basic amino acids in the flanking region (residues 26–39) were changed to Ser (data not shown), was not inserted into mitochondria. There might be the mechanism that assures post-translational targeting of newly synthesized mitochondrial outer membrane proteins (see Discussion).
SRP Recognizes the TMD of rTom20-GFP during Synthesis, but Positive Charges at the COOH-terminal Flanking Segment Inhibit Its Function
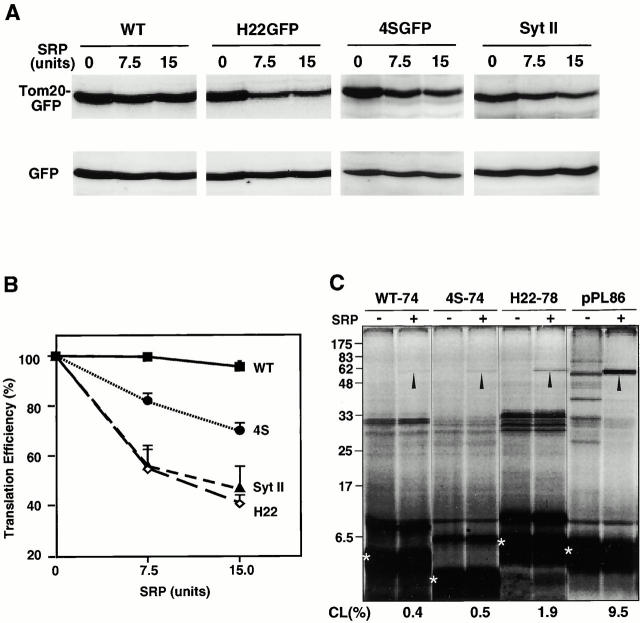
These results indicate that the TMD of rTom20-GFP by itself functions as an ER-targeting signal-anchor sequence, and suggest that basic amino acid residues within the COOH-terminal flanking region interfere with the function of SRP. We therefore examined several rTom20-GFP constructs using the SRP-induced translation arrest as a measure. mRNA for wild-type rTom20-GFP, H22GFP, 4SGFP, or Syt II was cotranslated with the control GFP mRNA in the presence or absence of SRP and the translation products were analyzed by Western blotting. Translation of H22GFP that was targeted to the ER-Golgi compartments in vivo was inhibited by SRP in a dose-dependent manner, whereas translation of GFP as an internal control was not affected (Fig. 9 A). SRP did not inhibit the translation of wild-type rTom20-GFP. In marked contrast, SRP did arrest, to a weak but significant extent, the translation of 4SGFP, the rTom20-GFP construct that does not carry positive charges in the COOH-terminal flanking region and that is transported exclusively to the Golgi compartment (Fig. 9A and Fig. B). Furthermore, synthesis of Syt II, a typical ER-targeted membrane protein (Kida et al. 2000), was inhibited by SRP to a similar extent as H22GFP (Fig. 9A and Fig. B), confirming that SRP was functional in the assay system.
Figure 9.
Interaction of the nascent rTom20-GFP constructs with SRP. (A) Translation arrest of rTom20-GFP constructs. mRNA for wild-type rTom20-GFP (WT), H22GFP, 4SGFP, or Syt II was cotranslated with the control GFP mRNA in the wheat germ lysate at 30°C for 40 min in the presence or absence of the indicated amounts of SRP. Translation products were analyzed by SDS-PAGE and subsequent Western blotting using anti–rTom20 IgG, anti–GFP IgG, or anti–Syt II IgG. (B) The Western blot images were analyzed by LAS1000 (Fuji Film Co.). Intensities of bands of the translated rTom20-GFP constructs were normalized by those of GFP, and then the translation efficiencies were calculated taking those in the absence of SRP as 100%. (C) Interaction of SRP with the nascent chains of rTom20-GFP constructs and preprolactin (pPL) as examined by photo–cross-linking. The truncated mRNAs indicated were translated in the wheat germ lysate system in the presence of 35S-methionine, TDBA-lys-tRNA, and, where indicated, SRP at 25°C for 20 min. After addition of 2 mM cycloheximide, the reaction mixtures were UV irradiated, and then subjected to SDS-PAGE and autoradiography. Arrowheads, cross-linked products; asterisks, truncated peptides synthesized. CL (%), efficiency of cross-linking = [signal in arrowhead/signal in asterisk] × 100.
We further examined the interaction of SRP with the truncated rTom20-GFP constructs in RNCs using photo–cross-linking. The truncated mRNA for rTom20-GFP, 4SGFP, H22GFP, or preprolactin was translated in the wheat-germ lysate system in the presence or absence of SRP. The nascent preprolactin, as a positive control, was cross-linked to SRP with significant efficiency, confirming validity of the assay system (Fig. 9 C). H22GFP, which is targeted to the ER-Golgi compartments in vivo and is translation-arrested in vitro to a similar extent as Syt II, indeed cross-linked to SRP. The truncated 4SGFP, which is targeted to the ER-Golgi compartments, was cross-linked to SRP to a weak but significant extent, coinciding with the extent of SRP-induced translation arrest. Thus, the results obtained by SRP-induced translation arrest were confirmed by the photo–cross-linking approach. The truncated wild-type rTom20-GFP was also cross-linked to SRP to a similar extent as 4SGFP. We presume that SRP remain associated with the nascent chain of wild-type rTom20-GFP, although this should be confirmed by different approaches.
Discussion
The mechanisms by which the mitochondrial outer membrane proteins are recognized and directed to the mitochondria remain mostly unknown. The present study analyzed the mitochondria-targeting signal of an NH2-terminal membrane-anchor protein, rTom20, using GFP as the reporter. We demonstrated that the TMD with a moderate hydrophobicity and at least one net positive charge within the COOH-terminal flanking region of five amino acid residues are both important determinants of the mitochondria-targeting signal. The TMD of rTom20 functioned by itself as a signal-anchor sequence to target GFP fusions to the ER. The TMD with increased hydrophobicity apparently counteracts the positive charges of the flanking region and functions as the ER-targeting signal-anchor sequence.
To our knowledge, this is the first demonstration of the importance of TMD hydrophobicity in addition to the net positive charge in the COOH-terminal flanking region in mitochondria targeting of the NH2-terminal membrane-anchor protein. This structural feature is conserved among Tom20 from various species; i.e., S. cerevisiae (Ramage et al. 1993), Neurospora crassa (Söllner et al. 1989), Caenorhabditis elegans (GenBank No. C46715), Drosophila melanogaster (AA990653), Xenopus laevis (AW638081), rats (Iwahashi et al. 1997), and humans (Goping et al. 1995; Seki et al. 1995; Hanson et al. 1996). A 23-kD protein of potato mitochondrial outer membrane, which is speculated to be a homologue of Tom20, is exceptional; however, it shares only 19% sequence identity with fungal Tom20 and, most notably, it carries a predicted TMD at the COOH terminus (Heins and Schmitz 1996). Furthermore, there are similar structural features for several other mitochondrial outer membrane proteins: Tom70 from N. crassa (Söllner et al. 1990) and rats (Suzuki and Mihara, unpublished results), and OM37 from rat liver mitochondria (Maeda and Mihara, unpublished results). We expect that these proteins are targeted to the mitochondria by the same mechanism as Tom20.
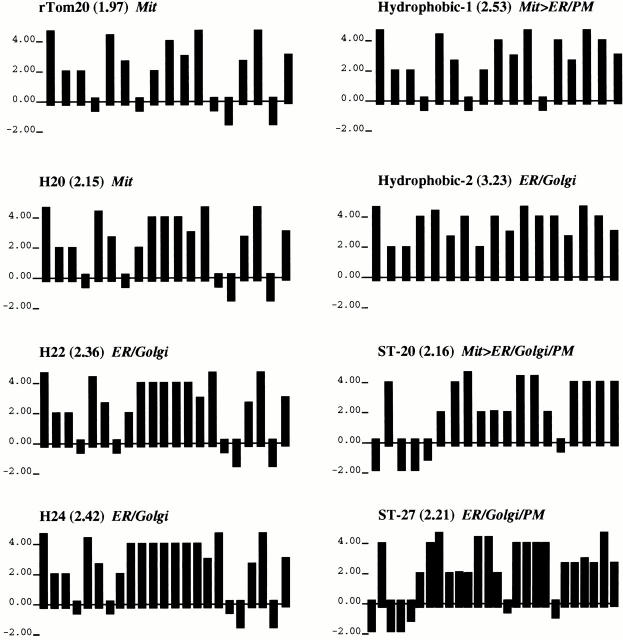
Although we demonstrated that the TMD with higher hydrophobicity acted dominantly over the positive charges of the flanking region and functioned as the ER-targeting signal-anchor sequence, we do not know whether the increased positive charges within the flanking region overcome the ER-targeting signal to direct the constructs to the mitochondria. Presumably this is not the case, since many ER membrane proteins carry enriched basic amino acid residues within the COOH-terminal flanking region of the signal-anchor sequence (Nelson and Strobel 1988; Li et al. 1995). Therefore, it is not a simple balance between the TMD hydrophobicity and the subsequent positive charges that determines the function of the mitochondria-targeting signal. Instead, the present data indicate that moderate TMD hydrophobicity and the subsequent positive charges are clearly required for proper function of the mitochondria-targeting signal. Of the TMDs examined to date, those with 18–20 residues and an average hydrophobicity (hydropathic index per residue) of 1.97–2.16 functioned as the mitochondria-targeting signal (Fig. 10). In contrast, TMDs with a higher hydrophobicity (average hydrophobicity, 2.21–3.23) functioned efficiently as the ER-targeting signal-anchor sequences, with the exception of Hydrophobic-1, which partly localized to the mitochondria in spite of its TMD having an average hydrophobicity of 2.53. In addition, the TMD length might be a critical determinant to match with the thickness of the mitochondrial outer membrane. More precise structural information in the TMD that serves as the mitochondria-targeting signal awaits analysis.
Figure 10.
Hydrophobicity plots of TMDs of the rTom20-GFP constructs. The algorithm of Kyte and Doolittle was used with a window size of one amino acid residue. The average hydrophobicity is shown in parentheses.
The TMD of rTom20 functioned, by itself, as an ER-targeting signal-anchor sequence. Because the ER-targeting signals are recognized by the SRP as they emerge from the ribosomes and are targeted to the ER by the SRP–SRP receptor system, the TMD of rTom20 should be recognized by the SRP in RNCs as it emerges from the ribosomes. How is rTom20 directed from the ribosomes to the mitochondrial outer membrane in an SRP-dependent ER-targeting system? Our results clearly demonstrated that positive charges at the proximal COOH-terminal flanking region of the TMD have a crucial role in this targeting; when hydrophobicity of the signal-anchor sequence is lower, net positive charges created within the flanking region interfere with the function of SRP, thus directing otherwise ER-targeted protein to the mitochondrial outer membrane. In this regard, a similar but clearly distinct example of switching the sorting signal has been reported. Cytochrome P4502B1 localizing predominantly in the ER contains a chimeric mitochondria- and ER-targeting signal at its NH2 terminus; the NH2-terminal 20 amino acid region with a hydrophobic helical structure is followed by a positively charged region that resembles the mitochondrial presequence. Phosphorylation of P4502B1 at Ser128 reduces the affinity of P4502B1 for SRP and directs otherwise ER-predominant protein to the mitochondrial matrix (Anandatheerthavarada et al. 1999).
The results obtained in the in vivo system were mostly confirmed by the in vitro membrane insertion systems. Some constructs (wild-type rTom20-GFP and H20GFP), however, which were targeted exclusively to the mitochondria in vivo, were also inserted cotranslationally into dog pancreas microsomes with significant efficiency. This is probably because the factors that prevent ER targeting and increase the fidelity of mitochondria targeting of the nascent precursors, such as the nascent polypeptide-associated complex (NAC), might be limiting in our in vitro assay system. NAC has been characterized as the heterodimeric, ribosome-associated chaperone that prevents promiscuous interactions between SRP and the nascent polypeptides destined for cellular compartments other than the secretory pathway (Wiedmann et al. 1994), although this function has been challenged (Raden and Gilmore 1998; Neuhof et al. 1998). The function of NAC in the targeting of preprotein to the mitochondria has been demonstrated in yeast using genetic and biochemical approaches. Depletion of the α-subunit of NAC leads to decreased mitochondria targeting and eventual loss of mitochondrial DNA (George et al. 1998). Purified NAC stimulates the import of ribosome-bound nascent preproteins by enhancing the productive binding of RNCs to mitochondria (Fünfschilling and Rospert 1999). Participation of NAC in mitochondria targeting of the NH2-terminal membrane-anchor proteins needs to be addressed in the future.
SRP-induced translation arrest and photo–cross-linking revealed that the TMD of rTom20-GFP constructs indeed interacted with SRP, although with low efficiency. The basic amino acid residues introduced into the COOH-terminal flanking region of the TMD inhibited the function of SRP. In addition, the truncated rTom20-GFP in RNCs was unable to be inserted into the mitochondria unless it is released from the ribosomes. The targeting factors that are present in the cytosol might release SRP from the TMD of rTom20 by binding to the positively charged COOH-terminal flanking region, thereby stabilizing the hydrophobic TMD and targeting rTom20 to the Tom machinery, probably to Tom40 directly (Schneider et al. 1991). Another possibility is that the factors do not induce dissociation of SRP from the TMD, but rather inhibit the function of SRP, such as translation arrest, and in conjunction with SRP, stabilize the polypeptide until its synthesis is completed, assuring post-translational integration of the nascent protein into the mitochondria. Thus, the mitochondrial outer membrane proteins with the NH2-terminal membrane-anchor might share part of the targeting system with the ER-targeted preproteins at the initial steps of translation in RNCs. It is thus intriguing that depletion of the SRP subunits, 7S RNA or SRP receptor, causes accumulation of a mitochondrial precursor and also causes respiration deficiency in yeast (Felici et al. 1989; Hann and Walter 1991; Ogg et al. 1992; Stirling and Hewitt 1992).
Acknowledgments
We thank Nobuhiro Nakamura for anti–rat GM130 and Golgin 97 antibodies and Katsuhiko Mikoshiba and Mitsunori Fukuda for Syt II cDNA and anti–mouse Syt II antibodies. We also thank Tom A. Rapoport and Sven U. Heinrich for teaching photo–cross-linking techniques, and Kazuhisa Ota for technical advice for cross-linking experiments. We are particularly grateful to the reviewers of a first version of the paper for their constructive criticism.
This study was supported by grants from the Ministry of Education, Science, and Culture of Japan, from the Human Frontier Science Program and from the Core Research for Evolutional Science and Technology (CREST).
Footnotes
Drs. Kanaji and Iwahashi contributed equally to this paper and are co–first authors.
Abbreviations used in this paper: GFP, green fluorescent protein; RNC, ribosome-nascent chain-complex; SRP, signal recognition particle; TMD, transmembrane domain.
References
- Anandatheerthavarada H.K., Biswas G., Mullick J., Sepuri N.B.V., Otvos L., Pain D., Avadhani N.G. Dual targeting of cytochrome P4502B1 to endoplasmic reticulum and mitochondria involves a novel signal activation by cyclic AMP–dependent phosphorylation as Ser128. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:5494–5504. doi: 10.1093/emboj/18.20.5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong L.C., Komiya T., Bergman B.E., Mihara K., Bornstein P. Metaxin is a component of a preprotein import complex in the outer membrane of the mammalian mitochondrion. J. Biol. Chem. 1997;272:6510–6518. doi: 10.1074/jbc.272.10.6510. [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Weissman A.M. Ubiquitin and the control of protein fate in the secretory and endocytotic pathways. Annu. Rev. Cell Dev. Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Douglas M.G. Biogenesis of ISP6, a small carboxyterminal anchored protein of the receptor complex of the mitochondrial outer membrane. J. Biol. Chem. 1995;270:5674–5679. doi: 10.1074/jbc.270.10.5674. [DOI] [PubMed] [Google Scholar]
- Dietmeier K., Hönlinger A., Bömer U., Dekker P.J.T., Eckerskorn C., Lottspeich F., Kübrich M., Pfanner N. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 1997;388:195–200. doi: 10.1038/40663. [DOI] [PubMed] [Google Scholar]
- Edward J.B., Roger G.S., Simon H.S., Manuel C. Interactions between multiple phosphorylation sites in the inactivation particle of a K+ channel. Insights into the molecular mechanism of protein kinase action. J. Gen. Physiol. 1998;112:71–84. doi: 10.1085/jgp.112.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan B., Beilharz T., George R., Isenmann S., Gratzer S., Wattenberg B., Lithgow T. Targeting of tail-anchored proteins to yeast mitochondria in vivo. FEBS Lett. 1999;451:243–248. doi: 10.1016/s0014-5793(99)00581-5. [DOI] [PubMed] [Google Scholar]
- Felici F., Cesareni G., Hughes J.M.X. The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol. Cell. Biol. 1989;9:3260–3268. doi: 10.1128/mcb.9.8.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Aruga J., Niinobe M., Aimoto S., Mikoshiba K. Inositol-1,3,4,5-tetrakisphosphate binding to C2B domain of IP4BP/synaptotagmin II. J. Biol. Chem. 1994;269:29206–29211. [PubMed] [Google Scholar]
- Fünfschilling U., Rospert S. Nascent-polypeptide–associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell. 1999;10:3289–3299. doi: 10.1091/mbc.10.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R., Beddoe T., Landl K., Lithgow T. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria. Proc. Natl. Acad. Sci. USA. 1998;95:2296–2301. doi: 10.1073/pnas.95.5.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goping I.S., Millar D.G., Shore G.C. Identification of the human mitochondrial protein import receptor, huMas20p. Complementation of Δmas20 in yeast. FEBS Lett. 1995;373:45–50. doi: 10.1016/0014-5793(95)01010-c. [DOI] [PubMed] [Google Scholar]
- Görlich D., Kurzchalia T.V., Wiedmann M., Rapoport T. Probing the molecular environment of translocating polypeptide chains by cross-linking. Methods Cell Biol. 1991;34:241–263. doi: 10.1016/s0091-679x(08)61684-2. [DOI] [PubMed] [Google Scholar]
- Hann B.C., Walter P. The signal recognition particle in S. cerevisiae . Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- Hanson B., Nuttall S., Hoogenraad N. A receptor for the import of proteins into human mitochondria. Eur. J. Biochem. 1996;235:750–753. doi: 10.1111/j.1432-1033.1996.t01-1-00750.x. [DOI] [PubMed] [Google Scholar]
- Heins L., Schmitz U.K. A receptor for protein import into potato mitochondria. Plant J. 1996;9:829–839. doi: 10.1046/j.1365-313x.1996.9060829.x. [DOI] [PubMed] [Google Scholar]
- Hurt E.C., Müller U., Schatz G. The first twelve amino acids of a yeast mitochondrial outer membrane protein can direct a nuclear encoded cytochrome oxidase subunit to the mitochondrial inner membrane. EMBO (Eur. Mol. Biol. Organ.) J. 1985;4:3509–3518. doi: 10.1002/j.1460-2075.1985.tb04110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenmann S., Khew-Goodall Y., Gamble J., Vadas M., Wattenberg B.W. A splice-isoform of vesicle-associated membrane protein-1 (VAMP-1) contains a mitochondria targeting signal. Mol. Biol. Cell. 1998;9:1649–1660. doi: 10.1091/mbc.9.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahashi J., Yamazaki S., Komiya T., Nomura N., Nishikawa S., Endo T., Mihara K. Analysis of the functional domain of the rat liver mitochondrial import receptor Tom20. J. Biol. Chem. 1997;272:18467–18472. doi: 10.1074/jbc.272.29.18467. [DOI] [PubMed] [Google Scholar]
- Keil P., Pfanner N. Insertion of MOM22 into the mitochondrial outer membrane strictly depends on surface receptors. FEBS Lett. 1993;321:197–200. doi: 10.1016/0014-5793(93)80107-6. [DOI] [PubMed] [Google Scholar]
- Kida Y., Sakaguchi M., Fukuda M., Mikoshiba K., Mihara K. Membrane topogenesis of a type I signal-anchor protein, mouse synaptotagmin II, on the endoplasmic reticulum. J. Cell Biol. 2000;150:719–730. doi: 10.1083/jcb.150.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda R., Ikenoue T., Honsho M., Tsujimoto S., Mitoma J., Ito A. Charged amino acids at the carboxy-terminal portions determine the intracellular locations of two isoforms of cytochrome b5. J. Biol. Chem. 1998;273:31097–31102. doi: 10.1074/jbc.273.47.31097. [DOI] [PubMed] [Google Scholar]
- Kutay U., Ahnert-Hilger G., Hartmann E., Wiedenmann B., Rapoport T.A. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Ullrich B., Zhang J.Z., Anderson R.G.W., Brose N., Südhof T.C. Ca2+-dependent and -independent activities of neural and non-neural synaptotagmins. Nature. 1995;375:594–599. doi: 10.1038/375594a0. [DOI] [PubMed] [Google Scholar]
- Lill R., Neupert W. Mechanisms of protein import across the mitochondrial outer membrane. Trends Cell Biol. 1996;6:56–61. doi: 10.1016/0962-8924(96)81015-4. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Donaldson J.G., Schweizer A., Berger E.G., Hauri H.P., Yuan L.C., Klausner R.D. Microtubule-dependent retrograde transport of proteins into the ER in the presence of Brefeldin A suggests an ER recycling pathway. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- Martoglio B., Dobberstein B. Signal sequencesmore than just a greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- Meijer M., Maarse A., Kübrich M., Pfanner N. Translocation of preproteins across the mitochondrial inner membraneTIMs and HSP70. Adv. Mol. Cell. Biol. 1996;17:127–148. [Google Scholar]
- Mannella C., Nuewald A., Lawrence C. Detection of likely beta-strand region in sequences of mitochondrial preproteins using Gibbs sampler. J. Bioenerg. Biomembr. 1996;28:163–169. doi: 10.1007/BF02110647. [DOI] [PubMed] [Google Scholar]
- McBride H.M., Millar D.G., Li J.M., Shore G.C. A signal-anchor sequence selective for the mitochondrial outer membrane. J. Cell Biol. 1992;119:1451–1457. doi: 10.1083/jcb.119.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara K. Targeting and insertion of nuclear-encoded preproteins into the mitochondrial outer membrane. Bioessays. 2000;22:364–371. doi: 10.1002/(SICI)1521-1878(200004)22:4<364::AID-BIES6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Mihara K., Omura T. Cytoplasmic chaperones in precursor targeting to mitochondriathe role of MSF and hsp70. Trends Cell Biol. 1996;6:104–108. doi: 10.1016/0962-8924(96)81000-2. [DOI] [PubMed] [Google Scholar]
- Mitoma J., Ito A. Mitochondrial targeting signal of rat liver monoamine oxidase B is located at its carboxy terminus. J. Biochem. 1992;111:20–24. doi: 10.1093/oxfordjournals.jbchem.a123712. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T., Warren G. Characterization of a cis-Golgi matrix protein, GM 130. J. Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.R., Strobel H.W. On the membrane topology of vertebrate cytochrome P-450 proteins. J. Biol. Chem. 1988;263:6038–6050. [PubMed] [Google Scholar]
- Neuhof A., Rolls M.M., Jungnickel B., Kalies K.-U., Rapoport T.A. Binding of signal recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction. Mol. Biol. Cell. 1998;9:103–115. doi: 10.1091/mbc.9.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Ng D.T.W., Brown J.D., Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S.C., Poritz M.A., Walter P. Signal recognition particle receptor is important for cell growth and protein secretion. Mol. Biol. Cell. 1992;3:895–911. doi: 10.1091/mbc.3.8.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T. Mitochondria-targeting sequence, a multi-role sorting sequence recognized at all steps of protein import into mitochondria. J. Biochem. 1998;123:1010–1016. doi: 10.1093/oxfordjournals.jbchem.a022036. [DOI] [PubMed] [Google Scholar]
- Ota K., Sakaguchi M., von Heijne G., Hamasaki N., Mihara K. Forced transmembrane orientation of hydrophilic polypeptide segments in multispanning membrane proteins. Mol. Cell. 1998;2:495–503. doi: 10.1016/s1097-2765(00)80149-5. [DOI] [PubMed] [Google Scholar]
- Raden D., Gilmore R. Signal recognition particle-dependent targeting of ribosomes to the rough endoplasmic reticulum in the absence and presence of the nascent polypeptide-associated complex. Mol. Biol. Cell. 1998;8:117–130. doi: 10.1091/mbc.9.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage L., Junne T., Hahne K., Lithgow T., Schatz G. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:4115–4123. doi: 10.1002/j.1460-2075.1993.tb06095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M., Mihara K., Sato R. Signal recognition particle is required for co-translational insertion of cytochrome P-450 into microsomal membranes. Proc. Natl. Acad. Sci. USA. 1984;81:3361–3364. doi: 10.1073/pnas.81.11.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi M., Tomiyoshi R., Kuroiwa T., Mihara K., Omura T. Functions of signal and signal-anchor sequences are determined by the balance between the hydrophobic segment and the N-terminal charge. Proc. Natl. Acad. Sci. USA. 1992;89:16–19. doi: 10.1073/pnas.89.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz G., Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Schneider H., Söllner T., Dietmeier K., Eckerskorn C., Lottspeich F., Trülzsch B., Neupert W., Pfanner N. Targeting of the master receptor MOM19 to mitochondria. Science. 1991;254:1659–1662. doi: 10.1126/science.1661031. [DOI] [PubMed] [Google Scholar]
- Schlossmann J., Neupert W. Assembly of the preprotein receptor Mom72/Mas70 into the protein import complex of the outer membrane of mitochondria. J. Biol. Chem. 1995;270:27116–27121. doi: 10.1074/jbc.270.45.27116. [DOI] [PubMed] [Google Scholar]
- Seki N., Moczko M., Nagase T., Zufall N., Ehmann B., Dietmeier K., Schäfer E., Nomura N., Pfanner N. A human homolog of the mitochondrial protein import receptor Mom19 can assemble with the yeast mitochondrial receptor complex. FEBS Lett. 1995;375:307–310. doi: 10.1016/0014-5793(95)01229-8. [DOI] [PubMed] [Google Scholar]
- Shore G.C., McBride H.M., Millar D.G., Steenaart N.A.Y., Nguyen M. Import and insertion of proteins into the mitochondrial outer membrane. Eur. J. Biochem. 1995;227:9–18. doi: 10.1111/j.1432-1033.1995.tb20354.x. [DOI] [PubMed] [Google Scholar]
- Söllner T., Griffiths G., Pfaller R., Pfanner N., Neupert W. MOM19, an import receptor for mitochondrial precursor proteins. Cell. 1989;59:1061–1070. doi: 10.1016/0092-8674(89)90762-9. [DOI] [PubMed] [Google Scholar]
- Söllner T., Pfaller R., Griffiths G., Pfanner N., Neupert W. A mitochondrial import receptor for the ADP/ATP carrier. Cell. 1990;62:107–115. doi: 10.1016/0092-8674(90)90244-9. [DOI] [PubMed] [Google Scholar]
- Stanley S.J., Davis A., D'Arcangelis D., Mannella C.A. Peptide-specific antibodies as probes of the topography of the voltage gated channel of the mitochondrial outer membrane of Neurospora crassa . J. Biol. Chem. 1995;270:16694–16700. doi: 10.1074/jbc.270.28.16694. [DOI] [PubMed] [Google Scholar]
- Stirling C.J., Hewitt E.W. The S. cerevisiae SEC65 gene encodes a component of yeast signal recognition particle with homology to human SRP19. Nature. 1992;356:534–537. doi: 10.1038/356534a0. [DOI] [PubMed] [Google Scholar]
- Voos W., Martin H., Krimmer T., Pfanner N. Mechanism of protein translocation into mitochondria. Biochim. Biophys. Acta. 1999;1422:235–254. doi: 10.1016/s0304-4157(99)00007-6. [DOI] [PubMed] [Google Scholar]
- Wada I., Rindress D., Cameron P.H., Ou W.-J., Doherty J.J., Louvard D., Bell A.W., Dignard D., Thomas D.Y., Bergeron J.J.M. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum. J. Biol. Chem. 1991;266:19599–19610. [PubMed] [Google Scholar]
- Walter P., Blobel G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 1980;77:7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Johnson A.E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Wiedmann B., Sakai H., Davis T.A., Wiedmann M. A protein complex required for signal sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]