Abstract
Hypoxia induces rapid and dramatic changes in cellular metabolism, in part through inhibition of target of rapamycin (TOR) kinase complex 1 (TORC1) activity. Genetic studies have shown the tuberous sclerosis tumor suppressors TSC1/2 and the REDD1 protein to be essential for hypoxia regulation of TORC1 activity in Drosophila and in mammalian cells. The molecular mechanism and physiologic significance of this effect of hypoxia remain unknown. Here, we demonstrate that hypoxia and REDD1 suppress mammalian TORC1 (mTORC1) activity by releasing TSC2 from its growth factor-induced association with inhibitory 14–3–3 proteins. Endogenous REDD1 is required for both dissociation of endogenous TSC2/14–3–3 and inhibition of mTORC1 in response to hypoxia. REDD1 mutants that fail to bind 14–3–3 are defective in eliciting TSC2/14–3–3 dissociation and mTORC1 inhibition, while TSC2 mutants that do not bind 14-3-3 are inactive in hypoxia signaling to mTORC1. In vitro, loss of REDD1 signaling promotes proliferation and anchorage-independent growth under hypoxia through mTORC1 dysregulation. In vivo, REDD1 loss elicits tumorigenesis in a mouse model, and down-regulation of REDD1 is observed in a subset of human cancers. Together, these findings define a molecular mechanism of signal integration by TSC1/2 that provides insight into the ability of REDD1 to function in a hypoxia-dependent tumor suppressor pathway.
[Keywords: Hypoxia, tuberous sclerosis, mTOR, REDD1, tumor suppressor]
The tuberous sclerosis tumor suppressor proteins TSC1 (hamartin) and TSC2 (tuberin) form a molecular complex that functions to integrate the cellular response to growth factor, nutrient, and oxygen availability (Pan et al. 2004). The essential role of the TSC1/2 complex in cellular homeostasis is evidenced by tuberous sclerosis syndrome, which results from germline mutations in the genes encoding either TSC1 or TSC2 and is characterized by benign tumors (hamartomas) of multiple tissues, including the kidney, brain, heart, lung, and skin (Crino et al. 2006). Genetic and biochemical data both in Drosophila and in mammalian cells have demonstrated that a major function of the TSC1/2 complex is to inhibit the activity of the checkpoint protein kinase complex TORC1 (target of rapamycin complex 1) (Hay and Sonenberg 2004). Inhibition of TORC1 activity results from TSC2 catalytic function as a GTPase-activating protein (GAP) toward the small GTPase Rheb, a positively acting upstream regulator of TORC1 (Inoki et al. 2003; Tee et al. 2003).
The mammalian TORC1 complex (mTORC1), composed of the proteins mTOR, raptor, and mLST8, functions as a key regulator of cell growth and cellular proliferation (Sarbassov et al. 2005). Two of the most well-studied targets of mTORC1 phosphorylation that mediate these effects are ribosomal S6 kinase 1 (S6K1) and the eukaryotic initiation factor 4E (eIF4E)-binding protein 4E-BP1 (Fingar and Blenis 2004). Phosphorylated S6K1 in turn phosphorylates ribosomal protein S6, an essential regulator of protein translation and cell growth (Ruvinsky et al. 2005). mTORC1-mediated phosphorylation of 4E-BP1 induces its dissociation from eIF4E, the rate-limiting factor for initiation of Cap-dependent translation (Sonenberg and Gingras 1998). Inappropriate control of mTORC1 activity is a hallmark of many benign and malignant human tumors, suggesting an important role in tumorigenesis (Guertin and Sabatini 2007).
A major mechanism for regulation of mTORC1 involves growth factor signaling, which promotes mTORC1 activity in large part through inhibition of the TSC1/2 complex (Gao and Pan 2001; Potter et al. 2001). TSC1/2 regulation in this setting occurs as a result of TSC2 phosphorylation through multiple growth factor signaling pathways, including PI3K/AKT, MEK/ERK/RSK, and MAPK/MK2 (Inoki et al. 2002; Manning et al. 2002; Li et al. 2003; Roux et al. 2004; Ma et al. 2005). In addition to inhibitory phosphorylation, TSC2 is activated by phosphorylation through the LKB1/AMPK pathway, which contributes to mTORC1 inhibition in the setting of energy deprivation and long-term hypoxic stress (Corradetti et al. 2004; Shaw et al. 2004; Liu et al. 2006).
The precise mechanism by which TSC2 phosphorylation regulates TSC1/2 complex activity remains controversial. One mechanism that is thought to contribute to TSC1/2 regulation involves phosphorylation-dependent association of TSC2 with 14–3–3 proteins. TSC2 binds 14–3–3 proteins in vitro (Nellist et al. 2002) and in vivo (Li et al. 2003; Zhang et al. 2003), and this interaction has been shown to inhibit TSC1/2 signaling to mTORC1 (Cai et al. 2006). Since TSC2 binding to 14–3–3 is enhanced by TSC2 phosphorylation through pathways including PI3K/AKT and MAPK (Li et al. 2003; Cai et al. 2006), a plausible model emerged whereby mTORC1 activation in response to growth factors is mediated at least in part through TSC2 phosphorylation and consequent 14–3–3 association (Cai et al. 2006). While substantial experimental evidence supports this model, studies have been hampered by the fact that the endogenous TSC2/14–3–3 interaction has proven difficult to characterize. Furthermore, recent studies have demonstrated that 14–3–3 proteins may also play a role in mTORC1 regulation downstream from the TSC1/2 complex (Sancak et al. 2007; Vander Haar et al. 2007). Thus, it remains to be determined to what extent the TSC2/14–3–3 interaction contributes to mTORC1 control, and under which physiologic circumstances this regulatory mechanism is most relevant.
Oxygen is an essential regulator of cellular metabolism. Under hypoxic conditions cells rapidly activate a variety of adaptive mechanisms that limit energy expenditure through inhibition of energy-intensive processes including protein translation (Wouters et al. 2005; Liu et al. 2006). A major mechanism for this effect involves the inhibition of mTORC1 activity that is observed following exposure to modest hypoxia (1% O2) (Arsham et al. 2003). Recent studies have demonstrated that regulation of mTORC1 activity under hypoxic conditions occurs through a novel pathway involving the TSC1/2 complex and the REDD1 gene (Brugarolas et al. 2004; Reiling and Hafen 2004). REDD1 (also know as RTP801/Dig1/DDIT4) is a member of a gene family that includes its paralog REDD2 (RTP801L, DDIT4L) and the Drosophila orthologs Scylla and Charybdis (Ellisen et al. 2002; Reiling and Hafen 2004; Sofer et al. 2005). Like the Drosophila orthologs, REDD1 expression is highly induced in response to hypoxia (Brugarolas et al. 2004). In mammalian cells, REDD1 overexpression is sufficient to potently inhibit mTORC1 activity, while genetic loss of REDD1 leads to a profound failure to appropriately down-regulate mTORC1 activity in response to hypoxia (Brugarolas et al. 2004; Corradetti et al. 2005; Sofer et al. 2005). Consistent with these findings, overexpression of Scylla and/or Charybdis produced smaller flies and smaller cell size, and loss of both genes was associated with increased cell size and larger flies (Reiling and Hafen 2004). Genetic data in flies and in mammalian cells demonstrate that REDD1 functions in a TSC2-dependent manner to regulate mTORC1 activity (Reiling and Hafen 2004; Sofer et al. 2005). Furthermore, REDD1 orthologs were shown to function in a dominant manner downstream from PI3K/AKT signaling, potentially suggesting an effect on the TSC1/2 complex itself (Reiling and Hafen 2004). Nevertheless, given that REDD1 exhibits neither homology with other known proteins nor recognizable functional domains, the mechanism by which REDD1 regulates mTORC1 activity has remained unresolved.
The link between mTORC1 dysregulation and human tumorigenesis suggests a potential role for hypoxia regulation of mTORC1 activity in tumor development. Given that a period of hypoxic stress is thought to occur virtually universally during human tumorigenesis, a failure to appropriately suppress mTORC1 activity under hypoxia could conceivably contribute to tumorigenesis. Here, we demonstrate the molecular mechanism by which REDD1 regulates TSC1/2-mTORC1 signaling, and we establish the relevance of this pathway in tumor suppression. We show that TSC1/2-dependent regulation of mTORC1 activity in response to hypoxia occurs through dissociation of inhibitory 14–3–3 from the TSC2 protein. This dissociation is mediated by the REDD1 protein, which binds 14–3–3 and is both necessary and sufficient to induce TSC2/14–3–3 dissociation and mTORC1 inhibition. We find that REDD1 specifically opposes PI3K/AKT-induced TSC2/14–3–3 association and mTORC1 activation, that loss of REDD1 drives tumor formation in an AKT-dependent genetic model, and that REDD1 is down-regulated in a subset of human carcinomas. Together, these results identify a molecular mechanism of positive and negative signal integration by the TSC1/2 complex. This mechanism provides an explanation for the ability of REDD1 to function in a hypoxia-mediated mTORC1 inhibitory pathway that contributes both to normal cellular homeostasis and tumor suppression.
Results
TSC2 and REDD1 are required for regulation of mTORC1 activity by hypoxia
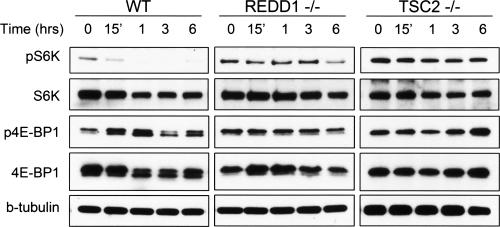
Studies in Drosophila and in mammalian cells have demonstrated that regulation of mTORC1 activity by hypoxia requires REDD1 and TSC2 (Brugarolas et al. 2004; Reiling and Hafen 2004). To analyze directly the requirement for these two genes in the hypoxia response we compared the timing of mTORC1 regulation in wild-type mouse embryo fibroblasts (MEFs), REDD1−/− MEFs, or TSC2−/− MEFs (the latter of which are derived on a p53−/− background in order to overcome premature senescence associated with TSC2 loss) (Fig. 1; Supplemental Fig. 1; Zhang et al. 2003). In agreement with previous observations (Arsham et al. 2003), we observed rapid inhibition of mTORC1 activity in wild-type cells under hypoxic conditions (1% O2), as assessed by examining phosphorylation of its two best-characterized direct substrates, S6K (Thr389, the major rapamycin-sensitive site) (Burnett et al. 1998), and 4E-BP1 (Thr70, one of several rapamycin-sensitive sites) (Fig. 1; Gingras et al. 2001). Down-regulation of mTORC1 substrate phosphorylation was maximal within 3 h in both wild-type cell populations. In contrast, both REDD1−/− and TSC2−/− MEFs showed a profound defect in mTORC1 substrate dephosphorylation at all time points in this analysis. These data demonstrate that regulation of mTORC1 activity by hypoxia requires both REDD1 and TSC2, consistent with genetic data demonstrating that REDD1 functions upstream of TSC2 to inhibit mTORC1 activity in response to hypoxia (Reiling and Sabatini 2006).
Figure 1.
REDD1 and TSC2 are required for inhibition of mTORC1 activity in response to hypoxia. Dephosphorylation of S6K (T389) and 4E-BP1 (T70) is observed in wild-type but not REDD1−/− or TSC2−/− cells. MEFs of each genotype growing in 10% serum were exposed to hypoxia (1% O2) for the indicated times. The same blot was stripped and reprobed for the respective total proteins. Note the prominence of hypophosphorylated 4E-BP1 (lower band) upon hypoxic exposure of wild-type cells. β-Tubulin (b-tubulin) serves as a loading control.
Hypoxia opposes serum-dependent association of endogenous 14–3–3 and TSC2
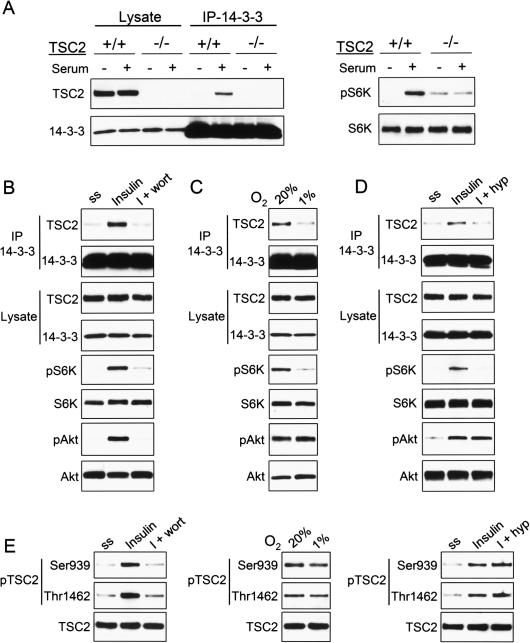
We observe that hypoxia inhibits mTORC1 activity in cells cultured in 10% serum (Fig. 1), and previous studies have demonstrated that hypoxia is sufficient to block insulin-induced mTORC1 activation (Arsham et al. 2003). Thus, hypoxia might function by opposing growth factor-mediated signaling to TSC1/2-mTORC1. Given that growth factors are thought to regulate TSC1/2 at least in part by promoting phosphorylation-dependent binding of TSC2 to inhibitory 14–3–3 proteins (Cai et al. 2006), we asked whether hypoxia had a measurable effect on the TSC2/14–3–3 interaction. To test this hypothesis we first established conditions under which we could readily detect the endogenous TSC2/14–3–3 complex. As predicted, serum stimulation robustly increased the association between endogenous TSC2 and 14–3–3 (Fig. 2A). Furthermore, insulin treatment stimulated TSC2/14–3–3 interaction coincident with AKT activation, while wortmannin treatment blocked insulin-induced complex formation and AKT activation (Fig. 2B). As expected, TSC2 inhibition through formation of the TSC2/14–3–3 complex was reflected in mTORC1 activation in response to serum and insulin, as assessed by phosphorylation of the mTORC1 substrate S6K (Fig. 2A,B). These findings are in agreement with previous studies demonstrating that PI3K signaling regulates TSC1/2 and promotes mTORC1 activation at least in part through regulation of the TSC2/14–3–3 association (Cai et al. 2006).
Figure 2.
Hypoxia opposes the PI3K-dependent 14–3–3/TSC2 association and mTORC1 activation. (A) The interaction of endogenous TSC2 and 14–3–3 is serum-inducible. Wild-type or TSC2−/− MEFs were serum-starved, then treated with serum (90 min) followed by IP for endogenous 14–3–3. (Right) pS6K (T389) indicating serum-induced activation of mTORC1. As expected, TSC2−/− cells exhibit elevated basal mTORC1 activity and reduced serum-responsiveness. (B) The endogenous TSC2/14–3–3 interaction is PI3K-dependent. MEFs were serum-starved, then treated with insulin (200 nM, 90 min) or pretreated with wortmannin (100 nM, 30 min) followed by insulin, prior to IP for endogenous 14–3–3. (C) Hypoxia induces TSC2/14–3–3 dissociation. MEFs in 10% serum were exposed to hypoxia (1% O2) for 3 h prior to IP as above. (D) Hypoxia opposes insulin-induced TSC2/14–3–3 association. Cells (293T) were serum-starved, then treated with insulin (200 nM, 5 h) or exposed to hypoxia (hyp) beginning 1 h prior to insulin treatment, followed by IP as above. Note that both wortmannin and hypoxia inhibit complex formation and mTORC1-dependent phosphorylation of S6K (T389), while only wortmannin inhibits AKT phosphorylation. (E) Hypoxia does not induce TSC2 dephosphorylation, unlike serum starvation or wortmannin. Lysates from experiments B–D were probed with the indicated phospho-specific TSC2 antibodies.
Next, we asked whether the TSC2/14–3–3 interaction was affected under hypoxic conditions. Remarkably, hypoxia consistently induced substantial dissociation of the endogenous TSC2/14–3–3 complex (Fig. 2C). As predicted this effect corresponded with inhibition of mTORC1 activity, evidenced by reduced S6K phosphorylation (Fig. 2C). Notably, hypoxia-induced dissociation of TSC2/14–3–3 and dephosphorylation of S6K were observed in cells grown in full (10%) serum, and these effects were not associated with any change in the levels of either TSC2 or 14–3–3 proteins. Furthermore, hypoxia also blocked the ability of insulin to induce endogenous TSC2/14–3–3 complex formation (Fig. 2D). Together, these findings demonstrate that hypoxia opposes serum and PI3K-induced association of endogenous TSC2 and 14–3–3. Potentially, hypoxia-induced release of TSC2 from inhibitory 14–3–3 might therefore provide a mechanism for rapid TSC2 activation and mTORC1 inhibition.
Wortmannin induces dissociation of TSC2 and 14–3–3 by inhibiting AKT activation, which results in dephosphorylation of TSC2 at critical residues required for 14–3–3 binding. Indeed, we observed that wortmannin treatment blocked the ability of insulin to induce phosphorylation of AKT, resulting in hypophosphorylation of TSC2 at S939 and T1462, two residues phosphorylated by AKT that are known to be important for 14–3–3 binding to TSC2 (Fig. 2B,E; Cai et al. 2006). Given that the effect of hypoxia in inducing dissociation of TSC2 and 14–3–3 was quantitatively similar to that of wortmannin on this protein complex (Fig. 2B,C), we asked whether hypoxia might also function by altering TSC2 phosphorylation. Interestingly, however, we observed no change in phosphorylation of AKT or TSC2 under hypoxic conditions that induced TSC2/14–3–3 dissociation (Fig. 2E). Thus, it appears that hypoxia opposes PI3K/AKT-induced TSC2/14–3–3 association through a mechanism that does not involve a change in AKT-dependent TSC2 phosphorylation.
Regulation of TSC2/14–3–3 association is important for hypoxia signaling to mTORC1.
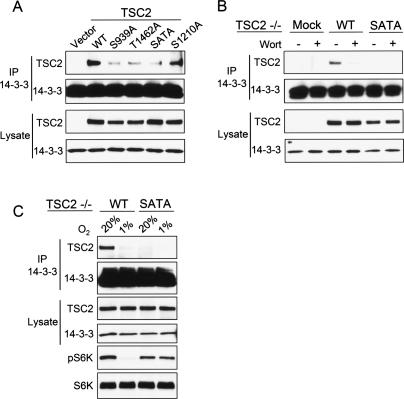
We next wished to establish whether the regulation of the TSC2/14–3–3 association we observed under hypoxic conditions was important for TSC1/2 signaling to mTORC1. Our initial approach to address this question was to generate a TSC2 point mutant that was defective for 14–3–3 binding. We reasoned that such a mutant should also be defective in hypoxia signaling to mTORC1. Given the ability of PI3K/AKT activation to stimulate endogenous TSC2/14–3–3 complex formation (Fig. 2), we focused on TSC2 residues S939 and T1462, which are direct sites for phosphorylation by AKT and are known to be critical for transducing growth factor signals from PI3K/AKT to mTORC1 (Manning et al. 2002). We first tested in a transfection assay the ability of each of these mutants or the double mutant (SATA) to bind endogenous 14–3–3. Each of these mutants demonstrated severely reduced binding to endogenous 14–3–3, with the SATA mutant exhibiting only a slightly greater defect in binding than either single-point mutant (Fig. 3A). Of note, mutation of S1210, a residue phosphorylated through the MAPK–MK2 pathway (Li et al. 2003), only minimally reduced binding of TSC2 to endogenous 14–3–3 (Fig. 3A). To explore the properties of the SATA mutant in a physiologic context, we stably reconstituted expression of either wild-type or SATA TSC2 at endogenous levels in TSC2−/− cells. As predicted, wild-type reconstituted TSC2 showed the expected binding to endogenous 14–3–3, which was regulated in a PI3K-dependent manner (Fig. 3B). In contrast, the SATA mutant showed little detectable interaction with 14–3–3, even in the presence of full serum. These findings are consistent with our data above that PI3K/AKT-dependent phosphorylation promotes the endogenous TSC2/14–3–3 interaction.
Figure 3.
TSC2/14–3–3 binding is important for hypoxia regulation of mTORC1. (A) AKT-dependent phosphorylation sites on TSC2 are required for 14–3–3 binding. Wild-type (WT) TSC2, the indicated point mutants, or the double mutant (SATA) were transfected into TSC2−/− cells, followed by IP for endogenous 14–3–3. S1210 is an MK2-phosphorylated site whose mutation does not dramatically affect 14–3–3 binding. (B) S939/T1462 are required for PI3K-dependent regulation of TSC2/14–3–3 binding. TSC2−/− cells were stably reconstituted with wild-type or SATA TSC2, then cells in full serum were treated with wortmannin (100 nM, 60 min) followed by IP for endogenous 14–3–3. (C) Absence of hypoxia regulation of mTORC1 correlates with defective TSC2/14–3–3 interaction. Stably reconstituted cells shown in B were exposed to hypoxia (3 h), followed by IP for endogenous 14–3–3 or Western blot analysis for phospho-S6K (T389) to assess mTORC1 activity. The same blot was stripped and reprobed for total S6K.
Although TSC2−/− cells exhibited little or no regulation of mTORC1 activity under hypoxic conditions (Fig. 1), reconstitution of wild-type TSC2 into these cells restored the ability of hypoxia to inhibit mTORC1 activity as evidenced by S6K dephosphorylation (Fig. 3C). More importantly, this effect correlated with the ability of hypoxia to induce dissociation of TSC2 and 14–3–3 (Fig. 3C). In contrast, however, cells reconstituted with TSC2 SATA showed little or no binding to endogenous 14–3–3 and essentially no regulation of mTORC1 substrate phosphorylation under hypoxic conditions (Fig. 3C). Together these findings suggest a model in which hypoxia opposes growth factor signaling to mTORC1 by inducing dissociation of the growth factor-induced TSC2/14–3–3 complex. As noted above (Fig. 2E), hypoxia-induced TSC2/14–3–3 dissociation involves a pathway independent of TSC2 dephosphorylation at the essential 14–3–3-binding residues.
A conserved 14–3–3-binding motif within REDD1 is required for signaling to TSC2/mTORC1
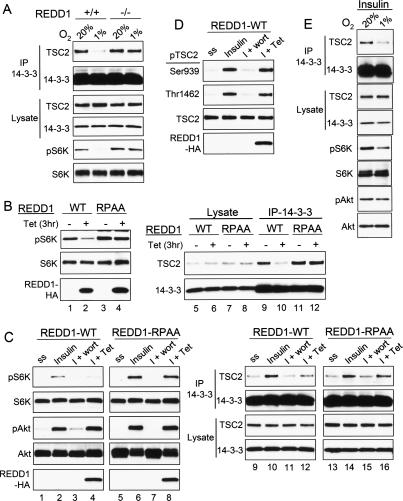
Since hypoxia signaling to mTORC1 is thought to require an effect of REDD1 on TSC1/2, we next sought to test whether REDD1-dependent signaling involved changes in TSC2/14–3–3 complex formation. Thus, we first used matched wild-type and REDD1−/− MEFs to establish whether endogenous REDD1 was required for hypoxia-mediated regulation of the TSC2/14–3–3 interaction. In wild-type MEFs, hypoxia induced the expected dissociation of endogenous TSC2 and 14–3–3, which correlated with significant inhibition of mTORC1 activity (Fig. 4A). In matched REDD1−/− cells, however, TSC2/14–3–3 dissociation was severely attenuated, and this effect coincided with defective mTORC1 inhibition (Fig. 4A). These data support the importance of TSC2/14–3–3 complex regulation in hypoxia signaling to mTORC1, and they demonstrate that endogenous REDD1 is required for this signaling mechanism.
Figure 4.
A conserved 14–3–3-binding motif within REDD1 is required for mTORC1 signaling and TSC2/14–3–3 regulation. (A) Endogenous REDD1 is required for hypoxia-induced TSC2/14–3–3 dissociation. MEFs of the indicated genotype were treated with hypoxia (3 h) followed by Western analysis or IP for endogenous 14–3–3. Hypoxia-induced TSC2/14–3–3 dissociation and S6K1 (T389) dephosphorylation are both absent in REDD1−/− MEFs. (B) Induction of REDD1 but not the REDD1-RPAA mutant inhibits mTORC1 activity and the endogenous TSC2/14–3–3 association. Expression of REDD1 was induced with tetracycline (Tet) for 3 h (hrs) in U2OS cells, followed by Western blot analysis or IP for endogenous 14–3–3. Dephosphorylation of S6K (T389) (lane 1 vs. lane 2) and TSC2/14–3–3 dissociation (lane 9 vs. lane 10) are only induced by wild-type REDD1. (C) Induction of REDD1 but not REDD1-RPAA opposes insulin-induced mTORC1 activation and endogenous TSC2/14–3–3 association. Cells were serum-starved, then treated with insulin (200 nM, 2 h), or were pretreated with wortmannin (100 nM, 30 min) or REDD1 induction (Tet, 60 min) prior to insulin (I) treatment. Lysates were analyzed by Western blot analysis and by IP for endogenous 14–3–3. Only wild-type REDD1 blocks insulin-induced S6K (T389) phosphorylation (lane 2 vs. lane 4) and opposes TSC2/14–3–3 association (lane 10 vs. lane 12). Note that REDD1 induction, unlike wortmannin treatment, does not affect AKT (S473) phosphorylation. (D) REDD1-mediated S6K dephosphorylation and TSC2/14–3–3 dissociation do not involve a change in AKT-dependent TSC2 phosphorylation. Lysates shown in C were probed for the indicated phosphorylated TSC2 residues. (E) Hypoxia replicates tetracycline-induced wild-type REDD1 in opposing insulin effects on TSC2/mTORC1 signaling. Serum-starved MEFs were treated with insulin (200 nM) prior to hypoxia (3 h).
To establish the mechanism of these REDD1-dependent effects, we first asked whether REDD1 was sufficient to induce TSC2/14–3–3 complex dissociation. We previously established that REDD1 expression is sufficient to inhibit endogenous mTORC1 activity in a TSC1/2-dependent manner (Li et al. 2003; Brugarolas et al. 2004; Sofer et al. 2005). Consistent with this finding, cells expressing tetracycline-inducible REDD1 demonstrated rapid dephosphorylation of S6K following induction of REDD1 (Fig. 4B). In order to ask whether REDD1 signaling to mTORC1 was associated with a change in the TSC2/14–3–3 complex, we immunoprecipitated the endogenous complex following tetracycline induction of REDD1. REDD1 induction consistently led to a substantial dissociation of the endogenous TSC2/14–3–3 complex, and this effect corresponded temporally with REDD1 signaling to mTORC1 (Fig. 4B).
Although the REDD1 primary amino acid sequence reveals no recognizable catalytic or other functional domains, we previously mapped an essential domain for REDD1 function to an evolutionarily conserved region between amino acids 96–153 (Li et al. 2003; Brugarolas et al. 2004; Sofer et al. 2005). Motif scanning within this domain revealed a consensus Arg-X-X-X-Ser/Thr-X-Pro-binding site for 14–3–3 proteins that is conserved among mammalian orthologs of both REDD1 and the REDD1-related protein REDD2 (Supplemental Fig. 2; Bridges and Moorhead 2005). This observation led us to hypothesize that REDD1 might oppose the TSC2/14–3–3 interaction through its ability to bind 14–3–3 via this conserved domain. To determine whether this putative 14–3–3-binding motif was important for REDD1 function, we mutated the conserved arginine (R) and proline (P) residues within this motif. Tetracycline induction of the mutant REDD1 (hereafter REDD1-RPAA) at comparable levels to wild-type REDD1 demonstrated that this protein was entirely inactive (Fig. 4B). Thus, REDD1-RPAA induction caused no regulation of the endogenous TSC2/14–3–3 complex and no inhibition mTORC1 activity (Fig. 4B).
We then asked whether REDD1 specifically opposed PI3K signaling to TSC1/2 and mTORC1. Indeed, induction of wild-type REDD1 was sufficient to block insulin-induced mTORC1 activation and TSC2/14–3–3 complex formation (Fig. 4C). In contrast, REDD1-RPAA affected neither mTORC1 induction nor endogenous complex formation following insulin simulation. As was observed with hypoxia itself, REDD1-mediated TSC2/14–3–3 dissociation and mTORC1 signaling did not involve a change in phosphorylation of AKT (Fig. 4C). Furthermore, REDD1 induced no change in TSC2 phosphorylation at the sites required for 14–3–3 binding (Fig. 4D). Taken together, these observations demonstrate that REDD1 functions to oppose serum and PI3K/AKT-induced TSC2/14–3–3 complex formation and thereby to inhibit mTORC1 activity. The requirement for the REDD1 14–3–3-binding motif suggests that REDD1 may regulate TSC2 and mTORC1 through its ability to bind directly to 14–3–3.
REDD1 binds 14–3–3 through the essential binding motif
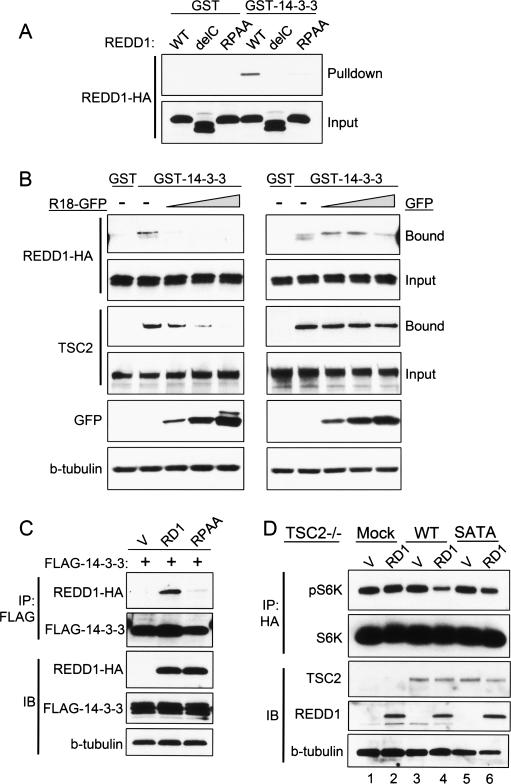
We next asked whether REDD1 could physically interact with 14–3–3 proteins, and whether this interaction was mediated through the putative REDD1 14–3–3-binding motif. We found that REDD1 was specifically affinity-purified using a GST-14–3–3 fusion protein but not GST alone (Fig. 5A). In contrast, the inactive REDD1-ΔC mutant (which lacks the domain containing the conserved 14–3–3-binding motif) bound neither GST-14–3–3 nor GST (Fig. 5A). Most notably, the inactive REDD1-RPAA mutant, which lacks the conserved arginine and proline residues within the putative 14–3–3-binding motif, was also severely defective in binding to GST-14–3–3 (Fig. 5A). Interestingly, mutation of the conserved serine residue within this motif (S137A) induced a partial loss of binding to GST-14–3–3, which correlated with a partial loss of function (data not shown). Thus, REDD1 function is correlated with its ability to bind 14–3–3.
Figure 5.
REDD1 binds 14–3–3 through the conserved binding motif. (A) REDD1 binding to 14–3–3 requires the conserved motif. 293T cells were transfected with wild-type (WT) REDD1, REDD1-delC (lacking the conserved central domain), or REDD1-RPAA, and lysates were purified by binding to GST or GST-14–3–3, followed by elution and Western blot analysis of bound protein. (B) Competition assay showing specific binding of REDD1 to 14–3–3. REDD1 was expressed as above, and lysates were incubated in the presence of increasing amounts of the 14–3–3-binding R18 decoy peptide fused to GFP (R18-GFP, left) or GFP control (right). Like REDD1, TSC2 also exhibits specific binding to 14–3–3, which can be competed out by R18-GFP but not GFP alone. (C) REDD1 but not REDD1-RPAA coimmunoprecipitates with 14–3–3. REDD1 and 14–3–3ζ were cotransfected into 293T cells, followed by IP for tagged 14–3–3. (D) REDD1 inhibition of mTORC1 activity requires TSC2/14–3–3 binding. TSC2−/− MEFs stably reconstituted with wild-type (WT) or mutant (SATA) TSC2 were cotransfected with REDD1 and HA-S6K, followed by IP for HA-S6K. REDD1-mediated dephosphorylation of S6K (T389) requires WT-TSC2 (lane 4) and is defective in SATA-TSC2-reconstituted cells (lane 6).
To further substantiate specific binding of REDD1 to 14–3–3, we performed a competition experiment using the well-characterized R18 decoy peptide that binds 14–3–3 with high affinity, resulting in disruption of substrate binding (Jin et al. 2004). We expressed this peptide as a GFP fusion protein and used it to compete with REDD1 for binding to 14–3–3 in the affinity purification assay as described above. As a control, we compared the ability of R18-GFP to compete for binding to 14–3–3 with TSC2. Expression of R18-GFP reduced to background levels the binding of both REDD1 and TSC2 to 14–3–3 in a dose-dependent manner (Fig. 5B). In contrast, equivalent levels of GFP alone had no effect on binding of either REDD1 or TSC2 to 14–3–3 (Fig. 5B). Specific binding of REDD1 but not REDD1-RPAA to 14–3–3 was also observed in coimmunoprecipitation assays (Fig. 5C). Together with the functional data above, these observations suggest that REDD1 binding to 14–3–3 is required for its ability to disrupt the endogenous 14–3–3 complex and to regulate mTORC1 activity.
Having shown that REDD1 binding to 14–3–3 is required for TSC1/2–mTORC1 regulation, we lastly wished to test whether TSC2 must bind 14–3–3 in order to be regulated by REDD1. We therefore asked whether REDD1 could signal to mTORC1 via the TSC2-SATA mutant, which is defective both in binding to 14–3–3 and in hypoxia signaling. We transfected REDD1 into TSC2−/− cells that had been stably reconstituted with either wild-type TSC2 or the TSC2-SATA mutant. In order to assay mTORC1 activity in the transfected cells we cotransfected HA-tagged S6K and examined phosphorylation of the immunoprecipitated S6K. As expected, REDD1 expression had little or no effect on S6K phosphorylation in TSC2−/− cells, while REDD1 substantially inhibited S6K phosphorylation in wild-type TSC2-reconstituted cells (Fig. 5D). In contrast, however, the effect of REDD1 was significantly attenuated in cells reconstituted with the TSC2-SATA mutant that exhibits reduced 14–3–3 binding (Fig. 5D). Thus, the REDD1 signaling pathway involves both REDD1 and TSC2 binding to 14–3–3. Together, our findings support a model whereby REDD1 and hypoxia-dependent mTORC1 regulation occurs through the ability of induced REDD1 to bind 14–3–3 and thereby disrupt binding of inhibitory 14–3–3 to TSC2. This model is further supported by our observation that endogenous REDD1 is colocalized with TSC2 in a cellular membrane compartment, which may function to increase the effective local concentration of REDD1 relative to 14–3–3, thereby enhancing its ability to sequester 14–3–3 from TSC2 (Supplemental Fig. 3).
Loss of hypoxia signaling to TSC2/14–3–3 promotes oncogenic properties
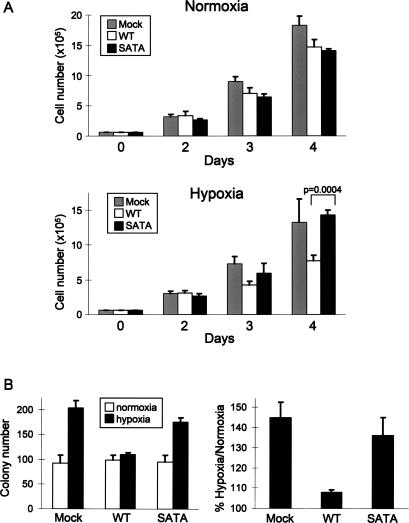
A key question remaining was the physiologic significance of REDD1 signaling to TSC2/14–3–3 in response to hypoxia. First, we sought to address the contribution of the TSC2/14–3–3 interaction in regulating proliferation under hypoxic conditions. We used the TSC2−/− cells described above (Fig. 3), which had been reconstituted with either wild-type TSC2 or the SATA mutant that does not bind 14–3–3 and that is defective for hypoxia-mediated suppression of mTORC1. We showed previously that TSC2−/− cells demonstrated a proliferation advantage specifically under hypoxic conditions compared with wild-type cells, and that this advantage was attributable to elevated mTORC1 activity (Brugarolas et al. 2004). In keeping with these results, we found that stable reconstitution of wild-type TSC2 into TSC2−/− cells substantially suppressed proliferation under hypoxic but not normoxic conditions (Fig. 6A). Reconstitution of the SATA mutant, however, failed to suppress proliferation in hypoxia, resulting in a consistent proliferation advantage compared with wild-type TSC2-reconstituted cells under hypoxic conditions (Fig. 6A). As expected, these proliferation differences in hypoxia were abolished by concurrent treatment of cells with rapamycin, confirming that mTORC1 dysregulation contributes to the growth advantage of TSC2−/− and TSC2-SATA-reconstituted cells (data not shown).
Figure 6.
Failure of hypoxia signaling through TSC2/14–3–3 promotes proliferation and anchorage-independent growth. (A) Proliferation advantage of TSC2−/− and TSC2-SATA-reconstituted cells (Fig. 3) specifically in hypoxia. Equal numbers of cells were plated in normoxia or hypoxia (1% O2), and viable cells were counted daily by trypan blue exclusion assay. Error bars show SD for triplicate plates in this representative experiment; P-value was derived from two-tailed Student’s t-test. (B) Hypoxia enhances colony formation when hypoxia signaling through TSC2/14–3–3 is defective. The indicated cells were transformed with H-Ras(V12), then plated in soft agar as described (see Materials and Methods) for 3 wk in either normoxia or hypoxia (1% O2). (Left) Colony counts performed in triplicate for a representative experiment. Error bars show SD. (Right) Summary of three independent experiments performed in triplicate. Error bars show SEM.
Given the strong link between mTORC1 dysregulation and tumorigenesis, we next asked whether the failure to suppress mTORC1 through TSC2/14–3–3 was also manifest as an advantage in anchorage-independent growth. We therefore performed a soft agar colony-forming assay in TSC2−/− or reconstituted cells following transformation with H-Ras(V12). Hypoxia had little effect on colony formation in transformed cells reconstituted with wild-type TSC2, with nearly equal numbers of colonies forming under either normoxic or hypoxic conditions (Fig. 6B). In contrast, both mock-transfected TSC2−/− and SATA TSC2-reconstituted cells consistently exhibited a higher frequency of colony formation under hypoxic compared with normoxic conditions (Fig. 6B), which correlated with a failure of hypoxia to suppress mTORC1 activity (Fig. 3) in these cells. These findings argue that regulation of mTORC1 activity through TSC2/14–3–3 is important for suppressing abnormal proliferation and anchorage-independent growth in response to hypoxia.
Endogenous REDD1 functions to suppress tumorigenesis in vivo
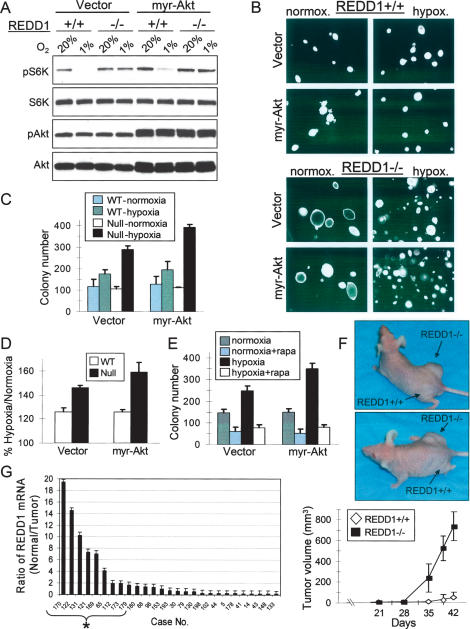
Finally, we wished to test directly whether REDD1-dependent signaling contributes to tumorigenesis. Our data suggest that the REDD1-dependent effect of hypoxia opposes PI3K/AKT activation of mTORC1. Therefore, we hypothesized that the ability of REDD1 to limit mTORC1 activity might be particularly relevant under conditions of elevated AKT activity, as is observed in many human cancers. We immortalized wild-type or REDD1−/− MEFs with SV40 large T-antigen (LTAg), then we examined the effect of hypoxia on mTORC1 activity and soft-agar colony formation in these cells following expression of either a retroviral vector or activated (myristoylated) AKT (myr-AKT). The ability of hypoxia to suppress mTORC1 activity was preserved even in the presence of constitutive AKT activation (Fig. 7A). This effect was REDD1-dependent, as REDD1−/− cells failed to suppress mTORC1 activity under hypoxia, either in the presence or absence of myr-AKT (Fig. 7A). REDD1-dependent dysregulation of mTORC1 activity was reflected in anchorage-independent growth, as REDD1−/− cells exhibited a substantially greater increase in colony number under hypoxic conditions than did wild-type cells (Fig. 7B–D). Under normoxic conditions, however, colony counts were essentially identical in wild-type and REDD1−/− cells, although REDD1−/− cells formed somewhat larger colonies (Fig. 7B,C). In addition, the increased colony number under hypoxia was enhanced in REDD1−/− cells, but not wild-type cells, expressing myr-AKT compared with the control vector (Fig. 7D). Treatment of either vector or myr-AKT-expressing cells with rapamycin substantially abrogated the increase in colony numbers observed in REDD1−/− cells (Fig. 7E). Thus, REDD1 is important for suppression of anchorage-independent growth in response to hypoxia due to its ability to inhibit mTORC1.
Figure 7.
REDD1-dependent hypoxia signaling suppresses tumorigenesis. (A) REDD1-dependent inhibition of mTORC1 in cells expressing myr-AKT. Fibroblasts expressing SV40 LTAg were infected with a control or myr-AKT-expressing retrovirus, then were treated with hypoxia for 3 h prior to Western analysis. (B) Photomicrographs demonstrating a substantially greater number of colonies under hypoxic conditions in the absence of REDD1. LTAg-expressing MEFs were infected with myr-AKT or a control retroviral vector and plated in soft agar under normoxic or hypoxic (1% O2) conditions. (C) Endogenous REDD1 suppresses anchorage-independent growth in hypoxia. Soft agar colony counts performed in triplicate for a representative experiment. Error bars equal SD. (D) Summary of three independent experiments, each performed in triplicate. Error bars equal SEM. (E) REDD1 suppresses anchorage-independent growth in hypoxia through mTORC1 regulation. Soft agar assay was performed in the presence or absence of rapamycin (25 nM) in LTAg-REDD1−/− MEFs infected with myr-AKT for 2 wk. Colony counts performed in triplicate for a representative experiment. Error bars equal SD. (F) Loss of REDD1 promotes tumorigenesis in vivo. Nude mice were injected with 2 × 106 myr-AKT-expressing cells described above. (Top) Representative mice showing large tumors in right (REDD1−/−-injected) but not left (wild-type-injected) flank. (Bottom) Mean tumor volume (N = 12 mice for each genotype). (G) REDD1 expression is down-regulated in primary breast carcinomas relative to specimen-matched normal epithelium. The ratio of REDD1 expression in normal/tumor pairs is shown, such that higher bars indicate lower tumor-specific REDD1 expression; asterisk (*) indicates tumors with more than twofold REDD1 down-regulation. Error bars show SD of three measurements.
To determine whether REDD1 signaling contributes to tumor suppression in vivo we injected either wild-type or REDD1−/− cells expressing myr-AKT subcutaneously into immunodeficient mice. Numerous studies have demonstrated that this tumorigenesis assay places cells under hypoxic stress, making this model a relevant one with which to test the consequences of defective REDD1 signaling (Blouw et al. 2003). As anticipated, tumor growth from wild-type myr-AKT-expressing cells was modest, as only very small tumors were evident in subset of injected mice 6 wk following injection. In dramatic contrast, REDD1−/− myr-AKT cells formed large, rapidly growing tumors that were manifest in a much shorter period of time (Fig. 7F). By 5 wk, for example, we observed a 36-fold difference in the mean volume of tumors derived from REDD1−/− versus wild-type cells (Fig. 7F). Loss of REDD1-dependent signaling therefore promotes in vivo tumorigenesis of murine cells.
If REDD1 function contributes to tumor suppression in humans, we anticipate that a subset of human tumors will exhibit loss of REDD1 function compared with normal tissues. To address this possibility we performed quantitative examination of REDD1 expression, using a panel of primary breast carcinoma specimens that had undergone laser-capture microdissection (LCM) in order to isolate distinct populations of carcinoma cells and specimen-matched normal epithelium (Ma et al. 2003). We assayed REDD1 expression by quantitative real-time RT–PCR (QRT–PCR) in 27 matched normal/tumor pairs. REDD1 expression was significantly down-regulated in eight of 27 carcinoma specimens relative to patient-matched control epithelium (Fig. 7G). We then confirmed these findings by REDD1 RNA in situ hybridization in the corresponding tissue specimens (data not shown). Thus, REDD1 is deficient in a subset of primary human breast carcinomas. It is notable that a similar frequency of REDD1 down-regulation was observed in an expression profiling study of 41 patient-matched normal prostate tissues versus invasive prostate carcinomas (Lapointe et al. 2004). Taken together, these findings support the view that the endogenous REDD1 pathway, which is essential for limiting mTORC1 activity under hypoxic stress, functions as a tumor suppressor mechanism in vivo (Fig. 8).
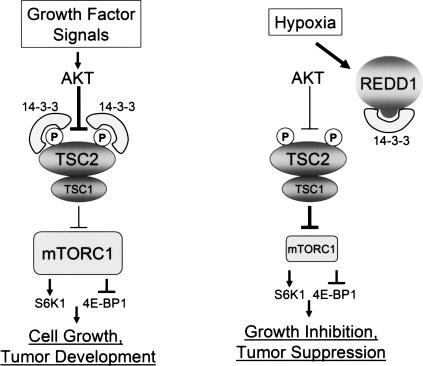
Figure 8.
Model for REDD1-dependent regulation of mTORC1 activity and cell growth. Insulin and other growth factors activate mTORC1 through AKT-mediated phosphorylation of TSC2, which promotes TSC2/14–3–3 association and thereby inhibits TSC1/2 function. In response to hypoxia, REDD1 is induced and binds 14–3–3, resulting in TSC2/14–3–3 dissociation, TSC1/2 activation, and mTORC1 inhibition. REDD1 can inhibit mTORC1 even in the presence of constitutive AKT activation. Consequently, loss of REDD1 in this setting induces further mTORC1 activation that drives tumorigenesis.
Discussion
Hypoxia-dependent regulation of TSC1/2–mTORC1 signaling
The TSC1/2 complex functions as both an essential integrator and gatekeeper of signaling to mTORC1. The profound consequences of TSC1/2 loss of function are evidenced biochemically by the elevation in basal mTORC1 activity observed in the absence of either of these proteins (Zhang et al. 2003). Physiologically, TSC1/2 haploinsufficiency leads to a tumor predisposition syndrome that is associated in most cases with somatic loss of the remaining TSC1/2 allele (Crino et al. 2006). Despite intensive efforts, the precise biochemical mechanisms regulating TSC1/2 activity are poorly understood. Here, we demonstrate that release of TSC2 from inhibitory 14–3–3 is an essential mechanism that restricts mTORC1 activity in response to hypoxic stress. We find that the TSC2/14–3–3 association correlates precisely with changes in mTORC1 activity, either positively in response to PI3K/AKT activity, or negatively in response to hypoxic stress. We tested directly the role of TSC2/14–3–3 binding in hypoxia signaling to mTORC1, by showing that a TSC2 point mutant that is unable to bind 14–3–3 is inert with respect to hypoxia-dependent mTORC1 regulation. Finally, we show that REDD1, an essential regulator of mTORC1 signaling through TSC1/2, is both necessary and sufficient to inhibit both the TSC2/14–3–3 interaction and mTORC1 activity in response to hypoxia. The critical role of TSC2/14–3–3 binding in this regulatory pathway is further underscored by our observation that REDD1 point mutants that are unable to bind 14–3–3, and therefore to inhibit the TSC2/14–3–3 interaction, are inactive for both TSC2/14–3–3 and mTORC1 regulation. Interestingly, recent studies suggest that 14–3–3-dependent pathways may regulate mTORC1 activity both upstream of and downstream from TSC1/2 (Sancak et al. 2007; Vander Haar et al. 2007). Our data, however, underscore the importance of 14–3–3 in the hypoxia-dependent regulation of mTORC1 through REDD1 and TSC1/2.
The TSC1/2 complex is positioned at the nexus of signaling pathways to mTORC1. TSC1/2 therefore functions as a pivotal integrator of positive and negative signals that control cellular metabolism. Nevertheless, the molecular mechanism by which the TSC1/2 complex integrates positive and negative signals has remained entirely elusive. In particular, an intriguing question has been the mechanism by which hypoxia signaling functions in a dominant manner vis-à-vis growth factor signaling to suppress mTORC1 activity (Arsham et al. 2003; Brugarolas et al. 2004). Our data suggest that the REDD1-mediated release of 14–3–3 from TSC2 explains how hypoxic stress acts to override growth factor inputs to TSC1/2. We show that like hypoxia itself, REDD1 induction is sufficient to abrogate both TSC2/14–3–3 binding and mTORC1 activation induced by PI3K/AKT. Dissociation of TSC2/14–3–3 thus serves as a critical switch that mediates the integration by TSC1/2 of growth factor and hypoxia signals to mTORC1. These observations are in keeping with studies in Drosophila, which demonstrated that the REDD1 orthlogs Scylla and Charybdis function in a TSC1/2-dependent manner as potent suppressors of cell growth induced by overexpression of the downstream PI3K-activated kinases PKB and PDK1 (Reiling and Hafen 2004). We find that the effects of REDD1 on TSC2/14–3–3 and mTORC1 require the ability of REDD1 itself to bind 14–3–3. In addition, these effects of hypoxia and REDD1 are not associated with a change in either AKT phosphorylation or TSC2 phosphorylation induced by AKT. This ability of REDD1 to disrupt TSC2/14–3–3 binding therefore provides a means to rapidly extinguish mTORC1 activity under hypoxia, even in the presence of abundant growth factors. The rapid REDD1-dependent effect on mTORC1 activity is evident as early as 15 min following exposure to hypoxia (Fig. 1) and precedes the increase in REDD1 levels, which begin to rise within 1 h (Supplemental Fig. 4). This observation is consistent with our finding that REDD1 is subject to complex post-translational modification that may contribute to its ability to regulate TSC2/14–3–3 and mTORC1. Presumably, the rapid and dominant efffect of hypoxia over growth factor signals in the regulation of mTORC1 activity evolved as a means to limit energy-intensive processes such as protein translation during the shift to anaerobic metabolism (Liu et al. 2006).
Subcellular localization of the TSC1/2 complex is thought to be critical to its signaling function (Cai et al. 2006). We find that REDD1 is colocalized with TSC1/2 within membranes, in keeping with the rapid and potent effect of REDD1 induction on TSC1/2 signaling. Colocalization of REDD1 and TSC2 is predicted to produce an effectively high local concentration of REDD1 relative to 14–3–3, which is only very weakly membrane-localized (Supplemental Fig. 3). Thus, REDD1/TSC2 colocalization likely contributes to the ability of REDD1 to sequester 14–3–3 proteins from TSC2 despite our observation that in total cellular lysates REDD1 is not in molar excess compared with abundant 14–3–3 proteins. Although the ability of REDD1 to bind 14–3–3 is essential for signaling, we find that the serine residue within the canonical Arg-X-X-X-Ser/Thr-X-Pro consensus 14–3–3-binding motif is partly dispensable for 14–3–3 binding and for REDD1 function. This finding implies that REDD1 phosphorylation at this site is not absolutely required for its ability to bind and sequester 14–3–3. Of note, both Drosophila REDD1 orthologs harbor a variant of this motif with the serine residue at the +2 position relative to arginine (Arg-X-Ser-X-X-X-Pro), suggesting evolutionary conservation of the essential arginine and proline residues and potentially of this 14–3–3-dependent mechanism.
REDD1 signaling and its implication for hypoxia as a tumor suppressor
Unlike loss of TSC2, germline loss of REDD1 does not result in dramatic elevation of basal mTORC1 activity in unstressed cells. This observation is likely to explain our observation that REDD1−/− mice, unlike TSC1−/− and TSC2−/− mice, are viable and not highly tumor prone (Sofer et al. 2005). Consistent with these findings, under normoxic conditions we observe similar rates of proliferation and soft agar colony formation in wild-type and REDD1−/− cells. In contrast, however, REDD1 nullizygosity was associated with a substantial increase in colony number under hypoxic conditions. Increased colonies correlated with mTORC1 dysregulation in REDD1−/− cells under hypoxia, and suppression of increased colony formation by rapamycin implicates mTORC1 directly in this phenotype. A particularly pronounced role for REDD1 was evident in cells engineered for constitutive AKT activation. Indeed, REDD1−/−, myr-AKT-expressing cells exhibited not only an enhanced increase in soft agar colonies under hypoxia, but they also rapidly formed tumors in immunodeficient mice compared with wild-type, myr-AKT cells. The ability of AKT expression to enhance the effect of REDD1 loss was also observed in Drosophila, as mutation of Scylla resulted in larger cell size only in the context of PKB/PDK1 overexpression (Reiling and Hafen 2004). The ability of REDD1 to function as a suppressor of tumorigenesis in this context is most likely directly related to the molecular mechanism of mTORC1 regulation that we uncovered. Thus, REDD1 is able to dominantly suppress oncogenic growth factor signals to mTORC1 through its effect on TSC1/14–3–3 (Fig. 7A), predicting that loss of REDD1 would result in unrestrained mTORC1 activity and potentiate tumor growth. Consistent with this hypothesis, our initial analysis of REDD1 expression in human tumors demonstrates that REDD1 is significantly down-regulated compared with normal tissues in a subset of cases (Fig. 7G). More detailed analysis of such tumors is likely to shed light on the tumor-specific genetic context in which REDD1 loss is observed most frequently.
While the role of hypoxia in tumorigenesis is the focus of intensive investigation, much of this work has emphasized the tumor-promoting effects of hypoxia. Indeed, the ability of cells to adapt to anaerobic metabolism and to elicit angiogenesis is important for the tumor cell phenotype. On the other hand, hypoxia is known to induce immediate metabolic demands that require cells to restrict growth-promoting activities (Wouters et al. 2005). Therefore, it seems plausible that hypoxia itself might function as a tumor suppressor mechanism. Nevertheless, little direct evidence has been provided to support this concept. We demonstrate directly that loss of hypoxia-dependent suppression of mTORC1 activity through REDD1 promotes anchorage-independent growth in vitro and tumorigenesis in vivo. These findings are in keeping with the many studies demonstrating the importance of hypoxia as a suppressor of mTORC1, and with the vast body of literature supporting the contribution of elevated mTORC1 activity to human cancer (Guertin and Sabatini 2007). Although our data suggest that little selective pressure may exist to inactivate the REDD1 pathway in normal cells, we speculate that hypoxic stress within nascent tumor cells provides an advantage to cells that have disabled the REDD1-dependent mechanism to limit mTORC1 activity. Once established, tumor cells that have evaded this metabolic checkpoint can still profit from the adaptations to hypoxia mentioned above, which are likely to contribute ultimately to tumor development and progression. Our identification of a hypoxia-dependent pathway linked to tumor suppression therefore provides new insight into potentially early events in tumor evolution.
Materials and methods
Cells and cell culture
Wild-type and REDD1−/− MEFs were derived from E14 litter-matched embryos, and matched wild-type and TSC2−/− (p53−/−) cells were the generous gift of Dr. David Kwiatkowski. To avoid any confounding due to p53 status, immortalized MEFs (SV40 LTAg) were used for data shown, although similar results were obtained with nonimmortalized cells. TSC2−/− cells were reconstituted with human wild-type or mutant TSC2 by transfection, and cells were selected and shown to express ectopic TSC2 at levels comparable to endogenous TSC2. All experiments were reproduced with at least two independently selected reconstituted lines. U2OS cells (T-REx-U2OS) expressing tetracycline-regulated REDD1 were described earlier (Sofer et al. 2005). All cells were propagated in DME/10% FCS/Pen/Strep, except as noted.
Immunoprecipitation (IP) and immunoblot analysis
Cells were lysed on ice in lysis buffer (0.75% NP-40, 1 mM DTT, protease inhibitors [Roche], and phosphatase inhibitors [Sigma] in PBS). Precleared lysates were incubated with either pan-14–3–3 mouse monoclonal antibody (Ab-4, LabVision) or mouse monoclonal HA.11 (Covance) for 2 h at 4°C, and immunocomplexes were precipitated using protein G Sepharose (Amersham Biosciences), then washed four times with lysis buffer prior to analysis by SDS-PAGE. For REDD1/14–3–3 coimmuniprecipitation, cells were harvested in hypotonic buffer (20 mM Tris at pH 7.5, 20 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 5 mM EGTA, 20 mM β-glycerophosphate, 1 mM DTT, protease and phosphatase inhibitors) and briefly sonicated. Precleared lysates were incubated with anti-Flag mouse monoclonal antibody (Sigma), then immunocomplexes were precipitated and washed three times in lysis buffer containing 0.5 M NaCl and 0.5% NP-40 and then washed twice with buffer two (10 mM HEPES at pH 6.0, 50 mM NaCl, 20 mM β-glycerophosphate, 20 mM NaF). Immunoblots were probed with the following antibodies: anti-REDD1 rabbit polyconal (Ellisen et al. 2002); anti-TSC2 (C-20), 4E-BP1 (R-113), GFP (B-2), and β-tubulin (D-10, all from Santa Cruz Biotechnology); anti-LDH (AB1222, Chemicon); anti-caveolin-1 (BD Biosciences); and phospho-S6K (Thr389), 4E-BP1 (Thr70), Akt (Ser473), TSC2 (Ser939 and Thr1462), total S6K, total Akt, and anti-Rheb (all from Cell Signaling Technology).
REDD1/14–3–3-binding and competition assays
For REDD1 affinity-purification assays, human embryonic kidney (HEK) 293T cells were transfected with pcDNA3-REDD1-HA wild-type, ΔC, or RPAA constructs, and lysates were prepared in detergent free lysis buffer (20 mM Tris-HCl at pH 7.5, 20 mM NaCl, 1 mM EDTA, 5 mM EGTA, 20 mM β-glycerolphosphate, 1 mM DTT, 1 mM PMSF, 10 μg/mL aprotinin, 10 μg/mL leupeptin, protease inhibitor cocktail, phosphatase inhibitor cocktail I and II) and sonicated. Lysates were cleared by centrifugation, and 250 μg of total protein were incubated with 50 μg of GST or GST-14–3–3β fusion protein linked to glutathione-Sepharose beads for 1 h at 4°C. Following the incubation, beads were washed twice in buffer 1 (0.1 M KCl, 20 mM Tris-HCl at pH 7.6, 0.1 mM EDTA, 10% glycerol, 0.2% NP-40, 1 mM DTT, 0.25 mM PMSF, 1 mM benzamidine, 2 μg/mL aprotinin) and twice in buffer 2 (buffer 1 with 0.5 M KCl). Protein elution was performed using 0.5% sarcosyl in buffer 1 for 2 × 15 min followed by Western blot analysis. For competition assays, the R18 peptide (PHCVPRDLSWLDLEANMCLP) was cloned C-terminally in-frame to GFP into the pcDNA3 vector. This construct or the GFP-containing control plasmid were transiently transfected into 293T cells and lysates were prepared as above. Lysates containing REDD1-HA were then combined with either R18-GFP or GFP-containing lysates in varying ratios (3:1, 1:1, 1:3), adjusting to a total volume of 1 mL using empty pcDNA3-transfected 293T cell lysate, followed by binding and washes as described above.
Cellular proliferation and anchorage-independent growth
TSC2−/− cells reconstituted with wild-type or SATA TSC2 were plated at a density of 6.5 × 104 cells per well in normoxia or hypoxia (1% O2) and growth was determined by counting viable cells daily using 0.4% trypan blue (Sigma) for 4 d. For soft agar assays, TSC2−/−;p53−/−MEFs or SV40 LTAg-expressing MEFs were infected with retroviruses expressing H-Ras(V12) or constitutively active Akt (myr-Akt), respectively, for 72 h prior to plating. High-titer amphotrophic retroviral stocks were generated by cotransfection with packaging vectors into 293T cells and viral supernatants were collected 48 h later. Cells were mixed in DMEM containing 10% FBS and 0.4% agar and were spread evenly onto a bottom layer of complete medium containing 0.5% agar in six-well plates in triplicate at two different dilutions (1.25 × 103 and 1.25 × 104). Cells were grown in normoxia or hypoxia (1% O2) for 2–3 wk. Each experiment was performed three times using independently derived MEF populations. At the end of the experiment, colonies were counted and photographed at a magnification of 40× under a phase contrast microscope.
Tumorigenicity studies
All animal studies were conducted according to protocols approved by the accredited MGH Subcommittee on Research Animal Care. Transformed MEFs retrovirally infected with myr-Akt (2 × 106) were subcutaneously injected into the dorsal flanks of nu/nu mice (REDD1+/+ MEFs on the left flank and REDD1−/− MEFs on the right flank). Animals were examined twice weekly and tumor volumes were determined by bidirectional measuring with calipers and calculated using the following formula: Tumor volume (cubic millimeters) = width2 × length/2.
Quantitative analysis of REDD1 expression in primary human tumors
LCM and analysis of primary human breast carcinomas were performed as described (Ma et al. 2003). All specimens underwent expert pathology review, and were collected prior to any treatment and with Human Subjects Institutional Review Board approval. Total RNA was extracted from the captured cells by using the Picopure RNA Isolation kit (Arcturus Engineering). T7-based RNA amplification was carried out by using the RiboAmp kit (Arcturus Engineering) according to the manufacturer’s instructions. Fidelity of amplification was verified, and QRT–PCR was carried out using internal cDNA standards as described (Ma et al. 2003). Sequences of the PCR primer pairs and fluorogenic probes are available on request.
Subcellular fractionation
Cells were rinsed and lifted in ice-cold PBS, pelleted, resuspended in hypotonic buffer (25 mM HEPES at pH 6.0, 150 mM NaCl, 10 mM NaF, 10 mM sodium pyrophosphate, 1 mM EDTA, protease inhibitors), and lysed using a dounce homogenizer. Nuclei were pelleted by centrifugation at 3000 rpm for 10 min at 4°C. The supernatant was further fractionated by ultracentrifugation at 25,000 rpm for 1 h at 4°C to collect the cytosolic fraction. The pellet (membrane fraction) was resuspended in 1% Triton X buffer (25 mM Tris at pH 8.0, 1% Triton X-100, 150 mM NaCl, 10 mM NaF, 10 mM sodium pyrophosphate, 1 mM EDTA, protease inhibitors) for 30 min at 4°C, followed by centrifugation at 13,000 rpm for 10 min at 4°C.
Acknowledgments
We thank David Kwiatkowski for the gift of matched wild-type and TSC2−/− cells; James Rocco for primary human fibroblasts; Brendan Manning, Kun-Liang Guan, and Yue Xiong for TSC2 constructs; Andre Bernards and Joseph Avruch for critical review of the manuscript; and Franziska Baenke and Nick Vidnovic for expert technical assistance. This work was supported by an American Cancer Society Research Scholar Award RSG-04-035 to L.W.E.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1617608
References
- Arsham A.M., Howell J.J., Simon M.C. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J. Biol. Chem. 2003;278:29655–29660. doi: 10.1074/jbc.M212770200. [DOI] [PubMed] [Google Scholar]
- Blouw B., Song H., Tihan T., Bosze J., Ferrara N., Gerber H.P., Johnson R.S., Bergers G. The hypoxic response of tumors is dependent on their microenvironment. Cancer Cell. 2003;4:133–146. doi: 10.1016/s1535-6108(03)00194-6. [DOI] [PubMed] [Google Scholar]
- Bridges D., Moorhead G.B. 14–3–3 proteins: A number of functions for a numbered protein. Sci. STKE. 2005;2005:re10. doi: 10.1126/stke.2962005re10. [DOI] [PubMed] [Google Scholar]
- Brugarolas J., Lei K., Hurley R.L., Manning B.D., Reiling J.H., Hafen E., Witters L.A., Ellisen L.W., Kaelin W.G. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes & Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett P.E., Barrow R.K., Cohen N.A., Snyder S.H., Sabatini D.M. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S.L., Tee A.R., Short J.D., Bergeron J.M., Kim J., Shen J., Guo R., Johnson C.L., Kiguchi K., Walker C.L. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J. Cell Biol. 2006;173:279–289. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradetti M.N., Inoki K., Bardeesy N., DePinho R.A., Guan K.L. Regulation of the TSC pathway by LKB1: Evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes & Dev. 2004;18:1533–1538. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradetti M.N., Inoki K., Guan K.L. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J. Biol. Chem. 2005;280:9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- Crino P.B., Nathanson K.L., Henske E.P. The tuberous sclerosis complex. N. Engl. J. Med. 2006;355:1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- Ellisen L.W., Ramsayer K.D., Johannessen C.M., Yang A., Beppu H., Minda K., Oliner J.D., McKeon F., Haber D.A. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol. Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- Fingar D.C., Blenis J. Target of rapamycin (TOR): An integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- Gao X., Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes & Dev. 2001;15:1383–1392. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Raught B., Gygi S.P., Niedzwiecka A., Miron M., Burley S.K., Polakiewicz R.D., Wyslouch-Cieszynska A., Aebersold R., Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes & Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin D.A., Sabatini D.M. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Hay N., Sonenberg N. Upstream and downstream of mTOR. Genes & Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Zhu T., Wu J., Guan K.L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Xu T., Guan K.L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes & Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Smith F.D., Stark C., Wells C.D., Fawcett J.P., Kulkarni S., Metalnikov P., O’Donnell P., Taylor P., Taylor L., et al. Proteomic, functional, and domain-based analysis of in vivo 14–3–3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr. Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Lapointe J., Li C., Higgins J.P., de van Rijn M., Bair E., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc. Natl. Acad. Sci. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Inoki K., Vacratsis P., Guan K.L. The p38 and MK2 kinase cascade phosphorylates tuberin, the tuberous sclerosis 2 gene product, and enhances its interaction with 14–3–3. J. Biol. Chem. 2003;278:13663–13671. doi: 10.1074/jbc.M300862200. [DOI] [PubMed] [Google Scholar]
- Liu L., Cash T.P., Jones R.G., Keith B., Thompson C.B., Simon M.C. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol. Cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.J., Salunga R., Tuggle J.T., Gaudet J., Enright E., McQuary P., Payette T., Pistone M., Stecker K., Zhang B.M., et al. Gene expression profiles of human breast cancer progression. Proc. Natl. Acad. Sci. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Chen Z., Erdjument-Bromage H., Tempst P., Pandolfi P.P. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- Manning B.D., Tee A.R., Logsdon M.N., Blenis J., Cantley L.C. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- Nellist M., Goedbloed M.A., de Winter C., Verhaaf B., Jankie A., Reuser A.J., van den Ouweland A.M., van der Sluijs P., Halley D.J. Identification and characterization of the interaction between tuberin and 14–3–3ζ. J. Biol. Chem. 2002;277:39417–39424. doi: 10.1074/jbc.M204802200. [DOI] [PubMed] [Google Scholar]
- Pan D., Dong J., Zhang Y., Gao X. Tuberous sclerosis complex: From Drosophila to human disease. Trends Cell Biol. 2004;14:78–85. doi: 10.1016/j.tcb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Potter C.J., Huang H., Xu T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell. 2001;105:357–368. doi: 10.1016/s0092-8674(01)00333-6. [DOI] [PubMed] [Google Scholar]
- Reiling J.H., Hafen E. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes & Dev. 2004;18:2879–2892. doi: 10.1101/gad.322704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling J.H., Sabatini D.M. Stress and mTORture signaling. Oncogene. 2006;25:6373–6383. doi: 10.1038/sj.onc.1209889. [DOI] [PubMed] [Google Scholar]
- Roux P.P., Ballif B.A., Anjum R., Gygi S.P., Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc. Natl. Acad. Sci. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I., Sharon N., Lerer T., Cohen H., Stolovich-Rain M., Nir T., Dor Y., Zisman P., Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes & Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Thoreen C.C., Peterson T.R., Lindquist R.A., Kang S.A., Spooner E., Carr S.A., Sabatini D.M. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Sabatini D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Shaw R.J., Kosmatka M., Bardeesy N., Hurley R.L., Witters L.A., DePinho R.A., Cantley L.C. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofer A., Lei K., Johannessen C.M., Ellisen L.W. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol. Cell. Biol. 2005;25:5834–5845. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Gingras A.C. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- Tee A.R., Manning B.D., Roux P.P., Cantley L.C., Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Vander Haar E., Lee S.I., Bandhakavi S., Griffin T.J., Kim D.H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Wouters B.G., van den Beucken T., Magagnin M.G., Koritzinsky M., Fels D., Koumenis C. Control of the hypoxic response through regulation of mRNA translation. Semin. Cell Dev. Biol. 2005;16:487–501. doi: 10.1016/j.semcdb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Zhang H., Cicchetti G., Onda H., Koon H.B., Asrican K., Bajraszewski N., Vazquez F., Carpenter C.L., Kwiatkowski D.J. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K–Akt signaling through downregulation of PDGFR. J. Clin. Invest. 2003;112:1223–1233. doi: 10.1172/JCI17222. [DOI] [PMC free article] [PubMed] [Google Scholar]