Abstract
Our understanding of the roles played by sex hormones in ovarian carcinogenesis has been limited by a lack of data concerning the mode of sex hormone action in human ovarian surface epithelial (HOSE) cells, the tissue of origin of >90% of ovarian cancers. We have compared the relative abundance of estrogen receptor (ER)α, ERβ, progesterone receptor (PR), and androgen receptor (AR) mRNA in four primary cultures of HOSE cells obtained from postmenopausal women to those found in late serous adenocarcinoma primary cell cultures and established ovarian cancer cell lines. We observed coexpression of ERα and ERβ mRNA along with AR and PR transcripts in normal HOSE cells and disruption of ERα mRNA expression as well as dramatic down-regulation of PR and AR transcript expression in most ovarian cancer cells. In contrast, levels of ERβ mRNA were unaffected by the malignant state. Additionally, a novel mutation involving a 32-bp deletion in exon 1 of ERα transcripts was detected in the SKOV3 cell line. This mutation would explain why SKOV3 was reported to be ER-positive but estrogen-insensitive. Taken together, these findings suggest that estrogens, signaling via either or both ER subtypes, may play an indispensable role in regulating normal HOSE cell functions. Therefore, loss of ERα, PR, and AR mRNA expression in HOSE cells may be responsible for neoplastic transformation in this cell type. In contrast, the roles played by ERβ in normal and malignant HOSE cells remain elusive. Finally, the coexistence of mutated ERα mRNA and normal ERβ transcripts in SKOV3 argues in favor of a dependency of ERβ action on functional ERαs.
Keywords: mutation, sequence deletion, estrogen action, androgen action, progesterone action
Ovarian carcinoma (OC) is the second most common and the most deadly malignancy of the female reproductive tract (for review, see refs. 1–4). Etiological factors involved in ovarian carcinogenesis remain poorly defined, and effective treatment protocols are limited (1–3). Epidemiological data suggest that endogenous and exogenous sex hormones may play important roles in the pathogenesis of the disease. In this regard, estrogens taken as oral contraceptives during premenopausal years offer protection, but when used postmenopausally as hormone replacement therapies elevate risk (1–6). The risk of developing invasive OC increases with ever-use of hormone replacement therapy and has been shown to depend on the duration of usage (5, 6). In addition to estrogens, other ovarian or adrenal steroids such as androstenedione, testosterone, and progestins have all been implicated as risk factors for OC (1–4). Androstenedione and progesterone are present at higher concentrations in the ovarian vein draining the affected ovary when compared with levels found in the vein draining the contralateral, disease-free gland (7, 8). Plasma levels of estradiol-17β (E2), progesterone, 20α-hydroxyprogesterone, testosterone, and androstenedione have all been shown to correlate with OC tumor masses (9–11). Taken together, these findings suggest that steroid hormones are likely involved in the genesis and progression of the disease, yet their mechanisms of action remain unclear.
The classical estrogen receptor (ER), recently renamed ERα (12), and the progesterone receptor (PR) were found in <50% of OC specimens, whereas androgen receptor (AR) was detected in most cases (>80%) (1–4). In recent studies, transcripts of the newly discovered ER subtype, ERβ (12), were found in normal human ovaries and benign and malignant ovarian tumors (13, 14), as well as in primary cultures of normal human ovarian surface epithelial (HOSE) cells (15). Unfortunately, none of the aforementioned studies demonstrated a strong association between ERα, PR, or AR status and OC histological types or grades. Furthermore, treatments of OCs with tamoxifen, antiandrogens, or progestins produced very dismal responses (for review, see refs. 1–4). Consequently, it is widely believed that levels of sex hormone receptors have little prognostic value and are poor predictors of hormone-manipulation outcomes for OCs.
A major challenge in assessing the significance of sex hormones and their receptors in ovarian carcinogenesis is the paucity of information about their expression levels in the normal HOSE. Over 90% of OCs arise from the HOSE, which shares a common embryonic origin with epithelia of Mullerian duct-derived tissues (Fallopian tube, endometrium, and endocervix) but is distinctly different from the granulosa–thecal cells of the ovary (1–4). In terms of tissue mass, this layer represents only a small fraction of the whole ovary. Thus, data generated from studies that compare levels of a molecular marker found in OCs with those observed in whole ovaries are difficult to interpret because expression pattern in HOSE could easily be masked by those in other ovarian cell types. In this regard, although previous studies have demonstrated localization of ERβ in ovarian granulosal cells and ERα throughout the ovary (12, 16, 17), only recently have both ER subtypes been found in normal HOSE cells (15). However, it remains unclear as to whether their expression levels are altered after neoplastic transformation. Importantly, little is known about the relationships between the expression patterns of the two ER subtypes and those of other steroid receptors in normal and malignant HOSE cells.
To fill this data gap, in the present study, we have compared the expression of ERα, ERβ, PR, and AR transcripts in primary cultures of normal HOSE cells, obtained from postmenopausal women, to those found in primary ovarian cancer cell cultures and in established OC cell lines. This approach has allowed us to observe that (i) ERα and ERβ mRNA, as well as AR and PR transcripts, are coexpressed in normal HOSE cells, (ii) whole exon-deletion variants of ERα and ERβ are commonly found in HOSE and ovarian cancer cells, and (iii) PR and AR mRNA expression are significantly down-regulated in ovarian cancer cells that exhibit altered ERα, but normal ERβ, message expression. A previously unknown ERα mutation, leading to a 32-bp deletion in exon 1, has been identified in an ovarian cancer cell line, SKOV3. This cell line has been shown to express ERα but is estrogen- and antiestrogen-resistant (18). Taken together, these findings implicate regulation of normal HOSE cell functions by estrogens, and possibly by progestins and androgens. Additionally, the emergence of sex hormone resistance, via down-regulation or mutational inactivation of receptors, may be a key feature of ovarian epithelial transformation.
MATERIALS AND METHODS
Cell Cultures and Cell Lines.
Four primary cultures of normal HOSE cells (HOSE27, HOSE20, HOSE17, and HOSE13), one primary culture of normal mesothelial cells (MesO13), four primary cultures of ovarian carcinoma cells (OVCA420, OVCA429, OVCA432, and OVCA433), and three ovarian cancer cell lines (DOV13, SKOV3, and CAOV3) were used in this study. Primary cultures of normal HOSE cells were initiated from surface scrapings of normal ovaries removed from postmenopausal women with benign gynecological diseases according to Tsao et al. (19). In each case, ovarian histology was performed by a pathologist, and only normal ovaries were used for normal HOSE cell collection. The one primary culture of normal mesothelial cells (MesO13) was obtained from peritoneal washing of a non-OC patient. Primary cultures of ovarian cancer cells (OVCA420, OVCA429, OVCA432, and OVCA433) were established from freshly isolated ascites or tumor explants obtained from patients with late stage serous ovarian adenocarcinomas according to Tsao et al. (19). All normal HOSE and ovarian cancer primary cell cultures were early passages in medium 199 and MCDB 105 (1:1) (Sigma) supplemented with 10% fetal calf serum (Sigma), 100 units/ml penicillin, and 100 μg/ml streptomycin under 5% CO2. Normal and malignant cells grew in this medium after two or more passages exhibited uniform epithelium-like morphology. Immunocytochemistry detection of cytokaratins (K7, K8, K18, and K19) and vimentin indicated little or no fibroblast contamination (19). Mok (19) showed that normal HOSE cells in primary cultures are susceptible to transforming growth factor-β (TGF-β) growth inhibition and secrete negligible amounts of CA125 (<5 units/ml). However, OC cells in primary cultures were resistant to TGF-β inhibition and secreted significant amounts of CA125 (65–461 units/ml) (19). All human tissues represented discarded tissues collected by the Laboratory of Gynecologic Oncology, Brigham and Women’s Hospital, Boston, Massachusetts. Three established ovarian carcinoma cell lines, CAOV3, DOV13, and SKOV 3, were purchased from American Type Culture Collection and maintained in culture media recommended by the organization.
RNA Isolation and Reverse Transcriptase–PCR (RT-PCR).
Total cellular RNA was isolated by using RNA Stat-60 reagent (Tel-Test, Friendswood, TX) according to protocols provided by the manufacturer. The quality of each total RNA sample was checked and controlled by using the following steps: (i) measurement of optical density, (ii) running of a denaturing RNA gel capable of detecting possible RNA degradation, and (iii) conducting a semiquantitative RT-PCR for glyceraldehyde-6-phosphate dehydrogenase (GAPDH), a housekeeping gene. Genomic DNA was eliminated by digestion with DNase I. One microgram of total RNA was reverse-transcribed by using the GeneAmp RNA PCR kit (Perkin–Elmer). Subsequently, 2 μl of the resulting cDNA samples was used in each PCR.
The primer sequences for GAPDH, ERα, ERβ, PR, and AR are given in Table 1 (20–22). Three pairs of primers were used to detect ERα wild-type and variant mRNAs, and two pairs of primers were used in the identification of wild-type and exon 5 deletion variant of ERβ (Table 1). Hot start PCR using AmpliTaq Gold DNA polymerase (Perkin–Elmer) was used in all amplification reactions. The enzyme was activated by preheating the reaction mixtures at 95°C for 6 minutes before thermal cycling. This protocol was chosen to minimize nonspecific product amplification. The routine PCR program was 30 cycles of 1 min at 94°C, 1 min at 60°C (annealing temperature), and 1 min at 72°C. mRNA-specific modifications included (i) an annealing temperature of 58°C used to amplify ERβ cDNA, (ii) an annealing temperature of 55°C in the amplification of ERα and AR cDNAs, (iii) a cycle number of 35 for ERα cDNA amplification, (iv) GAPDH cDNA amplified for 22–26 cycles, and its levels served as loading control. After PCR, the products were resolved on 2% agarose gel with ethidium bromide. The images was captured under UV transillumination. In initial experiments, after amplification, PCR products were excised, purified, and subjected to direct sequencing to verify amplification of the correct sequences. Serial dilutions of total RNA and cDNA were used in early experiments to establish the range of linearity between signal intensities and amounts of transcript in samples. The optimal PCR cycle number for each message was chosen to yield product levels at the linear portion of the serial dilution curve. All PCR conditions were optimized for quantification of relative message contents with respect to GAPDH product levels.
Table 1.
Primer sequences used for RT-PCR analysis
| Target gene | Primer sequence 5′ → 3′ | Location, nucleotide | Expected size, bp | Ref. |
|---|---|---|---|---|
| ERB | ||||
| Set 1 | ||||
| ERβ-1 | TGA AAA GGA AGG TTA GTG GGA ACC | 230-253 | 528 | |
| ERβ-2 | TGG TCA GGG ACA TCA TCA TGG | 737-757 | ||
| Set 2 | ||||
| ERβ-3 | GCC CAA GAG AAG TGG CGG CCA CG | 592-613 | 429 | 20 |
| ERβ-4 | AAA CCT TGA AGT AGT TGC CAG GAG C | 996-1020 | ||
| ERα | ||||
| Set 1 | ||||
| ERα-1 | TAC TGC ATC AGA TCC AAG GG | 41-60 | 650 | 22 |
| ERα-2 | ATC AAT GGT GCA CTG GTT GG | 671-690 | (exon 1–3) | |
| Set 2 | ||||
| ERα-3 | GTG GGA ATG ATG AAA GGT GG | 742-761 | 668 | |
| ERα-4 | TCC AGA GAC TTC AGG GTG CT | 1390-1409 | (exon 3–6) | |
| Set 3 | ||||
| ERα-5 | GTG CCT GGC TAG AGA TCC TG | 1142-1161 | 710 | |
| ERα-6 | TGG TGC ATG ATG AGG GTA AA | 1832-1851 | (exon 5–8) | |
| PR | ||||
| PR1 | GAT TCA GAA GCC AGC CAG AG | 1817-1836 | 533 | |
| PR2 | TGC CTC TCG CCT AGT TGA TT | 2330-2349 | ||
| AR | ||||
| AR1 | CTC TCT CAA GAG TTT GGA TGG CT | 2896-2918 | 342 | |
| AR2 | CAC TTG CAC AGA GAT GAT CTC TGC | 3214-3237 | ||
| GAPDH | ||||
| GAPDH-F | CCA CCC ATG GCA AAT TCC ATG GCA | 152-175 | 598 | 21 |
| GAPDH-R | TCT AGA CGG CAG GTC AGG TCC ACC | 726-749 |
Detection and Identification of ER Mutants and Variants by Using Direct DNA Sequencing Analyses.
RT-PCR products corresponding to different region of ERα or ERβ transcripts were amplified by using primer sets listed in Table 1. After amplification, products were analyzed by agar gel electrophoresis. Under UV transillumination, the DNA bands of interest were excised, reamplified, and resolved on agarose gel. The reamplified PCR products were eluted, purified by the Sephaglas BandPrep kit (Amersham Pharmacia), and subjected to radiolabeled primer cycle sequencing by using the ThermoSequenase cycle sequencing kit (Amersham Pharmacia). Two rounds of sequencing were performed to ensure reproducibility, and sequence data were compared with sequences published in GenBank.
RESULTS
ERα mRNA: Expression Levels and Identification of Exon 2, 4, 5, and 7 Deletion Variants.
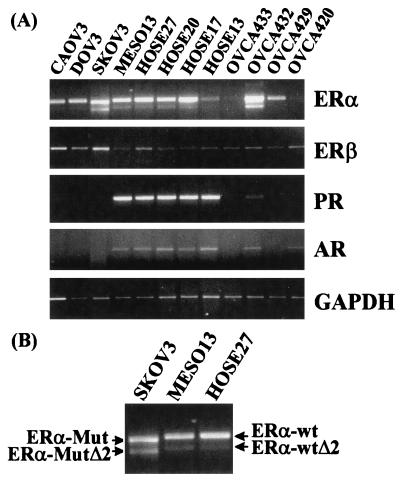
By using primer set 1 (Table 1), which amplified a 650-bp PCR product corresponding to nucleotides 41–690 (exons 1–3) of the ERα mRNA, we demonstrated expression of ERα transcript in all primary cultures of normal HOSE cells (HOSE27, HOSE20, HOSE 17, and HOSE13) and in a primary culture of mesothelial cells (MesO13) (Fig. 1). Among the primary cultures of ovarian cancer cells and the established OC lines, six of seven expressed ERα mRNA. The levels of this transcript in the three established cancer cell lines (CAOV3, DOV13, and SKOV3) were comparable to those found in HOSE cell cultures. However, ERα mRNA expression in primary cultures of ovarian cancer cells (OVCA433, OVCA432, OVCA429, and OVCA420) exhibited marked variability. Down-regulation of expression was noted in OVCA433 and OVCA420, enhanced expression was noted in OVCA432, and close-to-normal expression was noted in OVCA429.
Figure 1.
Relative abundance of ERα, ERβ, PR, and AR transcripts among normal and malignant human ovarian surface epithelial (HOSE) cells. Four HOSE cell primary cultures (HOSE13, HOSE17, HOSE20, and HOSE27), one mesoethelial cell culture (MesO13), four primary cell cultures established from late serous adenocarcinomas (OVCA420, OVCA429, OVCA432, OVCA433) and three established ovarian cancer cell lines (CAOV3, DOV13, SKOV3) were used to isolate total RNA. RT-PCRs were performed on 1 μg of RNA by using primer sets listed in Table 1. (A) A representative fluorograph of the RT-PCR products of one set of RNA samples is presented. Ethidium bromide-stained PCR products of ERα, ERβ, PR, AR, and GAPDH cDNA. (B) An excerpt from the ERα fluorograph, enlarged to illustrate that SKOV3 expressed the mutated ERα (ERα-Mut) and its exon 2 deletion variant (ERα-MutΔ2). Both contained a 32-bp deletion in exon 1 (see Fig. 2) and therefore exhibited higher mobilities than their corresponding wild-type transcripts (ERα-wt and ERα-wtΔ2).
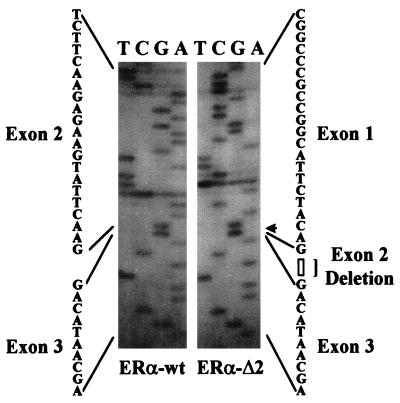
In addition to the predicted 650-bp PCR product, PCR with primer set 1 amplified an additional product of a smaller size. This variant product was present in all normal and some malignant ovarian cell lines or cultures (note the fainter band in HOSEs, MesO13, and OVCA429 in Fig. 1A). After excision, reamplification, and direct sequencing, it became apparent that this smaller size PCR product represented an ERα mRNA variant deleted in exon 2 (ER-αΔ2, Fig. 1 A and B; Fig. 2 Right). Further analysis showed additional ERα variants with exon 4, 5, or 7 deletion in cellular RNA samples isolated from all HOSE cell cultures and some cancerous ovarian epithelial cells (data not shown).
Figure 2.
Sequence analyses of the wild-type ERα transcript (ERα-wt) and its exon 2 deletion variant (ERα-Δ2). The PCR products of the respective cDNAs were excised, eluted, and reamplified. The reamplified products were subjected to radiolabeled primer cycle sequencing by using the ThermoSequenase cycle sequencing kit (Amersham Pharmacia). Exon 2 deletion is revealed by the sequencing results.
Identification of a ERα Exon 1 Deletion Mutant in SKOV3.
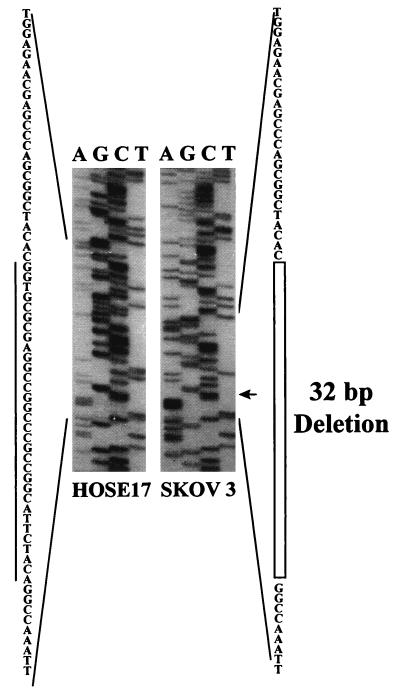
On closer examination, it was noted that PCR products derived from amplification of SKOV3 cellular RNA, using primer set 1, were of smaller than expected sizes (Fig. 1B). Subsequent DNA sequencing of these PCR products revealed the presence of a 32-bp deletion in exon 1 in both (ERα-Mut and ERα-MutΔ2, Fig. 1B and Fig. 3 Right). No normal ERα mRNAs were present in SKOV3 cells based on our analyses. Sequence analyses predicted a frameshift resulting in the production of a truncated ERα polypeptide of 145 aa (containing a partial A/B domain) from the mutated ERα transcript.
Figure 3.
Sequence analyses of PCR products of ERα cDNA derived from SKOV3 mRNA as compared with that derived from HOSE17 mRNA. A 32-bp deletion in exon 1 of the transcript was detected in transcripts of SKOV3 cells.
ERβ mRNA Expression.
ERβ mRNA expression was detected in all normal and malignant ovarian cell primary cultures/cell lines examined (PCRs were performed with primer pair Set 1, Fig. 1A). No dramatic differences in mRNA expression levels were observed among primary cultures of normal HOSE cells and ovarian cancer primary cell cultures/cell lines. A slight up-regulation of ERβ mRNA expression (≈2-fold of HOSE cells’ levels) was detected in SKOV3 and CAOV3, two established cancer cell lines. In light of a recent report of a ERβ exon 5 deletion variant (20), we used primer set 2 (Table 1) to demonstrate the presence of this variant in all normal HOSE cell cultures and in some ovarian cancer cell lines/cultures (data not shown).
PR mRNA Expression.
Expression of PR mRNA was demonstrated in all normal ovarian epithelial cells including the four primary cultures of normal HOSE cells and the one culture of mesoepithelial cells, MesO13 (Fig. 1, data from a 30-cycle PCR). PR mRNA expression in these cell cultures was strong. In contrast, most of the ovarian cancer cell lines/primary cultures (5/7) failed to express PR transcripts (negative expression was confirmed with 40-cycle PCRs, data not shown). Two ovarian cancer cell lines/cultures, OVCA432 and CAOV3, expressed extremely low levels of the transcript (<1/20 of HOSE cells’ levels).
AR mRNA Expression.
Thirty-cycle RT-PCR analyses revealed the presence of AR transcripts in all HOSE cell cultures and in the MesO13 culture (Fig. 1). Fairly uniform signal intensities were observed in these samples. Of the seven ovarian cancer cell lines/primary cultures, five did not express the AR transcript (40-cycle RT-PCR was used to confirm negativity, data not shown). Nevertheless, low levels of AR mRNA expression (<1/3 of HOSE cells’ levels) were found in OVCA420 and OVCA432.
DISCUSSION
In this study, semiquantitative RT-PCR and direct sequencing were used to investigate ERα, ERβ, PR, and AR mRNA expression in normal and malignant ovarian epithelial cells. Of significance to ovarian carcinogenesis, we report (i) coexpression of ERα and ERβ wild-type transcripts and their variants in four primary cultures of HOSE cells, the tissue of origin of >90% of OCs, and in one mesothelial cell culture (ii) loss of ERα, but not ERβ, mRNA expression in primary cultures of ovarian cancer cells and established OC cell lines, and (iii) expression of PR and AR mRNA in normal HOSE cells and marked down-regulation of these messages in ovarian cancer cells. Additionally, a novel ERα mRNA mutant, with a 32-bp deletion in exon 1, was identified in the estrogen-resistant ovarian cancer cell line, SKOV3 (18). Finally, several ER mRNA variants, commonly found in breast cancer specimens, were detected in HOSE cells as well as in ovarian cancer cells. These variants include ERα mRNA variants that lack exon 2, 4, 5, or 7, as well as an ERβ mRNA variant deleted in exon 5.
Our finding that ERα and ERβ transcripts are uniformly expressed in HOSE cells suggests that estrogens, via ER signaling, may play an important role in regulating normal HOSE cell functions. Because PR is a well recognized estrogen-regulated gene (23, 24) and high levels of PR mRNA are found in HOSE cells, we speculate that one or both ER isoforms are functional in these cells. Our data are in agreement with recent findings (15) that demonstrated expression of ERα and ERβ in 21-day cultured HOSE cells established from fresh ovarian scraping. However, when Brandenberger and coworkers (16) evaluated ERα and ERβ mRNA expression in normal human ovaries and in one immortalized HOSE cell line (IOSE-Van; ref. 25), they detected that ERβ as the predominant ER isoform in normal human ovary but found only ERα mRNA expression in IOSE-Van. Data from this latter report cannot be used for direct comparison with our findings because human ovaries are composed of many cell types, including granulosa cells, which express high levels of ERβ (12, 16, 17). Furthermore, IOSE-Van is a HOSE cell line immortalized by SV40 and therefore may exhibit receptor characteristics different from those found in normal HOSE cells.
Of particular interest to normal HOSE cell physiology is our observation that HOSE cells derived from postmenopausal women expressed substantial levels of ERα, ERβ, PR, and AR transcripts. These findings suggest that sex hormone responsiveness is retained in HOSE cells after menopause. It is well established that circulating estrogen levels are low after menopause but that the ovary continues to produce androgens and estrogens via aromatization (26, 27). Hence, ovarian estrogens and/or androgens may be responsible for maintained normal HOSE cell functions after menopause. In this connection, a logical question is whether loss of sex hormone regulation in HOSE cells is involved in the development of OCs that dramatically increase in incidence in women after the age of 45 (1–4).
Compared with HOSE cells, ovarian cancer cells express reduced levels of AR mRNA and no PR transcripts. Paradoxically, the loss of PR message expression in these malignant cells is not attended with concomitant down-regulation of ERα or ERβ mRNA expression. ERα mRNA expression is absent in only two ovarian cancer cell cultures (OVCA433 and OVCA420), whereas ERβ messages are present in all cancer cell cultures/lines at levels comparable to those found in HOSE cells. A lack of PR expression in ERα/ERβ-positive ovarian cancer cells may represent a state of estrogen resistance because, in most other estrogen-target tissues, PR expression is tightly associated with ERα (23, 24) or ERβ (28, 29) expression. The notion that ovarian cancer cells are estrogen-resistant is supported by clinical observations that tamoxifen treatment is a not an effective therapy for OCs and only 36% of OC specimens coexpress both ERα and PR (1–4).
The mechanisms underlying estrogen resistance have not been investigated in OCs. However, results from breast and endometrial cancer studies have identified reduction in ER expression, presence of dominant-negative ER variants, mutational inactivation of ER, and abnormal expression of coactivators/repressors of ER as probable causes for this phenomenon (30–34). ERα mutations are rare but have been reported in metastatic and tamoxifen-resistant malignancies of the breast (32, 33) and of the endometrium (34). Nevertheless, ERα mutations have not been reported in ovarian cancer specimens. In the present study, by using primers designed to amplify exon 1–3 of the ERα, we discovered an ERα mRNA mutation with a 32-bp deletion in exon 1 in SKOV3 cells. Based on sequence analyses, this deletion was predicted to cause a frameshift and result in the synthesis of a truncated, 145 aa protein. This protein, if synthesized in vivo, would lack both the DNA- and ligand-binding domains and likely be inactive. Previously, Hua and coworkers had reported SKOV3 cells to be estrogen-nonresponsive (18). These cells were shown to be insensitive to estrogen with respect to cell proliferation and induction of gene expression, and yet, they expressed ERα at both message and protein levels. Their latter findings do not contradict our results because, in their study, ERα messages were analyzed by using Northern hybridization, a method apt to miss a 32-bp mutation. Additionally, in their Western blot analyses, the mAb D75, instead of the ERα-specific H222 (35), was used. Hence, cross-reactivity with ERβ might have been a confounding factor. Our result clearly demonstrated expression of ERβ mRNA in SKOV3 cells, and if proteins were synthesized, immunoreactivity with D75 is conceivable. According to contemporary views (for review see ref. 36), if ERα and ERβ are coexpressed in the same cell, they may regulate different sets of cellular functions or play redundant roles. In the case of SKOV3 cells, mutated ERαs and wild-type ERβs likely coexist. It is therefore possible that estrogen resistance in this ovarian cancer cell line is because of mutational inactivation of the ERα. What remains to be explained is the roles played by ERβ in mediating estrogen responsiveness in SKOV3 cells, because this cell line expresses normal levels of ERβ mRNA. A comparable situation exists in ovarian granulosa cells of ERKO (ERα-knockout) mice (37). Disruption in folliculogenesis and abnormal granulosa cell functioning have been observed in these animals, and yet high levels of ERβ mRNA, but no ERα, are present in their granulosa cells (37). These findings in ERKO mice, together with our data, on SKOV3 suggest that the action of ERβ may depend on the presence of a functional ERα. If this supposition is verified, it would have significant implications in ERα/ERβ action.
ER mRNA variants are frequently expressed in many normal and malignant tissues (for review, see ref. 30). In breast cancers, variants of ERα are prevalent and they almost always coexist with wild-type ER (30). Even though little evidence has been presented in support of actual translation of these mRNA variants to protein products, in vitro studies suggest that they may have “outlaw functions,” i.e., dominant-positive or dominant-negative functions (30). Previously, one ERα variant with exon 4 deletion has been reported in OC specimens and in normal human ovaries (38). Our data now demonstrates prevalent occurrence of ER mRNA variants, coexisting with wild-type messages, in HOSE and ovarian cancer cells. Specifically, ERα mRNA variants that lack exon 2, 4, 5, or 7, as well as an ERβ mRNA variant deleted in exon 5 (20) were detected. Because the presence of ER mRNA variants is in both normal and malignant HOSE cells, this finding implies that mechanisms underpinning ER mRNA variant generation remain unaltered following neoplastic transformation.
Until recently, the biological significance of progesterone or androgen in ovarian carcinogenesis has remained unknown. Loss of estrogen responsiveness in HOSE cells likely leads to down-regulation of PR. Progesterone or progesterone responsiveness appears to offer protection against ovarian carcinogenesis. Previous epidemiological studies reported a reduction in OC risk in postmenopausal women using the combination (estrogens plus progestins) hormone replacement therapy (4, 39). In a recent epidemiological study, an increase in ovarian cancer incidence was observed among women with progesterone deficiency (40). In another study, 93% of malignant ovarian tumors exhibited PR immunonegativity, whereas PR immunopositivity was observed in the majority of borderline tumors (41). Exposure of ovarian cancer cells to progesterone up-regulated p53 and induced apoptosis in ovarian cancer cell lines (42). Similarly, testosterone and androstenedione were found to be effective in suppressing ovarian cancer cell proliferation (43). Of interest, a 75% rate of loss of heterozygosity at 11q23.3–24.3 that harbors the PR gene locus (44, 45) had been reported, and one of us (S.C.M.) recently detected a 40% loss of heterozygosity at Xq11.2–q12 that harbors the AR gene (46). In the present study, we observed significant down-regulation of PR and AR mRNA expression in several established ovarian cancer cell lines and in the small number of primary cell cultures established from patients with late serous adenocarcinoma. Taken together, these findings suggest that progesterone and testosterone signaling, via their cognate receptors, may have tumor suppressor function in ovarian carcinogenesis by induction of apoptosis and/or inhibition of proliferation. Hence, diminution in progesterone and/or androgen action/response may predispose HOSE cells to neoplastic transformation.
In summary, by comparing receptor mRNA levels in ovarian cancer cells to those found in HOSE cells, we have noted disruptions of ERα, PR, and AR mRNA expression in cancer cells. The association between loss of PR and/or AR expression and malignancy is especially apparent. It is conceivable that progesterone and/or androgen actions may protect HOSE cells from neoplastic transformation. If this premise can be verified, it may have significant clinical implications in postmenopausal hormone replacement therapy management. In contrast, expression of ERβ mRNA and those of several forms of ER mRNA variants are not affected by the malignant state. Future experiments are necessary to reveal the biological significance of these molecules in both normal HOSE cell physiology and ovarian tumorigenesis. Finally, of particular interest, the newly discovered ERα mutation involving a 32-bp deletion in exon 1 of the transcript in SKOV3 cells may explain the previously observed estrogen resistance in this ovarian cancer cell line.
Acknowledgments
This research is supported in part by National Institutes of Health–National Cancer Institute Grants CA-15776, CA62269, and AG13965 to S.M.H. and CA69453 and CA69291 to S.C.M.
ABBREVIATIONS
- HOSE
human ovarian surface epithilial
- ER
estrogen receptor
- PR
progesterone receptor
- AR androgen receptor
OC, ovarian carcinoma
- RT-PCR
reverse transcription–PCR
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
References
- 1.Roa B R, Slotman B S. Endocr Rev. 1991;12:14–26. doi: 10.1210/edrv-12-1-14. [DOI] [PubMed] [Google Scholar]
- 2.Clinton G M, Hua W. Crit Rev Oncol Hematol. 1997;25:1–9. doi: 10.1016/s1040-8428(96)00216-8. [DOI] [PubMed] [Google Scholar]
- 3.Kommoss F. In: Hormone-Dependent Cancer. Pasqualini J R, Katzenellenbogen B S, editors. New York: Dekker; 1996. pp. 541–572. [Google Scholar]
- 4.Risch H A. J Natl Cancer Inst. 1998;90:1774–1786. doi: 10.1093/jnci/90.23.1774. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez C, Calle E E, Coates R J, Miracle-McMahill H L, Thun M J, Heath C W., Jr Am J Epidemiol. 1995;141:828–835. doi: 10.1093/oxfordjournals.aje.a117518. [DOI] [PubMed] [Google Scholar]
- 6.Garg P P, Kerlikowske K, Subak L, Grady D. Obstet Gynecol. 1998;92:472–479. doi: 10.1016/s0029-7844(98)00139-2. [DOI] [PubMed] [Google Scholar]
- 7.Jeppsson S, Karlsson S, Kullarnder S. Acta Obstet Gynecol Scand. 1986;65:207–210. doi: 10.3109/00016348609155172. [DOI] [PubMed] [Google Scholar]
- 8.Heinonen P K, Koivula T, Pystynen P. Acta Obstet Gynecol Scand. 1985;64:649–652. doi: 10.3109/00016348509158207. [DOI] [PubMed] [Google Scholar]
- 9.Mahlck C G, Backstrom T, Kjellgren O. Gynecol Oncol. 1988;30:313–320. doi: 10.1016/0090-8258(88)90245-4. , 1988. [DOI] [PubMed] [Google Scholar]
- 10.Mahlck C G, Grankvist K, Backstrom T, Kjellgren O. Acta Obstet Gynecol Scand. 1986;65:533–538. doi: 10.3109/00016348609158381. [DOI] [PubMed] [Google Scholar]
- 11.Mahlck C G, Grankvist K, Kjellgren O, Backstrom T. Gynecol Oncol. 1990;36:219–225. doi: 10.1016/0090-8258(90)90178-n. [DOI] [PubMed] [Google Scholar]
- 12.Kuiper G G J M, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson J-Å. Proc Natl Acad Sci USA. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosselman S, Polman J, Dijkema R. FEBS Lett. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- 14.Pujol P, Rey J M, Nirde P, Roger P, Gastaldi M, Laffargue F, Rochefort H, Maudelonde T. Cancer Res. 1998;58:5367–5373. [PubMed] [Google Scholar]
- 15.Hillier S G, Anderson R A, Williams A R, Tetsuka M. Mol Hum Reprod. 1998;4:811–815. doi: 10.1093/molehr/4.8.811. [DOI] [PubMed] [Google Scholar]
- 16.Brandenberger A W, Tee M K, Jaffe R B. J Clin Endocrinol Metabol. 1998;83:1025–1028. doi: 10.1210/jcem.83.3.4788. [DOI] [PubMed] [Google Scholar]
- 17.Byers M, Kuiper G G J M, Gustafsson J-Å, Park-Sarge O K. Mol Endocrinol. 1997;11:172–182. doi: 10.1210/mend.11.2.9887. [DOI] [PubMed] [Google Scholar]
- 18.Hua W, Christianson T, Rougeot C, Rochefort H, Clinton G M. J Steroid Biochem Mol Biol. 1995;55:279–289. doi: 10.1016/0960-0760(95)00187-5. [DOI] [PubMed] [Google Scholar]
- 19.Tsao S W, Mok S C, Fey E G, Fletcher J A, Wan T S, Chew E C, Muto M G, Knapp R C, Berkowitz R S. Exp Cell Res. 1995;218:499–507. doi: 10.1006/excr.1995.1184. [DOI] [PubMed] [Google Scholar]
- 20.Vladusic E A, Hornby A E, Guerra-Vladusic F K, Lupu R. Cancer Res. 1998;58:210–214. [PubMed] [Google Scholar]
- 21.Maier J A, Voulalas P, Roeder D, Maciag T. Science. 1990;249:1570–1574. doi: 10.1126/science.2218499. [DOI] [PubMed] [Google Scholar]
- 22.Weigel R J, de Conick E C. Cancer Res. 1993;53:3472–3474. [PubMed] [Google Scholar]
- 23.Horwitz K B, Koseki Y, McGuire W L. Endocrinology. 1978;103:1742–1751. doi: 10.1210/endo-103-5-1742. [DOI] [PubMed] [Google Scholar]
- 24.Kruas W L, Montano M M, Katzenellenbogen B S. Mol Endocrinol. 1993;7:1603–1616. doi: 10.1210/mend.7.12.8145766. [DOI] [PubMed] [Google Scholar]
- 25.Maines-Bandiera S L, Kruk P A, Auersperg N. Am J Obstet Gynecol. 1992;167:729–735. doi: 10.1016/s0002-9378(11)91579-8. [DOI] [PubMed] [Google Scholar]
- 26.Lucisano A, Acampora M G, Russo N, Maniccia E, Montemurro A, Dell’Acqua S. Maturitas. 1984;6:45–53. doi: 10.1016/0378-5122(84)90064-1. [DOI] [PubMed] [Google Scholar]
- 27.Lucisano A, Russo N, Acampora M G, Fabiano A, Fattibene M, Parlati E, Maniccia E, Dell’Acqua S. Maturitas. 1986;8:57–65. doi: 10.1016/0378-5122(86)90008-3. [DOI] [PubMed] [Google Scholar]
- 28.Lau K M, Leav I, Ho S M. Endocrinology. 1998;139:424–427. doi: 10.1210/endo.139.1.5809. [DOI] [PubMed] [Google Scholar]
- 29.Shughrue P J, Lubahn D B, Negro-Vilar A, Korach K S, Merchenthaler E. Proc Natl Acad Sci USA. 1997;94:11008–11012. doi: 10.1073/pnas.94.20.11008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castles C G, Fuqua S A W. In: Hormone-Dependent Cancer. Pasqualini J R, Katzenellenbogen B S, editors. New York: Dekker; 1996. pp. 81–105. [Google Scholar]
- 31.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M J, O’Malley B W. Recent Prog Hormone Res. 1997;52:146–164. [PubMed] [Google Scholar]
- 32.Zhang Q X, Borg A, Wolf D M, Oesterreich S, Fuqua S A. Cancer Res. 1997;57:1244–1249. [PubMed] [Google Scholar]
- 33.Karnik P S, Kulkarni S, Liu X P, Budd G T, Bukowski R M. Cancer Res. 1994;54:349–353. [PubMed] [Google Scholar]
- 34.Assikis V J, Bilimoria M M, Muenzner H D, Lurain J R, Jordan V C. Gynecol Oncol. 1996;63:192–199. doi: 10.1006/gyno.1996.0305. [DOI] [PubMed] [Google Scholar]
- 35.Kuiper G G J M, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, Gustafsson J-Å. Endocrinology. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 36.Giguere V, Tremblay A, Tremblay G B. Steroids. 1998;63:335–339. doi: 10.1016/s0039-128x(98)00024-5. [DOI] [PubMed] [Google Scholar]
- 37.Couse J F, Korach K S. J Mol Med. 1988;76:497–511. doi: 10.1007/s001090050244. [DOI] [PubMed] [Google Scholar]
- 38.Park W, Choi J-J, Hwang E-S, Lee J-H. Clin Cancer Res. 1996;2:2029–2035. [PubMed] [Google Scholar]
- 39.Schneider H P, Birkhauser M. Int J Fertil Menopausal Stud. 1995;40, suppl. 1:40–53. [PubMed] [Google Scholar]
- 40.Modan B, Ron E, Lerner-Geva L, Blumstein T, Menczer J, Rabinovici J, Oelsner G, Freedman L, Mashiach S, Lunenfeld B. Am J Epidemiol. 1998;147:1038–1042. doi: 10.1093/oxfordjournals.aje.a009397. [DOI] [PubMed] [Google Scholar]
- 41.Noguchi T, Kitawaki J, Tamura T, Kim T, Kanno H, Yamamoto T, Okada H. J Steroid Biochem Mol Biol. 1993;44:657–660. doi: 10.1016/0960-0760(93)90275-2. [DOI] [PubMed] [Google Scholar]
- 42.Bu S-Z, Yin D-L, Ren X-H, Jiang L-Z, Wu Z-J, Gao Q-R, Pei G. Cancer. 1997;79:1944–1950. doi: 10.1002/(sici)1097-0142(19970515)79:10<1944::aid-cncr15>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 43.Thompson M A, Adelson M D. Adv Exp Med Biol. 1993;330:155–165. doi: 10.1007/978-1-4615-2926-2_12. [DOI] [PubMed] [Google Scholar]
- 44.Davis M, Hitchcock A, Foulkes W D, Campbell I G. Cancer Res. 1996;56:741–744. [PubMed] [Google Scholar]
- 45.Gabra H, Watson J E, Taylor K J, Mackay J, Leonard R C, Steel C M, Porteous D J, Smyth J F. Cancer Res. 1996;56:950–954. [PubMed] [Google Scholar]
- 46.Edelson M I, Lau C C, Colitt C V, Welch W R, Bell D A, Berkowitz R S, Mok S C. Oncogene. 1998;16:197–202. doi: 10.1038/sj.onc.1201479. [DOI] [PubMed] [Google Scholar]