Abstract
The linked Mrf4 and Myf5 genes encode two transcription factors essential for the determination and differentiation of skeletal muscle in the embryo. The locus is controlled by a multitude of interdigitated enhancers that activate gene expression at different times and in precisely defined progenitor cell populations. Manipulation of the enhancer–promoter composition of the locus reveals a novel mechanism for the regulation of such a gene cluster. Enhancers, promoters, and a new class of elements we call transcription balancing sequences, which can act as cryptic promoters, exist in a series of equilibria to ensure that enhancers and promoters together produce the highly dynamic and exquisitely specific expression patterns of the two genes. The proposed model depends upon nonproductive interactions between enhancers and both minimal and cryptic promoters, and is distinct from those developed for the β-globin and Hox clusters. Moreover, it provides an explanation for the unexpected phenotypes of the three Mrf4 knockout alleles.
[Keywords: Transcriptional regulation, cryptic promoter, Mrf4, Myf5, myogenesis]
Eukaryotes employ diverse mechanisms to control gene expression that can be categorized into transcriptional, post-transcriptional, and post-translational regulation. Of these, the rate of transcriptional initiation is probably the most important factor controlling the overall expression pattern of a gene. Basal promoters can initiate transcription on their own, but since most of the transcription factor-binding sites within them interact with ubiquitously expressed factors, transcription is not tissue-specific, and is usually driven at low levels. To increase transcription rates, and to confer specificity, enhancers interact with basal promoters by means that are not fully understood; the specificity is determined by the combination of binding sites in their sequences. In contrast, throughout mammalian genomes many cryptic promoters have been identified, sequences only able to initiate transcription following an alteration of their genomic environment, either experimentally or as the result of a translocation. These cryptic promoters must be prevented from interacting productively with the enhancers so that they remain transcriptionally silent. The expression pattern of any particular gene is established by the promoter/enhancer composition of the locus, the availability of cognate transcription factors, and the kinetics and mechanics affecting their interactions. The maintenance of the correct specificity of these interactions is crucial for orderly development and differentiation, and is a particular problem in loci containing multiple genes related by function and/or expression pattern. Of significant interest in this context is the regulation of genes encoding transcription factors that play key roles in a cascade controlling cell fate decisions.
During mouse embryogenesis, skeletal muscle arises from three different locations: segmented somitic paraxial mesoderm, unsegmented paraxial head mesoderm, and prechordal mesoderm. Trunk and limb muscles originate from the somitic epithelial dermomyotome (for review, see Kalcheim and Ben-Yair 2005). Progenitor cells delaminate from the four lips of the dermomyotome and move into the myotome, in which they develop in situ or subsequently travel to their final destination. At limb levels, cells from the ventrolateral lip (VLL) of the dermomyotome migrate into the developing limb buds, to form the appendicular musculature, while at more rostral positions VLL cells delaminate and follow a long-range migration along the hypoglossal chord to give rise to pharyngeal and tongue musculature. Facial musculature is derived from unsegmented head mesoderm and prechordal mesoderm cells, which either move through the branchial arches or directly to their final destination (for review, see Noden and Francis-West 2006).
In all of these locations the determination, migration, and terminal differentiation of skeletal muscle cells are controlled by a network in which the four myogenic regulatory factors (MRFs)—Myf5, Mrf4, MyoD, and myogenin—play key roles. Myf5 and MyoD can act as determination genes, while myogenin is essential for the terminal differentiation of committed myoblasts (for review, see Berkes and Tapscott 2005). Mrf4 has long been considered a differentiation gene, but it is expressed in undifferentiated cells and can act as a determination gene (Kassar-Duchossoy et al. 2004)
We have focused on the transcriptional regulation of Mrf4 and Myf5, which are separated by 8.7 kb in the mouse and are linked in all vertebrates. Myf5 is the first MRF gene expressed in the mouse; it is activated before 8.5 d post-coitum (dpc) in the dorsomedial lip of the somite and then extends into the newly formed myotome; by 9.5 dpc, expression begins in the ventral dermomyotome and branchial arches. As limbs develop, Myf5 is up-regulated in the central premuscle mass but not in migrating progenitors (Ott et al. 1991; Summerbell et al. 2000). During myoblast fusion expression is maintained in all skeletal muscle masses but it is down-regulated from late gestation, and only quiescent satellite cells and muscle spindles retain expression in the adult (Zammit et al. 2004). Mrf4 expression closely follows that of Myf5 but is triphasic. It begins in undifferentiated cells of the ventral somite coincident with the onset of Myf5 expression in the dorsomedial lip (Carvajal et al. 2001; Summerbell et al. 2002). It is then activated in the central myotome of rostral somites at ∼9.0 dpc, and by 10.0 dpc includes all somites but is absent from the limbs and branchial arch derivatives. This phase is down-regulated in a rostrocaudal direction in all somites from 12.5 dpc, followed by a third phase in which expression is up-regulated and maintained in all skeletal muscles in the adult.
The regulation of linked and functionally related genes has been studied in a number of systems. In the β-globin locus temporal switching from one gene to another in the same cell type is regulated by proximal sequences, insulators restrict interactions, while overall levels of expression are controlled by the distal locus control region (LCR), involved in chromatin architecture (for review, see de Laat and Grosveld 2003). In vertebrate Hox clusters global control regions regulate the expression of multiple genes, and locally acting elements shared between adjacent genes affect expression in particular cell types and tissues (Spitz et al. 2003). In Drosophila homeotic clusters, the precise structure of core promoter elements controls enhancer–promoter selectivity while a novel class of element (promoter tethers) facilitates interactions of enhancers with core promoters (Calhoun and Levine 2003).
Transcriptional regulation of Myf5 has been studied extensively in transgenic mice (Patapoutian et al. 1993; Zweigerdt et al. 1997; Hadchouel et al. 2000, 2003; Summerbell et al. 2000; Carvajal et al. 2001; Teboul et al. 2002; Buchberger et al. 2003; Zammit et al. 2004). A similar approach has been used in the analysis of the transcriptional regulation of Mrf4 (Patapoutian et al. 1993; Pin et al. 1997; Carvajal et al. 2001; Pin and Konieczny 2002). We have shown previously that a 250-kb BAC clone carrying 195 kb of sequence upstream of the Myf5 translational start site (BAC195) is able to fully recapitulate the expression patterns of both Mrf4 and Myf5 during embryonic development (Carvajal et al. 2001) and in the adult (Zammit et al. 2004). A complex picture has emerged (Fig. 1A) in which a multitude of enhancers regulate the activation of Myf5 at different times and anatomical locations during development, suggesting that the inductive signals for myogenesis are diverse, and that Myf5 may function to canalize these signals into a single skeletal muscle fate. Mrf4 is also differentially regulated in time and space, but the number of regulatory sequences involved is more limited (Carvajal et al. 2001). The majority of Myf5 enhancers are upstream of Mrf4 elements and closer to the Mrf4 promoter than to that of Myf5, and most are located in introns of the adjacent PTPRQ gene (Fig. 1A), which is not expressed in skeletal muscle precursors. The locus thus provides an ideal system to investigate the mechanisms by which enhancer–promoter specificity is controlled so as to precisely regulate the rapidly changing and exquisitely precise expression patterns of these determination genes.
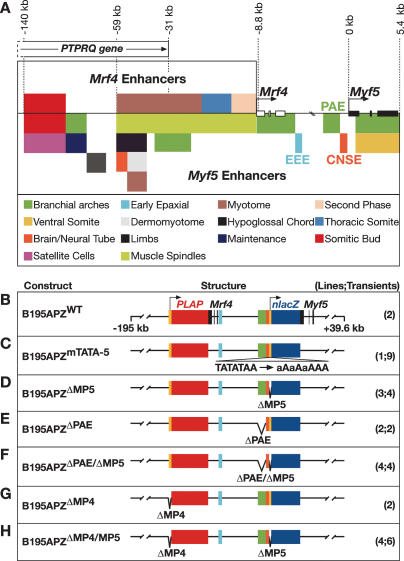
Figure 1.
Maps of the regulatory elements in the Mrf4/Myf5 locus and of the constructs generated for the study of global regulation. (A) Schematic representation of the regions involved in the transcriptional regulation of the Mrf4/Myf5 locus. Each colored box represents transcription occurring at either different times or anatomical locations in embryonic or adult skeletal muscle. The color key is included below the map. (B) Schematic representation of construct B195APZWT, showing the location of the reporter genes (red box, human placental alkaline phosphatase gene [PLAP]; dark blue box, nuclear localized lacZ gene [nlacZ]); the EEE, PAE, and CNSE (pale blue, green, and orange boxes, respectively); the minimal promoters (yellow boxes); and the end points of the BAC clone. (C–H) Schematic representation of the different modifications introduced into B195APZ, and the number of independent transgenic lines and individual transient embryos analyzed (in brackets). (MP5) Myf5 minimal promoter; (MP4) Mrf4 minimal promoter.
We propose the existence of a novel mechanism for the global transcriptional control of complex loci that depends on the maintenance of a series of equilibria between enhancers and promoters that are revealed by modification of the regulatory elements within the locus. Crucial to the establishment and fine tuning of the different equilibrium states are intergenic sequences that can initiate transcription when the locus is perturbed and can interact positively or negatively with the enhancers. Because of their activity, we term these elements transcription balancing sequences (TRABS).
Results
Deletion of the Myf5 minimal promoter allows some Myf5-specific elements to interact with the Mrf4 promoter
In Drosophila promoter–enhancer specificity can depend on the precise sequence of the TATA box (Ohtsuki et al. 1998; Butler and Kadonaga 2001). A putative TATA box is located between 183 and 177 base pairs (bp) upstream of the translational start site of Myf5. To identify the transcriptional start site we carried out 5′-rapid amplification of cDNA ends (RACE) amplification of total RNA from nontransgenic embryos and obtained a single amplification fragment. Multiple independent clones were sequenced that localized the transcriptional initiation site 155 bp upstream of the ATG, indicating that the putative TATA box is likely to be functional (data not shown).
We investigated whether the TATA box contributes to enhancer–promoter selectivity by mutating this element (TATATAA to aAaAaAA) in the context of BAC195APZ (constructs B195APZwt, B195APZmTATA-5) (Fig. 1B,C, respectively), which can fully recapitulate the expression patterns of Mrf4 and Myf5 (Carvajal et al. 2001). In B195APZmTATA-5 embryos these patterns were identical to those in B195APZwt embryos (data not shown).
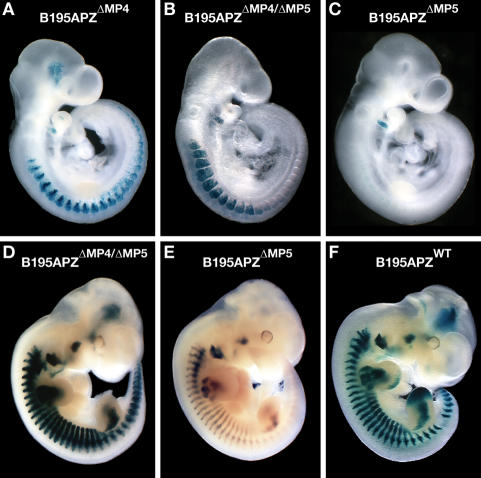
Since the TATA box is not required for correct enhancer–promoter specificity in this locus, we investigated the effects of deleting the entire Myf5 minimal promoter (as functionally defined by Summerbell et al. 2000; see Supplemental Fig. 1) to generate B195APZΔMP5 (Fig. 1D). The timing and pattern of expression of AP-Mrf4 is normal at 9.25 and 9.5 dpc (Fig. 2A,B) but then rapidly changes, incorporating aspects of the Myf5 pattern. At 10.5 dpc AP-Mrf4 expression is detected in both fore- and hindlimbs (Fig. 2C,D, respectively), 48 h earlier than endogenous Mrf4. At 11.0 dpc AP-Mrf4 is expressed in the brain (Fig. 2E), and is maintained in the limb buds and expressed weakly in the branchial arches (data not shown). The cells in the limbs and the arches in which this AP-Mrf4 expression is seen do not, at these times, express myogenin (Yee and Rigby 1993) and thus have not entered the terminal differentiation program. By 11.5 dpc, expression is still visible in the limb buds (Fig. 2F), and at 12.5 dpc (Fig. 2G), and later (data not shown), weak AP-Mrf4 expression is clearly seen in derivatives of the mandibular and hyoid arches, which are normally Myf5 but not Mrf4 expression domains. Thus, in the absence of the Myf5 minimal promoter, Myf5-specific elements are able to activate the remaining promoter, adding Myf5 expression domains to the normal pattern of Mrf4 (which is shown in Fig. 2H; also shown in Fig. 6 of Carvajal et al. 2001). It thus might appear that straightforward promoter competition operates for some enhancers; e.g., those functioning in the limbs, brain, and branchial arches. Their productive engagement is always with the Myf5 promoter but in its absence they engage with the Mrf4 promoter to direct transcription that is temporally and spatially correct for Myf5. However, it is particularly interesting to note that the early epaxial enhancer (EEE), which drives the earliest expression of Myf5 in the dorsomedial lip of the somite (Summerbell et al. 2000; Teboul et al. 2002), is not able to engage the Mrf4 promoter even in this context (Fig. 2A,B), indicating that enhancer–promoter specificity in this locus is controlled by a different mechanism.
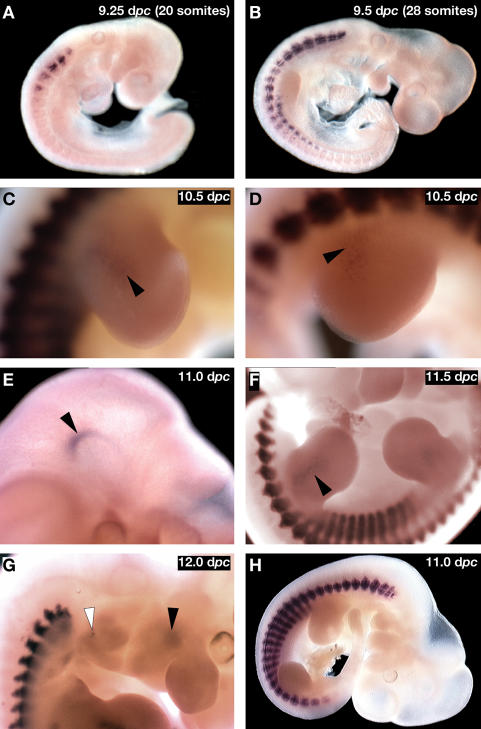
Figure 2.
In the absence of the Myf5 minimal promoter, some Myf5-specific enhancers drive AP-Mrf4 expression. In the absence of the Myf5 minimal promoter, expression of AP-Mrf4 at 9.25 dpc (A) and 9.5 dpc (B) is indistinguishable from that of the wild type. At 10.5 dpc, AP-Mrf4 is expressed in forelimbs (C, black arrowhead) and hindlimbs (D, black arrowhead). (E) At 11.0 dpc, AP-Mrf4 is expressed in Myf5-specific domains such as the brain (black arrowhead). (F) By 11.5 dpc, AP-Mrf4 expression is maintained in the limb buds, but is stronger in the hindlimb (black arrowhead). (G) By 12.0 dpc, AP-Mrf4 expression is activated in derivatives of the mandibular and hyoid arches (black and white arrowheads, respectively). (H) In the control embryo, carrying construct B195APZWT, AP-Mrf4 is not detected at these sites.
Myf5 expression is not abolished in the absence of its minimal promoter
To confirm that deletion of the minimal promoter abolishes nlacZ-Myf5 expression, we analyzed B195APZΔMP5 embryos for β-galactosidase activity. Expression is completely abolished at 8.5 dpc but, surprisingly, weak Myf5 expression is observed by 9.5 dpc in branchial arches and the caudal edges of the somites (Fig. 3A). By 10.5 dpc the intensity of expression is greatly increased and it is readily visible in arches, limb buds, and both rostral and caudal somitic edges, while still absent from the dorsomedial lip of the somite, the myotome, and the ventral somitic bud (Fig. 3B). At later stages, the expression pattern becomes virtually indistinguishable from that driven by the wild-type B195APZ (see Carvajal et al. 2001), except for the dorsomedial lip (Fig. 3C, 11.5 dpc) and the tail somites (Fig. 3D, 12.5 dpc) where it is missing. By 13.5 dpc the only difference from wild type is the missing expression in tail somites (Fig. 3E). Finally, from 14.5 dpc expression down-regulates (Fig. 3F), and it is abolished in all sites of normal Myf5 expression in adult muscle (data not shown).
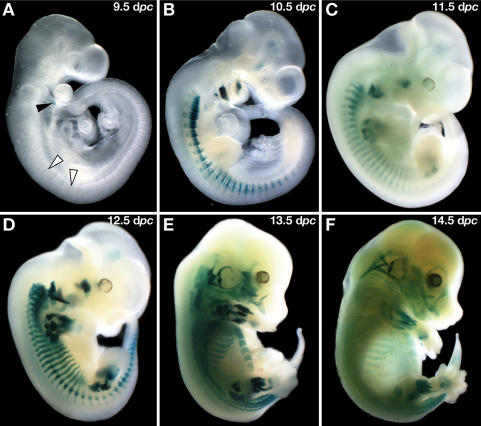
Figure 3.
Myf5 expression is not abolished in the absence of its minimal promoter. (A) nlacZ-Myf5 expression can be detected at 9.5 dpc in the hyoid arch and the caudal edges of interlimb somites (black and white arrowheads, respectively). (B) By 10.5 dpc, expression is up-regulated in the dermomyotome of cervical somites, the caudal lips of interlimb somites, and branchial arches. (C) By 11.5 dpc, expression is clearly visible in fore- and hindlimbs, and myotomal expression is up-regulated for the first time. By 12.5 dpc (D), expression increases in all premuscle masses, is maintained at 13.5 dpc (E), and then starts to down-regulate at 14.5 dpc (F).
The unexpected fact that Myf5 is widely expressed in the absence of its functionally defined minimal promoter means that the locus must contain at least one other, cryptic, promoter, and that many of the enhancers can activate it. This raises the question of whether the enhancers can engage with such sequences even when the minimal promoter is present. Moreover, it is clear that some of the enhancers that can activate the cryptic promoter—e.g., arch and limb enhancers—also activate the Mrf4 promoter in the same cell population at the same time, and are thus not making a simple competitive choice. We suggest that they participate in an equilibrium system in which they can engage with multiple other sequences within a short time period. It is again noteworthy that the EEE cannot activate the cryptic promoters in this context, nor can the element that normally maintains Myf5 expression after 14.5 dpc.
Modification or deletion of the Myf5 minimal promoter activates novel initiation sites in the locus
To map the cryptic promoter(s) we carried out 5′-RACE on mRNA extracted from 13.5-dpc B195APZwt, B195APZmTATA-5, and B195APZΔMP5 embryos. The TATA-box mutation alters the initiation site, leading to a variety of transcripts with 5′-ends ranging from −650 to −100 bp relative to the translational start site, and very little, if any, initiation at the normal site (Fig. 4A [lane 3], B; Supplemental Fig. 1), indicating that this consensus TATA box is responsible for specifying the transcriptional start site of Myf5 and that, under normal developmental conditions, its absence does not affect the expression pattern, while resulting in incorrect initiation. It also follows that transcription initiated at upstream start sites is correctly regulated by, as far as we can tell, all of the enhancers. For B195APZΔMP5 the nlacZ-specific nested primer set generates a ladder with fragments ranging from 50 to 1200 bp (Fig. 4A [lane 4], B; Supplemental Fig. 1), indicating that in the absence of the minimal promoter several transcripts are generated, starting within previously defined elements in the intergenic region: the proximal arch element (PAE), which in isolation is able to drive arch expression from 9.5 dpc to 12.5 dpc, and the CNS element (CNSE), which drives expression in the neural tube (Summerbell et al. 2000). We confirmed this using a different 5′-RACE protocol, which generated similar results (TaKaRa protocol) (data not shown). Because of the nature of 5′-RACE protocols we cannot rule out that some start sites are even further upstream.
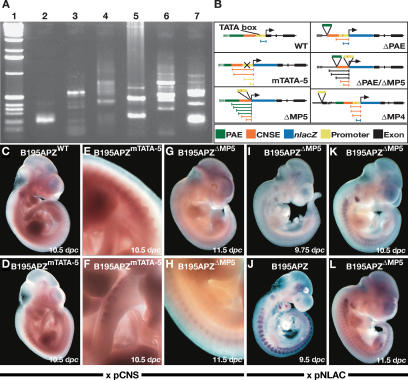
Figure 4.
Alterations in the composition of the locus result in modifications of the transcriptional start site of Myf5 as detected by 5′-RACE and in situ hybridization. The transcriptional start sites for the transgene change depending on the promoter/enhancer composition of the locus. Mutation of the TATA box in the Myf5 promoter results in the activation of multiple transcription initiation sites. (A) Deletion of the Myf5 minimal promoter, which activates a cryptic promoter, also results in multiple transcriptional starts, while deletion of the PAE or the Mrf4 promoter has a strong influence on transcription initiation at Myf5, resulting in the activation of additional cryptic sites. (Lane 1) One-kilobase DNA marker (GIBCO-BRL). (Lane 2) Wild type. (Lane 3) B195APZmTATA-5. (Lane 4) B195APZΔMP5. (Lane 5) B195APZΔPAE. (Lane 6) B195APZΔPAE/ΔMP5. (Lane 7) B195APZΔMP4. These putative initiation sites are represented in B, and have been shown to map to different enhancer elements upstream of the Myf5 promoter. These transcripts can be detected by using riboprobes against noncoding sequences immediately upstream of the promoter (D–H), but not in B195APZwt embryos (C). Hybridization with a nlacZ-specific riboprobe shows low overall levels of transgene transcription (I,K,L) compared with a transgenic line carrying the wild-type BAC (J).
To confirm the existence of these new transcripts we performed whole-mount in situ hybridization using a probe corresponding to the CNSE. Transcripts are not detected in nontransgenic (data not shown) or B195APZwt embryos (Fig. 4C), but transcripts containing this upstream sequence are detected in 10.5-dpc B195APZmTATA-5 embryos (Fig. 4D), in the somites and limbs and very faintly in the branchial arches (Fig. 4E,F). Transcripts containing CNSE sequences are also detected in 10.5- and 11.5-dpc B195APZΔMP5 embryos. At 10.5 dpc these transcripts are detected at low intensity in hyoid arch, somites, and limb buds (data not shown), while at 11.5 dpc transcripts are detected mainly in the dorsal somites (Fig. 4G,H). The low intensity of the signals indicates that deletion of the promoter reduces the levels of Myf5 transcription. Indeed, hybridization of B195APZΔMP5 embryos using an nlacZ-specific probe (pNLAC) shows that there is a significant reduction in transcription levels in all muscle precursors in early embryos (Fig. 4I) when compared with transgene expression in B195APZwt lines (Fig. 4J). At later stages (10.5 and 11.5 dpc) (Fig. 4K,L, respectively), there is an increase in nlacZ-Myf5 expression levels, particularly in derivatives of the hyoid arch, recapitulating some of the differences in intensity observed with β-galactosidase staining of these lines (Fig. 3B). The novel transcripts are found only where Myf5 is normally expressed; we saw no evidence for inappropriate expression.
The Mrf4 promoter is a key element in the maintenance of equilibria in the locus
We next generated a construct in which the Mrf4 minimal promoter (as functionally defined by Black et al. 1995; see Supplemental Fig. 2) was deleted (B195APZΔMP4) (Fig. 1G). The Myf5 expression pattern in such embryos (Fig. 5A) is indistinguishable from that of B195APZwt embryos (Fig. 6A), while AP-Mrf4 expression is completely abolished (data not shown), suggesting that all the cryptic promoters in the locus are located in the intergenic region. Because in the absence of the Myf5 minimal promoter some of the Myf5-specific enhancers are able to drive both nlacZ-Myf5 and AP-Mrf4 expression, we generated construct B195APZΔMP4/ΔMP5 (Fig. 1H), in which both promoters are absent, to ascertain if the nlacZ-Myf5 expression was a result of read-through from the Mrf4 promoter. At 9.5 dpc, construct B195APZΔMP4/ΔMP5 drives nlacZ-Myf5 expression in the dermomyotome and branchial arches but fails to drive dorsomedial lip or myotomal expression (Fig. 5B), showing that in the absence of the Myf5 minimal promoter nlacZ-Myf5 does not initiate at the Mrf4 promoter. More importantly, comparison of the nlacZ-Myf5 expression patterns of B195APZΔMP4/ΔMP5 and B195APZΔMP5 in the somites (Fig. 5B,C, respectively) shows that the Mrf4 promoter prevents a somitic enhancer from activating the cryptic promoters at this time, and only by removing the Mrf4 promoter is the enhancer able to drive nlacZ-Myf5 in the dermomyotome. We infer that the enhancer preferentially interacts nonproductively with the Mrf4 promoter when it is present. There is no expression in the dorsomedial lip, confirming that even if the Myf5 minimal promoter is not present the EEE is sequestered by other sequences and prevented from activating the Mrf4 promoter (see model in Fig. 7B). At later stages the double deletion results in up-regulation of nlacZ-Myf5 in all muscle precursors (Fig. 5D) when compared with B195APZΔMP5 (Fig. 5E), and the pattern became indistinguishable from that driven by B195APZwt (Fig. 5F). This shows that most, if not all, of the enhancers can activate the cryptic promoters if the Mrf4 promoter is absent. Surprisingly, this deletion alters the Myf5 initiation site, activating a site within the CNSE, and minor sites within the PAE in addition to the normal site (Fig. 4A [lane 7], B). The finding that deletion of the Mrf4 promoter affects transcriptional initiation at Myf5 indicates that both promoters are part of an equilibrium operating in the locus.
Figure 5.
The Mrf4 promoter is a key player in the enhancer–promoter equilibria in the locus. Deletion of the Mrf4 minimal promoter does not affect the nlacZ-Myf5 expression pattern (A), while the double MP4/MP5 deletion partially rescues the early (9.5 dpc) Myf5 expression pattern (B), lost in the single MP5 deletion (C). The double deletion also rescues the expression pattern at 11.5 dpc, resulting in up-regulation of the transgene in all premuscle masses (D), while the construct carrying the single MP5 deletion is unable to drive full expression in limbs, interlimb, or tail somites (E). (F) The B195APZΔMP4/ΔMP5 expression pattern is indistinguishable from that of B195APZwt at this stage.
Figure 6.
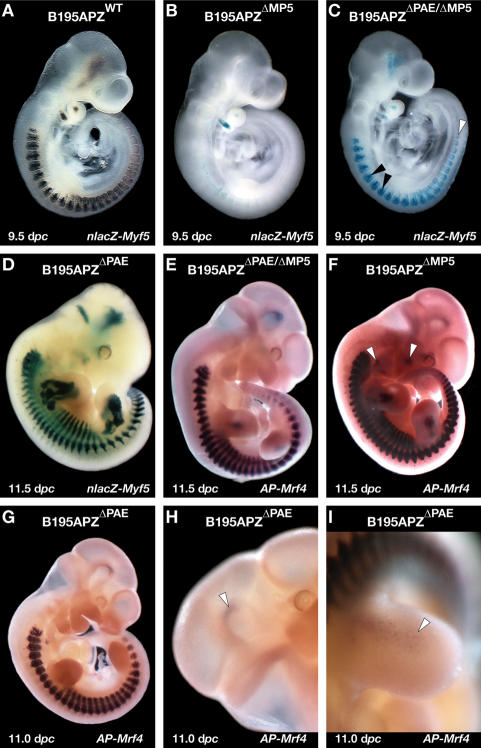
The PAE is essential for the maintenance of the equilibrium between enhancers and promoters in the locus. nlacZ-Myf5 expression patterns driven by B195APZwt (A) and B195ΔMP5 (B) constructs at 9.5 dpc. (C) Deletion of both the Myf5 minimal promoter and the PAE results in the rescue of the complete pattern of expression at this stage. (D) Single PAE deletion results in the loss of arch expression at 11.5 dpc, without any other effects on Myf5 expression. Double PAE/MP5 deletion abolishes AP-Mrf4 expression in the arches (E), which is readily detected in the single MP5 deletion (F). Single PAE deletion alters the expression of Mrf4 (G) by activating Myf5-specific expression in the brain (H) and early in the limbs (I).
Figure 7.
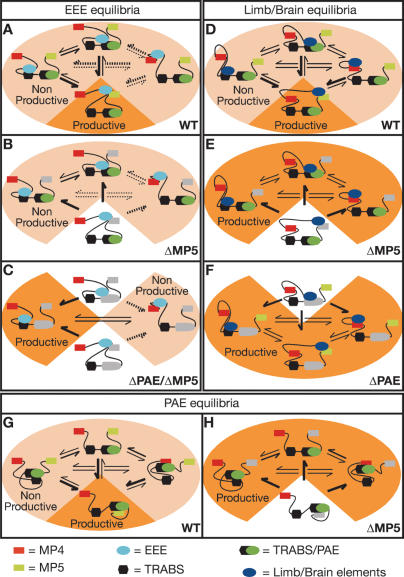
Schematic representation of the transcriptional equilibria in the Mrf4/Myf5 locus. The equilibria between the transcriptional elements in the locus control promoter–enhancer specificity in different ways depending on the enhancer required. (A–C) Early epaxial enhancer. In the wild-type situation, the equilibrium between the EEE and the other elements in the locus is displaced toward the Myf5 minimal promoter, which results in the only possible productive interaction. (B) Deletion of the Myf5 minimal promoter distorts the equilibrium, but none of the interactions are productive. (C) This distortion is detected when both the PAE and the Myf5 minimal promoter are removed, and the EEE can activate transcription from the TRABS but not from the Mrf4 promoter. (D–F) Brain and limb elements. (D) As with the EEE, in the wild-type situation the equilibrium is displaced toward the activation of the Myf5 minimal promoter. (E) Removal of this element results in the total reversion of the equilibrium generating productive interactions with the Mrf4 promoter and the TRABS in the locus. (F) Removal of a single TRABS (the one within the PAE interval) without modifying the promoter composition of the locus also results in reversion of the equilibrium, and these enhancers activate transcription at the Mrf4 and Myf5 promoters and at other TRABS. (G–H) Proximal arch element. (G) As in the other cases, in the wild-type situation the equilibrium favors the activation of the Myf5 promoter as a result of the only productive interaction. (H) Deletion of the promoter unbalances the equilibrium, making the different interactions productive. Arrows indicate the direction of the equilibria, while thickness indicates the favored state. Deleted elements are represented in gray. Dotted arrows indicate that the interaction of the EEE with the Mrf4 promoter, which we have not detected to date, may not form part of this equilibrium. Dark and pale orange represent productive and nonproductive states, respectively. Interaction and bending of the DNA is only used as a representation, and should not be taken as an indication of mechanism. We diagrammed only some of the possible interactions in order to exemplify the principles of our model.
The PAE is in equilibrium with other enhancers in the locus and with the Mrf4 minimal promoter
Expression in the branchial arches is subject to probably the most complex regulatory system in the locus, with at least five enhancers acting in the arches and their derivatives (Summerbell et al. 2000; Carvajal et al. 2001). Furthermore, we showed that in the absence of the Myf5 minimal promoter, nlacZ-Myf5 expression in the hyoid arch is maintained or up-regulated at the transcriptional level, suggesting that the PAE strongly activates the cryptic promoters in the locus. To investigate this and the contribution of the PAE to the normal regulation of Myf5, we analyzed nlacZ-Myf5 expression in B195APZΔPAE embryos, in which the PAE had been deleted (Fig. 1E; Supplemental Fig. 1). Early Myf5 arch expression was normal, but was down-regulated from 9.5 dpc (data not shown), although not completely abolished. We could not detect any other effects at any of the stages analyzed (9.5–13.5 dpc) (data not shown), indicating that sequences within the PAE are not required to activate Myf5 transcription at any other location. Surprisingly, deletion of the PAE alone results in the activation of an upstream start site, while the normal site downstream from the TATA box remains active (Fig. 4A [lane 5], B). The changes in the transcriptional start site of Myf5 in the absence of the PAE indicate that it has dual functions, to drive late arch expression and to ensure correct transcriptional initiation at the minimal promoter.
Our data suggest that sequences within the PAE may contribute to the regulation of the equilibria within the locus, and we showed that the EEE is subject to particularly stringent regulation. To further explore the role of the PAE we analyzed B195APZΔPAE/ΔMP5 animals, lacking both the Myf5 minimal promoter and the PAE (Fig. 1F). To our surprise, the full Myf5 expression pattern was restored at 9.5 dpc (Fig. 6C), including expression in the dorsomedial lip of the somite, driven by the EEE (Summerbell et al. 2000; Teboul et al. 2002). The pattern was indistinguishable from that of B195APZwt (Fig. 6A), showing that the enhancers accurately regulate the cryptic promoters. Deletion of both the PAE and the Myf5 minimal promoter allows the EEE to activate the other cryptic promoters to drive correct dorsomedial lip Myf5 expression. The simplest explanation of these data is that in the absence of the Myf5 minimal promoter the EEE interacts with sequences within the PAE interval in a way that prevents it from activating the cryptic promoters (see model in Fig. 7A–C).
By 11.5 dpc, Myf5 arch expression is down-regulated in B195APZΔPAE/ΔMP5 embryos (data not shown), reproducing the timing of arch expression driven by construct B195APZΔPAE (Fig. 6D). Because the down-regulation of Myf5 arch expression in B195APZΔPAE overlaps in time with the up-regulation of Mrf4 expression in B195APZΔMP5 lines, we hypothesized that in the absence of the Myf5 minimal promoter the PAE engages with the Mrf4 minimal promoter to drive this expression. Analyses of the AP-Mrf4 expression pattern at 11.5 dpc in B195APZΔPAE/ΔMP5 embryos (Fig. 6E) show that this is indeed the case, as these embryos lack the arch expression seen in B195APZΔMP5 embryos (Fig. 6F), while the remainder of the pattern was identical (data not shown). In the absence of both the PAE and the minimal promoter, transcription is still initiated within the CNSE and at locations upstream of the deleted PAE (Fig. 4A [lane 6], B), indicating that a variety of sequences in the locus are able to initiate transcription in the absence of the minimal promoter.
We showed that the PAE has a dual function, controlling the expression of Myf5 in the branchial arches and ensuring correct transcriptional initiation at the Myf5 promoter. In the absence of the Myf5 minimal promoter the PAE is involved in driving expression of AP-Mrf4 in the arches while double deletion of the PAE and the Myf5 minimal promoter reveals that sequences within the PAE interval sequester some Myf5-specific enhancers, thus preventing their interaction with other cryptic promoters in the locus. If the role of the PAE interval in the maintenance of the equilibria is crucial, deletion of this element in isolation should result in alteration of the enhancer–promoter specificity in the locus. While the changes in the pattern of nlacZ-Myf5 are restricted to the absence of early arch expression (Fig. 6D), there are important changes to the AP-Mrf4 pattern (Fig. 6G), which incorporates expression in the brain (Fig. 6H) and the limb buds (Fig. 6I). Therefore, in the absence of the PAE at least some enhancers are able to activate basal promoters that they cannot access when it is present, indicating that sequences within the PAE interval play a key role in regulating the overall enhancer–promoter equilibria (see model in Fig. 7D–H).
Discussion
Promoter–enhancer equilibria
Eukaryotes employ many different mechanisms to ensure that transcriptional units can respond appropriately to the surrounding environment in order to regulate the activity levels of the many proteins encoded in their genomes. Crucial to this response is the control of transcriptional regulators themselves, as downstream gene activation is mainly dependent on the correct control of transcription factor activity. In multicellular organisms complex regulatory mechanisms are required to ensure tight regulation of tissue-specific gene expression. These mechanisms must be even more complex when two or more genes, with distinct expression patterns, are linked in the genome, with the possibility of enhancer cross-talk even greater. Furthermore, it has been shown that gene duplication is usually accompanied by the sharing of enhancers belonging to the protogene, and the specialization of the duplicated genes for individual enhancers, usually resulting in nonoverlapping patterns that when combined cover the entire expression pattern of the original gene (Bruce et al. 2001; Locascio et al. 2002; de Souza et al. 2005). We propose that the regulation of such loci involves additional mechanisms to ensure the correct enhancer–promoter specificity.
In multigene loci, several mechanisms have been described to explain how promoter–enhancer specificity is established. These seem to rely on a hierarchical architecture in which promoters compete for the different enhancers, with sequences either within (TATA box, downstream promoter elements, initiator sequences) or outside (tethering elements) the promoters used to modify the strength of the competition, and/or block such interactions (silencers, insulators, boundary elements). Other elements involved are the LCRs, responsible for the initiation and/or maintenance of tissue-specific gene expression. The most widely studied LCR is that of the β-globin gene cluster, which has been shown to be required for high levels of β-globin expression (for reviews, see Fraser and Grosveld 1998; Li et al. 2002). Importantly, the individual globin genes compete for the LCR and, as with other forms of promoter competition, the LCR only interacts with one promoter at a time (Wijgerde et al. 1995). Finally, another cis-regulatory element, the promoter targeting sequence, has also been shown to contribute to promoter–enhancer specificity by selectively bypassing the effects of specific insulators in the Drosophila Abd-B gene (Zhou and Levine 1999).
By altering the regulatory element composition of the Mrf4/Myf5 locus we now reveal a new mechanism. In the absence of the Myf5 minimal promoter, some of the enhancers that interact with it in the normal context are recruited by the Mrf4 promoter to drive Myf5-specific expression. Strikingly, other sequences in the locus can also recruit these enhancers and assemble the transcriptional machinery to drive Myf5 expression in the absence of its promoter. Not all of the enhancers behave in the same manner, and thus, for example, expression is up-regulated in the hyoid arch at 9.5 dpc and myotomal expression is down-regulated at early stages, while dorsomedial lip expression mediated by the EEE is completely abolished. Because some of the enhancers drive expression of both AP-Mrf4 and nlacZ-Myf5 in the same location and at the same stage, this novel mechanism does not correspond to any previously described on/off-switch model. Deletion of the Mrf4 minimal promoter alters the expression of Myf5 at two time points during development. Early Myf5 expression is missing in the somitic bud, and late expression is abolished throughout the embryo and in the adult. Even more intriguingly, the deletion of both the PAE and the Myf5 minimal promoter results in pattern reversion of Myf5 expression to that of the wild type. Analyses at the RNA level indicate that deletion of either promoter results in modifications to the transcriptional start site for nlacZ-Myf5, further evidence of the cis-regulatory interactions between the two promoters in the locus. But these alterations in transcription initiation are not restricted to constructs missing the natural promoters. Indeed, deletion of the PAE also results in alteration of transcriptional initiation and changes in the pattern of Mrf4 expression. These observations indicate that the locus is not regulated by straightforward promoter competition, and that elements different from the core promoters are involved in enhancer–promoter selectivity, elements that we name TRABS.
Cryptic promoters or TRABS
The term cryptic promoter is commonly used in the literature to refer to a fragment of genomic DNA that is able to act as a promoter in certain circumstances but does not normally act. In this way, in several chromosomal translocations involving the c-myc locus, and giving rise to mouse plasmacytomas, it has been shown that the loss of the endogenous c-myc promoter in the derivative chromosome does not imply the silencing of c-myc, and that the generation of a truncated c-myc protein is due to the activation of bipolar cryptic promoters in the locus (Calabi and Neuberger 1985; Murphy et al. 1986). A cryptic promoter located 2 kb upstream of the Slp gene in the eIF4G locus, and originally defined as an internal ribosomal entry site (IRES), has been recently shown to be able to increase by 850 times the expression of an associated Renilla luciferase gene (Han and Zhang 2002). The authors also identified additional cryptic promoters in the 5′-untranslated region (UTR) of the Bad and Sno genes, previously defined as IRES, and concluded that cryptic promoter activities in the 5′-UTR sequences of cellular mRNAs are more prevalent than anticipated.
Deletion of the Myf5 minimal promoter in the context of B195APZ results in the activation of a cryptic promoter(s) in the locus. Furthermore, we showed that deletion of a single enhancer (the PAE) is enough to activate a cryptic promoter, as revealed by 5′-RACE analyses. It is not clear at the moment if these sequences are one or many, although the variation in the transcriptional start site for the nlacZ-Myf5 transgene in the different deletion constructs indicates that multiple sequences in the locus may be activated as cryptic promoters. Furthermore, we showed that the cryptic promoters are able to engage with most of the Myf5-specific enhancers in a context-dependent manner, changing their specificity according to the promoters in the locus, suggesting that the different sequences are in competition with the natural promoters and with each other. We propose that many of the sequences described as cryptic promoters in the literature are in fact TRABS, and that their function is to establish an equilibrium with the promoter(s) in the locus to ensure the fine control of the interactions between enhancers and natural promoters required for the correct transcriptional initiation of gene expression.
Analysis of the equilibrium
Deletion of the Myf5 minimal promoter leads to Mrf4 expression in a number of sites (brain, branchial arches, and limbs) that are normally Myf5-specific, which might be taken as indicating that for some enhancers there is straightforward promoter competition. However, other enhancers, most notably the EEE, cannot engage with the Mrf4 promoter even when the Myf5 promoter is absent, indicating that a different mechanism must operate. Many of the enhancers can thus engage with the cryptic promoters in an entirely specific fashion, but their ability to do so is modulated not only by the Myf5 promoter, which would be expected, but also by the Mrf4 promoter and the PAE. A key feature of the model that we propose (Fig. 7) is that these interactions between the enhancers and the cryptic promoters occur normally (Fig. 7A,D,G), and that they dynamically regulate the choices made as to which enhancers should engage with which promoter, in which progenitor cell population and at what time.
The second key observation is that in the absence of the Myf5 minimal promoter, enhancers operating in the arches (Fig. 7H), the limbs or the brain (Fig. 7E), engage with both the Mrf4 promoter and the cryptic promoters in the same progenitor cell population at the same time. It is clear that the ways in which the enhancers engage with the promoters are not controlled by an on/off switch. Rather, we propose that the enhancers are continuously interacting with a number of transcription initiation sites in the locus, and which interaction leads to the productive initiation of transcription is dictated by the instructive signals that the progenitor cell is receiving.
The model is further supported by the deletion of the minimal promoters of both Mrf4 and Myf5. While most of the early Myf5 somitic pattern is abolished in the absence of the Myf5 minimal promoter, deletion of the Mrf4 promoter results in partial rescue of the somitic pattern. We suggest that the enhancer driving this expression cannot engage with the cryptic promoters because it is sequestered nonproductively by the Mrf4 promoter. When the latter is removed, the enhancer is released and engages productively with the cryptic promoters. Similarly, the EEE cannot activate the cryptic promoters in this context because it is sequestered by sequences within the PAE interval. This is a third key feature of our model, the regulation of the locus depends on interactions between enhancers and promoters, both conventional and cryptic, that do not lead to the initiation of transcription.
It is striking that in our initial experiments the EEE was not seen to interact with anything other than the Myf5 minimal promoter, implying that there must be a mechanism that prevents its engagement with the Mrf4 and cryptic promoters (Fig. 7B). But deletion of the PAE and the Myf5 minimal promoter allows the EEE to activate one or more cryptic promoters (Fig. 7C). We thus infer that the EEE normally engages productively only with the Myf5 minimal promoter but that it is in an equilibrium between that promoter and the TRABS within the PAE interval. Only when both are removed is the EEE able to activate the cryptic promoters, although it is still, by an unknown mechanism, prevented from activating the Mrf4 promoter. It is important to mention that is not clear yet if the PAE interval contains two separable functional elements (the arch enhancer and the TRABS) or if they are interdigitated or even synonymous, and we are currently investigating this point. We suspect that if all the TRABS were removed, then the EEE would be able to activate the Mrf4 promoter, and we are examining this issue.
The Mrf4 ‘knockout’ and the Myf5 ‘knock-in’ revisited
Three different Mrf4 knockout alleles were generated more than a decade ago by the laboratories of Arnold, Olson, and Wold (for review, see Olson et al. 1996 and references therein). Although all three alleles abolished Mrf4 expression, variable cis-acting effects on the linked Myf5 gene were observed, resulting in homozygous phenotypes ranging from full viability to complete lethality. In a previous article (Olson et al. 1996), we and others hypothesized that the presence of the strong pgk promoter on the selectable marker was partially responsible for some of the cis-effects observed, but we did not have an explanation for the mechanism or for the apparent discrepancy in phenotypes between the two alleles where the construction of the targeted allele was very similar but the resulting phenotypes widely different. The Olson allele, in which the pgk-neo-selectable marker is transcribed in the opposite direction to Mrf4 and Myf5, presented the mildest of phenotypes, with some rib malformations and no overt problems with myogenesis. The Arnold and Wold alleles carried the pgk-neo-selectable marker in the same orientation as Mrf4 and Myf5, and while both presented myogenic defects, some animals carrying the Wold allele survived to adulthood while none of the animals carrying the Arnold mutation survived after birth.
The work presented here readily explains these results in that the presence of the strong pgk promoter would alter the transcriptional equilibria in the locus by competing with the endogenous promoters and TRABS, resulting in the misregulation of Myf5. Furthermore, in the Arnold allele, which resulted in a double Mrf4/Myf5 knockout, 102 bp of the Mrf4 minimal promoter were crucially removed, including the transcriptional start site (Braun and Arnold 1995). As we showed that the Mrf4 minimal promoter is a major player in establishing/maintaining the equilibria in the locus and is essential to determine the correct transcriptional initiation site for Myf5, it is likely that even its partial removal compromises Myf5 promoter function, as we showed with our B195APZΔMP4 lines, in which Myf5 expression in the somitic bud, late embryonic phases and the adult is abolished. Thus, the combination of the partial promoter deletion and the inclusion of an additional promoter in the locus could have resulted in the complete down-regulation of the linked Myf5 gene, resulting in the double knockout characteristic of this allele.
More recently, Kassar-Duchossoy et al. (2004) have shown other cis-effects in the locus in a series of Myf5 knock-in lines. The original Myf5nlacZ allele (Tajbakhsh et al. 1996a) and two new alleles, Myf5GFP-P and Myf5loxP, show differential effects on the levels of Mrf4. Expression of Mrf4 is abolished at 10.5 dpc in Myf5nlacZ/nlacZ embryos, and reduced to half of the normal levels in Myf5nlacZ/+ littermates. A similar knock-in allele, Myf5GFP-P also shows alteration in Mrf4 levels, in both heterozygous and homozygous animals, but this expression is not completely abolished. Finally, in Myf5loxP/loxP embryos, cis-effects were not observed. Although the three alleles abolish functional Myf5 protein and have similar genomic structures, the selectable markers and associated promoters used are different. In the Myf5nlacZ allele, the neo cassette is driven by the RNA pol II promoter (Tajbakhsh et al. 1996b), while in the Myf5GFP-P allele the puromycin cassette is driven by the pgk promoter (Kassar-Duchossoy et al. 2004). It is therefore possible to speculate that the phenotypes of these two alleles that introduce a foreign promoter into the locus are due to their ability to disrupt the delicate balance required to set the enhancer–promoter specificity. Differences in phenotypes between these two alleles would be caused by the different affinities of the foreign promoters for the existing transcriptional elements in the locus. Furthermore, Kaul et al. (2000) have shown that the rib phenotype observed in two of the original Myf5 knockout alleles is caused by the selectable marker, although in this case the mechanism is unclear.
Conclusion
There are now a number of examples of genes with regulatory elements located far from the promoter, and it is noteworthy that they tend to encode either signaling molecules (e.g., Shh) or transcription factors (e.g., Sox9) involved in the control of cell fate decisions (Sagai et al. 2005; Bien-Willner et al. 2007). The extremely complicated enhancer organization in the Mrf4/Myf5 locus is also not unique. Sox2 is expressed throughout the early nervous system, but this overall expression pattern is the summation of the activities of a number of dispersed enhancers, each of which operates in a particular population of progenitor cells at a particular time (Uchikawa et al. 2003). Structuring the regulatory elements in this way, together with the novel regulatory mechanism described here, would seem to be particularly appropriate for genes encoding transcription factors, like Myf5 and Sox2, which determine cell fate and have to be induced widely in the embryo in response to a variety of inductive signals.
Our data lead to a model that is quite distinct from those developed from analyses of the β-globin and Hox loci. They imply that the enhancers operate in what might be thought of as a neural network model. They are continuously interacting with not only the two conventional promoters in the locus but also with a number of sequences that can act as cryptic promoters, which we call TRABS. These interactions must be modulated by the inductive signals that determine skeletal muscle identity and lead to the activation of the two genes in an exquisite temporal and spatial pattern involving many different progenitor cell populations at many stages of development.
Materials and methods
Homologous recombination in Escherichia coli
Details on the construction of modifying cassettes can be found in the Supplemental Material.
We used the linear recombination method (Swaminathan et al. 2001) with some modifications. BAC clones were transferred into DY380 cells (kindly provided by Neil Copeland, National Cancer Institute-Frederick, Frederick, Maryland), which carry the temperature-inducible λ-recombinase system.
Single DY380 colonies carrying the desired BAC clone were isolated and grown overnight at 32°C. Two-hundred-twenty microliters of these cultures were used to seed 11 mL of LB and were incubated at 32°C with vigorous shaking (>300 rpm) until OD600 was 0.6 ± 0.05. Cultures were then transferred to a 42°C waterbath and incubated for 15 min. Cultures were transferred to ice, incubated for 20 min, made electrocompetent by standard methods, and resuspended in a final volume of 30 μL.
Modifying cassettes (2 nmol for TATA box mutation; 1 μg for other cassettes) were denatured, ethanol-precipitated, and resuspended in 20 μL of ice-cold H2O. The denatured cassettes were mixed with 30 μL of electrocompetent DY380 cells carrying the BAC clone to be modified. Electroporations were carried out using the Bio-Rad GenePulser II system (1.75 kV, 100 Ω, 25 μF). Electroporated cells were diluted 1:106 into a final volume of 50 mL of LB and aliquoted into a single 96-well plate (500 μL per well), resulting in an average of 20–50 cells per well. After overnight incubation at 32°C, pools were screened using the appropriate primers (see the Supplemental Material). Two to six positive pools were plated to obtain single colonies, and a second round of screening was carried out to identify single positives. Clones were sequenced using primers outside the homology arms.
Generation of transgenic mice
All in vivo experimentation was performed according to United Kingdom Home Office Regulations. BAC DNA was prepared using the QIAgen Maxiprep kit (Qiagen) as described previously (Carvajal et al. 2001). After dialysis against microinjection buffer (10 mM Tris-HCl at pH 7.5, 0.1 mM EDTA at pH 8.0, 100 mM NaCl), DNA was diluted to 1.5 ng/mL in microinjection buffer and used to inject fertilized mouse eggs from CBA/Ca × C57Bl/6 crosses, as described previously (Yee and Rigby 1993).
Histochemical staining
β-Galactosidase: Embryos were fixed overnight in Mirsky’s fixative (National Diagnostics) at 4°C, washed three times in PBSA (Ca2+, Mg2+-free phosphate-buffered saline)/0.02% Nonidet P-40 for 20 min at room temperature, placed in 10 mL of X-gal solution [5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6 ⋅ 3H2O, 2 mM MgCl2, 0.02% Nonidet P-40, 0.4 mg/mL X-Gal in PBSA] for 2–20 h (depending on stage) at 37°C, and post-fixed in Mirsky’s fixative.
Alkaline phosphatase: Embryos were fixed in 4% paraformaldehyde in PBSA overnight at 4°C, rinsed twice in PBSA + 2 mM MgCl2, and washed in PBSA + 2 mM MgCl2 for 10 min. Endogenous phosphatases were inactivated by incubation for 1 h in PBSA + 2 mM MgCl2 at 65°C. Embryos were equilibrated in AP-buffer (100 mM Tris-HCl at pH 9.5, 100 mM NaCl, 50 mM MgCl2, 0.1% Tween 20). Embryos were washed once in ice-cold AP-buffer, transferred to AP-staining buffer (AP-buffer + 0.1 mg/mL of BCIP; 0.5 mg/mL NBT), and stained for 3–10 h (depending on stage) in the dark at 4°C. The reaction was stopped by washing in ice-cold PBSA containing 2 mM EDTA in the dark for 24 h at 4°C. Embryos were post-fixed in Mirsky’s fixative adjusted to pH 5.0.
RACE
Embryos were obtained from timed matings, taking the day of vaginal plug as 0.5 dpc, and dissected in PBSA. For the identification of transgenic embryos, left forelimbs were transferred to single tubes containing 1 mL of X-gal solution (see above) and incubated at 37°C until coloration was detected. Single 13.5-dpc embryos were homogenized in 1 mL of TRI reagent (Sigma) using disposable plastic pestles and RNA isolated following the manufacturer’s recommendations. RNA was resuspended in a final volume of 500 μL of DEPC-treated H2O. We used the SMART RACE Amplification Kit (BD Biosciences) following manufacturer’s recommendations. Single-stranded cDNA was amplified by PCR using the Advantage 2 PCR Kit (BD Biosciences) using either Myf5- or nlacZ-specific primers (Supplemental Material). Single amplification bands were gel-purified and cloned into pCR-TOPO (Invitrogene) for sequencing.
Acknowledgments
We are grateful to all members of the Rigby laboratory for helpful discussions, and the anonymous referees for their insightful comments. This work was supported by The Institute of Cancer Research, Medical Research Council Programme Grant G040119, and the MYORES Network of Excellence, contract 511978, from the European Commission 6th Framework Programme. A.K. held a Medical Research Council Studentship.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.442408
References
- Berkes C.A., Tapscott S.J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Bien-Willner G.A., Stankiewicz P., Lupski J.R. SOX9cre1, a cis-acting regulatory element located 1.1Mb upstream of SOX9, mediates its enhancement through the SHH pathway. Hum. Mol. Genet. 2007;16:1143–1156. doi: 10.1093/hmg/ddm061. [DOI] [PubMed] [Google Scholar]
- Black B.L., Martin J.F., Olson E.N. The mouse MRF4 promoter is trans-activated directly and indirectly by muscle-specific transcription factors. J. Biol. Chem. 1995;270:2889–2892. doi: 10.1074/jbc.270.7.2889. [DOI] [PubMed] [Google Scholar]
- Braun T., Arnold H.H. Inactivation of Myf-6 and Myf-5 genes in mice leads to alterations in skeletal muscle development. EMBO J. 1995;14:1176–1186. doi: 10.1002/j.1460-2075.1995.tb07101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A.E.E., Oates A.C., Prince V.E., Ho R.K. Additional hox clusters in the zebrafish: Divergent expression patterns belie equivalent activities of duplicate hoxB5 genes. Evol. Dev. 2001;3:127–144. doi: 10.1046/j.1525-142x.2001.003003127.x. [DOI] [PubMed] [Google Scholar]
- Buchberger A., Nomokonova N., Arnold H.H. Myf5 expression in somites and limb buds of mouse embryos is controlled by two distinct distal enhancer activities. Development. 2003;130:3297–3307. doi: 10.1242/dev.00557. [DOI] [PubMed] [Google Scholar]
- Butler J.E., Kadonaga J.T. Enhancer–promoter specificity mediated by DPE or TATA core promoter motifs. Genes & Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabi F., Neuberger M.S. Chromosome translocation activates heterogeneously initiated, bipolar transcription of a mouse c-myc gene. EMBO J. 1985;4:667–674. doi: 10.1002/j.1460-2075.1985.tb03681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun V.C., Levine M. Long-range enhancer–promoter interactions in the Scr–Antp interval of the Drosophila Antennapedia complex. Proc. Natl. Acad. Sci. 2003;100:9878–9883. doi: 10.1073/pnas.1233791100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal J.J., Cox D., Summerbell D., Rigby P.W.J. A BAC transgenic analysis of the Mrf4/Myf5 locus reveals interdigitated elements that control activation and maintenance of gene expression during muscle development. Development. 2001;128:1857–1868. doi: 10.1242/dev.128.10.1857. [DOI] [PubMed] [Google Scholar]
- de Laat W., Grosveld F. Spatial organization of gene expression: The active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- de Souza F.S.J., Bumaschny V.F., Low M.J., Rubinstein M. Subfunctionalization of expression and peptide domains following the ancient duplication of the proopiomelanocortin gene in teleost fishes. Mol. Biol. Evol. 2005;22:2417–2427. doi: 10.1093/molbev/msi236. [DOI] [PubMed] [Google Scholar]
- Fraser P., Grosveld F. Locus control regions, chromatin activation and transcription. Curr. Opin. Cell Biol. 1998;10:361–365. doi: 10.1016/s0955-0674(98)80012-4. [DOI] [PubMed] [Google Scholar]
- Hadchouel J., Tajbakhsh S., Primig M., Chang T.H., Daubas P., Rocancourt D., Buckingham M. Modular long-range regulation of Myf5 reveals unexpected heterogeneity between skeletal muscles in the mouse embryo. Development. 2000;127:4455–4467. doi: 10.1242/dev.127.20.4455. [DOI] [PubMed] [Google Scholar]
- Hadchouel J., Carvajal J.J., Daubas P., Bajard L., Chang T., Rocancourt D., Cox D., Summerbell D., Tajbakhsh S., Rigby P.W.J., et al. Analysis of a key regulatory region upstream of the Myf5 gene reveals multiple phases of myogenesis, orchestrated at each site by a combination of elements dispersed throughout the locus. Development. 2003;130:3415–3426. doi: 10.1242/dev.00552. [DOI] [PubMed] [Google Scholar]
- Han B., Zhang J.T. Regulation of gene expression by internal ribosome entry sites or cryptic promoters: The eIF4G story. Mol. Cell. Biol. 2002;22:7372–7384. doi: 10.1128/MCB.22.21.7372-7384.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalcheim C., Ben-Yair R. Cell rearrangements during development of the somite and its derivatives. Curr. Opin. Genet. Dev. 2005;15:371–380. doi: 10.1016/j.gde.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L., Gayraud-Morel B., Gomes D., Rocancourt D., Buckingham M., Shinin V., Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Kaul A., Koster M., Neuhaus H., Braun T. Myf-5 revisited: Loss of early myotome formation does not lead to a rib phenotype in homozygous Myf-5 mutant mice. Cell. 2000;102:17–19. doi: 10.1016/s0092-8674(00)00006-4. [DOI] [PubMed] [Google Scholar]
- Li Q., Peterson K.R., Fang X., Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A., Manzanares M., Blanco M.J., Nieto M.A. Modularity and reshuffling of Snail and Slug expression during vertebrate evolution. Proc. Natl. Acad. Sci. 2002;99:16841–16846. doi: 10.1073/pnas.262525399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W., Sarid J., Taub R., Vasicek T., Battey J., Lenoir G., Leder P. A translocated human c-myc oncogene is altered in a conserved coding sequence. Proc. Natl. Acad. Sci. 1986;83:2939–2943. doi: 10.1073/pnas.83.9.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noden D.M., Francis-West P. The differentiation and morphogenesis of craniofacial muscles. Dev. Dyn. 2006;235:1194–1218. doi: 10.1002/dvdy.20697. [DOI] [PubMed] [Google Scholar]
- Ohtsuki S., Levine M., Cai H.N. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes & Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E.N., Arnold H.H., Rigby P.W.J., Wold B.J. Know your neighbours: Three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- Ott M.O., Bober E., Lyons G., Arnold H., Buckingham M. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development. 1991;111:1097–1107. doi: 10.1242/dev.111.4.1097. [DOI] [PubMed] [Google Scholar]
- Patapoutian A., Miner J.H., Lyons G.E., Wold B. Isolated sequences from the linked Myf-5 and MRF4 genes drive distinct patterns of muscle-specific expression in transgenic mice. Development. 1993;118:61–69. doi: 10.1242/dev.118.1.61. [DOI] [PubMed] [Google Scholar]
- Pin C.L., Konieczny S.F. A fast fibre enhancer exists in the muscle regulatory factor 4 gene promoter. Biochem. Biophys. Res. Commun. 2002;299:7–13. doi: 10.1016/s0006-291x(02)02571-8. [DOI] [PubMed] [Google Scholar]
- Pin C.L., Ludolph D.C., Cooper S.T., Klocke B.J., Merlie J.P., Konieczny S.F. Distal regulatory elements control MRF4 gene expression in early and late myogenic cell populations. Dev. Dyn. 1997;208:299–312. doi: 10.1002/(SICI)1097-0177(199703)208:3<299::AID-AJA2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Sagai T., Hosoya M., Mizushina Y., Tamura M., Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- Spitz F., Gonzalez F., Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Summerbell D., Ashby P.R., Coutelle O., Cox D., Yee S., Rigby P.W.J. The expression of Myf5 in the developing mouse embryo is controlled by discrete and dispersed enhancers specific for particular populations of skeletal muscle precursors. Development. 2000;127:3745–3757. doi: 10.1242/dev.127.17.3745. [DOI] [PubMed] [Google Scholar]
- Summerbell D., Halai C., Rigby P.W.J. Expression of the myogenic regulatory factor Mrf4 precedes or is contemporaneous with that of Myf5 in the somitic bud. Mech. Dev. 2002;117:331–335. doi: 10.1016/s0925-4773(02)00208-3. [DOI] [PubMed] [Google Scholar]
- Swaminathan S., Ellis H.M., Waters L.S., Yu D., Lee E.C., Court D.L., Sharan S.K. Rapid engineering of bacterial artificial chromosomes using oligonucleotides. Genesis. 2001;29:14–21. doi: 10.1002/1526-968x(200101)29:1<14::aid-gene1001>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Bober E., Babinet C., Pournin S., Arnold H., Buckingham M. Gene targeting the myf-5 locus with nlacZ reveals expression of this myogenic factor in mature skeletal muscle fibres as well as early embryonic muscle. Dev. Dyn. 1996a;206:291–300. doi: 10.1002/(SICI)1097-0177(199607)206:3<291::AID-AJA6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Rocancourt D., Buckingham M. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature. 1996b;384:266–270. doi: 10.1038/384266a0. [DOI] [PubMed] [Google Scholar]
- Teboul L., Hadchouel J., Daubas P., Summerbell D., Buckingham M., Rigby P.W.J. The early epaxial enhancer is essential for the initial expression of the skeletal muscle determination gene Myf5 but not for subsequent, multiple phases of somitic myogenesis. Development. 2002;129:4571– 4580. doi: 10.1242/dev.129.19.4571. [DOI] [PubMed] [Google Scholar]
- Uchikawa M., Ishida Y., Takemoto T., Kamachi Y., Kondoh H. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell. 2003;4:509–519. doi: 10.1016/s1534-5807(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Wijgerde M., Grosveld F., Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- Yee S.P., Rigby P.W.J. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes & Dev. 1993;7:1277–1289. doi: 10.1101/gad.7.7a.1277. [DOI] [PubMed] [Google Scholar]
- Zammit P.S., Carvajal J.J., Golding J.P., Morgan J.E., Summerbell D., Partridge T.A., Rigby P.W.J., Beauchamp J.R. Myf5 expression in adult muscle spindles and satellite cells is controlled by separate genetic elements. Dev. Biol. 2004;273:454–465. doi: 10.1016/j.ydbio.2004.05.038. [DOI] [PubMed] [Google Scholar]
- Zhou J., Levine M. A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell. 1999;99:567–575. doi: 10.1016/s0092-8674(00)81546-9. [DOI] [PubMed] [Google Scholar]
- Zweigerdt R., Braun T., Arnold H.H. Faithful expression of the Myf-5 gene during mouse myogenesis requires distant control regions: A transgene approach using yeast artificial chromosomes. Dev. Biol. 1997;192:172–180. doi: 10.1006/dbio.1997.8759. [DOI] [PubMed] [Google Scholar]