Abstract
Caulobacter crescentus divides asymmetrically into a swarmer cell and a stalked cell, a process that is governed by the imbalance in phosphorylated levels of the DivK cell fate determinant in the two cellular compartments. The asymmetric polar localization of the DivJ kinase results in its specific inheritance in the stalked daughter cell where it phosphorylates DivK. The mechanism for the polar positioning of DivJ is poorly understood. SpmX, an uncharacterized lysozyme homolog, is shown here to control DivJ localization and activation. In the absence of SpmX, DivJ is delocalized and dysfunctional, resulting in developmental defects caused by an insufficiency in phospho-DivK. While SpmX stimulates DivK phosphorylation in the stalked cell, unphosphorylated DivK in the swarmer cell activates an intricate transcriptional cascade that leads to the production of the spmX message. This event primes the swarmer cell for the impending transition into a stalked cell, a transition that is sparked by the abrupt accumulation and localization of SpmX to the future stalked cell pole. Localized SpmX then recruits and stimulates DivJ, and the resulting phospho-DivK implements the stalked cell fate. The dynamic interplay between SpmX and DivK is at the heart of the molecular circuitry that sustains the Caulobacter developmental cycle.
[Keywords: Asymmetric cell division, cell fate determinant, polar protein localization, muramidase, kinase]
Asymmetric division is fundamental to differentiation in eukaryotes and prokaryotes. A signature of this process is the generation of an imbalance in the distribution and/or activity of cell fate determinants in daughter cells, endowing them with distinctive features (Horvitz and Herskowitz 1992; Shapiro and Losick 1997). For example, in the asymmetric division of Drosophila neuroblasts, a protein kinase (aPKC) within the Par complex is polarized to the apical membrane. This asymmetric disposition of aPKC, along with its cell cycle activation, governs the timely phosphorylation of a cell fate determinant (Lgl) in one of the two daughter cells (Wirtz-Peitz and Knoblich 2006). A remarkably similar principle operates at the heart of the developmental program that instructs stem cell-like division in the crescent-shaped bacterium Caulobacter crescentus (Fig. 1). A protein kinase (DivJ) that is asymmetrically localized and activated during the cell cycle phosphorylates a cell fate determinant (DivK) specifically in one of the two daughter cells (Skerker and Laub 2004). Here we describe an uncharacterized muramidase homolog (SpmX) that acts as spatiotemporal regulator of DivJ and DivK.
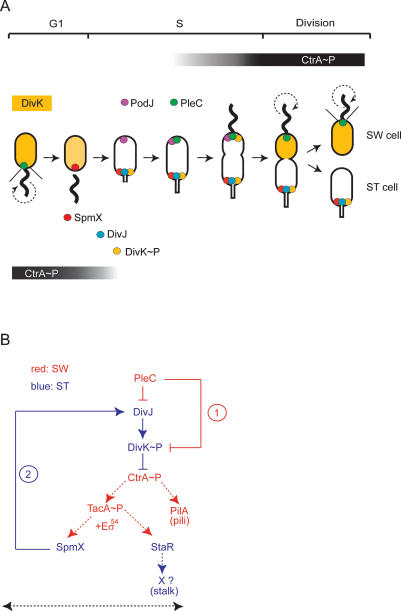
Figure 1.
Graphical representation of the cell fate regulators in Caulobacter and the genetic circuit they constitute. (A) Subcellular localization of DivJ, DivK, PleC, and the localization factors PodJ and SpmX during the Caulobacter asymmetric division cycle: SpmX (red dot) localizes to the pole previously occupied by PleC (green dot), which eventually morphs into the stalked pole. SpmX recruits DivJ (blue dot), initiating the phosphorylation and localization of DivK (yellow), eventually resulting in bipolar DivK∼P (yellow dot) in predivisional cells. Predivisional cells also display PleC at the flagellated pole. PleC has been recruited previously to that site by PodJ (purple dot), which localizes to that pole in late stalked cells (Viollier et al. 2002b; Hinz et al. 2003). Coincident with the compartmentalization of the late predivisional cell, localized DivK∼P and diffuse (dephosphorylated) DivK are present in the stalked (ST) cell chamber and the swarmer (SW) cell chamber, respectively. Pili and the flagellum are indicated by the thin lines (black) and the thick wavy line, respectively. The circular dashed arrow denotes a rotating flagellum. The graded black bar indicates the time during the cell cycle that CtrA∼P is present. In late predivisional cells, CtrA∼P accumulates in the swarmer compartment and is eliminated from the stalked cell compartment. (B) In the swarmer cell compartment (SW, red), PleC reduces DivK∼P levels by direct dephosphorylation of DivK∼P (rectangular arrow, event 1) (Matroule et al. 2004), and possibly indirectly by inhibiting the DivJ kinase (Sommer and Newton 1991; Wheeler and Shapiro 1999) by the production of an inhibitor or modulator of DivJ activity. High levels of DivK∼P inhibit CtrA∼P-mediated transcriptional activation of tacA and pilA by a mechanism that is poorly understood (Biondi et al. 2006a). The tacA translation product along with σ54-containing RNA polymerase (Eσ54) catalyzes transcription of spmX and staR, the gene for a transcriptional regulator of stalk biogenesis (Biondi et al. 2006b). At the swarmer-to-stalked cell transition, SpmX accumulates to localize and activate DivJ (event 2), thereby producing a surge in DivK∼P that signals the removal of CtrA∼P by a complex phosphosignaling cascade (Biondi et al. 2006a; Iniesta et al. 2006). This event, coupled with the disappearance of TacA at the swarmer-to-stalked cell transition (see Fig. 4A), leads to the shut-down of SpmX transcription in stalked cells (ST, blue). The dotted lines indicate transcriptional regulation; bold lines indicate the regulation at the level of protein localization or activity. The black dashed arrow indicates the new connections of the circuit uncovered herein.
The Caulobacter predivisional cell features a cylindrical extension of the cell envelope, the stalk, at the old cell pole and a rotating flagellum along with the pili synthetic apparatus at the new pole (Fig. 1A). A stalked daughter cell that replicates its chromosome and a swarmer daughter cell that is maintained in a (G1-like) nonreplicative state are the products of each cell division. DivJ is localized to the stalked pole in the predivisional cell (Fig. 1A) and is inherited by the stalked daughter cell where it phosphorylates DivK, an essential single domain response regulator. The PleC phosphatase is localized to the flagellated pole and, directly and possibly also indirectly, antagonizes DivJ to maintain DivK∼P levels low in the swarmer cell compartment (Fig. 1A; Matroule et al. 2004; Skerker and Laub 2004).
DivK implements the fate of the two daughter cells through CtrA (Fig. 1B; Wu et al. 1998), a DNA-binding response regulator that is stimulated by phosphorylation to interact with its target sites (CtrA boxes) (Quon et al. 1996). The stability and phosphorylation of CtrA are cell cycle-regulated (Fig. 1A; Domian et al. 1997), and DivK controls both these events. In the stalked cell, DivK∼P triggers the removal of phosphorylated CtrA (CtrA∼P) (Hung and Shapiro 2002; Biondi et al. 2006a; Iniesta et al. 2006). In the swarmer cell, CtrA∼P represses DNA replication by binding to five target sites within the chromosomal origin of replication (Cori) (Quon et al. 1998). In addition, CtrA∼P regulates transcription at promoters of developmental genes, including pilA, which encodes the structural subunit of the pilus filament that is extruded from the flagellated pole of swarmer cells (Skerker and Shapiro 2000; Laub et al. 2002). A hallmark of the Caulobacter division cycle is that the swarmer cell morphs into a replicative stalked cell in response to an unknown cell cycle cue. In doing so, a stalk elaborates from the pole previously vacated by ejecting the flagellum and retracting the pili. Coincident with these morphological changes, CtrA phosphorylation subsides and the protein is degraded. This relieves repression of Cori and DNA replication commences in the nascent stalked cell (Domian et al. 1997), events bearing functional resemblance to the eukaryotic G1–S transition.
The inactivation of CtrA∼P is signaled by a surge in DivK∼P, caused by the abrupt localization and activation of DivJ at the pole previously occupied by PleC (Fig. 1A; Wheeler and Shapiro 1999; Hung and Shapiro 2002). Mutations in divJ and divK result in elevated CtrA activity and aberrant cell division, and a mutation in ctrA can overcome the requirement of DivK for viability (Wu et al. 1998; Hung and Shapiro 2002; Pierce et al. 2006). In the absence of PleC, CtrA∼P levels are reduced, DivK∼P levels are dramatically elevated, and DivJ is delocalized yet still able to efficiently phosphorylate DivK (Wheeler and Shapiro 1999; Biondi et al. 2006a). The finding that DivK∼P levels are lower, but not higher, in the pleC divJ double mutant compared with the divJ single mutant (Wheeler and Shapiro 1999) begs the question of whether in addition to controlling DivJ localization, PleC plays a role in preventing precocious activation of DivJ (Fig. 1B). PleC is know to promote stalk elongation, flagellum rotation, and pilA transcription (Ohta et al. 1992; Wang et al. 1993; Viollier and Shapiro 2003).
Since the PleC–DivJ–DivK phosphosignaling cascade impinges on motility, we explored if additional components of this pathway can be identified in a screen for motility mutants. Using such a screen (S. Pritchard, D. Matteson, E. Huitema, S.K. Radhakrishnan, and P.H. Viollier, in prep.), we here unveil the uncharacterized muramidase homolog SpmX as a PleC-dependent protein that promotes localization and activation of DivJ, and that therefore indirectly stimulates the surge of DivK∼P at the G1–S transition. We also found that DivK feeds back on SpmX by way of a transcriptional cascade that relies on the sequential action of CtrA and the recently characterized transcriptional activator TacA that directs stalk elongation at the G1–S interface (Biondi et al. 2006b). We propose that SpmX and DivK represent critical nodes of the regulatory circuitry that sustains the Caulobacter asymmetric division cycle.
Results
In the absence of the SpmX muramidase homolog, cell division is perturbed
To identify additional regulators of DivK, we screened our comprehensive library of transposon motility mutants for developmental defects by light and transmission electron microscopy (TEM). Strains NS190 and NS349 frequently have bipolar stalks and are perturbed in cell division, a phenotype resembling that of a strain with a cold-sensitive mutation in divK (divKcs) grown under permissive conditions (Fig. 2C; Supplemental Table S1). Moreover, they were found to be flagellated, but to swim poorly. NS190 and NS349 harbor a Himar1 and an EZ-Tn5 insertion, respectively, in the uncharacterized CC2173 gene (henceforth referred to as spmX for stalked pole muramidase homolog) (Fig. 2A). The spmX gene encodes a polytopic membrane protein of 431 residues with two membrane-spanning domains (residues 366–388 and 398–420) near the C terminus and a lysozyme/muramidase-like domain (cd00737; residues 7–143) near the N terminus (Fig. 2A). In lysozyme, this domain hydrolyzes the β-1,4-glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine of the peptidoglycan cell wall (Holtje 1996). An in-frame deletion in spmX (ΔspmX) (Fig. 2A) gave rise to a motility and division defect as did the spmX transposon insertions in strains NS190 and NS349 (Fig. 2B; data not shown). These results, along with complementation experiments (see Supplemental Fig. S1), indicate that the observed defects are due to the loss of SpmX.
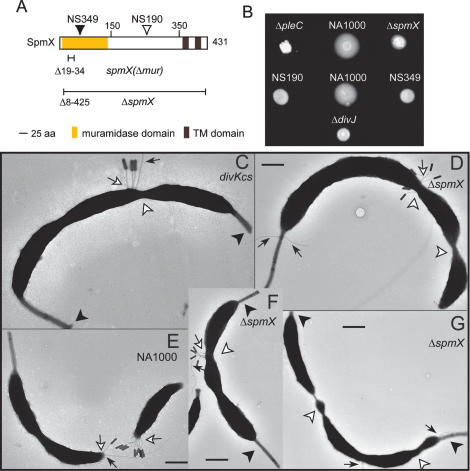
Figure 2.
A motility mutant screen identifies spmX, an uncharacterized gene encoding a muramidase homolog required for proper cell division. (A) Domain organization of SpmX showing the predicted transmembrane domains (brown), the muramidase domain (yellow), the position of the Himar1 (white triangle), and the EZ-Tn5 (black triangle) insertion in strains NS349 and NS190, respectively. The line below the domain architecture shows the deleted coding region of the ΔspmX strain. (B) Motility assay of NA1000 (wild type), NS190, NS349, ΔspmX, ΔpleC, and ΔdivJ strains. Overnight cultures (2.5 μL) were placed on PYE swarm (0.3%) agar plates and incubated for 60 h at 30°C. Motility defects can be seen as swarms with a compact appearance, whereas those from wild type are diffuse and enlarged. (C–G) Transmission electron micrographs of wild-type (E), ΔspmX (D,F,G), and divKcs (C) cells grown at 30°C. Pili (visualized indirectly by staining with pilus-specific bacteriophage ΦCbK) and flagella are indicated by white and black arrows, respectively. Arrowheads indicate bipolar stalks (black) or abnormal constrictions (white). Bars, ∼200 nm.
SpmX mutant cells have a strong propensity to elongate into either smooth (unconstricted) filamentous cells or cells with multiple constrictions that are often found at aberrant positions (Fig. 2D,F,G; Supplemental Fig. S2). These aberrant divisions likely underlie the occurrence of minicell-like particles that are occasionally observed in spmX cultures (Supplemental Fig. S2B). The motility defect of spmX mutants (Fig. 2B) could be due to inefficient flagellar rotation or the presence of ectopic flagella that are often observed at the division plane, where pili are also located. The abnormal location of these organelles at the division plane can be reconciled with the observed cell separation defect of ΔspmX mutants. If sister cells remain associated with one another and the developmental program proceeds unhindered, this unseparated “post-divisional” cell will grow a stalk at each end. Similarly, pili and flagella will eventually emerge from constrictions where functional, albeit physically associated, poles are located. Based on the results shown below, we propose that the phenotypes of spmX mutants are primarily due to improper regulation of DivK and DivJ.
SpmX controls the localization and activity of DivJ and DivK
The similarity of divKcs and ΔspmX mutants prompted us to investigate if SpmX impinges on DivK activity. Phosphorylation of DivK affects its activity as well as its ability to localize to the cell poles (Lam et al. 2003). A low-copy plasmid harboring divK-gfp under the control of the native divK promoter was transformed into wild-type and spmX− cells to determine if DivK is localized and phosphorylated in the absence of SpmX. Live-cell fluorescence microscopy revealed that, in contrast to the intense polar foci of DivK-GFP in wild-type cells, only diffuse fluorescence was observed in cells lacking SpmX (Fig. 3A; Supplemental Fig. S3A). Immunoblotting using a DivK-specific antiserum demonstrated that the absence of SpmX had no impact on DivK or DivK-GFP steady-state levels (Supplemental Fig. S3B). Thus, SpmX is required for DivK localization.
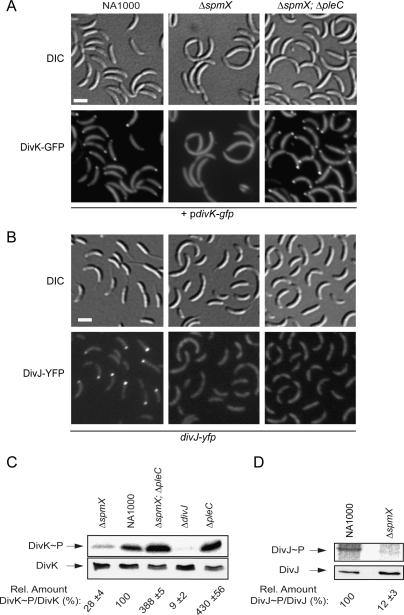
Figure 3.
Localization and in vivo phosphorylation of DivK and DivJ are impaired in the ΔspmX mutant. (A,B) Differential interference contrast (DIC) and fluorescence micrographs of NA1000, ΔspmX, and ΔspmX ΔpleC cells expressing either DivK-GFP expressed from a low-copy plasmid (pdivK-gfp) under the control of the native divK promoter (A) or DivJ-YFP from the endogenous divJ locus (B). (C) Determination of relative DivK∼P/DivK levels in NA1000, ΔspmX, ΔpleC ΔspmX, ΔpleC, and ΔdivJ cells. (D) Measurement of relative DivJ∼P/DivJ levels in NA1000 and ΔspmX cells. Bars, ∼2 μm.
To test if SpmX affects DivK phosphorylation, we determined DivK∼P levels in ΔspmX and ΔdivJ cells relative to wild-type cells (Fig. 3C). Consistent with previous results (Wheeler and Shapiro 1999), DivK∼P levels are severely reduced in ΔdivJ cells, because they lack DivJ, the principal kinase for DivK (Figs. 1B, 3C). Similarly, in ΔspmX cells, DivK∼P levels were 72% lower than in wild-type cells. This result is consistent with the idea that inefficient phosphorylation of DivK underlies its inability to localize to the poles of ΔspmX cells. Moreover, this result raised the possibility that, akin to divJ mutations, the perturbation in cytokinesis of ΔspmX cells is a consequence of low DivK∼P. If so, then a pleC mutation, which causes a dramatic increase in cellular DivK∼P levels (Wheeler and Shapiro 1999), might ameliorate the cytokinetic abnormalities by raising DivK∼P levels. Microscopic examination revealed that ΔspmX ΔpleC double-mutant cells localize DivK-GFP to the cell poles and have lost the characteristic cell division phenotype of spmX single mutants (Fig. 3A,B). Instead, the cells had acquired the hallmarks of ΔpleC cells (stalkless and pililess) (Fig. 3A; data not shown). The relative levels of DivK∼P were elevated in ΔspmX ΔpleC double-mutant cells and were comparable with those observed in the ΔpleC single mutant (Fig. 3C). To confirm that this effect was caused by the loss of activity rather than the physical absence of PleC, we deleted spmX in the pleC(H610A) strain, in which the pleC gene bears a point mutation encoding catalytically inactive PleC (PleCH610A) (Viollier et al. 2002a). The phenotype of the resulting ΔspmX pleC(H610A) double mutant was indistinguishable from that of the ΔspmX ΔpleC mutant (data not shown), confirming that the loss PleC activity can ameliorate the ΔspmX phenotype.
Histidine kinases like DivJ first undergo a transient trans-autophosphorylation event at a conserved histidine residue before the phosphate is passed on to a response regulator, such as DivK (Stock et al. 1990). Because the results above suggested that DivJ kinase activity is compromised in ΔspmX cells, we measured the relative levels of autophosphorylated DivJ (DivJ∼P) in wild-type and ΔspmX mutant cells. DivJ∼P levels were dramatically reduced (88%) (Fig. 3D) in ΔspmX cells compared with wild-type cells. This result, along with the requirement of SpmX for efficient DivK phosphorylation, indicates that DivJ activity is compromised in the ΔspmX mutant. Epistasis experiments provided additional evidence that SpmX lies in a genetic pathway with DivJ and DivK. The phenotype of ΔspmX ΔdivJ double-mutant cells is indistinguishable from that of the ΔdivJ single mutant (data not shown), showing that the effects of ΔdivJ and ΔspmX deletions, each of which compromise DivK phosphorylation on their own, are not additive.
Because DivJ is localized to the stalked pole, we tested the idea that SpmX controls both the polar positioning and activation of DivJ. Accordingly, we constructed ΔspmX strains in which the divJ gene was replaced with a variant encoding either DivJ-YFP (Fig. 3B) or DivJ-GFP (Supplemental Fig. S3E) under control of the divJ promoter. Analysis of these strains by fluorescence microscopy along with a spmX+ isogenic parent revealed only diffuse fluorescence in ΔspmX cells or in ΔspmX ΔpleC cells (Fig. 3B). Immunoblot analysis using antibodies to DivJ showed DivJ-YFP or DivJ-GFP steady-state levels were maintained in the ΔspmX mutant (Supplemental Fig. S3D,F). Based on these results we conclude that SpmX is required (1) to direct DivJ to the stalked pole, (2) to stimulate autophosphorylation of DivJ, and, therefore, (3) to catalyze efficient phosphotransfer from DivJ to DivK. Because DivJ kinase activity is dispensable for DivJ localization to the stalked pole (Lam et al. 2003), we surmise that the effect of SpmX on DivJ localization is not a secondary consequence of a primary effect on kinase activity. Instead, we propose that the principal role of SpmX is to direct DivJ to the stalked pole and to activate it upon its recruitment to the cell pole.
SpmX localizes to the nascent stalked pole during the G1–S transition
To explore if SpmX is localized to the same subcellular site as DivJ, we visualized SpmX by live-cell fluorescence microscopy in cells producing SpmX translationally fused to the C terminus or the N terminus of the red fluorescent protein mCherry. Strains bearing these modified alleles in place of wild-type spmX appear phenotypically normal as determined by differential interference contrast (DIC) microscopy, indicating that these SpmX chimeras are functional. Live-cell fluorescence microscopy of these strains revealed a bright fluorescent focus of SpmX-mCherry or mCherry-SpmX at the stalked pole (Fig. 4B). To corroborate this localization data, we also visualized SpmX in fixed cells by indirect immunofluorescence microscopy (IFM) using a polyclonal antibody raised against SpmX (Fig. 4C). Unipolar foci derived from SpmX were observed in wild-type cells, but not in ΔspmX mutant cells. Because stalk integrity is compromised by the cell permeabilization procedure used during IFM, we were unable to unambiguously assign the location of the SpmX signal to the stalked pole. To resolve this ambiguity, SpmX was immunolocalized in cells expressing the stalked pole marker DivJ-GFP and was found to colocalize with DivJ-GFP (Fig. 4C). While cells that had polar DivJ-GFP always contained overlapping SpmX signals, the converse was not true: Occasionally cells with polar SpmX were observed in which DivJ is not localized. This observation, along with the finding that SpmX is required for DivJ localization, suggested that polar localization of SpmX precedes that of DivJ. This prompted us to construct a divJ-yfp spmX-mCherry strain to colocalize SpmX and DivJ in live cells. Consistent with the IFM results, SpmX-mCherry and DivJ-YFP colocalize in cells of an unsynchronized culture (Fig. 4D). Time-lapse fluorescence microscopy of synchronized swarmer cells was used to determine the temporal sequence of localization of SpmX and DivJ. Initially, neither SpmX nor DivJ was localized. Subsequently, polar SpmX-mCherry foci appeared, followed by the emergence of DivJ-YFP foci and subsequently the stalk (Fig. 4E) at the same pole. To determine whether or not SpmX is present in swarmer cells, we probed blots containing extracts prepared from wild-type cells at different stages of the cell cycle with the anti-SpmX antibody. As shown in Figure 4A, the SpmX protein is at first barely detectable in swarmer cells (t = 0 min), but accumulates sharply during the transition into stalked cells (t = 20 min) and is present henceforth. An identical pattern of abundance was observed in spmX-mCherry cells (Supplemental Fig. S4). Thus, the accumulation and localization of SpmX is an early event during the G1–S transition that is required for and precedes the recruitment of DivJ to the nascent stalked pole.
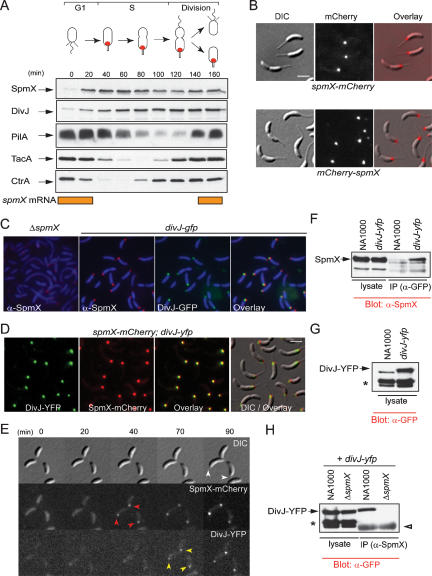
Figure 4.
Colocalization of SpmX and DivJ. (A) Immunoblots showing cellular levels of SpmX, DivJ, PilA, TacA, and CtrA during various stages of the wild-type (NA1000) cell cycle. The orange bars indicate the time when spmX mRNA is present during the cell cycle as determined previously (Laub et al. 2000; McGrath et al. 2007). (B) Polar localization of SpmX-mCherry and mCherry-SpmX, expressed from the endogenous chromosomal locus in place of wild-type SpmX in strains MT237 and MT272, respectively. (C) Immunofluorescence micrographs using anti-SpmX antiserum in LS3200 (divJ-gfp) cells. SpmX-derived signals (red) colocalize (yellow) with DivJ-GFP signals (green). Cells were visualized by staining in 1 μg/mL DAPI (4′,6-diamidino-2-phenylindole). No SpmX-derived foci were seen at the poles in ΔspmX cells. (D) DivJ-YFP (green) and mCherry-SpmX (red) colocalize (yellow) to the stalked pole of NR3601 (spmX-mCherry; divJ-yfp) cells in which the modified alleles replaced the native spmX and divJ genes. (E) Time-lapse fluorescence microscopy of purified NR3601 swarmer cells growing on a cushion of 1% M2G agarose on a microscope slide, showing that SpmX-mCherry (red arrowheads) localizes to the pole first, followed by that of DivJ-YFP (yellow arrowheads), and finally the stalk (white arrow heads) elongates from the same pole. Bars, ∼2 μm. (F) Immunoprecipitation (IP) analysis of membrane-solubilized extracts of the divJ-yfp and NA1000 strain with a monoclonal antibody to GFP (α-GFP). Precipitated samples were analyzed by immunoblotting (Blot) using specific polyclonal antibodies to SpmX (α-SpmX). (G) Immunoblot analysis of extracts from NA1000 and divJ-yfp to detect DivJ-YFP using a monoclonal antibody to GFP. The asterisk (*) marks two nonspecific signals reacting with the anti-GFP antibody. (H) Immunoprecipitation experiments using membrane solubilized extracts of the divJ-yfp and ΔspmX; divJ-yfp strains and polyclonal antibodies to SpmX. DivJ-YFP was detected by immunoblotting using a monoclonal antibody to GFP. Note that the two nonspecific signals (*) do not coprecipitate. The white arrowhead marks the position of the signals derived from the IgG heavy chains.
Prompted by the findings above, we explored whether SpmX and DivJ reside in the same complex at the stalked pole using coimmunoprecipitation experiments. A monoclonal antibody to GFP was used to precipitate DivJ-YFP from membrane-solubilized extracts of the divJ-yfp strain. Immunoblotting showed that SpmX was present in this sample, but not in the sample in which NA1000 solubilized extracts were processed in an identical manner (Fig. 4F). In a reciprocal experiment, DivJ-YFP could be coimmunoprecipitated with the anti-SpmX antibodies from extracts of the divJ-yfp strain, but not from those of the ΔspmX; divJ-yfp strain (Fig. 4H). In contrast, two other proteins that are detected by the anti-GFP antibody (Fig. 4G) and the polarly localized McpA chemoreceptor (Supplemental Fig. S5) were not present in the precipitates from both extracts. These experiments indicate that DivJ and SpmX reside in the same protein complex, providing a mechanistic basis for the notion that DivJ is attracted to the stalked cell pole via the prior localization of SpmX. DivJ is predicted to have six TM domains and to lack major segments that protrude into the periplasm (Sommer and Newton 1991), suggesting that this complex is membrane-anchored.
Role of the muramidase domain in SpmX and DivJ localization
To explore if the muramidase domain is required for SpmX function and/or localization, we engineered alleles encoding SpmX(1–150), SpmX(1–350), and SpmX(Δmur) under control of the vanillate-inducible promoter, Pvan, on a low-copy plasmid (Thanbichler et al. 2007). SpmX(1–150) just encompasses the muramidase domain, SpmX(1–350) lacks the two TM segments, and SpmX(Δmur) is deleted for the active site residues from E19-T34 (see Fig. 2A). Compared with wild-type SpmX, none of the mutants was able to support DivJ localization or wild-type morphology when expressed in the ΔspmX; divJ-yfp strain (Fig. 5A). Immunoblotting showed that this deficiency was not due to a failure to produce SpmX(1–350) or SpmX(Δmur) (Fig. 5B). To investigate if these mutants were localized to the stalked pole, we created translational fusions to the N terminus of mCherry and localized the fusion proteins in NA1000 (Fig. 5C) and the ΔspmX mutant (data not shown). The localization results were similar in both backgrounds and revealed that the muramidase domain contains information sufficient to direct mCherry to the stalked pole. In contrast, SpmX(Δmur)-mCherry was expressed (Fig. 5D) but delocalized, suggesting that an intact muramidase active center is critical for localization. Because SpmX(1–350) (Fig. 5E), but not the cytoplasmic protein CtrA (Fig. 5F), was released into the supernatant upon spheroblasting, we surmise that SpmX(1–350) is found in the periplasmic compartment and that the inability to recruit DivJ to the stalked pole is not due to its absence from the periplasm. The fact that SpmX(1–150) retains the ability to localize to the stalked pole suggests that it is also exported to the periplasmic space (Fig. 5B).
Figure 5.
Role of the muramidase domain in the localizing of SpmX and DivJ to the stalked pole. (A) Fluorescence and DIC micrographs of ΔspmX; divJ-yfp cells expressing SpmX derivatives from a vanillate-inducible promoter (Pvan) on low-copy plasmid pRVMCS-5 (Thanbichler et al. 2007) after growth in PYE supplemented with tetracycline (1 μg/mL) and induced with vanillate (50 nM) for 2 h. (B) Immunoblot analysis of SpmX and CtrA steady-state levels present in equal amounts of cellular extracts from strains in A. Note that the SpmX(1–150) derivative is not efficiently detected by the anti-SpmX antibody because a recombinant SpmX derivative from residues 120–351 was used as immunogen. (C) Localization of SpmX-mCherry derivatives expressed from pRVMCS-5 in NA1000 cells grown as in A. Note that as a result of ectopic expression from Pvan, SpmX-mCherry can often be seen to localize to the pole opposite the stalk. (D) Immunoblot experiments to determine SpmX-mCherry steady-state levels present in cell extracts from strains in C. Equal amounts of cell extracts were loaded in each lane. Similar localization results and abundance patterns as in C and D, respectively, were observed when the SpmX-mCherry derivatives were expressed in a ΔspmX background (data not shown). (E) NA1000 cells expressing the SpmX-mCherry or SpmX(1–350)-mCherry (as in D) were converted to spheroblasts (SB) and gently centrifuged. The cell (SB Cells) and supernatant (SB Sup.) fraction, along with a control lysate of untreated cells (UT Lysate), were examined for the presence of SpmX-mCherry or CtrA by immunoblotting. The presence of SpmX(1– 350)-mCherry in the spheroblast supernatant indicates that it is exported into the periplasm in untreated cells. Note that spheroblasting is incomplete, explaining why a substantial amount of SpmX(1–350)-mCherry remains associated with cells. Similar experiments with SpmX(1–150)-mCherry were inconclusive because spheroblasts were fragile, undergoing lysis during centrifugation (data not shown). (F) The immunoblot shown in E was reprobed for the presence of a cytoplasmic protein (CtrA) as a control for cell lysis. In B, D, E, or F, SpmX-mCherry derivatives and CtrA were detected using polyclonal antibodies to SpmX (α-SpmX), CtrA (α-CtrA), and mRFP (α-mRFP). Note that the anti-mRFP antibody reacts with mCherry.
Together, our results support a model in which an intact muramidase active center serves to target SpmX to the stalked pole. The subsequent recruitment of DivJ into a complex at the stalked pole by SpmX is likely mediated via membrane interactions requiring the SpmX transmembrane domains. We propose that in this complex, activated DivJ will then catalyze efficient phosphotransfer to DivK.
PleC regulates DivJ localization via transcriptional control of spmX
It is known that PleC is required for polar localization of DivJ (Wheeler and Shapiro 1999), but whether or not this is due to a direct or an indirect mechanism is unclear. Based on two lines of evidence we speculated that the latter is true and that SpmX is the centerpiece of this pathway. First, SpmX is necessary to localize DivJ (Fig. 3), and second, mRNA profiling experiments indicated that the abundance of the spmX message is positively dependent on PleC (Chen et al. 2006). The prospect that PleC impinges on the spatiotemporal regulation of DivJ by controlling spmX transcription prompted us to investigate the underlying mechanism in detail.
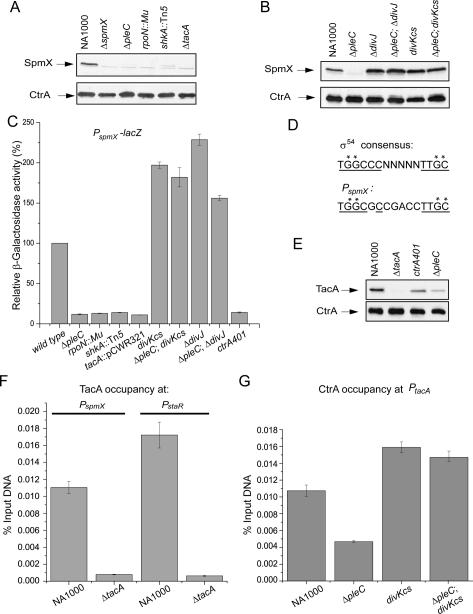
First, we confirmed that SpmX levels are dramatically reduced in ΔpleC cells by immunoblot analysis of extracts from wild-type and ΔpleC mutant cells (Fig. 6A), and that this effect requires the phosphatase activity of PleC (Supplemental Fig. S6). Next, we tested if spmX transcription is impaired in ΔpleC mutant cells. To this end, we engineered a transcriptional fusion of the spmX promoter (PspmX) to a promoterless lacZ gene on a low-copy plasmid and introduced the PspmX-lacZ reporter construct into the wild-type and ΔpleC mutant strain. Quantification of lacZ-encoded β-galactosidase activity revealed that the PspmX-lacZ reporter is only 10% as active in ΔpleC mutant cells as in wild-type cells (Fig. 6C). Thus, PleC is required to activate transcription of spmX as well as that of the pilA pilin gene. Incidentally, the messages of spmX and pilA exhibit nearly identical abundance patterns during the cell cycle: Both are highly abundant in swarmer cells, are down-regulated in stalked cells, and reaccumulate coincident with the compartmentalization of the predivisional cell (Laub et al. 2000; McGrath et al. 2007). This pattern is no surprise for the pilA message since the abundance profile of the PilA protein essentially reflects that of the message (Fig. 4A). However, for the spmX mRNA this pattern is unexpected, since the SpmX translation product accumulates during the G1–S transition when spmX transcript levels begin to decline. This suggests that SpmX accumulation is regulated at the post-transcriptional level. Since the spmX and pilA messages are both dependent on PleC, and since CtrA∼P levels are markedly reduced in the pleC mutant (Biondi et al. 2006a), the possibility existed that CtrA∼P also directly activates transcription from the spmX promoter, along with that of pilA (Fig. 1B). In support of this, PspmX-lacZ was only 10% as active in cells with a temperature-sensitive loss-of-function mutation in ctrA (ctrA401) as in wild-type cells (Fig. 6C). Moreover, SpmX was barely detectable by immunoblot analysis in extracts from ctrA401 cells (Fig. 7B). To determine whether CtrA binds to PspmX in vivo, we conducted quantitative chromatin immunoprecipitation (qChIP) experiments using a polyclonal antibody to CtrA. Control qChIP experiments showed that CtrA interacts efficiently with the promoters of pilA and fliL (Supplemental Fig. S7A), both promoters that CtrA∼P binds to in vitro (Wu et al. 1998; Skerker and Shapiro 2000). However, the spmX promoter was at least an order of magnitude less abundant in these immunoprecipitated fragments than the fliL or pilA promoters. In fact, precipitation of spmX promoter DNA was in the same range as the background signal obtained when pilA promoter DNA was immunoprecipitated with antibodies to the CpaE structural protein of the pilus assembly machinery (data not shown), a protein that is not known to bind promoters (Skerker and Shapiro 2000). Based on these results, we surmise that regulation of PspmX by CtrA is indirect (Fig. 1B).
Figure 6.
Transcriptional regulation of spmX. (A) Immunoblots showing SpmX steady-state levels in wild-type (NA1000), ΔpleC, rpoN∷Tn5, shkA∷Tn5, and ΔtacA mutants. (B) Immunoblots showing that ΔdivJ and divKcs mutations restore SpmX production to ΔpleC mutant cells. (C) β-Galactosidase assays using the PspmX-lacZ reporter plasmid to determine spmX promoter activity in NA1000 and various mutants. Temperature-sensitive mutants were grown at the permissive temperature. (D) Comparison of the spmX promoter sequence with the Eσ54 consensus sequence. Asterisks (*) indicate nucleotides required for Eσ54 binding. Underlined are the −24 and −12 consensus sequences for Eσ54 promoters. (E) Immunoblot showing TacA steady-state levels in NA1000, ΔtacA, ctrA401, and ΔpleC mutants. CtrA was used as a loading control for the immunoblots in A, B, and E. (F) Measurement of TacA occupancy at the spmX and staR promoters in vivo using qChIP analysis. (G) qChIP experiments showing the reduction in CtrA occupancy at the tacA promoter in ΔpleC mutant cells, and an increase in the divKcs single mutant and the ΔpleC divKcs double mutant relative to NA1000.
Figure 7.
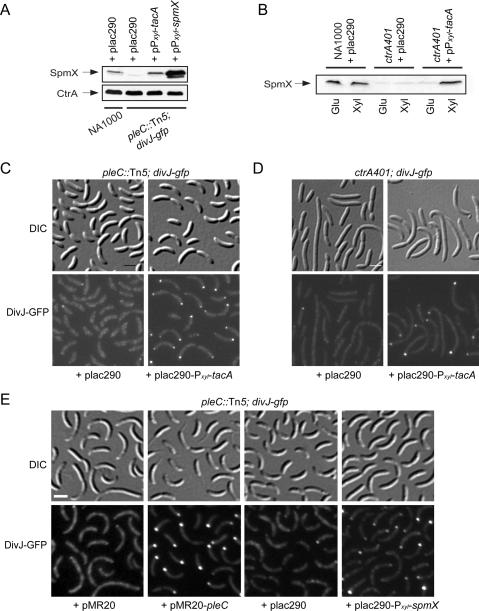
The absence of SpmX underlies the DivJ localization defect in pleC and ctrA401 mutants. (A,B) Immunoblots of pleC∷Tn5 divJ-gfp and ctrA401 divJ-gfp cells upon expression of TacA or SpmX from a xylose-inducible promoter on a low-copy plasmid (pPxyl-tacA). In A, cells were grown in PYE containing tetracycline (1 μg/mL) and 20 mM xylose, except for the strain containing pPxyl-spmX that was grown in 2 mM xylose and tetracycline. In B, cells were grown in PYE containing tetracycline (1 μg/mL) as well as 20 mM xylose (Xyl) or 20 mM glucose (Glu). (C,D) Fluorescence micrographs showing DivJ-GFP in pleC∷Tn5 divJ-gfp (C) and ctrA401 divJ-gfp (D) cells that were grown in PYE containing tetracycline (1 μg/mL) and 20 mM xylose and harbored either plasmid pPxyl-tacA or the empty vector. (E) Localization of DivJ-GFP in a pleC∷Tn5 mutant that expresses SpmX from a xylose-inducible promoter on a low-copy plasmid (pPxyl-spmX) or with a complementing plasmid harboring the pleC gene. No localization is observed in strains with the empty vector. Cells were grown in PYE containing tetracycline (1 μg/mL). Bars, ∼2 μm.
A spmX transcriptional cascade is active in swarmer cells
There are several arguments pointing to the TacA transcriptional activator as the unidentified PleC- and CtrA-dependent transcription factor that binds to PspmX. First, since the accumulation of the tacA transcript slightly precedes that of spmX (Laub et al. 2000; McGrath et al. 2007), the temporal pattern of tacA expression is consistent with the proposed role of TacA as transcriptional regulator of spmX. Second, the abundance of the tacA message is dramatically reduced in the absence of PleC, a feature tacA shares with pilA and spmX (Chen et al. 2006). Third, the spmX promoter sequence suggests that an RNAP holoenzyme containing a σ54 factor (Eσ54) transcribes spmX (Fig. 6D; McGrath et al. 2007). Eσ54-catalyzed transcription normally requires a coactivator of a specialized clade of response regulators (Popham et al. 1989), of which there are only four encoded in the Caulobacter genome and TacA is one of them (Marques et al. 1997).
Immunoblot analysis and PspmX-lacZ reporter assays were used to test if TacA regulates spmX expression. A marked reduction of cellular SpmX levels was observed in strains mutant for TacA or σ54 (RpoN) (Fig. 6A). Moreover, the PspmX-lacZ reporter was only 10% as active in these mutants compared with the wild-type strain (Fig. 6C). Like other response regulators, TacA is also subject to regulation by phosphorylation at a conserved aspartic acid residue (at position 54) (Stock et al. 1990). The ShkA hybrid histidine kinase was shown recently to initiate a phosphorelay that activates TacA by phosphorylation (Biondi et al. 2006b). Consistent with the idea that active TacA stimulates transcription of spmX, disruption of the shkA gene severely impairs SpmX accumulation and PspmX-lacZ activity (Fig. 6A,C). To determine if regulation of spmX transcription by TacA is direct, we conducted qChIP experiments using a polyclonal antibody to TacA. The spmX promoter precipitated specifically and efficiently from lysates of wild-type cells, but not from those of ΔtacA cells (Fig. 6F; Supplemental Fig. S7B). The staR gene that encodes a regulator of stalk elongation shares promoter sequence motifs with spmX (McGrath et al. 2007) and is thought to be directly regulated by TacA (Biondi et al. 2006b). Our qChIP experiments also demonstrated that TacA interacts efficiently in vivo with the promoter of staR (Fig. 6F), but not with that of two other PleC-dependent genes (CC1695 and CC0167) (Supplemental Fig. S7B). These findings, along with the immunoblots showing that TacA is present in the swarmer cell compartment (Fig. 4A), support the view that TacA binds to PspmX in swarmer cells to stimulate transcription by Eσ54 (Fig. 1B).
Why is spmX transcription impaired in ΔpleC cells? Previous experiments implicated that tacA transcription is under the control of CtrA (Marques et al. 1997), suggesting that ΔpleC cells might be unable to activate spmX transcription because they lack sufficient amounts of TacA. As expected, immunoblotting revealed that TacA levels are markedly reduced in extracts from ΔpleC and ctrA401 cells compared with those from wild-type cells (Fig. 6E). The tacA promoter features a CtrA box that overlaps the −35 promoter motif (Marques et al. 1997), providing further support for the hypothesis that a decrease in CtrA∼P is responsible for the reduction of TacA in ΔpleC cells. Whether or not CtrA binds the tacA promoter in vivo is unknown. Our qChIP experiments revealed that the tacA promoter is indeed an in vivo target of CtrA (Fig. 6G). The tacA promoter precipitated with efficiency comparable with the fliL promoter (Supplemental Fig. S7A). Next, we used qChIP analysis to explore if CtrA occupancy at the tacA promoter is compromised in ΔpleC cells compared with wild-type cells. As shown in Figure 6G, CtrA binding to the tacA promoter is reduced by 60% when PleC is absent, thus explaining the reduction of TacA in ΔpleC mutant cells. Based on this data, we propose the following sequence of events. First, low CtrA∼P levels caused by the loss of PleC prevent efficient tacA transcription, which in turn causes a reduction in TacA steady-state levels below the threshold necessary to support spmX transcription. Since spmX is not transcribed, cells are devoid of SpmX, the localization determinant for DivJ, and, thus, DivJ remains dispersed.
If our interpretation of this transcriptional hierarchy is correct, then SpmX expression should be restored to pleC or ctrA mutant cells when TacA is expressed from a xylose-inducible Pxyl-tacA construct on a low-copy-number plasmid. In support of our model, Figure 7, A and B, shows that inducing expression of TacA upon administering xylose restored wild-type levels of SpmX to ctrA401 and ΔpleC mutant cells. Based on these data, we surmise that spmX transcription in the swarmer cell involves an intricate transcriptional regulatory cascade following the order PleC → CtrA → TacA → SpmX.
SpmX couples PleC signaling to DivJ localization
Since ctrA401 cells are devoid of SpmX, they should be unable to direct DivJ to the pole. As expected, only diffuse fluorescence was observed in ctrA401 divJ-gfp cells (Fig. 7D). Since the Pxyl-tacA construct endows both ctrA401 and pleC mutant cells with SpmX, we asked if this plasmid could remedy the DivJ localization defect of these cells. Indeed, ctrA401 divJ-gfp or pleC∷Tn5 divJ-gfp cells bearing the Pxyl-tacA plasmid sequestered DivJ-GFP to the pole (Fig. 7C,D) in a xylose-dependent manner. Interestingly, the Pxyl-tacA plasmid also restored stalk biogenesis to the pleC mutant (Supplemental Fig. S8), indicating that DivJ localization and stalk biogenesis at the G1–S transition rely on TacA whose production is dependent on PleC signaling. To test if SpmX is sufficient to direct DivJ to the pole in the absence of PleC, we expressed SpmX from a Pxyl-spmX plasmid in pleC∷Tn5 divJ-gfp cells. As expected, Pxyl-spmX induced the formation of polar DivJ-GFP clusters (Fig. 7E) in the pleC∷Tn5 mutant, but not stalks. Thus, as already suggested by the ΔspmX phenotype, SpmX is not the TacA-dependent gene that promotes stalk synthesis. Instead, SpmX plays a pivotal role in reprogramming the G1 cell into an S-phase cell by the recruitment of DivJ to the nascent stalked pole at the G1–S transition, an event that is prepared by PleC through its transcriptional regulation of spmX.
SpmX and DivK define critical nodes of an intricate genetic circuit
The data presented above outline two branches of a putative autoregulatory circuit in which PleC facilitates SpmX production, whereas SpmX promotes the accumulation of DivK∼P, which PleC acts against. If these dependencies indeed form the basis of a closed genetic circuit, PleC must signal spmX transcription through DivK. Immunoblotting and PspmX-lacZ reporter experiments confirmed this prediction, as the divKcs mutation overcame the reduction in cellular SpmX levels and the spmX transcriptional defect of ΔpleC cells (Fig. 6B,C). Moreover, qChIP analysis showed that CtrA is again efficiently associated with the tacA promoter in ΔpleC divKcs cells (Fig. 6G). Based on these results, we position DivK between PleC and CtrA in the spmX transcriptional cascade (Fig. 1B). The view that dephosphorylated DivK signals the onset of spmX transcription in the swarmer cell compartment stems from the result that the ΔdivJ mutation, which causes DivK to predominate over DivK∼P, recapitulates the effects of the divKcs mutation on PspmX-lacZ activity and SpmX accumulation in ΔpleC cells (Fig. 6B,C). Thus, high DivK∼P levels that occur at the G1–S transition or upon inactivating PleC will indirectly shut down PspmX.
On the basis of these results, we propose the following sequence of events that lead to the accumulation of the spmX transcript in G1 (swarmer) cells. (1) The PleC phosphatase directly and/or indirectly lowers DivK∼P levels; (2) dephosphorylated DivK promotes to CtrA∼P accumulation; (3) CtrA∼P subsequently activates the tacA promoter; and (4) with the synthesis and phosphorylation of TacA, transcription of spmX is initiated by Eσ54 upon stimulation by TacA. (5) Early at the G1–S transition, either regulated translation or an increase in SpmX stability promotes its accumulation, whereupon it localizes to the developing stalked pole. (6) Polar SpmX then recruits and activates the DivJ kinase. (7) DivK is phosphorylated and (8) indirectly turns off transcription of spmX in nascent stalked cells by down-regulating CtrA∼P, which (9) eliminates de novo production of TacA, while (10) pre-existing TacA is proteolyzed (Fig. 4A).
Discussion
The spmX gene can be considered a focal point of regulation of the Caulobacter asymmetric division cycle, since it is intricately regulated at multiple levels both in time and space. Commensurate with its sophisticated regulation, the SpmX protein performs functions that act at the heart of this developmental program. SpmX stimulates DivJ at the G1–S transition and, thus, triggers the surge of DivK∼P. Moreover, SpmX is a localization factor that directs DivJ and, indirectly, DivK to the pole. Finally, transcription of spmX in the incipient swarmer cell compartment is activated through DivK as part of a PleC-dependent signal transduction cascade. Based on these data, we propose that SpmX and DivK act as critical nodes of an autoregulatory loop interwoven into the genetic circuitry that implements and sustains Caulobacter asymmetric division. DivK activates the transcriptional branch of this loop to prime swarmer cells for the impending transition into stalked cells, while SpmX that accumulates in stalked cells promotes DivK∼P production, which shuts off spmX transcription.
In the absence of SpmX, DivJ is delocalized and its kinase function is impaired. As a result, DivK is poorly phosphorylated and its ability to execute cell specification is compromised. Our result that the requirement for SpmX to stimulate DivJ activity can be overcome with a ΔpleC mutation illustrates another nuance of the PleC regulatory pathway that is required to inhibit DivJ (Sommer and Newton 1991; Wheeler and Shapiro 1999). It is conceivable that a negative regulatory activity that is induced by PleC inhibits premature activation of residual DivJ found in swarmer cells. If that is true, we hypothesize that SpmX is required to eliminate this activity at the G1–S transition, thereby relieving repression of DivJ. This hypothesis accounts for the observed requirement of SpmX to stimulate DivJ in wild-type cells and also explains the observation that DivJ activity is unrestrained in ΔpleC cells, in which both SpmX and this putative inhibitor are absent. Based on this idea, we posit two concurrent functions of SpmX to exert spatiotemporal control over DivJ. First, by recruiting DivJ to the emerging stalked pole at the G1–S transition, SpmX ensures that the bulk of DivJ partitions with the stalked daughter cell at the next cytokinetic event. Thus, this function of SpmX is central for daughter cell-specific distribution of DivJ. Second, SpmX stimulates DivJ activity, possibly by relieving inhibition of the hypothetical negative regulator outlined above. We cannot exclude an alternative model involving a positive regulator of DivJ that is stimulated by SpmX, but that is hyperactivated by another pathway in pleC mutants where SpmX is absent. Intriguingly, DivK was found recently to act as a positive allosteric regulator of the DivJ kinase (R. Paul and U. Jenal, pers. comm.), raising the possibility that DivK itself might represent the aforementioned hypothetical regulator of DivJ.
In addition to the known battery of effectors, at least one unknown phosphodonor of DivK still exists (Wheeler and Shapiro 1999; Ohta and Newton 2003). The regulatory network converging on DivK bears a remarkable resemblance to that regulating Spo0F, a single domain response regulator that plays an important role in committing Bacillus subtilis cells to the sporulation pathway (Stragier and Losick 1996). Spo0F is subject to regulation by phosphorylation and dephosphorylation by multiple kinases and phosphatases (Jiang et al. 2000a, b; Smits et al. 2007). While it is certainly conceivable that another histidine kinase is the source of DivK∼P in cells lacking both PleC and DivJ (Wheeler and Shapiro 1999), DivK∼P might also acquire its phosphate from a high-energy phosphometabolite like acetylphosphate (Wolfe 2005). The regulatory pathways controlling DivK are central for the G1–S transition and likely act in concert with others to complete the underlying developmental changes that remodel a swarmer cell into a replicative stalked cell. These include pathways specified by the master transcriptional regulators GcrA and DnaA as well as the SsrA (tmRNA) quality control system, all of which are active during the G1–S transition and facilitate the onset of DNA replication (Keiler and Shapiro 2003a, b; Holtzendorff et al. 2004; Collier et al. 2006).
A fundamental developmental principle that has found precedence in asymmetric cell division in eukaryotes and in prokaryotes is the targeting of cell fate determinants to specific subcellular positions as exemplified in budding yeast, in Drosophila neuroblasts, and in Caulobacter. The localization of SpmX early during the G1–S transition to recruit DivJ to the stalked pole can be compared with the asymmetrically localized Par complex in Drosophila neuroblasts, a complex that includes aPKC along with other proteins that are likely required for its recruitment to the apical membrane (Wirtz-Peitz and Knoblich 2006). As for the Par complex, the current challenge for SpmX is to elucidate how this asymmetric positioning is achieved. SpmX might recognize a structural and/or chemical modification in the polar peptidoglycan layer that attracts it to the old cell pole, a view that is supported by our result that the muramidase domain is necessary and sufficient for the localization of SpmX to the stalked pole. Indeed, the cell division apparatus that directs the biosynthesis of the polar peptidoglycan during cytokinesis has been implicated in the deposition of a spatial mark at the newborn cell pole to cue protein localization (MacAlister et al. 1987; Huitema et al. 2006; Lam et al. 2006). Like SpmX, the localization factor PodJ that recruits the PleC phosphatase to the newborn pole contains a motif thought to interact with peptidoglycan (Viollier et al. 2002b; Hinz et al. 2003). This raises the intriguing possibility that the ability of selected proteins to discriminate between the old and the new cell pole could be based on unique features present in the polar peptidoglycan. If that is true, cellular asymmetry in Caulobacter or other bacteria might be founded on this principle. Even bacteria that appear morphologically symmetrical feature an old and a new cell pole that might promote the formation of unique molecular microdomains that can be exploited for specialized polar functions (Shapiro et al. 2002), predicting that such a localization concept could be widespread among prokaryotes.
Another remarkable aspect unearthed in this work is the complex transcriptional cascade that activates spmX in swarmer cells. It is an essential part of a new genetic circuit in which SpmX promotes the production of DivK∼P, and DivK∼P in turn terminates the pathway that activates spmX transcription at the G1–S transition. This is achieved by halting transcription to prevent de novo synthesis of TacA and degrading pre-existing TacA at the G1–S transition. Later in the cell cycle, once the predivisional cell is compartmentalized and DivK∼P levels fall in the swarmer cell chamber, the spmX transcriptional pathway is once again activated. Launching the pathway for spmX transcription before cell separation has been completed ensures that the daughter swarmer cell is born with the spmX message in anticipation of the impending transition into stalked cells. The synthesis of a transcript in a compartment long before the function of its translation product is required is a fundamental regulatory strategy that also underlies development of the Drosophila embryo (Micklem 1995). Transcripts like nanos, which codes for a key developmental regulator, are deposited in the egg by the mother before fertilization has occurred (Wang and Lehmann 1991). Once fertilization occurs, the newly formed embryo is already endowed with maternal-derived messages that can implement developmental changes. Many of these maternal mRNAs are later degraded and replaced with other de novo synthesized embryonic transcripts during the midblastula (also known as the oocyte–zygote) transition (Schier 2007). In a remarkable analogy to the fate of these developmental messages, transcripts like spmX that are synthesized before the birth of the swarmer cell begin to disappear during the swarmer-to-stalked cell transition, the time when the synthesis of stalked cell-specific transcripts commences. Thus, the localization of SpmX as well as the underlying spmX transcriptional cascade both represent striking manifestations of remarkably similar fundamental developmental mechanisms that operate in both eukaryotes and prokaryotes.
Materials and methods
Strains and growth conditions
Caulobacter NA1000 (Evinger and Agabian 1977) and derivatives were grown at 30°C in PYE, M2G, or M5G, except divKcs derivatives that were grown at 32°C. Escherichia coli S17-1 (Simon et al. 1983) and EC100D (Epicentre Technologies) were grown at 37°C in LB. Motility assays, swarmer cell isolation, intergeneric conjugations, electroporations, and bacteriophage ΦCr-30-mediated generalized transductions were performed as described (Ely 1991; Viollier and Shapiro 2003; Chen et al. 2005).
Isolation of motility mutants NS190, NS349, NS229, and NS217
From a library of 20,000 transposon mutants that were tested for reduced motility on semisolid PYE (0.3%) agar plates, >400 mutants were isolated and the transposon insertion site was determined (S. Pritchard D. Matteson, E. Huitema, S.K. Radhakrishnan, and P.H. Viollier, in prep.). To uncover unidentified components of the PleC–DivJ–DivK signaling pathway, we screened all mutants with insertions in uncharacterized genes by TEM for pleiotropic phenotypes resembling those exhibited by divJ, divK, or pleC strains, such as abnormalities in cell division and/or polar development (Sommer and Newton 1991). As described in the Results section, NS190 and NS349 shared several properties with the divKcs mutant.
NS190 contains a mariner-derived Himar1 insertion at nucleotide position 6336 of AE005889 delivered by conjugation from E. coli S17-1 harboring pHPV414 (Viollier et al. 2004). In NS349, an EZ-Tn5 transposon (Epicentre Technologies) that had been introduced by electroporation was inserted at nucleotide position 5805 of AE005889. Strain NS229 has a HyperMu (Epicentre Technologies) insertion in rpoN (nucleotide position 4011 of AE006018). An EZ-Tn5 insertion at nucleotide 3762 of AE005688 disrupted shkA in strain NS217.
β-Galactosidase assays and immunoblots
β-Galactosidase assays were performed at 30°C as described earlier (Viollier and Shapiro 2003; Huitema et al. 2006). For the production of anti-SpmX and anti-TacA antibodies, the sequences coding for residues 120–351 of SpmX and 226–488 of TacA were overexpressed in E. coli Rosetta (DE3)/pLysS using pET28a (Novagen) as N-terminal His6-tagged variants and were purified using Ni-NTA agarose (Qiagen). Purified proteins were excised from a 12.5% SDS polyacrylamide gel and used to immunize rabbits (Josman LLC). Antisera were used at the following dilutions: anti-SpmX (1:50,000), anti-TacA (1:30,000), anti-McpA (1:10;000), anti-CtrA (1:10,000), anti-DivJ (1:10,000), anti-mRFP1 (1:5000), anti-DivK (1:10,000), and anti-PilA (1:5000) (Alley et al. 1992; Domian et al. 1997; Wheeler and Shapiro 1999; Jacobs et al. 2001; Viollier et al. 2002a; Chen et al. 2005).
Microscopy
TEM was performed as described earlier (Skerker and Shapiro 2000; Huitema et al. 2006). For fluorescence and DIC imaging, a Nikon Eclipse 90i microscope fitted with a 100× oil TIRF (1.45 numerical aperture) objective and a CoolSnap HQ2 camera (Photometrics) with suitable filters was used. Images were acquired and processed using MetaMorph version 7.0 (Molecular Devices). IFM was conducted as described (Viollier et al. 2002a) using the anti-SpmX antiserum at a dilution of 1:2000.
In vivo 32P labeling
In vivo phosphorylation experiments were performed with certain modifications of a previously described method (Domian et al. 1997). A single colony of cells picked from a PYE agarose plate was washed with M5G medium lacking phosphate and was grown overnight in M5G with 0.05 mM phosphate to an optical density of 0.3 at 660 nm. One milliliter of culture was labeled for 4 min at 28°C using 30 μCi of γ-[32P]ATP. Following lysis, proteins were immunoprecipitated with 3 μL of anti-DivJ or anti-DivK antiserum. The precipitates were resolved by SDS-PAGE, and [32P]-labeled DivK or DivJ were quantified using a Storm 820 PhosphorImager and ImageQuant software version 4.0 (Molecular Dynamics) and were normalized to the relative cellular content as determined by immunoblotting of lysates.
Release of periplasmic proteins
A previously described procedure for releasing periplasmic protein upon spheroblasting was used (Judd et al. 2005) with minor modifications. Ready-Lyse lysozyme solution (Epicentre) was used at a concentration of 9 U/μL, and the cells were incubated for 10 min at room temperature. Spheroblasts were centrifuged at 4500g for 5 min, and samples were collected and then subjected to immunoblot analysis.
Sequence analysis
The DAS (http://www.sbc.su.se/~miklos/DAS/maindas.html) server was used for the prediction of transmembrane domains in SpmX.
Immunoprecipitations, strain, and plasmid constructions
Details for these procedures can be found in the Supplemental Material.
Acknowledgments
We thank Sean Pritchard for excellent technical assistance; Piet de Boer and Arne Rietsch for critical reading of the manuscript; and Mike Laub, Christine Jacobs-Wagner, and Kathleen Ryan for materials. Supported by funds from the School of Medicine, the Mount Sinai Health Care Foundation, and the U.S. Department of Energy Office of Science (BER, grant no. DE-FG02-05ER64136) to P.V., and start-up funds from the Max Planck Society to M.T.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1601808
References
- Alley M.R., Maddock J.R., Shapiro L. Polar localization of a bacterial chemoreceptor. Genes & Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- Biondi E.G., Reisinger S.J., Skerker J.M., Arif M., Perchuk B.S., Ryan K.R., Laub M.T. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006a;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- Biondi E.G., Skerker J.M., Arif M., Prasol M.S., Perchuk B.S., Laub M.T. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol. Microbiol. 2006b;59:386–401. doi: 10.1111/j.1365-2958.2005.04970.x. [DOI] [PubMed] [Google Scholar]
- Chen J.C., Viollier P.H., Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol. Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- Chen J.C., Hottes A.K., McAdams H.H., McGrath P.T., Viollier P.H., Shapiro L. Cytokinesis signals truncation of the PodJ polarity factor by a cell cycle-regulated protease. EMBO J. 2006;25:377–386. doi: 10.1038/sj.emboj.7600935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J., Murray S.R., Shapiro L. DnaA couples DNA replication and the expression of two cell cycle master regulators. EMBO J. 2006;25:346–356. doi: 10.1038/sj.emboj.7600927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian I.J., Quon K.C., Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- Evinger M., Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz A.J., Larson D.E., Smith C.S., Brun Y.V. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol. Microbiol. 2003;47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- Holtje J.V. Bacterial lysozymes. EXS. 1996;75:65–74. doi: 10.1007/978-3-0348-9225-4_4. [DOI] [PubMed] [Google Scholar]
- Holtzendorff J., Hung D., Brende P., Reisenauer A., Viollier P.H., McAdams H.H., Shapiro L. Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science. 2004;304:983–987. doi: 10.1126/science.1095191. [DOI] [PubMed] [Google Scholar]
- Horvitz H.R., Herskowitz I. Mechanisms of asymmetric cell division: Two Bs or not two Bs, that is the question. Cell. 1992;68:237–255. doi: 10.1016/0092-8674(92)90468-r. [DOI] [PubMed] [Google Scholar]
- Huitema E., Pritchard S., Matteson D., Radhakrishnan S.K., Viollier P.H. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Hung D.Y., Shapiro L. A signal transduction protein cues proteolytic events critical to Caulobacter cell cycle progression. Proc. Natl. Acad. Sci. 2002;99:13160–13165. doi: 10.1073/pnas.202495099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta A.A., McGrath P.T., Reisenauer A., McAdams H.H., Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc. Natl. Acad. Sci. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C., Hung D., Shapiro L. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc. Natl. Acad. Sci. 2001;98:4095–4100. doi: 10.1073/pnas.051609998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Grau R., Perego M. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J. Bacteriol. 2000a;182:303–310. doi: 10.1128/jb.182.2.303-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Shao W., Perego M., Hoch J.A. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 2000b;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- Judd E.M., Comolli L.R., Chen J.C., Downing K.H., Moerner W.E., McAdams H.H. Distinct constrictive processes, separated in time and space, divide Caulobacter inner and outer membranes. J. Bacteriol. 2005;187:6874–6882. doi: 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K.C., Shapiro L. tmRNA in Caulobacter crescentus is cell cycle regulated by temporally controlled transcription and RNA degradation. J. Bacteriol. 2003a;185:1825–1830. doi: 10.1128/JB.185.6.1825-1830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K.C., Shapiro L. tmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J. Bacteriol. 2003b;185:573–580. doi: 10.1128/JB.185.2.573-580.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H., Matroule J.Y., Jacobs-Wagner C. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev. Cell. 2003;5:149–159. doi: 10.1016/s1534-5807(03)00191-6. [DOI] [PubMed] [Google Scholar]
- Lam H., Schofield W.B., Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Laub M.T., McAdams H.H., Feldblyum T., Fraser C.M., Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- Laub M.T., Chen S.L., Shapiro L., McAdams H.H. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. 2002;99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister T.J., Cook W.R., Weigand R., Rothfield L.I. Membrane-murein attachment at the leading edge of the division septum: A second membrane-murein structure associated with morphogenesis of the Gram-negative bacterial division septum. J. Bacteriol. 1987;169:3945–3951. doi: 10.1128/jb.169.9.3945-3951.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques M.V., Gomes S.L., Gober J.W. A gene coding for a putative σ54 activator is developmentally regulated in Caulobacter crescentus. J. Bacteriol. 1997;179:5502–5510. doi: 10.1128/jb.179.17.5502-5510.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matroule J.Y., Lam H., Burnette D.T., Jacobs-Wagner C. Cytokinesis monitoring during development: Rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell. 2004;118:579–590. doi: 10.1016/j.cell.2004.08.019. [DOI] [PubMed] [Google Scholar]
- McGrath P.T., Lee H., Zhang L., Iniesta A.A., Hottes A.K., Tan M.H., Hillson N.J., Hu P., Shapiro L., McAdams H.H. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat. Biotechnol. 2007;25:584–592. doi: 10.1038/nbt1294. [DOI] [PubMed] [Google Scholar]
- Micklem D.R. mRNA localisation during development. Dev. Biol. 1995;172:377–395. doi: 10.1006/dbio.1995.8048. [DOI] [PubMed] [Google Scholar]
- Ohta N., Newton A. The core dimerization domains of histidine kinases contain recognition specificity for the cognate response regulator. J. Bacteriol. 2003;185:4424–4431. doi: 10.1128/JB.185.15.4424-4431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N., Lane T., Ninfa E.G., Sommer J.M., Newton A. A histidine protein kinase homologue required for regulation of bacterial cell division and differentiation. Proc. Natl. Acad. Sci. 1992;89:10297–10301. doi: 10.1073/pnas.89.21.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce D.L., O’Donnol D.S., Allen R.C., Javens J.W., Quardokus E.M., Brun Y.V. Mutations in DivL and CckA rescue a divJ null mutant of Caulobacter crescentus by reducing the activity of CtrA. J. Bacteriol. 2006;188:2473–2482. doi: 10.1128/JB.188.7.2473-2482.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popham D.L., Szeto D., Keener J., Kustu S. Function of a bacterial activator protein that binds to transcriptional enhancers. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- Quon K.C., Marczynski G.T., Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- Quon K.C., Yang B., Domian I.J., Shapiro L., Marczynski G.T. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier A.F. The maternal–zygotic transition: Death and birth of RNAs. Science. 2007;316:406–407. doi: 10.1126/science.1140693. [DOI] [PubMed] [Google Scholar]
- Shapiro L., Losick R. Protein localization and cell fate in bacteria. Science. 1997;276:712–718. doi: 10.1126/science.276.5313.712. [DOI] [PubMed] [Google Scholar]
- Shapiro L., McAdams H.H., Losick R. Generating and exploiting polarity in bacteria. Science. 2002;298:1942–1946. doi: 10.1126/science.1072163. [DOI] [PubMed] [Google Scholar]
- Simon R., Priefer U., Puhler A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram negative bacteria. Biotechnology (N.Y.) 1983;1:784–790. [Google Scholar]
- Skerker J.M., Laub M.T. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat. Rev. Microbiol. 2004;2:325–337. doi: 10.1038/nrmicro864. [DOI] [PubMed] [Google Scholar]
- Skerker J.M., Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits W.K., Bongiorni C., Veening J.W., Hamoen L.W., Kuipers O.P., Perego M. Temporal separation of distinct differentiation pathways by a dual specificity Rap–Phr system in Bacillus subtilis. Mol. Microbiol. 2007;65:103–120. doi: 10.1111/j.1365-2958.2007.05776.x. [DOI] [PubMed] [Google Scholar]
- Sommer J.M., Newton A. Pseudoreversion analysis indicates a direct role of cell division genes in polar morphogenesis and differentiation in Caulobacter crescentus. Genetics. 1991;129:623–630. doi: 10.1093/genetics/129.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J.B., Stock A.M., Mottonen J.M. Signal transduction in bacteria. Nature. 1990;344:395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- Stragier P., Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- Thanbichler M., Iniesta A.A., Shapiro L. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 2007;35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier P.H., Shapiro L. A lytic transglycosylase homologue, PleA, is required for the assembly of pili and the flagellum at the Caulobacter crescentus cell pole. Mol. Microbiol. 2003;49:331–345. doi: 10.1046/j.1365-2958.2003.03576.x. [DOI] [PubMed] [Google Scholar]
- Viollier P.H., Sternheim N., Shapiro L. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J. 2002a;21:4420–4428. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier P.H., Sternheim N., Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc. Natl. Acad. Sci. 2002b;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier P.H., Thanbichler M., McGrath P.T., West L., Meewan M., McAdams H.H., Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Lehmann R. Nanos is the localized posterior determinant in Drosophila. Cell. 1991;66:637–647. doi: 10.1016/0092-8674(91)90110-k. [DOI] [PubMed] [Google Scholar]
- Wang S.P., Sharma P.L., Schoenlein P.V., Ely B. A histidine protein kinase is involved in polar organelle development in Caulobacter crescentus. Proc. Natl. Acad. Sci. 1993;90:630–634. doi: 10.1073/pnas.90.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler R.T., Shapiro L. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol. Cell. 1999;4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Knoblich J.A. Lethal giant larvae take on a life of their own. Trends Cell Biol. 2006;16:234–241. doi: 10.1016/j.tcb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Wolfe A.J. The acetate switch. Microbiol. Mol. Biol. Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Ohta N., Newton A. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc. Natl. Acad. Sci. 1998;95:1443–1448. doi: 10.1073/pnas.95.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]