Abstract
Neural networks are balanced by inhibitory and excitatory neuronal activity. The formation of these networks is initially generated through neuronal subtype specification controlled by transcription factors. The basic helix–loop–helix (bHLH) transcription factor Ptf1a is essential for the generation of GABAergic inhibitory neurons in the dorsal spinal cord, cerebellum, and retina. The transcription factor Rbpj is a transducer of the Notch signaling pathway that functions to maintain neural progenitor cells. Here we demonstrate Ptf1a and Rbpj interact in a complex that is required in vivo for specification of the GABAergic neurons, a function that cannot be substituted by the classical form of the bHLH heterodimer with E-protein or Notch signaling through Rbpj. We show that a mutant form of Ptf1a without the ability to bind Rbpj, while retaining its ability to interact with E-protein, is incapable of inducing GABAergic (Pax2)- and suppressing glutamatergic (Tlx3)-expressing cells in the chick and mouse neural tube. Moreover, we use an Rbpj conditional mutation to demonstrate that Rbpj function is essential for GABAergic specification, and that this function is independent of the Notch signaling pathway. Together, these findings demonstrate the requirement for a Ptf1a–Rbpj complex in controlling the balanced formation of inhibitory and excitatory neurons in the developing spinal cord, and point to a novel Notch-independent function for Rbpj in nervous system development.
[Keywords: Ptf1a, Notch, cerebellum, dorsal spinal cord, neuronal specification, bHLH transcription factor]
Normal nervous system function requires the correct balance of inhibitory and excitatory neurons. Alterations in this balance are thought to underlie diverse neurological disorders from epilepsy to autism to hyperalgesia. Although a handful of transcription factors have been shown to initiate the formation of these two major classes of neurons in multiple CNS regions, we have an incomplete understanding of the mechanisms that regulate this balance. In the cerebellum, dorsal spinal cord, and retina, the basic helix–loop–helix (bHLH) transcription factor Ptf1a (p48) plays a central role in the generation of inhibitory GABAergic neurons while suppressing the generation of excitatory glutamatergic neurons (Glasgow et al. 2005; Hoshino et al. 2005; Fujitani et al. 2006; Nakhai et al. 2007; Pascual et al. 2007). Here we demonstrate that Ptf1a utilizes a nonclassical bHLH transcription factor complex for these functions.
Inhibitory and excitatory interneurons in the dorsal horn of the spinal cord require the combined actions of bHLH and homeodomain (HD) transcription factors (Gowan et al. 2001; Gross et al. 2002; Müller et al. 2002; Cheng et al. 2004, 2005; Glasgow et al. 2005; Helms et al. 2005; Mizuguchi et al. 2006; Wildner et al. 2006). Neurogenesis in this CNS region is characterized by two waves: an early wave (embryonic day 10–11.5 [E10–E11.5]) that generates sensory relay and local interneurons that settle in the deep dorsal horn (dI1–dI6), and a late wave (E11.5–E13) that generates interneurons in the superficial laminae (dILA and dILB) (for reviews, see Caspary and Anderson 2003; Helms and Johnson 2003). Specification of the major GABAergic inhibitory interneurons dI4 and dILA requires Ptf1a (Glasgow et al. 2005) and the HD factors Lbx1 and Pax2 (Cheng et al. 2004, 2005). In contrast, Ptf1a represses the HD factor Tlx3, a factor required for specification of the major glutamatergic excitatory interneurons dI5 and dILB. Another bHLH factor, Ascl1 (previously Mash1), has a more complex role in specifying these neurons. In early stages it is required with the HD factors Gsh1 and Gsh2 for dI5 glutamatergic neurons (Helms et al. 2005; Mizuguchi et al. 2006), but in the late-born dIL populations it is required for Ptf1a expression and, thus, for the dILA GABAergic neurons (Mizuguchi et al. 2006; Wildner et al. 2006). The central role played by Ptf1a in guiding inhibitory versus excitatory neuronal specification makes it an attractive target for studies revealing the molecular mechanisms controlling neuronal network formation.
Insight into the mechanism of Ptf1a function has come from studies of transcriptional control of pancreatic digestive enzymes in the adult pancreas (Beres et al. 2006) and the mapping of human mutations in the PTF1A gene (Sellick et al. 2004). The classical form for a class II bHLH factor such as Ptf1a is as a heterodimer with E-protein (for review, see Massari and Murre 2000). However, Ptf1a was identified as a component of the pancreas transcription factor complex (PTF1) that is required for expression of pancreatic enzymes such as Elastase1 (Cockell et al. 1989; Krapp et al. 1996, 1998). This protein complex includes Ptf1a with any one of the E-proteins, Tcfe2a (E12 and E47), Tcf4 (E2.2), or Tcf12 (HEB), and Rbpjl, a homolog of Drosophila Suppressor of Hairless (Beres et al. 2006). Human mutations in PTF1A that resulted in cerebellar and pancreatic agenesis (Hoveyda et al. 1999; Sellick et al. 2004) were identified as truncations that left the bHLH domain intact but deleted the Rbpjl interaction domain (Beres et al. 2006). Together, these results suggest the trimeric complex with Rbpjl is required for normal pancreas. However, it is Rbpj (RBP-Jk, CBF1, CSL, Rbpsuh), not Rbpjl, that is present in the developing neural tube and cerebellum. In vitro studies have shown Rbpj can also form a complex with Ptf1a and E-protein (Beres et al. 2006).
Rbpj is the vertebrate ortholog of the Drosophila gene Suppressor of Hairless, and is a DNA-binding factor essential in transducing Notch signals during neural development. Rbpj binds to its consensus site DNA in regulatory regions of target genes and mediates the transcriptional effects of Notch signaling by associating with the protease-cleaved Notch intracellular domain (NICD) and coactivators such as Maml (Mastermind-like) (for review, see Louvi and Artavanis-Tsakonas 2006). For example, Rbpj binds and activates transcription from the promoter of the Hes1 gene in the presence of activated Notch, a mechanism for inhibiting neuronal differentiation (Jarriault et al. 1995). In contrast, in the absence of Notch signaling, Rbpj can recruit corepressors such as the LIM-protein KyoT2, or SMRT and N-CoR, components of histone deacetylase corepressor complexes (Kao et al. 1998; Taniguchi et al. 1998). Extensive studies in vertebrates and invertebrates have demonstrated Rpbj functions via Notch signaling in many processes during neural development, including neural progenitor cell maintenance and promoting the generation of astrocytes (Furukawa et al. 2000; Gaiano et al. 2000; Hitoshi et al. 2002).
In this study, we test the requirement for a transcription complex containing both Ptf1a and Rbpj in determining the neuronal identity of GABAergic neurons in the dorsal spinal cord. We demonstrate that a mutant, Ptf1aW298A, is capable of forming a heterodimer with the E-protein Tcf12 but is incapable of forming the heterotrimer with the E-protein plus Rbpj. This mutant has no activity in inducing Ptf1a-dependent neurons when overexpressed in the chick neural tube, an activity normally detected with wild-type Ptf1a (Wildner et al. 2006). Furthermore, in mice with Ptf1aW298A replacing the wild-type protein, the phenotype in the dorsal neural tube and cerebellum is identical to the Ptf1a null. Conditional knockout of Rbpj in the dorsal neural tube phenocopies the Ptf1aW298A/W298A in specifying dorsal neuronal identity. Thus, we provide in vivo evidence implicating a protein complex of Ptf1a with Rbpj as the essential transcription complex for GABAergic neuronal development in multiple regions of the nervous system.
Results
Ptf1aW298A efficiently forms a heterodimer with the E-protein Tcf12 but does not interact with Rbpj
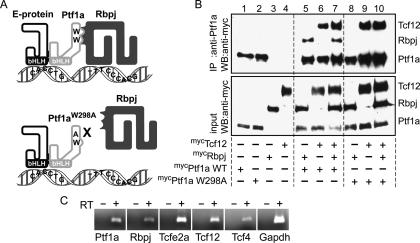
PTF1 is a transcription factor complex identified in adult pancreas (Roux et al. 1989; Beres et al. 2006). In pancreas, this complex consists of Ptf1a, Rbpjl, and any one of the E-proteins such as Tcf12. However, although Ptf1a and E-proteins are present in the developing neural tube, Rbpjl is not (R.M. Henke, unpubl.). Biochemical studies demonstrated that a PTF1 complex could contain Rbpj in place of Rbpjl (Fig. 1A; Beres et al. 2006). Using RT–PCR from isolated Ptf1a lineage cells, we show that Rbpj is present with the other PTF1 complex components, including Ptf1a, Tcf12, Tcf4, and Tcfe2a, in E12.5 neural tube (Fig. 1C). To begin to address whether the functional form of Ptf1a in the nervous system requires its interaction with Rbpj, we took advantage of a mutant form of Ptf1a that disrupts this interaction. Ptf1aW298A contains a single amino acid substitution of a conserved tryptophan in the C terminus of the protein, and it fails to function efficiently with Rbpj to activate transcription in reporter assays (Beres et al. 2006). Furthermore, in contrast to the wild-type Ptf1a (Fig. 1B, lanes 5–7), Ptf1aW298A does not coimmunoprecipitate Rbpj when these proteins are coexpressed in COS cells (Fig. 1B, lanes 8–10). Notably, Ptf1aW298A does retain its ability to interact with E-protein (Fig. 1A,B). These results indicate that the single amino acid substitution of alanine for tryptophan at position 298 specifically impairs the association of Ptf1a with Rbpj without impairing its ability to heterodimerize with E-protein.
Figure 1.
Ptf1aW298A mutant protein efficiently forms a heterodimer with an E-protein (Tcf12) but not with Rbpj. (A) The diagram depicts the PTF1 trimer on DNA containing an E-box and T/C-box. When the tryptophan (W) at position 298 in Ptf1a is mutated to an alanine (A), there is a loss of the trimer complex, but the dimer with an E-protein can form. (B) Immunoprecipitation (IP) of COS cells transfected with Ptf1a wild type, Ptf1aW298A mutant, Rbpj, or Tcf12 alone (lanes 1–4); cells cotransfected with Ptf1a wild type or Ptf1aW298A mutant with Rbpj (lanes 5,8), or Tcf12 (lanes 6,9); or cells triple-transfected (lanes 7,10). All transfected proteins were myc-tagged. The top panel shows cell lysates immunoprecipitated with anti-Ptf1a antibodies and immunoblotted (WB) with anti-myc antibodies. Equal input of proteins in each sample is shown in Western blot with anti-myc antibodies in the bottom panel. (C) RT–PCR from RNA isolated by FACS from E12.5 Ptf1aCre/+/ROSA26YFP E12.5 neural tubes demonstrating coexpression of the PTF1 trimer components in dorsal neural tube. (RT) Reverse transcriptase.
Ptf1aW298A fails to induce Pax2 (dILA neurons) and suppress Tlx3 (dILB neurons) in chick neural tube
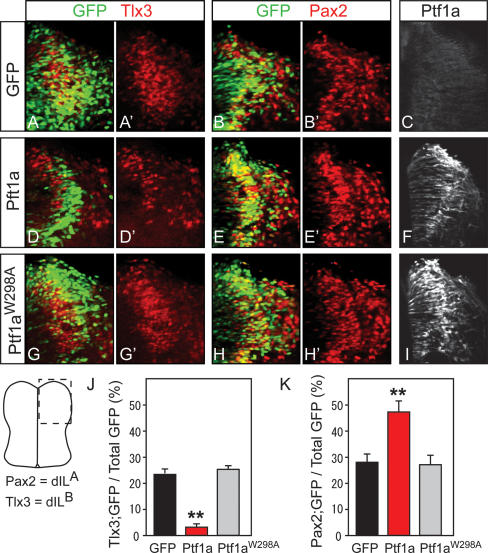
Ectopic expression of Ptf1a can induce the generation of dILA (Pax2+) and suppression of dILB (Tlx3) neurons in chick neural tube (Wildner et al. 2006). To test if these functions of Ptf1a depend on the interaction of Ptf1a and Rbpj, we performed in ovo electroporation experiments with the mutant Ptf1aW298A. Electroporation of HH26 chick neural tubes with vectors expressing GFP and GFP plus Ptf1a or Ptf1aW298A were analyzed at HH29–HH30 (48 h later) for Pax2 and Tlx3 as markers for dILA and dILB neurons, respectively (Fig. 2). Cells expressing the ectopic wild-type Ptf1a were more likely to express Pax2 (dILA) (48% vs. 29%) and less likely to express Tlx3 (dILB) (2% vs. 23%) when compared with control electroporated cells (Fig. 2A–F, quantified in J,K). In contrast, expression of Ptf1aW298A did not alter marker expression from that seen in controls (28% vs. 29% for Pax2 and 24% vs. 23% for Tlx3), indicating loss of activity of the protein (Fig. 2G–I). Ectopic expression of Ptf1a and Ptf1aW298A was confirmed by immunostaining the electroporated chick neural tubes using Ptf1a-specific antibodies (Fig. 2F,I). These results demonstrate the requirement for the tryptophan residue in the ability of Ptf1a to alter neuronal fate, and suggest that the interaction of Ptf1a and Rbpj is required for Ptf1a function in inducing dILA and suppressing dILB neurons in the neural tube.
Figure 2.
Ptf1a induction of Pax2 and suppression of Tlx3-expressing neurons in chick neural tube requires the interaction with Rbpj. Chick neural tubes were electroporated at HH26 with constructs expressing GFP (A–C), Ptf1a and GFP (D–F), or Ptf1aW298A and GFP (G–I), and the effects on neural specification were assessed at HH29–HH30 using GFP fluorescence (green) and antibodies against Tlx3 (red in A,A′,D,D′,G,G′) and Pax2 (red in B,B′,E,E′,H,H′). (C,F,I) Ectopic mouse Ptf1a was detected with guinea pig anti-Ptf1a antibody that does not recognize chick Ptf1a. The proportion of Tlx3+;GFP+ cells (J) and Pax2+;GFP+ cells (K) after electroporation of expression constructs for control GFP only (black), Ptf1a plus GFP (red), or Ptf1aW298A plus GFP (gray) was determined in the dorsal half of the electroporated side of the neural tube (see diagram for region where images were taken). Four embryos were analyzed per construct, and selected sections were between fore- and hindlimb levels in regions with GFP+ cells. (**) P < 0.001.
A Ptf1a that can interact with Rbpj is required in vivo for the formation of GABAergic neurons in the dorsal horn of the spinal cord and the cerebellum
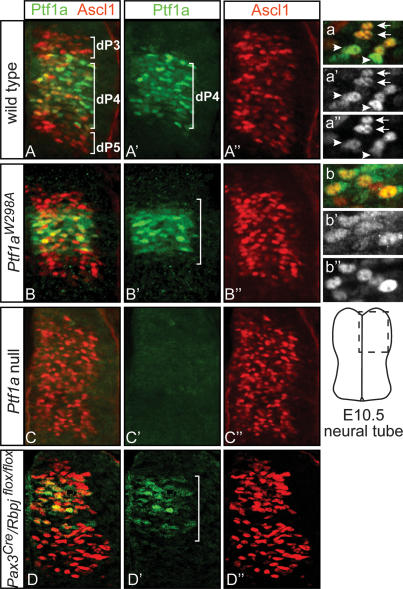
To identify the contribution in vivo of the Ptf1a–Rbpj interaction to normal dorsal spinal cord development, we examined mutant mice that express the mutant form Ptf1aW298A (Masui et al. 2007). Similar to the null mutant Ptf1aCre/Cre (Kawaguchi et al. 2002), these animals die as neonates and lack a pancreas (Masui et al. 2007). In our initial characterization of the neural tube in these mutants, we used coimmunofluorescence with anti-Ptf1a and anti-Ascl1 antibodies to examine if the mutant protein was made and present in the correct domain. In E10.5 neural tubes, Ptf1a is restricted to progenitors of dI4 interneurons (dP4), whereas Ascl1 is present in progenitors to dI3, dI4, and dI5 interneurons (Fig. 3A–A″, dP3, dP4, dP5). The Ptf1aW298A protein in mutants homozygous for the Ptf1aW298A allele is detected, and although it appears more diffuse within the cells and is at slightly lower levels, it is present in the dI4 progenitor domain similar to the wild-type protein (Fig. 3B, B′). The absence of detectable Ptf1a in the Ptf1a-null neural tubes demonstrates the specificity of the antibody (Fig. 3C, C′).
Figure 3.
Ptf1aW298A is detected in the progenitors to dI4 neurons but fails to repress Ascl1 levels. Immunofluorescence on neural tube transverse sections of wild-type (A–A″), homozygous Ptf1aW298A (B–B″), Ptf1a-null (C–C″), and Pax3Cre/Rbpjflox/flox (D–D″) mouse E10.5 embryos. Wild-type Ptf1a and Ptf1aW298A protein (green) is present in dI4 progenitors (dP4), although the number of positive cells in Ptf1aW298A/W298A and Pax3Cre/ Rbpjflox/flox neural tubes (B′,D′) appears slightly decreased relative to that of wild type (A′). (A″–D″) Ascl1 (red) levels appear to increase in the dI4 progenitor domain in all the mutant neural tubes. (a′,b′) In high-magnification views, Ptf1aW298A is localized mainly to the nucleus, though its pattern is slightly disrupted compared with wild type. (a–a″) The level of Ptf1a in a cell is complementary to the level of Ascl1 in wild type. (b–b″) This pattern is disrupted between Ptf1aW298A and Ascl1. Arrows and arrowheads indicate Ascl1high/Ptf1alowest cells and Ascl1lowest/Ptf1ahigh cells, respectively.
A first indication of a neural tube phenotype was the apparent increase in Ascl1 levels in the dI4 progenitor domain relative to that seen in wild type. Normally at this stage, Ascl1 levels are complementary to Ptf1a levels in that they are generally low in the dI4 progenitors but high in the dI3 and dI5 progenitors (Glasgow et al. 2005). Within individual cells in the dI4 progenitor domain, cells with highest Ptf1a have the lowest levels of Ascl1 (Fig. 3a–a″, arrowheads), and vice versa (Fig. 3a–a″, arrows). In contrast, in both the null Ptf1a and the variant Ptf1aW298A, Ascl1 levels were more uniform across dI3, dI4, and dI5 progenitor domains (Fig. 3B″–C″), and individual cells with or without Ptf1aW298A protein have indistinguishable levels of Ascl1 (Fig. 3b–b″). This increase in Ascl1 levels does not appear to be a consequence of loss of Notch signaling, since we detect no decrease in the levels of the Notch target Hes5 (data not shown). The results suggest Ptf1aW298A has lost the ability to modulate Ascl1 levels.
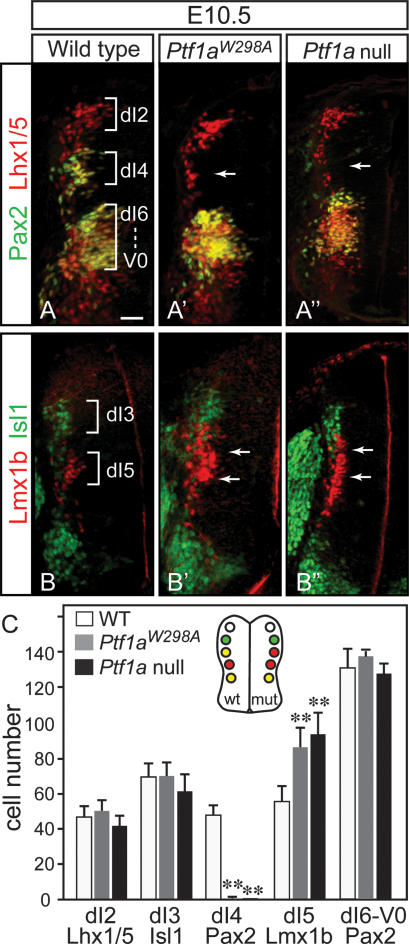
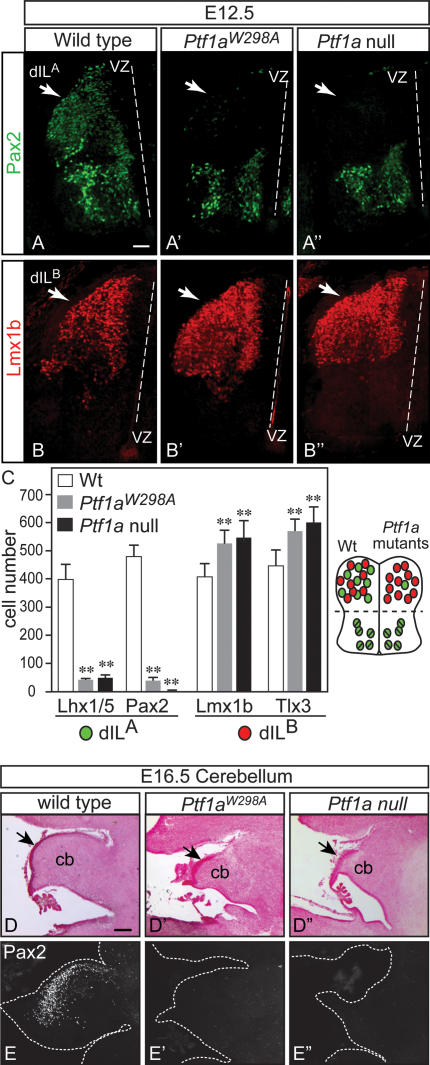
The E10.5 dorsal neural tube has six populations of neurons defined by combinations of HD transcription factor expression (Helms and Johnson 2003). We analyzed five of these populations, comparing the wild type, the homozygous Ptf1aW298A, and the Ptf1a null. Just as for the Ptf1a null (Glasgow et al. 2005), in Ptf1aW298A/W298A dorsal neural tube, the dI4 cells defined by location and coexpression of Pax2 and Lhx1/5 were absent (Fig. 4A–A″, arrows), whereas the dI5 population, marked by Lmx1b, expanded dorsally into the dI4 domain (Fig. 4B–B″, arrows). In contrast, no significant change in cell number was observed in the dI2 (Lhx1/5), dI3 (Isl1), or dI6-V0 (Pax2/Lhx1/5) domains (see cell counts in Fig. 4C). Thus, the heterodimer of Ptf1a with E-protein is not sufficient to specify dI4 neurons and suppress dI5, implicating the requirement for the Ptf1a complex with Rpbj.
Figure 4.
Dorsal interneurons dI4 are lost in mutants homozygous for Ptf1aW298A. Immunofluorescence on transverse sections of E10.5 neural tubes from wild-type (A,B), homozygous Ptf1aW298A (A′,B′), and Ptf1a-null (A″,B″) embryos (only left half of neural tube is shown). (A–A″) dI4 neurons (yellow) identified by location and colabeling with Pax2 (green) and Lhx1/5 (red) are lost in the Ptf1aW298A mutant and the Ptf1a null (arrows). There is no significant change in dI2 neurons identified by Lhx1/5 (red) or dI6-V0 neurons Pax2;Lhx1/5 (yellow ventral to dI4). (B–B″) dI5 neurons identified by Lmx1b (red) are expanded dorsally into the dI4 domain in Ptf1aW298A mutants, as is seen in Ptf1a null (arrows), whereas dI3 and motor neurons marked by Isl1 (green) are not affected. These data are quantified in C from sections in the upper limb level in at least four embryos of each genotype. The neural tube diagram summarizes the changes in neuronal populations detected in the Ptf1a mutants relative to wild type. (**) P < 0.001. Bar, 50 μm.
In the second wave of neurogenesis in the dorsal neural tube, two major interneuron populations arise out of a common progenitor domain. At E12.5, these neurons, dILA and dILB, are marked by Pax2 and Lhx1/5 (dILA), and Tlx3 and Lmx1b (dILB) (Helms and Johnson 2003). At E12.5, Ptf1aW298A/W298A mutant neural tubes were found to have a dramatic reduction in dILA neurons and an increase in dILB compared with wild type (Fig. 5). This is illustrated as a loss of Pax2- and Lhx1/5-positive cells (Fig. 5A, A′, counts in C), and an increase in Lmx1b- and Tlx3-positive cells (Fig. 5B, B′, counts in C). Furthermore, it has been demonstrated that the dILA neurons identified by Pax2 and Lhx1/5 become GABAergic neurons in the dorsal horn and that dILB neurons identified by Lmx1b and Tlx3 become glutamatergic (Cheng et al. 2004). Thus, we examined E16.5 mutant spinal cords for expression of GABAergic (Pax2, GABA) and glutamatergic (Tlx3,VGLUT2, GluR2/3) markers. Again, as was reported for the Ptf1a null (Glasgow et al. 2005), there was a dramatic loss of cells expressing the neurotransmitter GABA and the HD transcription factor Pax2 in the dorsal horn of the Ptf1aW298A/W298A compared with wild-type spinal cords (Supplemental Fig. 1A–B′). Conversely, these mutants have an increase over wild type in cells with markers associated with the glutamatergic phenotype (Supplemental Fig. 1C–E′). These results demonstrate the Ptf1aW298A/W298A mutant phenotype is nearly identical to the Ptf1a null in E10.5, E12.5, and E16.5 spinal neural tube. Thus, the reported role of Ptf1a as a switch between major GABAergic and glutamatergic neurons in the dorsal spinal cord is mediated through a Ptf1a protein that requires the ability to complex with Rbpj. The Ptf1a/E-protein heterodimer is not sufficient to control GABAergic identity.
Figure 5.
Dorsal interneurons dILA are lost in Ptf1aW298A mutants. Immunofluorescence on transverse sections of E12.5 neural tubes from wild-type (A,B), homozygous Ptf1aW298A (A′,B′), and Ptf1a-null (A″,B″) embryos. Only the left half of the neural tube is shown (dashed line indicates the position of the ventricle). (A–A″) Pax2 identifies dILA neurons (green), which are essentially absent in Ptf1aW298A and Ptf1a-null embryos specifically in the dorsal neural tube. (B–B″) dILB neurons, marked by Lmx1b (red), increase in both the Ptf1aW298A and Ptf1a-null embryos. These data plus those for Lhx1/5 (dILA marker) and Tlx3 (dILB marker) are quantified in C from sections in the upper limb level in six embryos of each genotype. (**) P < 0.001. The neural tube diagram summarizes the changes in neuronal populations detected in the Ptf1a mutants relative to wild type. (D–E″) Sagittal sections of wild-type (D,E), Ptf1aW298A (D′,E′), or Ptf1a-null (D″,E″) mouse E16.5 cerebellar primordium (cb) are shown with nuclear fast red histology demonstrating the decreased size of the cerebellum (cb), the thickening of the EGL (arrow) in the mutants, and immunofluorescence detecting the GABAergic marker Pax2 (E–E″). Dashed white line indicates tissue border. Bars: A–B″, 50 μm; D–E″, 150 μm.
Ptf1a is also required for GABAergic neurons in cerebellum in mouse and human (Sellick et al. 2004; Hoshino et al. 2005; Pascual et al. 2007). Histological analysis of sagittal sections at E16.5 showed aberrant morphology of the cerebellar primordia in Ptf1aW298A mutant mice reminiscent of that reported for the null allele of Ptf1a (Fig. 5D–D″). The E16.5 Ptf1aW298A/W298A mutant cerebellum is smaller than wild type with a thickening of the Atoh1-expressing external germinal layer that contains progenitors to the major excitatory interneuron population in the cerebellum, the granule neurons (Supplemental Fig. 2A–B″, arrows). The decreased size of the E16.5 cerebellum is largely due to a dramatic reduction of GABAergic neurons as determined by decreased immunoreactivity using antibodies to the transcription factor Pax2, to the neurotransmitter GABA, and to Calbindin, a marker for Purkinje cells, just as seen in the Ptf1a null (Fig. 5E–E″; Supplemental Fig. 2C–E″). In contrast to the loss of GABAergic neurons in both mutants, cells expressing Tbr1 (Supplemental Fig. 2F–F″), a marker for progenitors of glutamatergic neurons in deep cerebellar nuclei, are still present (Fink et al. 2006). Taken together, the ability of Ptf1a to interact with Rbpj appears critical for the specification of GABAergic neurons not only in the developing spinal cord but also the cerebellum.
Rbpj is required specifically for GABAergic interneurons dI4 and dILA independent of its function as the effector of Notch signaling
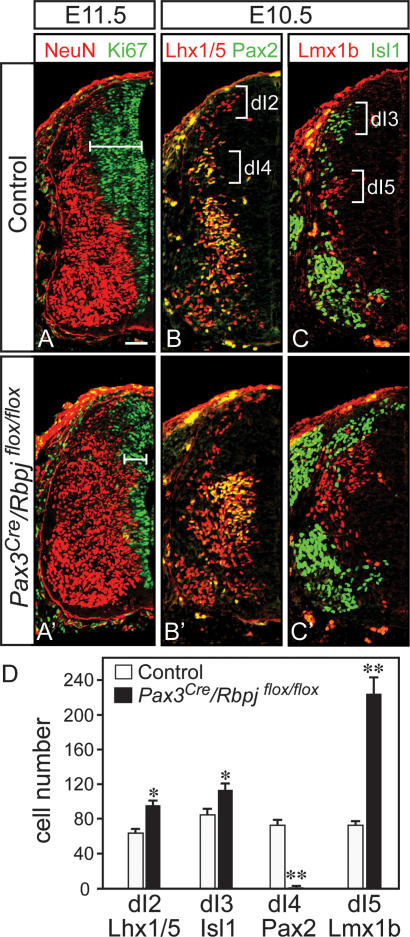
The phenotype in the Ptf1aW298A/W298A embryos demonstrates that the tryptophan 298 residue is essential for all described function of Ptf1a, and implicates Rbpj as an obligate cofactor. However, the known function of Rbpj is as the transcriptional effector of Notch signaling, which acts in the nervous system to inhibit neuronal differentiation (de la Pompa et al. 1997). To address the role of Rbpj in determining the identity of GABAergic interneurons in the dorsal spinal cord, we employed conditional mutagenesis of Rbpj (Tanigaki et al. 2002). A Pax3Cre driver line that induces efficient recombination in the dorsal neural tube from E9 was used to delete Rbpj prior to the early neurogenic phase (Engleka et al. 2005). Pax3Cre/Rbpjflox/flox embryos have a decreased progenitor domain in the dorsal neural tube visualized with anti-Ki67, and precocious neuronal differentiation visualized with NeuN, relative to control embryos at E10.5 and E11.5 (Fig. 6A, A′). This result is in accordance with previous reports that demonstrate an important role for Notch signaling in the maintenance of neural progenitors (de la Pompa et al. 1997). To ascertain whether Rbpj also functions in specification of dI4 neurons, we examined the composition of the various dorsal neuron types at E10.5 in the Pax3Cre/Rbpjflox/flox embryos. Mutant mice suffer a complete loss of dI4 interneurons, which were identified by their position in the dorsal mantle zone and by their coexpression of Pax2 and Lhx1/5 (Fig. 6B, B′, see D for quantification). This was accompanied by an increase in the number of dI2 (Lhx1/5), dI3 (Isl1/2), and dI5 (Lmx1b/Tlx3) neurons (Fig. 6B–D). The number of dI2 and dI3 neurons was slightly increased in mutant mice, reflecting an overall increase in neurogenesis in the dorsal spinal cord. In contrast, the increase in the number of dI5 neurons was dramatic in the mutant embryos relative to control (Fig. 6C,D). This excess in the dI5 population accompanied by the absence of dI4 suggests the dI4 neurons assume dI5 identity in the absence of Rbpj. The combination of the loss of dI4 neurons with the increase in dI5 mimics the phenotype seen in Ptf1a mutants (Fig. 4; Glasgow et al. 2005). The loss of dI4 neurons in Pax3Cre/Rbpjflox/flox embryos occurred even though Ptf1a was present in dorsal progenitors (Fig. 3D). Notably, the changes in Ptf1a and Ascl1 levels in the Pax3Cre/Rbpjflox/flox mutants appear similar to those detected in the Ptf1aW298A/W298A mutants (Fig. 3, cf. B–B″ and D–D″). Taken together, the similarity in phenotype between the Ptf1a and Rbpj mutants provides in vivo evidence supporting the requirement of the Ptf1a–Rbpj complex in determining neuronal identity in dI4 versus dI5 interneurons.
Figure 6.
Rbpj is required to determine the identity of dI4 neurons. Immunofluorescence on transverse sections of E10.5 or E11.5 neural tubes from Control (Pax3Cre/Rbpjflox/+) (A–C) and mutant Pax3Cre/Rbpjflox/flox (A′–C′) embryos (only the left half of neural tube is shown). (A,A′) A decrease in the progenitor domain (indicated by the bar) and an increase in the neuronal domain in the dorsal neural tube of the Pax3Cre/Rbpjflox/flox mutants were detected using Ki67 (green) and NeuN (red), respectively. (B,B′) dI2 (Lhx1/5, red) are increased while dI4 (Pax2/Lhx1/5, yellow) are lost in the Pax3Cre/Rbpjflox/flox mutant. (C,C′) dI3 (Isl1, green) and dI5 (Lmx1b, red) neurons are both increased in the Pax3Cre/Rbpjflox/flox relative to controls. The dramatic increase in dI5 neurons and the loss of dI4 neurons suggests that dI4 assume the identity of dI5 neurons in the absence of Rbpj. These data are quantified in D from three embryos each using sections from the upper limb level. Error bars represent standard error. (*) P < 0.05; (**) P < 0.001. Bar, 50 μm.
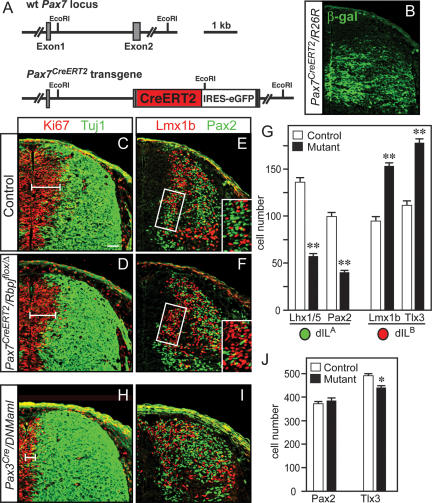
The dorsal progenitor domain of Pax3Cre/Rbpjflox/flox mice was depleted by E12, which precluded further investigation of the role of Rbpj in the generation of dILA and dILB neurons (data not shown). To assess the function of Rbpj during this second neurogenic phase, we generated a BAC transgenic mouse strain that expresses a tamoxifen-inducible Cre recombinase under the control of the Pax7 locus (Fig. 7A). Recombination induced by the Pax7CreERT2 transgene was investigated using the ROSA26R reporter line that expresses β-galactosidase after Cre-mediated recombination (Soriano 1999). After administration of tamoxifen at E10.5, efficient recombination was observed in the ventral part of the alar plate by E12.5 (Fig. 7B). We detected no obvious depletion of the progenitor domain in the Pax7CreERT2/RbpjΔ/flox mutant mice at E12.5, 2 d after tamoxifen administration (Fig. 7C,D), although depletion of progenitors was observable at E13.5 (data not shown). However, there was a significant reduction in the number of newborn dILA neurons (Pax2 and Lhx1/5), while the number of dILB neurons (Lmx1b and Tlx3) was increased (Fig. 7E,F, see G for quantification). Taken together, these results demonstrate a role for Rbpj in determining the identity of the GABAergic neurons dI4 and dILA, while suppressing the identity of the glutamatergic neurons dI5 and dILB, with little effect on the other dorsal interneuron cell types.
Figure 7.
Rbpj is required to determine the identity of dILA neurons independent of its role in Notch signaling. (A) Diagram of the Pax7CreERT2-BAC transgenic mouse strain used to delete Rbpj during the second neurogenic phase of neural tube development. (B) β-gal expression in a Pax7CreERT2/ROSA26R E12.5 embryo treated with tamoxifen at E10.5 demonstrates active Cre recombination in the dorsal neural tube. Immunofluorescence images on E12.5 neural tubes after 10.5-d-post-coitum (dpc) tamoxifen administration of control (C,E) and Pax7CreERT2/Rbpjflox/Δ (D,F), or Pax3Cre/DNMaml (H,I) mutants. Antibodies used include Ki67 (red) and Tuj1 (green) to distinguish progenitor and differentiation domains (C,D,H), or Pax2 (green) and Lmx1b (red) to distinguish dILA and dILB populations (E,F,I). (E,F) The boxed areas are shown at higher magnification in the insets and indicate the domain that was quantified for the number of dILA and dILB neurons (shown in G). This domain was used since Cre recombination induced by tamoxifen at 10.5 dpc was efficient in this region (see B) and it contains newly born neurons. (J) The number of dILA (Pax2) and dILB (Tlx3) cells in Pax3Cre/DNMaml embryos relative to control was quantified in the entire dorsal and neural tube, reflecting the recombination efficiency by Pax3Cre. Data quantified in G and J are from three embryos each using sections from the upper limb level. Error bars represent standard error. (*) P < 0.05; (**) P < 0.001. Bars: 50 μm; insets in E,F, 20 μm.
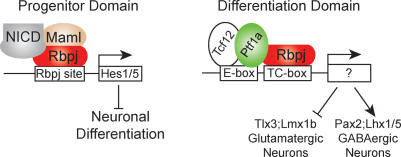
In order to separate the role of Rbpj in the Ptf1a complex versus its role in the Notch signaling pathway, we used a mouse strain that expresses, after Cre-induced recombination, a dominant-negative variant of Mastermind-like (DNMaml) under the control of the ROSA26 locus (Tu et al. 2005). Maml factors act with NICD to form a transcription activation complex with Rbpj. The dominant-negative Maml lacks its activation domain and is thought to block Notch signaling by forming an inactive complex with NICD and Rbpj (Tu et al. 2005). We analyzed the dorsal spinal cord of control and mutant mice (Pax3Cre/DNMaml) at E12.5 with Ki67 and the neuronal marker Tuj1, and observed a partial depletion of the progenitor domain in mutants when compared with control, consistent with a reduction of Notch signaling (Fig. 7C,H). However, in contrast to changes seen in the Rbpj mutants, there was no loss of dILA (Pax2) neurons. Instead, there was a minor decrease in the dILB population (Fig. 7I, quantified in J). This decrease in dILB is consistent with a previous report that demonstrated Notch signaling biases the dIL progenitor to a dILB fate (Mizuguchi et al. 2006). These results demonstrate two functions for Rbpj in dorsal neural tube development. Rbpj acts both in a complex with Ptf1a and an E-protein to specify dI4 and dILA neurons, and independently, in a complex with Maml, to transduce Notch signals that maintain neural progenitors and suppress neuronal differentiation (Fig. 8).
Figure 8.
Distinct functions for the two independent Rbpj-containing transcription complexes. Diagram depicting the two Rbpj-containing complexes. Rbpj with the NICD and Maml enhance expression of Hes1 and Hes5, which in turn inhibit neuronal differentiation and maintain the progenitor domain. DNA binding is through a consensus binding site for Rbpj. The PTF1 complex containing Rbpj with Ptf1a and an E-protein such as Tcf12 is distinct and binds a bipartite sequence that includes an E-box plus a TC-box. The TC-box is similar to the Rbpj consensus site, but not all TC-boxes serve as Rbpj-binding sites (Beres et al. 2006). The PTF1 transcription factor complex determines neuronal identity by inducing GABAergic-specific genes and suppressing glutamatergic-specific genes.
Discussion
Requirement for a Ptf1a–Rbpj complex in development of specific GABAergic neuronal populations
Ptf1a is an unusual bHLH transcription factor. Like other class II bHLH factors, it heterodimerizes with an E-protein such as Tcf12 and binds the hexameric E-box DNA sequence. What is unique to Ptf1a is that it also can form a trimeric complex by direct binding to Rbpj or Rbpjl, forming a complex that binds a compound DNA sequence that includes the E-box plus a TC-box, a consensus Rbpj site (Fig. 1A; Beres et al. 2006). In this study, we provide in vivo evidence that a Ptf1a–Rbpj complex is essential in neural development for determining the identity of specific populations of GABAergic neurons while suppressing neighboring glutamatergic neurons. First, embryos homozygous for the Ptf1aW298A allele that encodes a Ptf1a variant deficient in Rbpj binding fail to generate GABAergic neurons in the dorsal spinal cord (dI4 and dILA) and cerebellum. Second, in contrast to wild-type Ptf1a, the Ptf1aW298A mutant protein cannot induce the generation of dILA (Pax2) and the suppression of dILB (Tlx3) neurons when overexpressed in the chick neural tube. Third, conditional loss of Rbpj demonstrates its requirement in determining the identity of dI4 and dILA neurons but not the other dorsal neuronal populations. And fourth, in both the Ptf1aW298A mutants and the conditional loss of Rbpj, the absence of GABAergic neurons is countered by an increase in the neighboring glutamatergic populations. Thus, we demonstrate that a unique transcription complex containing Ptf1a and Rbpj is the functional complex in vivo for determining the balance of inhibitory and excitatory neurons in the dorsal spinal cord. The classic form of the bHLH heterodimer with E-protein is not sufficient to replace the Ptf1a–Rbpj function. And finally, the requirement for Rbpj in the Ptf1a–Rbpj complex is unique for Rbpj biology in that it is distinct from its role in transducing Notch signals.
Human mutations in PTF1A support the conclusion that in vivo the functional complex requires PTF1A–Rbpj protein–protein interaction. The human mutations of PTF1A result in pancreatic agenesis and cerebellar hypoplasia (Hoveyda et al. 1999; Sellick et al. 2004). These mutations resulted in C-terminal-truncated PTF1As that leave the bHLH domain intact but delete the tryptophan-containing peptide region shown in mouse to be required for Ptf1a–Rbpj complex formation. Originally, it was suggested that the human phenotype was due to the loss of bHLH heterodimer formation or stability. However, it was subsequently shown that the truncated human proteins could still form heterodimers with E-protein but not heterotrimers with Rbpj or Rbpjl, and they were devoid of transcriptional activity in cell transfection assays (Beres et al. 2006). Therefore, as in the Ptf1aW298A mouse mutant analyzed here (and in Masui et al. 2007), the human mutations disrupt formation of the PTF1A–RBPJ complex, resulting in the pancreatic and cerebellar phenotypes reported.
Transcription factor control of neuronal cell type identity in the dorsal spinal cord
We demonstrate that Ptf1a and Rbpj are essential for the transcriptional control of genes that generate the balance of inhibitory and excitatory neurons. The Ptf1a–Rbpj complex along with Ascl1 controls neuronal specification upstream of HD transcription factors such as Pax2, Lhx1/5, Tlx3, and Lmx1b. In particular, Ptf1a is essential for Pax2 expression (dI4) in the dorsal neural tube, whereas Ascl1, with Gsh1/2, is required for Tlx3 (dI5) (Glasgow et al. 2005; Helms et al. 2005; Mizuguchi et al. 2006). In the absence of Ptf1a, the dI4 neurons assume a dI5-like identity (Glasgow et al. 2005). This increase in dI5-like neurons may be a consequence of the up-regulation of Ascl1 that occurs in cells that have lost Ptf1a–Rpbj function, shown here in the Ptf1aW298A and Pax3Cre/Rbpjflox/flox mutants (Fig. 3). Thus, in this early phase of dorsal spinal cord development, part of Ptf1a–Rbpj function is to cell-autonomously suppress Ascl1 levels while inducing dI4-specific genes.
In the second neurogenic phase in the dorsal neural tube, neurons are generated from a progenitor domain that has Ptf1a- and Ascl1-expressing cells dispersed throughout the dorsal progenitor zone. The current model for balancing the generation of excitatory and inhibitory neurons in this region places Ascl1 in progenitor cells where it cell-autonomously regulates Ptf1a to promote the dILA lineage, or non-cell-autonomously to activate the Notch pathway that biases cells to the dILB lineage (Mizuguchi et al. 2006). Inhibition of Notch signaling through Rbpj and Maml in the dorsal neural tube resulted in a subtle decrease specifically in the dILB population, consistent with this model (Fig. 7J). However, the most dramatic effect on the balance of dILA and dILB populations was through a Notch-independent role for Rbpj in a complex with Ptf1a. In this case, dILA neurons take on the identity of dILB-like neurons and thus completely disrupt the balance of inhibitory and excitatory neurons. So although Notch signaling modulates the balance of the dILA and dILB lineages, Rbpj in a Notch-independent complex is essential for determining identity of the dorsal horn GABAergic neurons.
Ptf1a functions with Rbpj in multiple developmental pathways
Cerebellar development serves as another model for uncovering the molecular logic used to generate the correct balance of excitatory and inhibitory neurons in a neuronal network. Ptf1a plays a central role here as well, and similar to the dorsal spinal cord, Ptf1a is required for generation of the major populations of GABAergic neurons including Purkinje, basket, stellate, and golgi cells (Hoshino et al. 2005; Pascual et al. 2007). Ptf1a suppresses the glutamatergic phenotype, because in the Ptf1a-null mouse, Ptf1a lineage cells are at least partially misspecified to granule-like cells in the external granule layer (Pascual et al. 2007). The cerebellum in Ptf1aW298A mutant mice is similar in phenotype to the Ptf1a null. While we did not demonstrate a similar requirement for Rbpj in this region of the developing brain, we propose that the Ptf1a–Rbpj complex is a common mechanism for maintaining the balance of inhibitory and excitatory neurons wherever Ptf1a is expressed. However, the Ptf1a–Rbpj complex cannot be required for specification of all GABAergic neurons, because Ptf1a is not present in all brain regions containing these neurons (e.g., ventral spinal cord, tectum, and cortex [Glasgow et al. 2005]). Thus, other mechanisms for transcriptional control of the GABAergic phenotype must be in play. For example, Ascl1 functioning with the Hes-related bHLH factor Helt has been implicated in the GABAergic phenotype in the midbrain and cortex (Fode et al. 2000; Miyoshi et al. 2004; Guimera et al. 2006; Nakatani et al. 2007).
Ptf1a is not restricted to the developing nervous system; in fact, Ptf1a was originally identified as a key transcriptional regulator in pancreas (Krapp et al. 1996; Rose et al. 2001). In mature pancreas, Ptf1a forms a complex with an E-protein and Rbpjl that is required in acini for expression of secretory digestive enzymes (Beres et al. 2006). However, mouse knockouts of Ptf1a revealed that it functions earlier in the developing pancreas at a stage when Rbpjl is not present (Krapp et al. 1998; Kawaguchi et al. 2002). In a study performed in parallel with the study reported here, Ptf1a was shown to be in a complex with Rbpj and an E-protein early in pancreas development (Masui et al. 2007). The Ptf1aW298A variant was used to demonstrate that the interaction between Ptf1a and Rbpj is required for early functions controlling pancreatic growth, morphogenesis, and lineage fate decisions. Taken together, in each Ptf1a-expressing tissue examined, the functional form for this transcription factor is in a nonclassical bHLH complex, one that includes Rbpj or Rbpl, and is independent of Notch.
Two distinct roles for Rbpj in dorsal neural tube development
Rbpj has been described as a bifunctional protein that acts both as a transcriptional activator and repressor by associating with various cofactors, and it plays a pivotal role in control of cell fate decisions in the developmental programs of a variety of tissues (Honjo 1996). Although autoactivation of the Drosophila ortholog of Rbpj, Suppressor of Hairless, during development of mechanosensory organs has been reported as a Notch-independent function for Suppressor of Hairless, known molecular functions of Rbpj in vertebrates are through its function as the transcriptional regulator of Notch signaling (Barolo et al. 2000). In this role, Rbpj functions as a DNA-binding factor and mediates the transcriptional effects of Notch by associating with the protease-cleaved NICD and its coactivator Maml. Notch signaling through an Rbpj-dependent mechanism activates expression of the inhibitory bHLH factors Hes1 and Hes5 to suppress neuronal differentiation and maintain cells in a progenitor state (Fig. 8; Ohtsuka et al. 1999; Gaiano et al. 2000). Our data confirm that Rbpj, through transducing Notch signals, is required in this process since blocking the activation of the complex with dominant-negative Maml resulted in depletion of the progenitor domain and precocious neuronal differentiation (Figs. 6, 7).
Our data also describe a novel role for Rbpj in neural development distinct from its function in maintaining the neural progenitor state. Rbpj is required to determine the identity of GABAergic neurons over glutamatergic neurons in specific CNS regions, and this function is independent of the Notch signaling cascade. Although this function of Rbpj could involve its role as a repressor, our data with the Ptf1aW298A variant strongly implicate a Ptf1a–Rbpj1 complex. PTF1, the transcription complex comprising Ptf1a, Rbpj, and an E-protein, functions through regulating the expression of specific targets by recognition of its compound DNA-binding consensus containing an E-box and a TC-box (Beres et al. 2006). This transcription complex induces expression of GABAergic-specific genes such as Pax2, Lhx1/5, and GABA, while inhibiting expression of glutamatergic genes such as Tlx3, Lmx1b, and Vglut (Fig. 8). Which, if any, of these genes are direct targets is not yet known. Future studies defining target genes directly regulated by the PTF1 transcription complex will begin to provide a molecular framework for understanding function of this novel complex, and in turn address the important question of how neuronal networks are generated during early development.
Materials and methods
Mouse strains
Ptf1aCre (p48Cre) mutant mice have been described previously (Kawaguchi et al. 2002) and are used here as the Ptf1a null. Rbpjflox (Tanigaki et al. 2002), Pax3Cre (Engleka et al. 2005), DNMAML (Tu et al. 2005), and ROSA26R (Soriano 1999) mice have been described previously. Rbpjflox/+ mice were used to generate the RbpjΔ allele by the use of a Cre-deleter strain (Schwenk et al. 1995).
Ptf1aW298A mutant mice were generated by recombinase-mediated cassette exchange (Feng et al. 1999) using a two-step staggered selection strategy (Long et al. 2004) to insert the single-codon W298A mutation into the endogenous Ptf1a locus using a mouse embryonic stem (ES) cell line that was previously engineered to contain a loxed cassette acceptor allele (M.A. Magnuson, unpubl.). ES cells containing the mutated Ptf1a gene sequences were then injected into blastocysts (Masui et al. 2007). A Frt-flanked hygromycin resistance cassette was removed by crossing to a FlpE-expressing transgenic mouse (Rodriguez et al. 2000) prior to analysis. Genotyping for Ptf1aW298A mutant mice was performed using a PCR protocol that distinguishes the 2-base-pair (bp) difference (TGG) to (GCG) using the following primers: 5′-TCATCCGTACAGCTAAAGTGTG-3′ and 5′-TCTGTCAAAGGTGCTTCAGG-3′ for the wild-type Ptf1a locus (∼300 bp), and 5′-TCATCCGTACAGCTAAAG TGGC-3′ and 5′-CGGAGTTTCCTGGACAGAGT-3′ for the Ptf1aW298A mutant locus (∼300 bp).
The Pax7CreERT2 mice were generated as BAC transgenics using pronuclear injection. A 185-kb BAC clone RP23-387L10 (RZPD) containing the Pax7 locus was modified by homologous recombination in bacteria (Lee et al. 2001) using a cassette encoding the entire sequence of CreERT2 (containing the initiating ATG codon) and IRES-eGFP-pA designed to replace the second exon of Pax7. Exon 1 was modified to remove possible ATG codons. The linearized Pax7CreERT2-BAC was injected into pronuclei of fertilized eggs. Four transgenic founders were obtained and screened for Cre-induced recombination. The transgenic founder that induced the highest recombination rate in the spinal cord was selected. Genotyping of Pax7CreERT2 transgenic mice was performed using the following primers: 5′-CTC CCCCACACTAACTGCAT-3′ and 5′-ATGTTTAGCTGGCC CAAATG-3′ giving a PCR product of 380 bp.
Embryos were staged based on assumed copulation at E0, halfway through the dark cycle. Tamoxifen (Sigma-Aldrich) was dissolved at a concentration of 20 mg/mL in sunflower oil and administered to pregnant females at 100 mg/kg (Joyner and Zervas 2006). Embryos were dissected for analysis 48 h after tamoxifen treatment.
Plasmid construction
All chick electroporations and COS cell coimmunoprecipitation experiments used the expression vector pMiWIII, which derives expression through a chick β-actin promoter (Suemori et al. 1990; Matsunaga et al. 2001). Mouse Ptf1a and Ptf1aW298A, human RBPJ, and human TCF12 coding regions (Beres et al. 2006) were subcloned as PCR products with NcoI and XbaI ends into a modified pMiWIII vector that results in fusion of five myc tags at the N terminus of the proteins. Plasmids were sequence-verified.
Immunofluorescence
Embryos at E10.5 and E12.5 were dissected in ice-cold 0.1 M sodium phosphate buffer (pH 7.4), fixed in 4% formaldehyde for 2 h at 4°C, and washed three times in 0.1 M sodium phosphate buffer (pH 7.4) for 2 h. For E16.5 spinal cords and brains, the tissues were dissected out, fixed in 4% formaldehyde overnight at 4°C, and washed overnight with 0.1 M sodium phosphate buffer (pH 7.4). Embryos and dissected tissues were sunk overnight in 30% sucrose in 0.1 M sodium phosphate buffer (pH 7.4), embedded in OCT, and cryosectioned at 12–30 μm. All sections of neural tubes and spinal cords are from the upper limb level.
Immunofluorescence was performed using the following primary antibodies: rabbit anti-calbindin (1:500; Sigma), rabbit anti-GABA (1:1000; Sigma), rabbit anti-GluR2/3 (1:100; Chemicon), rabbit anti-Islet1/2 (1:5000; Tsuchida et al. 1994), guinea pig anti-Islet1/2 (1:20,000; gift from T. Jessell), mouse anti-Lhx1/5 (1:100; 4F2; Developmental Studies Hybridoma Bank), guinea pig anti-Lmx1b (1:5000; Müller et al. 2002), mouse anti-Mash1 (1:1000; Lo et al. 1991), rabbit anti-Math1 (1:100; Helms and Johnson 1998), rabbit anti-Pax2 (1:1000; Zymed), rabbit anti-Tlx3 (1:20,000; gift from T. Müller and C. Birchmeier), guinea pig anti-VGLUT2 (1:2500; Chemicon), goat anti-β-Gal (1:500; Biogenesis), mouse anti-NeuN (1:1000; Chemicon), rat anti-Ki67 (1:50; DakoCytomation), and mouse anti-TuJ1 (1:1000; Covance). Guinea pig polyclonal antisera to Ptf1a (1:5000) were raised against amino acids 11–235 of recombinant mouse Ptf1a (GenBank NM_018809) fused with glutathione-S-transferase (Ptf1a-GST expression plasmid was a gift from H. Edlund). The specificity of the antisera was confirmed by staining E10.5 neural tube sections from wild-type and Ptf1a-null embryos (Fig. 3). Fluorescence imaging and cell counts of specific neuronal cell types were carried out on a Bio-Rad MRC 1024 or Zeiss LSM Pascal confocal microscope. For each experiment, multiple sections from at least three different animals were analyzed.
Chicken in ovo electroporation
Fertilized White Leghorn eggs were obtained from the Texas A&M Poultry Department and incubated for 5 d at 39°C. Solutions of supercoiled plasmid DNA (0.5 μg/μL) in PBS/0.02% Trypan Blue were injected into the lumen of the closed neural tube at stage HH26, and embryos were electroporated as described previously (Muramatsu et al. 1997; Funahashi et al. 1999; Nakada et al. 2004; Wildner et al. 2006). An expression vector with nuclear localized GFP (pCIG) (Megason and McMahon 2002) was coinjected as a control to monitor efficiency and extent of electroporation. Embryos were harvested 48 h later at HH29–HH30, fixed with 4% formaldehyde for 1 h, and processed as above for cryosectioning and immunofluorescence.
COS transfection and coimmunoprecipitation
COS cells were transiently transfected individually with Ptf1a, Ptf1aW298A, Rbpj, or TCF12, or cotransfected with the combinations indicated in Figure 1 using Fugene 6 Transfection Reagent (Roche). All proteins expressed were myc-tagged and expressed using pMiWIII expression vector. After 48 h, the cells were washed with ice-cold PBS, and homogenized in lysis buffer containing 50 mM Tris-Cl (pH 7.5), 5 mM MgCl2, 100 mM KCl, 1 mM DTT, 0.2% NP-40, 1 mM PMSF, and protease inhibitor cocktail. After centrifugation at 20,000g for 20 min, the supernatant was incubated with guinea pig anti-Ptf1a antibody-conjugated protein G Sepharose 4B (Sigma) for 12 h at 4°C. After being washed with lysis buffer, the proteins bound to Sepharose beads were solubilized with SDS sample buffer. Immunoprecipitants were immunoblotted with rabbit polyclonal anti-Myc antibody (A-14, Sigma) and visualized using peroxidase-conjugted secondary antibody followed by SuperSignal West Pico (Pierce).
RT–PCR
cDNA was prepared from 20 ng of total RNA from YFP fluorescence-activated cells sorted from E12.5 neural tubes from Ptf1Cre/+/ROSA26YFP embryos using 16-mer oligo-dT primers and Sensiscript Reverse Transcriptase (Qiagen). One-twentieth of the cDNA solution was used for each PCR reaction. Samples not including reverse transcriptase were used to control for genomic DNA contamination. PCR was performed for 35 cycles of 30 sec at 94°C, 30 sec at 58°C, and 30 sec at 68°C. The following primers were used to detect the expression of mRNA of PTF1 components and the control Gapdh: Ptf1a-sense, 5′-TCATCC GTACAGCTAAAGTGTG-3′; Ptf1a-antisense, 5′-TCTGTCA AAGGTGCTTCAGG-3′; Rbpj-sense, 5′-TGGCACTGTTCAA TCGCCTT-3′; Rbpj-antisense, 5′-AATCTTGGGAGTGCCAT GCCA-3′; Tcfe2a-sense, 5′-AGGTCCCACGCACGCGCACC 3′; Tcfe2a-antisense, 5′-CGCCTGCTGCAGGATGAGCA-3′; Tcf12-sense, 5′-CCATCCCCAAATTCTGACGAT-3′; Tcf12-antisense, 5′-CTGGACATTGGCGGAAGACTT-3′; Tcf4-sense, 5′-TGTACCCAATCACGACAGGA-3′; Tcf4-antisense, 5′-GCCAGCTCGTAGTATTTTGCC-3′; Gapdh-sense, 5′-GT GAGGCCGGTGCTGAGTAT-3′; Gapdh-antisense, 5′-TCAT GAGCCCTTCCACAATG-3′.
Acknowledgments
We are grateful to Dr. W. Pear for generating and sharing the DNMaml mouse strain. We thank Susan Hipkens for performing the recombinase-mediated cassette exchange procedure for Ptf1aW298A, and the Vanderbilt Transgenic Mouse/ES Cell Shared Resource for performing the blastocyst microinjection experiments. We thank D. Anderson, T. Jessell, and T. Müller for antibodies; H. Edlund for the Ptf1a-GST plasmid; and Drs. H. Lai and K. Zimmerman for critical reading of this manuscript. This work was funded by NIH R01 HD037932 to J.E.J., DK072473 to M.A.M., and DK61220 and DK55266 to R.J.M.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1628008
References
- Barolo S., Walker R.G., Polyanovsky A.D., Freschi G., Keil T., Posakony J.W. A notch-independent activity of suppressor of hairless is required for normal mechanoreceptor physiology. Cell. 2000;103:957–969. doi: 10.1016/s0092-8674(00)00198-7. [DOI] [PubMed] [Google Scholar]
- Beres T., Masui T., Swift G.H., Shi L., Henke R.M., MacDonald R.J. PTF1 is an organ-specific and Notch-independent bHLH complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue RBP-L. Mol. Cell. Biol. 2006;26:117–130. doi: 10.1128/MCB.26.1.117-130.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T., Anderson K.V. Patterning cell types in the dorsal spinal cord: What the mouse mutants say. Nat. Rev. Neurosci. 2003;4:289–297. doi: 10.1038/nrn1073. [DOI] [PubMed] [Google Scholar]
- Cheng L., Arata A., Mizuguchi R., Qian Y., Karunaratne A., Gray P.A., Arata S., Shirasawa S., Bouchard M., Luo P., et al. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat. Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- Cheng L., Abdel-Samad O., Xu Y., Mizuguchi R., Luo P., Shirasawa S., Goulding M., Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat. Neurosci. 2005;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- Cockell M., Stevenson B.J., Strubin M., Hagenbuchle O., Wellauer P.K. Identification of a cell-specific DNA-binding activity that interacts with a transcriptional activator of genes expressed in the acinar pancreas. Mol. Cell. Biol. 1989;9:2464–2476. doi: 10.1128/mcb.9.6.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Pompa J.L., Wakeham A., Correia K.M., Samper E., Brown S., Aguilera R.J., Nakano T., Honjo T., Mak T.W., Rossant J., et al. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- Engleka K.A., Gitler A.D., Zhang M., Zhou D.D., High F.A., Epstein J.A. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev. Biol. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Feng Y.Q., Seibler J., Alami R., Eisen A., Westerman K.A., Leboulch P., Fiering S., Bouhassira E.E. Site-specific chromosomal integration in mammalian cells: Highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- Fink A.J., Englund C., Daza R.A., Pham D., Lau C., Nivison M., Kowalczyk T., Hevner R.F. Development of the deep cerebellar nuclei: Transcription factors and cell migration from the rhombic lip. J. Neurosci. 2006;26:3066–3076. doi: 10.1523/JNEUROSCI.5203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C., Ma Q., Casarosa S., Ang S.L., Anderson D.J., Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes & Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y., Fujitani S., Luo H., Qiu F., Burlison J., Long Q., Kawaguchi Y., Edlund H., MacDonald R.J., Furukawa T., et al. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Funahashi J., Okafuji T., Ohuchi H., Noji S., Tanaka H., Nakamura H. Role of Pax-5 in the regulation of a mid-hindbrain organizer’s activity. Dev. Growth Differ. 1999;41:59–72. doi: 10.1046/j.1440-169x.1999.00401.x. [DOI] [PubMed] [Google Scholar]
- Furukawa T., Mukherjee S., Bao Z.Z., Morrow E.M., Cepko C.L. rax, Hes1, and notch1 promote the formation of Müller glia by postnatal retinal progenitor cells. Neuron. 2000;26:383–394. doi: 10.1016/s0896-6273(00)81171-x. [DOI] [PubMed] [Google Scholar]
- Gaiano N., Nye J.S., Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- Glasgow S.M., Henke R.M., Macdonald R.J., Wright C.V., Johnson J.E. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- Gowan K., Helms A.W., Hunsaker T.L., Collisson T., Ebert P.J., Odom R., Johnson J.E. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Gross M.K., Dottori M., Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34:535–549. doi: 10.1016/s0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Guimera J., Weisenhorn D.V., Wurst W. Megane/Heslike is required for normal GABAergic differentiation in the mouse superior colliculus. Development. 2006;133:3847–3857. doi: 10.1242/dev.02557. [DOI] [PubMed] [Google Scholar]
- Helms A.W., Johnson J.E. Progenitors of dorsal commissural interneurons are defined by MATH1 expression. Development. 1998;125:919–925. doi: 10.1242/dev.125.5.919. [DOI] [PubMed] [Google Scholar]
- Helms A.W., Johnson J.E. Specification of dorsal spinal cord interneurons. Curr. Opin. Neurobiol. 2003;13:42–49. doi: 10.1016/s0959-4388(03)00010-2. [DOI] [PubMed] [Google Scholar]
- Helms A.W., Battiste J., Henke R.M., Nakada Y., Simplicio N., Guillemot F., Johnson J.E. Sequential roles for Mash1 and Ngn2 in the generation of dorsal spinal cord interneurons. Development. 2005;132:2709–2719. doi: 10.1242/dev.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitoshi S., Alexson T., Tropepe V., Donoviel D., Elia A.J., Nye J.S., Conlon R.A., Mak T.W., Bernstein A., van der Kooy D. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes & Dev. 2002;16:846–858. doi: 10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo T. The shortest path from the surface to the nucleus: RBP-J κ/Su(H) transcription factor. Genes Cells. 1996;1:1–9. doi: 10.1046/j.1365-2443.1996.10010.x. [DOI] [PubMed] [Google Scholar]
- Hoshino M., Nakamura S., Mori K., Kawauchi T., Terao M., Nishimura Y., Fukuda A., Fuse T., Matsuo N., Sone M. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Hoveyda N., Shield J.P., Garrett C., Chong W.K., Beardsall K., Bentsi-Enchill E., Mallya H., Thompson M.H. Neonatal diabetes mellitus and cerebellar hypoplasia/agenesis: Report of a new recessive syndrome. J. Med. Genet. 1999;36:700–704. [PMC free article] [PubMed] [Google Scholar]
- Jarriault S., Brou C., Logeat F., Schroeter E.H., Kopan R., Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Joyner A.L., Zervas M. Genetic inducible fate mapping in mouse: Establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev. Dyn. 2006;235:2376–2385. doi: 10.1002/dvdy.20884. [DOI] [PubMed] [Google Scholar]
- Kao H.Y., Ordentlich P., Koyano-Nakagawa N., Tang Z., Downes M., Kintner C.R., Evans R.M., Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes & Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y., Cooper B., Gannon M., Ray M., MacDonald R.J., Wright C.V.E. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Krapp A., Knofler M., Frutiger S., Hughes G.J., Hagen-buchle O., Wellauer P.K. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix–loop–helix protein. EMBO J. 1996;15:4317–4329. [PMC free article] [PubMed] [Google Scholar]
- Krapp A., Knofler M., Ledermann B., Burki K., Berney C., Zoerkler N., Hagenbuchle O., Wellauer P.K. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes & Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.C., Yu D., de Martinez Velasco J., Tessarollo L., Swing D.A., Court D.L., Jenkins N.A., Copeland N.G. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Lo L.-C., Johnson J.E., Wuenschell C.W., Saito T., Anderson D.J. Mammalian achaete-scute homolog 1 is transiently expressed by spatially-restricted subsets of early neuroepithelial and neural crest cells. Genes & Dev. 1991;5:1524–1537. doi: 10.1101/gad.5.9.1524. [DOI] [PubMed] [Google Scholar]
- Long Q., Shelton K.D., Lindner J., Jones J.R., Magnuson M.A. Efficient DNA cassette exchange in mouse embryonic stem cells by staggered positive–negative selection. Genesis. 2004;39:256–262. doi: 10.1002/gene.20053. [DOI] [PubMed] [Google Scholar]
- Louvi A., Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Massari M.E., Murre C. Helix–loop–helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui T., Long Q., Beres T.M., Magnuson M.A., MacDonald R.J. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes & Dev. 2007;21:2629–2643. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga E., Araki I., Nakamura H. Role of Pax3/7 in the tectum regionalization. Development. 2001;128:4069–4077. doi: 10.1242/dev.128.20.4069. [DOI] [PubMed] [Google Scholar]
- Megason S.G., McMahon A.P. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- Miyoshi G., Bessho Y., Yamada S., Kageyama R. Identification of a novel basic helix–loop–helix gene, Heslike, and its role in GABAergic neurogenesis. J. Neurosci. 2004;24:3672–3682. doi: 10.1523/JNEUROSCI.5327-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R., Kriks S., Cordes R., Gossler A., Ma Q., Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat. Neurosci. 2006;9:770–778. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- Müller T., Brohmann H., Pierani A., Heppenstall P.A., Lewin G.R., Jessell T.M., Birchmeier C. The homeodomain factor Lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron. 2002;34:551–562. doi: 10.1016/s0896-6273(02)00689-x. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Mizutani Y., Ohmori Y., Okumura J. Comparison of three nonviral transfection methods for foreign gene expression in early chicken embryos in ovo. Biochem. Biophys. Res. Commun. 1997;230:376–380. doi: 10.1006/bbrc.1996.5882. [DOI] [PubMed] [Google Scholar]
- Nakada Y., Hunsaker T.L., Henke R.M., Johnson J.E. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus cell-type specification. Development. 2004;131:1319–1330. doi: 10.1242/dev.01008. [DOI] [PubMed] [Google Scholar]
- Nakatani T., Minaki Y., Kumai M., Ono Y. Helt determines GABAergic over glutamatergic neuronal fate by repressing Ngn genes in the developing mesencephalon. Development. 2007;134:2783–2793. doi: 10.1242/dev.02870. [DOI] [PubMed] [Google Scholar]
- Nakhai H., Sel S., Favor J., Mendoza-Torres L., Paulsen F., Duncker G.I., Schmid R.M. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development. 2007;134:1151–1160. doi: 10.1242/dev.02781. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T., Ishibashi M., Gradwohl G., Nakanishi S., Guillemot F., Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M., Abasolo I., Mingorance-Le Meur A., Martinez A., Del Rio J.A., Wright C.V., Real F.X., Soriano E. Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proc. Natl. Acad. Sci. 2007;104:5193–5198. doi: 10.1073/pnas.0605699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Rose S.D., Swift G.H., Peyton M.J., Hammer R.E., MacDonald R.J. The role of PTF1-P48 in pancreatic acinar gene expression. J. Biol. Chem. 2001;276:44018–44026. doi: 10.1074/jbc.M106264200. [DOI] [PubMed] [Google Scholar]
- Roux E., Strubin M., Hagenbuchle O., Wellauer P.K. The cell-specific transcription factor PTF1 contains two different subunits that interact with the DNA. Genes & Dev. 1989;3:1613–1624. doi: 10.1101/gad.3.10.1613. [DOI] [PubMed] [Google Scholar]
- Schwenk F., Baron U., Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick G.S., Barker K.T., Stolte-Dijkstra I., Fleischmann C., Coleman R.J., Garrett C., Gloyn A.L., Edghill E.L., Hattersley A.T., Wellauer P.K., et al. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat. Genet. 2004;36:1301–1305. doi: 10.1038/ng1475. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Suemori H., Kadodawa Y., Goto K., Araki I., Kondoh H., Nakatsuji N. A mouse embryonic stem cell line showing pluripotency of differentiation in early embryos and ubiquitous β-galactosidase expression. Cell Differ. Dev. 1990;29:181–186. doi: 10.1016/0922-3371(90)90120-l. [DOI] [PubMed] [Google Scholar]
- Tanigaki K., Han H., Yamamoto N., Tashiro K., Ikegawa M., Kuroda K., Suzuki A., Nakano T., Honjo T. Notch–RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y., Furukawa T., Tun T., Han H., Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol. Cell. Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T., Ensini M., Morton S.B., Baldassare M., Edlund T., Jessell T.M., Pfaff S.L. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Tu L., Fang T.C., Artis D., Shestova O., Pross S.E., Maillard I., Pear W.S. Notch signaling is an important regulator of type 2 immunity. J. Exp. Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildner H., Muller T., Cho S.H., Brohl D., Cepko C.L., Guillemot F., Birchmeier C. dILA neurons in the dorsal spinal cord are the product of terminal and non-terminal asymmetric progenitor cell divisions, and require Mash1 for their development. Development. 2006;133:2105–2113. doi: 10.1242/dev.02345. [DOI] [PubMed] [Google Scholar]