Abstract
Natural selection acts on variation within populations, resulting in modified organ morphology, physiology, and ultimately the formation of new species. Although variation in orthologous proteins can contribute to these modifications, differences in DNA sequences regulating gene expression may be a primary source of variation. We replaced a limb-specific transcriptional enhancer of the mouse Prx1 locus with the orthologous sequence from a bat. Prx1 expression directed by the bat enhancer results in elevated transcript levels in developing forelimb bones and forelimbs that are significantly longer than controls because of endochondral bone formation alterations. Surprisingly, deletion of the mouse Prx1 limb enhancer results in normal forelimb length and Prx1 expression, revealing regulatory redundancy. These findings suggest that mutations accumulating in pre-existing noncoding regulatory sequences within a population are a source of variation for the evolution of morphological differences between species and that cis-regulatory redundancy may facilitate accumulation of such mutations.
[Keywords: Chiroptera, Carollia perspicillata, Prx1, Prrx1, limb development, evolution]
Changes in DNA sequence that alter existing noncoding regulatory elements or create new ones have been hypothesized to be enabling events for natural selection and consequently morphological evolution (Stern 2000; Wray 2003; Carroll et al. 2004; Carroll 2005). These types of mutations can modify specific aspects of the patterns and/or levels of gene expression, in contrast to coding sequence mutations that cause global alterations of gene function in all expressing tissues. Regulatory mutations that cause alterations in the expression of developmental control genes can lead to changes in organogenesis, resulting in morphological and/or physiological modification. For example, a single base change in a limb-specific enhancer of the Sonic hedgehog (Shh) gene causes preaxial polydactyly in mice by directing Shh expression ectopically in the anterior limb bud during development (Maas and Fallon 2005). Descriptive and correlative evidence supports this hypothesis, and in some cases genetic evidence suggests that cis-regulatory divergence of specific loci may be the cause of morphological change (Stern 1998; Sucena and Stern 2000; Sucena et al. 2003; Wittkopp et al. 2003; Shapiro et al. 2004; Wang et al. 2004; Gompel et al. 2005; Prud’homme et al. 2006). No studies have been reported that experimentally test these ideas between different species of mammals.
Results
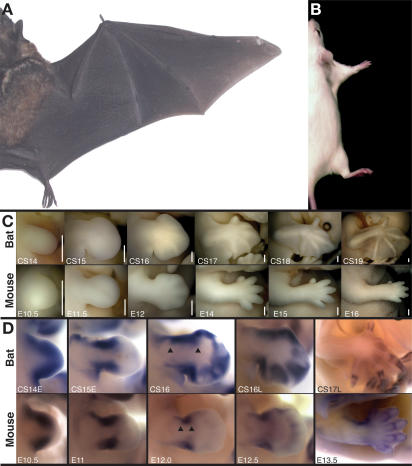
We compared limb morphogenesis between the mouse (Mus musculus) and our model Chiropteran, the short-tailed fruit bat (Carollia perspicillata) (Rasweiler and Badwaik 1996; Cretekos et al. 2005). These animal models represent the two largest orders of mammals (Rodentia and Chiroptera, respectively) in terms of numbers of extant species and are thought to have shared their most recent common ancestor ∼80–100 million years ago (Murphy et al. 2004; Springer et al. 2004). There are striking morphological differences between the forelimbs of adult bats and mice (Fig. 1A,B). These include differences in the relative length of anterior and posterior digits, presence or absence of tissue between the digits and between the limbs, and differences in the proximal–distal length of all three forelimb segments relative to body length. While the relative timing and positioning of the initial formation of forelimb buds appears to be conserved between bat and mouse (Theiler 1989; Cretekos et al. 2005), all of the striking differences in adult limb morphology arise from differences in limb morphogenesis that appear during embryonic development (Fig. 1C)
Figure 1.
Comparison of bat and mouse forelimb morphogenesis and Prx1 expression. Dorsal view of the right side of an adult Carollia (A) and mouse (B) scaled for equivalent body length. (C) Developmental series of Carollia (bat) and mouse forelimbs at equivalent stages, anterior at top and distal to right. Staging is according to Cretekos et al. (2005) for bat and in days post-coitum (E) for mouse and is indicated at the bottom left of each panel. Bars (bottom right of each panel), 0.5 mm. (D) Prx1 RNA expression in the developing forelimb detected by whole-mount in situ hybridization for Carollia (bat) and mouse. Staging is according to Cretekos et al. (2005) for bat and in days post-coitum (E) for mouse and is indicated at the bottom left of each panel. Views are not to scale.
Of particular significance to the experiments described here, all three segments of the forelimb skeleton elongate in the proximal–distal axis to a much greater extent relative to body length during development of the bat compared with mouse. For example, the ratio of forelimb length to crown–rump length (FL:CR) is about the same for mouse or bat at embryonic day 11.5 (E11.5)/Carollia stage 15 (CS15), before skeletal development begins (FL:CR of embryos pictured in Fig. 1C is 0.19 for mouse and 0.20 for bat). However, for the most advanced specimens pictured in Figure 1C, this ratio has increased almost fivefold for bat (from 0.20 to 0.98), compared with a less than twofold increase for mouse (from 0.19 to 0.34). Interestingly, the roughly 1:3 FL:CR length ratio persists through postnatal development to adulthood for mouse, while for the bat limb elongation continues to outpace body growth until the adult FL:CR is approximately 2:1 (Fig. 1A,B).
One developmental control gene known to promote limb skeletal elongation is Prx1, also called MHox or Prrx1 (Martin et al. 1995). Prx1 is a paired-related homeobox gene expressed in somites, craniofacial mesenchyme, and limb mesoderm during mouse development (Cserjesi et al. 1992; Martin et al. 1995). Prx1-null mutants die at birth with severe craniofacial defects and significantly shortened limbs (Martin et al. 1995). Early events of limb development are normal in Prx1-null mutants as the limb skeleton forms and is patterned correctly. However the long bones of the limbs are significantly shortened, showing that this transcription factor is an essential regulator of long bone elongation during limb development. Given that the forelimb long bones are strikingly elongated during bat development (Fig. 1C; Cretekos et al. 2005; Sears et al. 2006), and mouse Prx1-null mutants show a specific defect in the elongation of these elements (Martin et al. 1995), we hypothesized that differences in Prx1 activity between mouse and bat may contribute to the morphological differences that develop in the forelimbs of these two divergent mammals.
Bat and mouse Prx1 gene organization and expression
We isolated and sequenced the Carollia Prx1 locus and found that its structure is highly conserved with mouse (Supplemental Fig. 1A). The mouse locus produces two isoforms by RNA splicing between two alternative fourth exons, encoding proteins that differ only in the C-terminal region. Alignment of the deduced amino acid sequences of Carollia PRX1A and B shows >99% identity with the orthologous isoforms of mouse PRX1 (Supplemental Fig. 1B). Only two amino acid differences are found between bat and mouse PRX1 proteins, and these occur outside of the conserved homeodomain, PRX and OAR motifs, thus there is no evidence to suggest that these orthologous proteins differ in biochemical activity.
Prx1 expression was examined by whole-mount RNA in situ hybridization of stage-equivalent bat and mouse embryos from early limb bud stage (CS12; Theiler stage 15 [TS15]/E9.5) through the onset of skeletal elongation (CS18; TS22/E13.5). The overall expression pattern of Prx1 is similar in bat embryos compared with mouse but some differences are apparent (Fig. 1D). Prx1 is initially expressed uniformly throughout the limb bud mesoderm in both bat and mouse (Cserjesi et al. 1992; Martin et al. 1995; data not shown). As the limb bud grows out, expression persists in anterior and posterior domains in both species, but these domains appear expanded in the bat relative to mouse in subsequent stages (Fig. 1D, arrowheads). Expression is not detected in the early handplate of either species (Fig. 1D, E11.5, CS15). In subsequent stages however, Prx1 is up-regulated in the bat handplate compared with mouse, where expression is detected but at significantly lower levels compared with the more proximal domains of expression (Fig. 1D, E12–E12.5, CS16–CS16L). As the digital condensations begin to differentiate into chondrogenic elements, Prx1 is expressed in the perichondrium surrounding these elements in both species (Fig. 1D, E13.5, CS17L). Thus, there are similarities and differences in Prx1 limb expression between bat and mouse, and the differences correlate with morphological differences in limb development between these species. The expression differences observed could be a direct result of differences in the sequences in and around the Prx1 locus that regulate transcription and/or indirect results of differences in the expression of the transcription factors that bind these sequences. Moreover, the differences in expression could be either a cause or an effect of the correlated morphological differences. To address these issues, we focused next on the cis-regulation of Prx1.
Identification of bat and mouse Prx1 limb transcriptional enhancers
Previously, deletion mapping identified a region upstream of the start of Prx1 transcription capable of directing reporter gene expression in the limbs of transgenic mice (Martin and Olson 2000). Comparison of the upstream region of bat Prx1 with the orthologous sequence of mouse revealed regions of noncoding homology corresponding to conserved region 1 (CR1) and CR2 proposed by Martin and Olson (2000). Carollia CR1 and CR2 show 94.1% and 93.2% identity to mouse, respectively (Supplemental Fig. 1C).
To investigate the developmental expression pattern governed by this cis-regulatory region in greater detail, we constructed transgenes combining 1-kb genomic fragments centered on the CR1/2 homology of either mouse or bat (Supplemental Fig. 1C) with a basal promoter-lacZ-polyA reporter cassette and generated transgenic mouse lines. The mouse and bat 1-kb CR1/2-containing regions share 67% overall DNA sequence identity. Four mouse- and 6 bat-enhancer transgenic lines express β-galactosidase (β-gal) activity in a similar Prx1-like pattern during development at similar transgenic frequencies, as assayed by whole-mount X-gal staining (Supplemental Fig. 1D). Differences were seen between each line in the relative levels of reporter gene expression and in specific details of expression patterns at later stages of development (Supplemental Fig. 1D; data not shown). To demonstrate that this region contains a bona fide enhancer activity (Atchison 1988), we also generated transgenes inserting a smaller fragment spanning CR1 and CR2 flanked by minimal additional sequence (581-base-pair [bp] fragment in Supplemental Fig. 1C) in either orientation upstream of the reporter cassette. This fragment directed expression of β-gal activity in E11.5 G0 transgenic embryos in a Prx1-like pattern regardless of orientation (data not shown).
Replacing the endogenous mouse Prx1 limb enhancer with the corresponding bat enhancer
To directly address the functional relevance of limb enhancer divergence between bat and mouse, we generated mice that express the endogenous mouse Prx1 gene under control of the bat limb enhancer (Prx1BatE) using an enhancer replacement gene targeting strategy in mouse embryonic stem (ES) cells (see Fig. 2; Materials and Methods for details). Because regions flanking the CR1 and CR2 homologies might contain relatively less well-conserved species-specific regulatory information (Gompel et al. 2005), we substituted the 1-kb bat enhancer described above for the equivalent 1 kb of mouse genomic sequence. The analyses presented in this study for all of the Prx1 alleles generated by gene targeting were performed exclusively on a 129/SvEv inbred genetic background to eliminate genetic background variation.
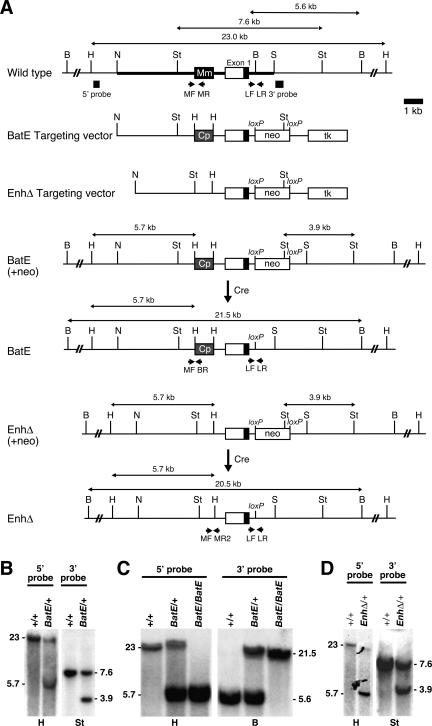
Figure 2.
Prx1 limb enhancer replacement and deletion by gene targeting. (A) Strategy for generating targeted alleles. The genomic region surrounding the Prx1 limb enhancer is diagrammed schematically. The targeting vector possesses 7.05 kb of total homology in three parts: 3.8 kb 5′, 2.2 kb central, and 1.05 kb 3′ homology (indicated by the thick line in wild-type allele). A PGKneobpA-lox neo expression cassette (neo) was introduced into the BamHI site of intron 1 separating the central and 3′ homologies. The neo cassette introduces a StuI restriction site used for Southern analysis. A MCTK1 expression cassette (tk) was included for negative selection. The Carollia limb enhancer (Cp) was introduced on a 1-kb HindIII restriction fragment replacing the equivalent 1-kb segment surrounding the endogenous enhancer (Mm) and separating the 5′ and central homologies (BatE targeting vector). An identical vector was also constructed but with the 1-kb enhancer segment deleted and replaced with a HindIII restriction site for Southern analysis (EnhΔ targeting vector). The positions of the external 5′ and 3′ probes used for Southern analysis to screen for targeted ES cell clones are shown below the wild-type allele. The sizes of the expected restriction fragments recognized by the probes are indicated above each allele. The neo cassettes were removed in vivo by Cre expressed from an ectopic male germline-specific Prm1-Cre transgene. (B) Southern analysis of bat enhancer replacement targeted ES cell clones. Targeted clones digested with HindIII produce 23-kb wild-type and 5.7-kb targeted bands when hybridized with an external 5′ probe. Targeted clones digested with StuI produce 7.6-kb wild-type and 3.9-kb targeted bands when hybridized with an external 3′ probe. (C) Southern analysis of Prx1BatE mice: Genomic DNA extracted from Prx1BatE/+ mice digested with HindIII and hybridized with an external 5′ probe produces 23-kb wild-type and 5.7-kb targeted bands; Prx1BatE/BatE mice produce only the 5.7-kb targeted band. Genomic DNA extracted from Prx1BatE/+ mice digested with BamHI and hybridized with an external 3′ probe produces 5.6-kb wild-type and 21.5-kb targeted bands when hybridized with an external 3′ probe; Prx1BatE/BatE mice produce only the 21.5-kb targeted band. (D) Southern analysis of enhancer deletion targeted ES clones. Targeted clones digested with HindIII produce 23-kb wild-type and 5.7-kb targeted bands when hybridized with an external 5′ probe. Targeted clones digested with StuI produce 7.6-kb wild-type and 3.9-kb targeted bands when hybridized with an external 3′ probe. (B) BamHI; (H) HindIII; (N) NsiI; (S) SalI; (St) StuI.
To select for targeted alleles, a loxP-flanked neomycin (flox-neo) resistance cassette was inserted into a nonconserved region of intron 1, ∼500 bp 3′ of the exon 1 splice donor. The flox-neo cassette was later excised using a Cre recombinase-expressing transgene, leaving a single loxP site. Our targeting strategy also produced a control Prx1 allele, Prx1loxP, that differs from wild type only in the presence of this intronic loxP site. Morphometric analysis of the skeletons of Prx1loxP/loxP mice showed no significant difference in forelimb length at E18.5 compared with wild-type controls, demonstrating that the residual loxP site in the first intron of Prx1 does not influence forelimb length (Fig. 3A; Supplemental Table 1).
Figure 3.
Forelimb length of Prx1 mutants. Limb morphometry at E18.5 (A) and E15.5 (B). An example of the right forelimb of a Prx1BatE/BatE, a Prx1+/+ sibling, and a Prx1−/−-null mutant of comparable mass, stained with alizarin red for minerized bone and alcian blue for cartilage (A) or alcian blue alone (B). (A) Mass-normalized limb measurements for E18.5 pups are shown as percentage difference relative to average wild type. Prx1BatE/BatE forelimbs are on average 6% longer than wild type at E18.5 (Student’s t-test P = 0.01); mass-normalized average forelimb length of E18.5 Prx1loxP/loxP controls; Prx1EnhΔ/EnhΔ and Prx1−/− mutants processed identically are shown for comparison. (B) Mass-normalized limb measurements for E15.5 pups are summarized: Average length of Prx1BatE/BatE forelimbs is not significantly different from wild-type siblings at E15.5. Genotype, sample size (n), and Student’s t-test P value are indicated above. (**) P ≤ 0.01; (‡) P > 0.05. Error bars show standard error for each sample set.
The bat Prx1 limb enhancer increases forelimb length in mice during development
Mice heterozygous or homozygous for the Prx1BatE allele segregate in the expected Mendelian ratio, are viable and fertile, and show no overt morphological phenotypes. However, morphometric analysis of skeletal preparations reveals that the forelimbs of Prx1BatE/BatE mutants are on average ∼6% longer than their wild-type littermates at E18.5 (Fig. 3A; Supplemental Table 1). Like the Prx1-null mice, limb pattern is unchanged even though length is altered (Fig. 4A; Martin et al. 1995). Statistical analysis of these data using Student’s t-test, Kruskal-Wallis test, and Wilcoxon rank sum test reveals that this difference is highly significant (t-test P = 0.01; Kruskal-Wallis P < 0.0001; Wilcoxon P = 0.02) (see Materials and Methods for details of statistical analysis). We chose this stage for morphometric analysis because preliminary examination of newborn pups indicated Prx1BatE/BatE mutant limbs appeared proportionally longer than wild-type littermates, but variability in overall pup size made this difficult to judge accurately. Morphometric analysis at E18.5, just before birth, allowed us to control for the normal variance in size using mass without introducing differences in feeding as an additional mass variable. These and all subsequent skeletal length measurements are therefore normalized against embryo mass to control for the normal variance in pup size within and between litters.
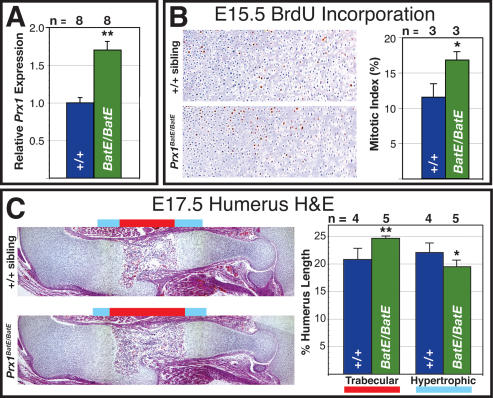
Figure 4.
Quantification of Prx1 expression and long bone growth in Prx1BatE/BatE mutant forelimbs. (A) Average Prx1 expression level in developing Prx1BatE/BatE forelimb long bones is 1.7-fold higher than wild type, quantified by real-time QRT–PCR analysis of total RNA purified from humeri dissected from eight Prx1BatE/BatE compared with eight Prx1+/+ siblings at E17.5 and shown graphically. (B) Long bone chondrocyte proliferation is elevated by ∼6% at E15.5 in Prx1BatE/BatE mutants. Immunohistochemical detection of BrdU incorporation in equivalent sections of the proximal humerus growth plate in pulse-labeled E15.5 Prx1BatE/BatE and Prx1+/+ sibling. Average increase in mitotic index of three Prx1BatE/BatE compared with three Prx1+/+ siblings is shown graphically at the right. (C) Equivalent sections of the stylopod of E17.5 Prx1BatE/BatE and Prx1+/+ sibling stained with hematoxylin/eosin. The proportion of overall humerus longitudinal length comprised of trabecular bone and hypertrophic cartilage in five Prx1BatE/BatE and four Prx1+/+ siblings is shown graphically at the right. Error bars show standard error for each sample set. (**) P ≤ 0.01; (*) P ≤ 0.05.
The forelimbs of Prx1-null homozygotes are ∼12.5% shorter than controls at E18.5 (Fig. 3A; Martin et al. 1995). Thus, the change in forelimb length shown for Prx1BatE/BatE mutants is about half the magnitude (in the opposite direction) of the length change produced by the complete elimination of Prx1 function. Although a 6% increase may seem to be a modest change in forelimb length in absolute terms, it is a highly significant change in terms of Prx1 gene function. As Prx1-null homozygotes also show significantly shortened hindlimbs (Supplemental Table 1; Martin et al. 1995), we performed morphometric analysis on Prx1BatE/BatE mutant hindlimbs and found no significant difference in either length or pattern at E18.5 compared with wild-type littermates (Supplemental Table 1; data not shown).
To investigate the developmental basis of the phenotype, we analyzed skeletal preparations of E15.5 embryos. At this stage of development, all of the elements of the limb skeleton are present in the form of cartilaginous precursors. The chondrocytes that make up these elements have begun to undergo differentiation, and some hypertrophic chondrocytes are seen in the central region of more proximal elements (Fig. 3B), but little if any mineralized trabecular bone has yet formed. We find no significant differences between Prx1BatE/BatE mutants and their wild-type littermates in either morphology or limb skeletal element length at this stage (Fig. 3B; data not shown). Thus, the Prx1BatE mutant limb skeleton shows no indication of precocious formation, and the significant length difference seen 3 d later at E18.5 is unlikely to result from developmental heterochrony.
At the developmental stages when the mutant phenotype is detected, between E15.5 and birth, Prx1 is expressed predominantly in the perichondrium surrounding the limb skeletal elements and in some chondrocytes. We find no obvious difference in the pattern of Prx1 expression between Prx1BatE/BatE mutants and their wild-type littermates either by whole-mount in situ hybridization of RNA expression or immunohistochemical analysis of protein expression in this developmental interval (Supplemental Fig. 2A; data not shown). By design, the only difference between the BatE and wild-type alleles is the replacement of a mouse Prx1 noncoding regulatory sequence with the orthologous element from bat, so the alteration of Prx1 expression responsible for the phenotype may be a difference in expression level, perhaps in a subset of the overall expression pattern. Consistent with this idea, we find that LacZ reporter expression driven by either the bat or mouse Prx1 limb enhancer in the transgenic mouse lines described above is most abundant in a subset of perichondrium and chondrocyte cells at the ends of the long bones at E17.5 (Supplemental Fig. 2B). To test this hypothesis we conducted real-time quantitative PCR analysis of RNA purified from long bones dissected from mutant and wild-type forelimbs at E17.5. We found that on average mutant long bones express ∼70% more Prx1 than wild-type siblings relative to endogenous glyceraldehyde-3-phosphate dehydrogenase (Gapdh) control expression (Fig. 4A).
At E17.5, we find that the trabecular zone of the humerus of the Prx1BatE/BatE mutants is significantly expanded relative to wild-type siblings, apparently at the expense of the hypertrophic chondrocyte zone (Fig. 4C), suggesting that chondrocyte differentiation is accelerated in the mutants relative to controls. Mouse models of chondrodysplasia syndromes involving elevated or constitutive fibrobast growth factor signaling in developing long bones show a similar acceleration in chondrocyte differentiation (for review, see Ornitz and Marie 2002). These mouse mutants develop shorter than normal limbs, however, because increased differentiation rate depletes the pool of chondrocyte precursors. We find an average 6% increase in chondrocyte mitotic index at E15.5 in the Prx1BatE/BatE mutants relative to controls, as assayed by BrdU pulse-labeling (Fig. 4B). Finally, we examined apoptosis by TUNEL analysis at E15.5 and E17.5 and found no apparent difference between Prx1BatE/BatE mutants and controls at either time point (data not shown).
Taken together, these data provide a cellular mechanism for the observed forelimb lengthening by a coordinated increase of both chondrocyte proliferation and differentiation without any change in cell death. These changes in limb long bone growth are detectable at perinatal stages of development but apparently regulate during postnatal skeletal growth as adult mutants show no difference in limb length compared with wild-type at 6 wk of age (Supplemental Table 1; data not shown).
Prx1 limb enhancer deletion mutant mice have normal limb length and expression
To test whether the Prx1 limb enhancer is essential for limb-specific transcription, we generated mice lacking it using gene targeting. The targeting strategy is similar to the one described above for the enhancer replacement except that the 1-kb genomic region that includes the enhancer was deleted. The Prx1EnhΔ allele was analyzed on a 129/SvEv inbred genetic background.
Mice homozygous for the Prx1EnhΔ allele segregate in the expected Mendelian ratio, are viable and fertile, and surprisingly do not exhibit the limb defects of Prx1-null mice (Martin et al. 1995). Morphometric analysis of Prx1EnhΔ/EnhΔ skeletal preparations shows that there is no significant difference in comparison with controls in forelimb length at E18.5 (Fig. 3A; Supplemental Table 1). Interestingly, Prx1 expression at the RNA level, assayed by whole-mount in situ hybridization between E10.5 and E13.5 and real-time PCR analysis of RNA generated from long bones dissected from mutant and wild-type forelimbs at E17.5, or at the protein level, assayed by immunohistochemisty between E16.5 and E18.5 in Prx1EnhΔ/EnhΔ mice and their heterozygote and wild-type siblings, show no significant differences (Supplemental Fig. 2C–E). These results demonstrate that the Prx1 locus contains additional enhancers whose activity is redundant with the limb enhancer that we deleted or replaced in this study.
Discussion
The fact that Prx1 expression level is elevated and a forelimb elongation phenotype develops in the Prx1BatE/BatE mutants shows that the bat enhancer responds distinctly from the corresponding mouse enhancer even in the context of a mouse transcription factor environment. This limb elongation arises at stages of mouse gestation when Prx1 expression is limited to the perichondrium of the developing limb skeletal elements. It is known that the perichondrium is an important regulator of endochondrial bone growth (Eames et al. 2003; Colnot et al. 2004). We have shown previously that Bmp expression and signaling are increased relative to mouse in the perichondrium of bat forelimb skeletal elements during limb development (Sears et al. 2006). Our findings indicate that the perichondrium is an important tissue for modifying skeletal morphologies between species.
To our surprise, Prx1EnhΔ/EnhΔ mutants did not reproduce the shortened limb phenotype of Prx1-null mice, and expression in the limb bud, hand plate, and perichondrium was overtly normal, indicating the presence of an additional limb enhancer. The existence of another Prx1 limb enhancer could also account for the observation that LacZ reporter protein expression driven by the known enhancer in the transgenic mouse lines described here is restricted to a subset of the overall Prx1 expression pattern. Experiments to identify additional Prx1 limb enhancers are currently in progress. Multiple enhancers regulating fully or partially overlapping expression patterns have been identified for a variety of developmental control genes (Helms et al. 2000; Pappu et al. 2005; Uemura et al. 2005; Jeong et al. 2006), and functional enhancer redundancy has been demonstrated for immunoglobulin and T-cell receptor genes (Wiersma et al. 1999; Xiong et al. 2002). Maintenance of redundant enhancers for essential developmental control genes would allow changes in expression pattern to arise from mutations that alter regulatory activity while preserving the required gene function (Stern 2000; Wray 2003; Carroll et al. 2004; Carroll 2005). The presence of additional endogenous mouse Prx1 limb enhancers may buffer the effects of the bat enhancer on forelimb lengthening. In addition, the related Prx2 gene has been shown to be partially redundant with Prx1 during limb development (ten Berge et al. 1998), thus endogenous Prx2 expression might also dilute the activity of the bat limb enhancer on forelimb lengthening.
In On the Origin of Species (Darwin 1859), Darwin discussed how divergent limb morphologies evolved through “successive slight modifications” of a conserved fundamental pattern. He stated “there will be little or no tendency to alter the original pattern” and that “the bones of a webbed foot might have all its bones, or certain bones, lengthened and the membrane connecting them increased so as to serve as a wing” (Chapter 13, subheading Morphology, paragraph 3). Here we show that targeted replacement of an endogenous mouse Prx1 limb enhancer with the bat equivalent produces mice with increased forelimb length though pattern is maintained, identifying transcriptional regulatory sequence differences between species as a source of variation for organ modification. Since the limb modification reported here accounts for a portion of the relative increase in forelimb length of bat compared with mouse, this may be one of Darwin’s predicted “successive slight modifications” required to evolve a mammalian wing. Because the forelimb skeletal structure of the oldest bat fossils known is similar in structure and proportion to extant bats, it has been proposed that the morphological changes required for powered flight in bats evolved relatively rapidly and the underlying genetic changes might therefore be few in number (Simmons and Geisler 1998; Speakman 2001; Teeling et al. 2005; Sears et al. 2006). Alternatively, the proposed rapid change from arm to wing morphology may have resulted from many independently arising predispositions toward longer limbs accumulating in a population over a long period of time. Each of these modifications would individually cause minor or transient effects on limb phenotype due to the buffering effect on phenotypic output by canalization of developmental mechanisms. Once combined in a bat ancestor, however, the cumulative effect could override genetic capacitance to produce a large and seemingly instantaneous change at the morphological level. A general mechanism for the advent of morphological variation among mammals might therefore require divergence of many regulatory elements for several developmental control genes.
Materials and methods
Isolation and analysis of the Carollia Prx1 locus
A probe for the first exon of Carollia Prx1 was generated by PCR using degenerate primers (Prx1dF, ATGRCSTCCAGCTAYG SSCACG; Prx1dR, GACACCCCKCAGCAGGACAATG) designed by aligning the first exon sequences of mouse, rat, human, and chicken Prx1 using ClustalW. The resulting 245-bp product was cloned and confirmed by sequencing. The same primers were used to screen a fractionated and arrayed Carollia genomic λ library (Cretekos et al. 2001) by PCR. Prx1-positive phage pools were plated, lifted, and screened by hybridization using a radiolabeled Carollia exon1 probe as described (Ausubel 1989). Six independent recombinants were identified and subjected to insert end sequencing. Sequences were aligned to the human genome by BLAST (NCBI), and the clone whose end sequence matched the human sequence furthest 5′ of PMX1 (human Prx1 ortholog) was selected for subcloning and sequencing (CPGL-Prx1#71; 17,257-bp insert; GenBank accession no. DQ396876). The Carollia first exon probe was subsequently used to screen a genomic BAC library (E.D. Green, unpubl.) to identify Carollia clone 484G18 (diagrammed in Fig. 2A). This 132,080-bp clone was subjected to shotgun sequencing by the NIH Intramural Sequencing Center (http://www.nisc.nih.gov) (GenBank accession no. AC148270).
Whole-mount in situ hybridization and immunohistochemistry
Whole-mount in situ hybridization was performed as described (Wilkinson 1992) on mouse embryos and on Carollia embryos with the following modifications: Proteinase K (Roche) digestion of bat embryos was 15 min for CS14, 20 min for CS15, 35 min for CS16, and 50 min for CS17. The Carollia embryos used in this study were collected from the wild on the island of Trinidad, as described previously (Cretekos et al. 2005). Digoxigenin-labeled antisense riboprobe against Carollia Prx1 was generated by digesting a 4.8-kb XhoI genomic subclone of CPGL-Prx1#71 in pBluescript(-)KS with DraI, followed by transcription with T7 RNA polymerase. An equivalent riboprobe against mouse Prx1 was generated by digesting a 4.5-kb XbaI genomic subclone of Prx1 in pBluescript SK+ (pRP4-2, provided by J.F. Martin) with BglII, followed by transcription with T3 RNA polymerase. Both the bat and mouse riboprobes are ∼1.1 kb in length and correspond to ∼700 bp of 5′ untranslated leader and 244 bp of coding sequence from the first exon, with the remainder intron sequence. Immunohistochemical detection of PRX1 protein expression was performed using an anti-PRX1 polyclonal antibody (Chesterman et al. 2001) and Vectastain anti-rabbit HRP kit (Vector). Immunohistochemical detection of LacZ protein was performed using an anti-LacZ polyclonal antibody (MP Biomedical) and Vectastain fluorescein anti-rabbit kit (Vector). Tissues were fixed overnight in 4% paraformaldehyde and embedded in paraffin, and 7-μm-thick sections were counterstained with hematoxylin/eosin or DAPI, respectively.
Generation and analysis of transgenic mice
Carollia and mouse genomic sequences upstream of the first exon of Prx1 were aligned using ClustalW. Species-specific primer pairs incorporating HindIII restriction sites were designed to amplify ∼1-kb fragments centered on the noncoding homology elements CR1 and CR2 proposed by Martin and Olson (2000) as limb-specific cis-regulatory elements (Fig. 2C) by PCR. Carollia primers were CpPrxEnHF2 (CCCAAGCTTCCCCCTCGGCCTTCCACCTTAG) and CpPrxEnHR2 (CCCAAGCTTCCCGCCCCACAAACCCCAGTCC) and mouse primers were MmPrxEnHF2 (CCCAAGCTTGAAAGGGTGTCTCCT TTCCC) and MmPrxEnHR2 (CCCAAGCTTCCCCTACCTTT GCCAATGT); HindIII restriction sites are indicated by bold type and sequences complementary to target are underlined. A 13-kb NotI genomic subclone (mouse strain 129/SvEv) in pBluescript SK+-containing sequence upstream of and including part of exon1 (pRP4-1, provided by J.F. Martin) was used as a template to amplify the mouse enhancer, and CPGL-Prx1#71 was used as a template to amplify the bat enhancer. The ∼1-kb PCR product for each species was digested with HindIII, cloned into pBluescript KS−, verified by sequencing, then subcloned into the HindIII site of phspPTlacZpA (Kothary et al. 1989). Transgenic founder mice were generated from both mouse (11 of 41 G0 founders transgenic by tail biopsy/lacZ PCR) and bat (19 of 65 G0 founders transgenic by tail biopsy/lacZ PCR) Prx1 putative enhancer transgenes by pronuclear microinjection as described (Brinster et al. 1985). Transgenic lines were propagated from all founders and analyzed for β-gal activity by X-gal staining (Nagy et al. 2003). Six of 19 (32%) lines express β-gal activity in a Prx1-like pattern during development under the control of the bat 1-kb region, and four of 11 (36%) lines express β-gal activity in a Prx1-like pattern under control of the mouse 1-kb region, as assayed by whole-mount X-gal staining (Fig. 2L; data not shown).
Generation of gene targeting vectors
HindIII sites were engineered by PCR into the mouse genomic DNA flanking the limb enhancer in positions corresponding to the ends of the 1-kb mouse enhancer described above. For the enhancer replacement targeting vector (BatE), the 1-kb Carollia limb enhancer HindIII fragment described above was inserted between the engineered HindIII sites. For the enhancer deletion targeting vector (EnhΔ), the 1-kb enhancer-containing sequence between the engineered HindIII sites was deleted. A loxP-flanked (flox) neo expression cassette (PGKneobpA-lox) was inserted, in reverse orientation relative to the direction of Prx1 transcription, into a Klenow-blunted BamHI site in the first intron of the targeting vector (Fig. 3A). A herpes simplex virus-thymidine kinase expression cassette (MCTK1) was inserted into a Klenow-blunted SalI site at the end of the 3′ arm of homology with enrich for homologous recombinants by negative selection (Mansour et al. 1988). The vectors were sequenced to confirm the enhancer replacement or deletion and the insertion of the flox-neo cassette, and to ensure that no other changes were introduced. The presence of an exogenous selectable marker can influence the activity of a targeted locus (Pham et al. 1996). Therefore, we flanked the neomycin resistance cassette with loxP sites (flox-neo) and used PC3, a 129/SvEv-derived ES cell line that carries an ectopic Cre transgene driven by the male germline-specific Prm1 promoter, for gene targeting (O’Gorman et al. 1997). Germline transmission of targeted alleles through male chimeras results in in vivo excision of the flox-neo cassette, leaving a single loxP site at the position of the destroyed BamHI site.
The region of vector homology begins at an NsiI site 5′ of the first exon of the Prx1 gene and extends 3′ to a SalI site in the first intron, providing 7.05 kb of total homology in three segments. The 5′ arm of vector homology contains 3.8 kb from the NsiI site to the 5′ introduced HindIII site. The central region of homology contains 2.2 kb from the 3′ introduced HindIII site to the destroyed BamHI in intron 1 and contains the entire first exon of Prx1. The 3′ arm contains 1.05 kb of homology from the destroyed BamHI site in intron 1 to the destroyed SalI site. The targeting vectors each contain the same three segments of homology with the target locus. Homologous recombination in the 3′ homology arm along with either the 5′ arm or the central region of homology results in two different gene targeting events, distinguishable by Southern analysis with external probes. Recombination in the 5′ homology arm results in replacement or deletion of the limb enhancer, depending on which targeting vector was used, as well as insertion of the flox-neo cassette. Recombination in the central homology region results in the insertion of the flox-neo cassette with no other changes using either targeting vector. After Cre-mediated excision of the neo cassette, the former becomes the experimental Prx1BatE or Prx1EnhΔ alleles and the latter serves as a control for the residual intronic loxP site (Prx1loxP). Insertion of the neo cassette brought in a new StuI restriction site that was exploited during Southern blot analysis.
Generation of targeted ES cell clones and germline alleles
The targeting vectors were linearized with SalI outside the 5′ homology. PC3 ES cells (O’Gorman et al. 1997) were electroporated in duplicate with 25 μg of vector each and were selected for 10 d in medium with G418 and FIAU (Behringer et al. 1994). Eight-hundred-forty and 768 G418, FIAU-resistant ES cell colonies were picked, respectively, for the BatE and EnhΔ targeting vectors and were analyzed by Southern blot (Fig. 3A,B,D; Ramirez-Solis et al. 1992). Correctly targeted ES cell clones were identified by the presence of additional 3.9-kb bands when digested with StuI and hybridized with a 3′ external probe (0.3-kb SalI–HincII) (Fig. 3B). Correctly 3′ targeted ES cell clones could have recombined either in the 5′ homology arm (BatE or EnhΔ) or in the central homology region (floxneo): These two possibilities were resolved by Southern analysis with a 5′ external probe (0.4-kb HindIII–EcoRV). Correctly targeted enhancer replacement (Prx1BatE), deletion (Prx1EnhΔ), and floxneo (Prx1floxneo) alleles were identified by the presence or absence of additional 5.7-kb bands when digested with HindIII and hybridized with 5′ probe (Fig. 2B,D). A targeting frequency of 1.1% (14 of 1287) G418, FIAU-resistant ES cell colonies screened for 3′ and 5′ targeting was obtained, four Prx1BatE, two Prx1EnhΔ and eight Prx1floxneo, respectively. Correctly targeted ES cell lines were karyotyped (Nagy et al. 2003) and three Prx1BatE, two Prx1EnhΔ and three Prx1floxneo with normal karyotype were injected into C57BL/6 (B6) blastocysts and transferred into 2.5-d-post-coitum pseudopregnant Swiss foster mothers. High-percentage chimeras (70%–90% agouti pigmentation) were obtained for all of the clones. The PC3 ES cells possess a protamine-Cre transgene expressing Cre recombinase in the male germline that results in in vivo excision of the PGKneobpA-lox cassette (O’Gorman et al. 1997) and loss of the internal StuI site (Prx1loxP). For all subsequent 3′ Southern analyses, DNA prepared from tail biopsies was digested with BamHI and scored for the presence of an additional 21.5-kb (Prx1BatE) or 20.5-kb (Prx1EnhΔ) band resulting from the destruction of the intronic BamHI site. Male chimeras were bred to B6 females, and all agouti pups were genotyped by tail biopsy DNA/Southern analysis using the 5′ and 3′ external probes to confirm germline transmission of the targeted alleles (Fig. 3C). After confirmation of genotype by Southern analysis, transmitting male chimeras were bred to 129/SvEv females (Taconic Farms) to propagate lines on a 129/SvEv inbred background. The F1 progeny of these crosses were genotyped by PCR of DNA prepared from tail biopsies (see Fig. 3A for primer positions). PCR with (1) MF (TAGGAACATGGGTAACCAGC), MR1 (GAGATGCCA ACGGTGAGACTCC), and BR (CTCGACTTACTCCTGCGG AGAGG) produces a 492-bp product from wild-type or Prx1loxP alleles and a 427-bp product from Prx1BatE; (2) MF, MR1, and MR2 (GTCCTGATAAGCTTCTCTCTTC) produces a 492-bp product from wild-type or Prx1loxP alleles and a 370-bp product from Prx1EnhΔ; (3) LF (GGCAGCGACACCCCTCAGCAGG) and LR (GGAGATGCAGCCCTGGCGGCTGG) produces a 336-bp product from the wild-type allele and a 390-bp product from any of the targeted alleles; and (4) Cre-Fw1 (GGACATGT TCAGGGATCGCCAGGC) and Cre-Rv2 (CGACGATGAAG CATGTTTAGCTG) produces a 219-bp product from mice carrying the Prm1-Cre transgene. F1 progeny that gave both 427- and 492-bp products in PCR1 and both 336- and 392-bp products in PCR3, and were negative for the Cre transgene in PCR4 were selected to propagate the Prx1BatE allele. F1 progeny that gave both 370- and 492-bp products in PCR2 and both 336- and 392-bp products in PCR3, and were negative for the Cre transgene in PCR4 were selected to propagate the Prx1EnhΔ allele. F1 progeny that gave only the 492-bp product in PCR1 or PCR2 and both 336- and 392-bp products in PCR3, and were negative for the Cre transgene in PCR4 were selected to propagate the Prx1loxP allele. For each of these, the PCR products were sequenced to confirm enhancer replacement or deletion and excision of the neo cassette (data not shown).
Limb morphometry
Prx1BatE/+, Prx1LacZ(null)/+, Prx1EnhΔ/+, or Prx1loxP/loxP 129/SvEv coisogenic mice were intercrossed and pregnant females were sacrificed at E15.5 and/or E18.5 time points. The E18.5 litters were dissected, weighed, and processed for whole-mount alcian blue and alizarin red staining of cartilage and bone, respectively (Nagy et al. 2003). A skin biopsy was saved from each pup to prepare DNA for PCR genotyping. The E15.5 litters were dissected, weighed, and processed for whole-mount alcian blue staining of cartilage (Nagy et al. 2003). The yolk sac of each conceptus was saved to prepare DNA for PCR genotyping. The right fore- and hindlimb of each stained specimen was dissected from the rest of the skeleton, mounted flat, and imaged using a stereo dissecting photomicroscope with digital image capture. The stylopod, zeugopod, and autopod of each limb were measured using the Measure Tool of Adobe Photoshop. For the purposes of these measurements, the stylopod is defined as the longest axis of the humerus or femur, the zeugopod is defined as longest axis of the radius or fibula, and the autopod is defined as the longest axis from the most proximal metacarpal or metatarsal element to the most distal element of the third digit. Imaging and PCR genotyping were conducted separately on specimens identified by an alphanumeric code, and all measurements were made in numbered sequence without prior knowledge of genotype (single blind). Measured limb length was normalized against embryo mass to control for the normal variance in body size within and between litters. For adult limb measurements, pups were genotyped by tail biopsy PCR, separated by sex at weaning, and held until they were 6-wkold adults. Adults were sacrificed and weighed, and all four limbs were dissected and mounted flat for limb skeleton imaging. Skeletons were imaged by small animal X ray with digital capture (Faxitron model MX-20D) and measured as described above.
Normalized limb measurements for each group of homozygous mutants were compared with those of their wild-type siblings using the two-tailed Student’s t-test assuming unequal variance. In addition to t-tests, nonparametric tests for the significance of the difference between the distributions of normalized E18.5 forelimb lengths among Prx1-null, bat enhancer, enhancer deletion, loxP-only controls, and the wild-type controls (+/+ siblings of the null, bat enhancer, and enhancer deletion homozygotes) were performed. Data from the three wild-type groups were combined, and a Kruskal-Wallis test was used to compare FL among the three wild-type mouse groups: No statistically significant difference was detected (P = 0.77). The Kruskal-Wallis test was also used to compare FL among the combined wild-type, BatE, EnhΔ, loxP, and null groups. The result indicates that FL in at least one group was significantly different from that of the other groups (P < 0.0001). The Wilcoxon rank sum test was then used for comparison of each of the genetically manipulated groups with the wild-type group. The result suggests that FL in bat enhancer group is significantly longer than that in the wild type (P = 0.02), and FL in null group was significantly shorter than that in wild type (P = 0.001).
Quantitative real-time PCR analysis
Prx1BatE/+ or Prx1EnhΔ/+ 129/SvEv coisogenic mice were intercrossed, and pregnant females were sacrificed at E17.5. The humerus was dissected from each forelimb, and connective tissue was removed by treatment with 0.1 mg/mL collagenase D (Roche) in PBS. The remaining limb tissue was used to prepare DNA for PCR genotyping. Total RNA was prepared from each pair of dissected humeri using a Qiagen RNeasy miniprep kit; yield and purity were assayed by UV spectrophotometry at 260 and 280 nm. QRT–PCR on oligo-dT-primed cDNA generated from the equivalent of 100 ng of total RNA from Prx1BatE/BatE, Prx1EnhΔ/EnhΔ or wild-type sibling humeri (n = 5–8 per geno type) was performed in duplicate on an ABI 7900 Real-Time PCR System (Applied Biosystems). Primer sequences for Prx1 and Gapdh, our endogenous control, are as follows: Prx1Exon2F2 (GATCTCGCACGTCGGGTGAACC), Prx1Exon3R1 (GCCAG CATGGCTCGCTCATTC), GAPDH-F (TTGTCTCCTGCGA CTTCAACA), and GAPDH-R (ACCAGGAAATGAGCTTGA CAAA). Relative Prx1 expression was quantified using the comparative CT method, normalized to Gapdh endogenous reference, and calibrated to wild-type Prx1 expression level as described by the instrument manufacturer (Applied Biosystems publication no. 4303859B).
Cell proliferation and cell death analysis
To examine cell proliferation, a BrdU staining kit (Calbiochem) was used to stain BrdU-incorporated cells fixed 2 h after intraperitonial injection of pregnant female mice with 0.02 mL/g body weight of BrdU/FdU labeling reagent (Amersham). Tissues were fixed in 4% paraformaldehyde and embedded in paraffin. Seven-micron-thick sections were counterstained with hematoxylin/eosin. The central two-thirds of the proximal humerus growth plate was used for analysis; the lateral portions of the growth plate adjacent to the perichondrium were excluded. The chondrocyte mitotic index was defined as the number of BrdU-labeled nuclei divided by the total number of nuclei. The mitotic index of each sample was based on at least 500 cells, using the manual count function of Metamorph software (Universal Imaging). To examine cell death, a TUNEL immunofluorescent staining kit (Roche Applied Science) was used to identify apoptotic cells. Tissues were fixed in 4% paraformaldehyde and embedded in paraffin. Seven-micron-thick sections were counterstained with DAPI.
Acknowledgments
We gratefully acknowledge Lianchun Xiao and the Department of Biostatistics and Applied Mathematics at University of Texas M.D. Anderson Cancer Center for assistance with statistical analyses. We thank George Adebayo for transgenic mouse production, Jian Min Deng for tissue culture support, Hank Adams for advice on microscopic imaging, Rui Zhu for advice on QRT–PCR, Michael Kern for anti-PRX1 polyclonal antibody, and Simeon Williams for field assistance. We are grateful to Terence Capellini, Eric Davidson, Georg Halder, Robin Hiesinger, Susan Mackem, Lee Niswander, Scott Weatherbee, and anonymous reviewers for helpful comments on the manuscript. We also thank the Department of Life Sciences, University of the West Indies, Trinidad, for generous assistance and the use of departmental facilities. We are also grateful to the Wildlife Section, Forestry Division, Agriculture Land and Marine Resources of the Republic of Trinidad and Tobago for providing collecting and export permits. These studies were supported by National Science Foundation grant IBN 0220458 and endowments from the Barnts family and Ben F. Love to R.R.B., and National Institutes of Health (NIH) grant DE 16329 to J.F.M.. C.J.C. was supported by NIH training grants CA09299 and HD07325. Blastocyst injection and veterinary resources were supported by the NIH Cancer Center Support Grant CA16672. This work was supported in part by the Intramural Research Program of the National Human Genome Research Institute, NIH.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1620408
References
- Atchison M.L. Enhancers: Mechanisms of action and cell specificity. Annu. Rev. Cell Biol. 1988;4:127–153. doi: 10.1146/annurev.cb.04.110188.001015. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M. Current protocols in molecular biology. John Wiley & Sons; New York: 1989. [Google Scholar]
- Behringer R.R., Finegold M.J., Cate R.L. Mullerian-inhibiting substance function during mammalian sexual development. Cell. 1994;79:415–425. doi: 10.1016/0092-8674(94)90251-8. [DOI] [PubMed] [Google Scholar]
- Brinster R.L., Chen H.Y., Trumbauer M.E., Yagle M.K., Palmiter R.D. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc. Natl. Acad. Sci. 1985;82:4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.B. Evolution at two levels: On genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.B., Grenier J.K., Weatherbee S.D. From DNA to diversity: Molecular genetics and the evolution of animal design. Blackwell Science; Malden, MA: 2004. [Google Scholar]
- Chesterman E.S., Gainey G.D., Varn A.C., Peterson R.E., Kern M.J. Investigation of Prx1 protein expression provides evidence for conservation of cardiac-specific posttranscriptional regulation in vertebrates. Dev. Dyn. 2001;222:459–470. doi: 10.1002/dvdy.1198. [DOI] [PubMed] [Google Scholar]
- Colnot C., Lu C., Hu D., Helms J.A. Distinguishing the contributions of the perichondrium, cartilage, and vascular endothelium to skeletal development. Dev. Biol. 2004;269:55–69. doi: 10.1016/j.ydbio.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Cretekos C.J., Rasweiler J.J., Behringer R.R. Comparative studies on limb morphogenesis in mice and bats: A functional genetic approach towards a molecular understanding of diversity in organ formation. Reprod. Fertil. Dev. 2001;13:691–695. doi: 10.1071/rd01115. [DOI] [PubMed] [Google Scholar]
- Cretekos C.J., Weatherbee S.D., Chen C.H., Badwaik N.K., Niswander L., Behringer R.R., Rasweiler J.J. Embryonic staging system for the short-tailed fruit bat, Carollia perspicillata, a model organism for the mammalian order Chiroptera, based upon timed pregnancies in captive-bred animals. Dev. Dyn. 2005;233:721–738. doi: 10.1002/dvdy.20400. [DOI] [PubMed] [Google Scholar]
- Cserjesi P., Lilly B., Bryson L., Wang Y., Sassoon D.A., Olson E.N. MHox: A mesodermally restricted homeodomain protein that binds an essential site in the muscle creatine kinase enhancer. Development. 1992;115:1087–1101. doi: 10.1242/dev.115.4.1087. [DOI] [PubMed] [Google Scholar]
- Darwin C. On the origin of species by means of natural selection, or the preservation of favored races in the struggle for life. John Murray; London, UK: 1859. [Google Scholar]
- Eames B.F., de la Fuente L., Helms J.A. Molecular ontogeny of the skeleton. Birth Defects Res. C Embryo Today. 2003;69:93–101. doi: 10.1002/bdrc.10016. [DOI] [PubMed] [Google Scholar]
- Gompel N., Prud’homme B., Wittkopp P.J., Kassner V.A., Carroll S.B. Chance caught on the wing: Cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Helms A.W., Abney A.L., Ben-Arie N., Zoghbi H.Y., Johnson J.E. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Jeong Y., El-Jaick K., Roessler E., Muenke M., Epstein D.J. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133:761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- Kothary R., Clapoff S., Darling S., Perry M.D., Moran L.A., Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- Maas S.A., Fallon J.F. Single base pair change in the long-range Sonic hedgehog limb-specific enhancer is a genetic basis for preaxial polydactyly. Dev. Dyn. 2005;232:345–348. doi: 10.1002/dvdy.20254. [DOI] [PubMed] [Google Scholar]
- Mansour S.L., Thomas K.R., Capecchi M.R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: A general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Martin J.F., Olson E.N. Identification of a prx1 limb enhancer. Genesis. 2000;26:225–229. [PubMed] [Google Scholar]
- Martin J.F., Bradley A., Olson E.N. The paired-like homeo box gene MHox is required for early events of skeletogenesis in multiple lineages. Genes & Dev. 1995;9:1237–1249. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Pevzner P.A., O’Brien S.J. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein M., Vintersten K., Behringer R. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2003. [Google Scholar]
- O’Gorman S., Dagenais N.A., Qian M., Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz D.M., Marie P.J. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes & Dev. 2002;16:1446–1465. doi: 10.1101/gad.990702. [DOI] [PubMed] [Google Scholar]
- Pappu K.S., Ostrin E.J., Middlebrooks B.W., Sili B.T., Chen R., Atkins M.R., Gibbs R., Mardon G. Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development. 2005;132:2895–2905. doi: 10.1242/dev.01869. [DOI] [PubMed] [Google Scholar]
- Pham C.T., MacIvor D.M., Hug B.A., Heusel J.W., Ley T.J. Long-range disruption of gene expression by a selectable marker cassette. Proc. Natl. Acad. Sci. 1996;93:13090–13095. doi: 10.1073/pnas.93.23.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme B., Gompel N., Rokas A., Kassner V.A., Williams T.M., Yeh S.D., True J.R., Carroll S.B. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- Ramirez-Solis R., Rivera-Perez J., Wallace J.D., Wims M., Zheng H., Bradley A. Genomic DNA microextraction: A method to screen numerous samples. Anal. Biochem. 1992;201:331–335. doi: 10.1016/0003-2697(92)90347-a. [DOI] [PubMed] [Google Scholar]
- Rasweiler J.J., Badwaik N.K. Improved procedures for maintaining and breeding the short-tailed fruit bat (Carollia perspicillata) in a laboratory setting. Lab. Anim. 1996;30:171–181. doi: 10.1258/002367796780865718. [DOI] [PubMed] [Google Scholar]
- Sears K.E., Behringer R.R., Rasweiler J.J., Niswander L.A. Development of bat flight: Morphologic and molecular evolution of bat wing digits. Proc. Natl. Acad. Sci. 2006;103:6581–6586. doi: 10.1073/pnas.0509716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M.D., Marks M.E., Peichel C.L., Blackman B.K., Nereng K.S., Jonsson B., Schluter D., Kingsley D.M. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- Simmons N.B., Geisler J.H. Phylogenetic relationships of Icaronycteris, Archaeonycteris, Hassianycteris, and Palaeochiropteryx to extant bat lineages, with comments on the evolution of echolocation and foraging strategies in Microchiroptera. Bull. Am. Mus. Nat. Hist. 1998;235:4–182. [Google Scholar]
- Speakman J.R. The evolution of flight and echolocation in bats: Another leap in the dark. Mammal Rev. 2001;31:111–130. [Google Scholar]
- Springer M.S., Scally M., Madsen O., de Jong W.W., Douady C.J., Stanhope M.J. The use of composite taxa in supermatrices. Mol. Phylogenet. Evol. 2004;30:883–884. doi: 10.1016/j.ympev.2003.09.019. [DOI] [PubMed] [Google Scholar]
- Stern D.L. A role of Ultrabithorax in morphological differences between Drosophila species. Nature. 1998;396:463–466. doi: 10.1038/24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.L. Evolutionary developmental biology and the problem of variation. Evolution Int. J. Org. Evolution. 2000;54:1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- Sucena E., Stern D.L. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc. Natl. Acad. Sci. 2000;97:4530–4534. doi: 10.1073/pnas.97.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucena E., Delon I., Jones I., Payre F., Stern D.L. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature. 2003;424:935–938. doi: 10.1038/nature01768. [DOI] [PubMed] [Google Scholar]
- Teeling E.C., Springer M.S., Madsen O., Bates P., O’Brien S.J., Murphy W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- ten Berge D., Brouwer A., Korving J., Martin J.F., Meijlink F. Prx1 and Prx2 in skeletogenesis: Roles in the craniofacial region, inner ear and limbs. Development. 1998;125:3831–3842. doi: 10.1242/dev.125.19.3831. [DOI] [PubMed] [Google Scholar]
- Theiler K. The house mouse: An atlas of embryonic development. Springer-Verlag; New York: 1989. [Google Scholar]
- Uemura O., Okada Y., Ando H., Guedj M., Higashijima S., Shimazaki T., Chino N., Okano H., Okamoto H. Comparative functional genomics revealed conservation and diversification of three enhancers of the isl1 gene for motor and sensory neuron-specific expression. Dev. Biol. 2005;278:587–606. doi: 10.1016/j.ydbio.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Wang X., Greenberg J.F., Chamberlin H.M. Evolution of regulatory elements producing a conserved gene expression pattern in Caenorhabditis. Evol. Dev. 2004;6:237–245. doi: 10.1111/j.1525-142X.2004.04029.x. [DOI] [PubMed] [Google Scholar]
- Wiersma E.J., Ronai D., Berru M., Tsui F.W., Shulman M.J. Role of the intronic elements in the endogenous immunoglobulin heavy chain locus. Either the matrix attachment regions or the core enhancer is sufficient to maintain expression. J. Biol. Chem. 1999;274:4858–4862. doi: 10.1074/jbc.274.8.4858. [DOI] [PubMed] [Google Scholar]
- Wilkinson D.G. In situ hybridization: A practical approach. Oxford University Press; New York: 1992. [Google Scholar]
- Wittkopp P.J., Williams B.L., Selegue J.E., Carroll S.B. Drosophila pigmentation evolution: Divergent genotypes underlying convergent phenotypes. Proc. Natl. Acad. Sci. 2003;100:1808–1813. doi: 10.1073/pnas.0336368100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray G.A. Transcriptional regulation and the evolution of development. Int. J. Dev. Biol. 2003;47:675–684. [PubMed] [Google Scholar]
- Xiong N., Kang C., Raulet D.H. Redundant and unique roles of two enhancer elements in the TCRγ locus in gene regulation and γδ T cell development. Immunity. 2002;16:453–463. doi: 10.1016/s1074-7613(02)00285-6. [DOI] [PubMed] [Google Scholar]