Abstract
In approximately 20% of cases of severe congenital neutropenia (SCN), mutations are found in the gene encoding the granulocyte colony-stimulating factor receptor (G-CSF–R). These mutations introduce premature stop codons, which result in truncation of 82–98 COOH-terminal amino acids of the receptor. SCN patients who develop secondary myelodysplastic syndrome and acute myeloid leukemia almost invariably acquired a GCSFR mutation, suggesting that this genetic alteration represents a key step in leukemogenesis. Here we show that an equivalent mutation targeted in mice (gcsfr-Δ715) results in the selective expansion of the G-CSF– responsive progenitor (G-CFC) compartment in the bone marrow. In addition, in vivo treatment of gcsfr-Δ715 mice with G-CSF results in increased production of neutrophils leading to a sustained neutrophilia. This hyperproliferative response to G-CSF is accompanied by prolonged activation of signal transducer and activator of transcription (STAT) complexes and extended cell surface expression of mutant receptors due to defective internalization. In view of the continuous G-CSF treatment of SCN patients, these data provide insight into why progenitor cells expressing truncated receptors clonally expand in vivo, and why these cells may be targets for additional genetic events leading to leukemia.
Keywords: neutropenia, severe congenital neutropenia, granulocyte colony-stimulating factor receptor, mutations, acute myeloid leukemia
Severe congenital neutropenia (SCN)1 is a heterogeneous hematopoietic disorder characterized by a profound neutropenia due to a maturation arrest of granulocytic progenitor cells in the bone marrow (BM). As a result, SCN patients suffer from recurrent, often life-threatening, bacterial infections (1). The disease presents with a variable inheritance, and the mechanisms underlying its pathogenesis are largely unknown (2–5). Granulocyte colony-stimulating factor (G-CSF) is a major regulator of neutrophil production (6, 7). However, although defective production of G-CSF has been suggested as a possible cause of SCN in isolated cases (8, 9), in the majority of SCN patients G-CSF serum levels are normal or elevated (10–12). In >90% of SCN patients, daily administration of pharmacological doses of G-CSF increases neutrophil levels and significantly reduces infection-related events (1, 13). In contrast, treatment with GM-CSF is rarely effective (13), suggesting that specifically the G-CSF signal transduction pathway is affected in SCN.
G-CSF mediates its effects through activation of its cognate receptor (G-CSF–R), a single-transmembrane protein of the hematopoietin receptor superfamily which forms homooligomeric complexes upon ligand binding (14–16). Like all members of this group of receptors, G-CSF–R lacks intrinsic tyrosine kinase activity but can activate cytoplasmic tyrosine kinases of the Jak and Src families (17–23). These kinases tyrosine phosphorylate substrates, including the receptor, to provide docking sites for other proteins, which, in turn, can be phosphorylated as well. Proteins docking to the G-CSF–R complex include members of the signal transducer and activator of transcription (STAT) family. Upon tyrosine phosphorylation, STATs form dimeric complexes and translocate to the nucleus, where they influence gene transcription (24, 25). Of the six members of the STAT family identified in mammalian cells, STAT1, STAT3, and STAT5 have been implicated in G-CSF signaling (19, 26–29). STAT3 has recently been linked with IL-6–induced macrophage differentiation and G-CSF–induced neutrophilic differentiation (30–32), whereas STAT5 has been implicated in the control of hemopoietic cell proliferation by some cytokines, including G-CSF (33, 34).
In a subgroup of SCN patients, acquired mutations in the GCSFR gene are found in BM and peripheral blood neutrophils (18, 35–39). These mutations introduce premature stop codons that result in the truncation of 82–98 COOH-terminal amino acids, a region previously implicated in the control of neutrophilic differentiation (40, 41). In recent studies of a total of 78 SCN patients, 18 cases with point mutations in the critical region in the GCSFR gene were found (38, 42). Among those patients with GCSFR mutations, eight developed acute myeloid leukemia (AML), whereas only one of the patients without a mutation has thus far showed clinical or cytogenetic evidence of leukemic progression (38). This latter patient represents a rare case of autosomal dominant SCN affecting a mother and four siblings. Up to now, no GCSFR mutations have been detected in patients with idiopathic neutropenia, chronic myeloid leukemia (CML), or myelodysplastic syndrome (MDS) without history of neutropenia. However, mutations occur at very low frequency in de novo AML (18, 43–45). These clinical observations suggest that acquisition of a GCSFR mutation may be a critical step in the leukemic transformation of progenitor cells from SCN patients.
Recently, we generated mice with a mutation in their gcsfr gene (Δ715) equivalent to one of the nonsense mutations found in SCN patients (46). Both homozygous and heterozygous Δ715 mice have reduced numbers of circulating neutrophils compared with their wt/wt littermates, supporting the notion that the G-CSF–R COOH terminus has a role in granulopoiesis in vivo. Strikingly, after injection with G-CSF, gcsfr-wt/Δ715 and gcsfr-Δ715/Δ715 mice showed increased peripheral neutrophil counts compared with their wt/wt littermates (46). Analysis of cells labeled with the thymidine analogue 5-bromo-2′-deoxyuridine in vivo indicated that the rise was due to de novo generation of neutrophils, although specific mechanistic details were not addressed.
In this study, we show that the gcsfr-Δ715 mutation results in an approximately twofold increase in granulocyte colony-forming cell (G-CFC) content in the BM. In contrast, the numbers of more primitive hematopoietic progenitors, as assessed in a cobblestone area–forming cell (CAFC) assay (47), are slightly decreased. Sustained G-CSF treatment results in a continuous neutrophilia in gcsfr-Δ715 mice, suggesting a persistent increase in G-CSF– induced proliferation of neutrophilic progenitors due to the G-CSF–R truncation. Finally, we demonstrate altered levels and kinetics of G-CSF–induced activation of STAT proteins in gcsfr-Δ715 mice, including a significantly extended activation of STAT proteins that correlated with a delay in internalization of the truncated G-CSF–R proteins. These altered signaling properties provide potential explanations for the preleukemic nature of GCSFR mutations in SCN.
Materials and Methods
Mice.
gcsfr-Δ715 mice (46) and control littermates were housed in a specific pathogen–free environment. F1 and F2 mice were generated from gcsfr-wt/Δ715 (129/OLA × FVB/N) parents, and mice were used at the age of 2–3 mo. The gcsfr-Δ715 mutation was back-crossed on an FVB/N background. Progeny of gcsfr-wt/Δ715 mice of the third back-cross generation were used in the in vivo G-CSF treatment experiment, in two out of four in vitro colony assays, and in one of the two CAFC assays.
Blood and BM Sampling.
Blood samples were collected from the tail vein at fixed time points to avoid variation due to circadian rhythms (48). Blood cell counts were performed on a Sysmex-K1000X automated counter (Toa Medical Electronics Co. Ltd.). To obtain BM cell suspensions, femurs and tibias were crushed in a mortar in HBSS with 5% FCS (HBSS/FCS). BM cells were passed through a 100-μm sieve, spun down, and resuspended. The resulting monocellular suspensions contained 98–99% viable cells, as determined by trypan blue exclusion. For differential countings, blood smears were fixed in methanol, May-Grunwald-Giemsa (MGG) stained. At least 300 blood cells were analyzed in a Zeiss Axioscope microscope.
Isolation of BM Progenitor Cells.
BM cells of two mice were pooled and suspended in HBSS/FCS and subsequently incubated in a 162-cm2 cell culture flask (Costar) in a humidified atmosphere of 5% CO2 in air at 37°C for 1 h. Nonadherent cells were spun down and resuspended in 45% (ρ = 1.058) Percoll (Pharmacia Fine Chemicals) solution in Ca2+/Mg2+-free HBSS. Cells were then loaded on a Percoll density gradient as described (49). BM progenitor cells were collected from the interface between the layers with densities of 1.058 and 1.064. Cells were washed twice in Ca2+/Mg2+-free HBSS supplemented with 0.2% BSA (HBSS/BSA). The cell numbers recovered in these fractions from wt/wt, wt/Δ715, and Δ715/Δ715 mice were similar.
In Vitro Colony Assays.
BM progenitor cells were plated in triplicate in 35-mm Petri dishes (Falcon; Becton Dickinson) containing 1 ml of methylcellulose medium (Methocult M3230; Stem-cell Technologies, Inc.) containing 30% fetal bovine serum, 1% BSA, 0.1 mM 2-ME, 2 mM l-glutamine, without additional growth factor or with 100 ng/ml G-CSF (Amgen, Inc.), 20 U/ml murine GM-CSF (provided by Dr. Steven Neben, Genetic Institute, Cambridge, MA), 100 U/ml murine macrophage (M)-CSF, 4 U/ml human erythropoietin (EPO; a gift of Janssen-Cilag) plus 10 μg/ml murine stem cell factor (SCF) and 0.2 mM hemin (bovine type 1; Sigma). Colonies (50 cells or more) were counted on day 7 or 8 of culture. CAFC assays were performed as described (47). In this limiting dilution assay, phase-dark hematopoietic colonies (cobblestone areas) are formed under a preestablished stromal layer. Early appearing cobblestone areas (week 1–2) represent clones from the relatively mature stem/progenitor cell subset, while the most primitive hemopoietic stem cells, i.e., those with long-term repopulating abilities in vivo, are assessed at week 4–5.
Preparation of Nuclear Extracts.
BM cells isolated as described above were washed once with HBSS, resuspended in RPMI/ 10% FCS, and transferred to a tissue culture flask. After 1.5 h, nonadherent cells were transferred to a fresh flask for another 3.5 h before stimulation with murine IL-3 (100 ng/ml) or human G-CSF (100 ng/ml). Stimulation was terminated by adding ice-cold PBS containing 0.1 mM Na3VO4. Cells were lysed in 200 μl hypotonic buffer (20 mM Hepes, 20 mM NaF, 1 mM Na3VO4, 1 mM Na4P2O7, 0.125 μM okadaic acid, 1 mM EDTA, 1 mM EGTA, 0.2% NP-40, 1 mM dithiothreitol, 0.5 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.5 μg/ml bacitracin, 0.5 μg/ml iodoacetamide, and 1 mM Pefabloc). Nuclei were spun down at 15,000 g for 30 s, and proteins were extracted by rocking for 30 min at 4°C in 20 μl high-salt buffer (hypotonic buffer with 420 mM NaCl, and 20% glycerol). Insoluble materials were removed by centrifugation at 20,000 g at 4°C for 20 min.
Electrophoretic Mobility Shift Assays with STAT-binding Oligonucleotides.
Nuclear extracts were incubated with double-stranded [γ-P32]ATP end-labeled STAT1- and STAT3-binding oligonucleotide m67 (5′-CATTTCCCGTAAATC-3′; reference 50) or STAT1- and STAT5-binding oligonucleotide (5′-AGATTTCTAGGAATTCAATCC-3′) derived from the 5′ regulatory region of the β-casein gene (51). Binding reactions were performed for 30 min at room temperature in a 20-μl reaction volume containing 3 μl nuclear extract, 1 μl dIdC (2 mg/ml), 11 μl H2O, 4 μl 5× binding buffer (50 mM Hepes, pH 7.6–7.8, 85 mM NaCl, 15 mM NaMoO4, 4.25 mM dithiothreitol, and 25% glycerol), and 1 μl labeled double-stranded oligonucleotides (10.2 ng/μl). The reaction mixtures were fractionated by electrophoresis on 5% polyacrylamide gels containing 5% glycerol in 0.25× TBE (Tris-boric acid-EDTA). In supershift experiments, 2 μl of either anti-STAT1, anti-STAT3 (Santa Cruz Biotechnology), or anti-STAT5 antibodies (provided by B. Groner, Institute of Experimental Cancer Research, Tumor Biology Center, Freiburg, Germany) was added to the reaction mixture and incubated for 30 min on ice, before the addition of the radiolabeled oligonucleotides. The gels were dried and subsequently analyzed by autoradiography. For quantitation, gels were exposed to PhosphorImager screens and analyzed with ImageQuant software (Molecular Dynamics).
Flow Cytometric Analysis of G-CSF–R Internalization.
G-CSF–R expression on BM cells was measured by flow cytometry. To this end, G-CSF was biotinylated using d-biotinoyl-ε-aminocaproic acid-N-hydroxy-succinimide ester (Biotin-7-NHS; Boehringer Mannheim). Free biotin was removed by gel filtration on Sephadex G-25. BM cells (106) were incubated for various times at 37°C with 0.2 μg/ml biotinylated G-CSF. Subsequently, cells were incubated for 30 min at 4°C with PE-conjugated streptavidin (SA-PE; Caltag Labs) in the presence of 0.05% NaN3. Cells were subjected to flow cytometric analysis on a FACS®. A window was set on the basis of forward and side light scatter to restrict the analysis to myeloid cells.
Results
Numbers of Myeloid Progenitors Are Increased in gcsfr-Δ715 Mice.
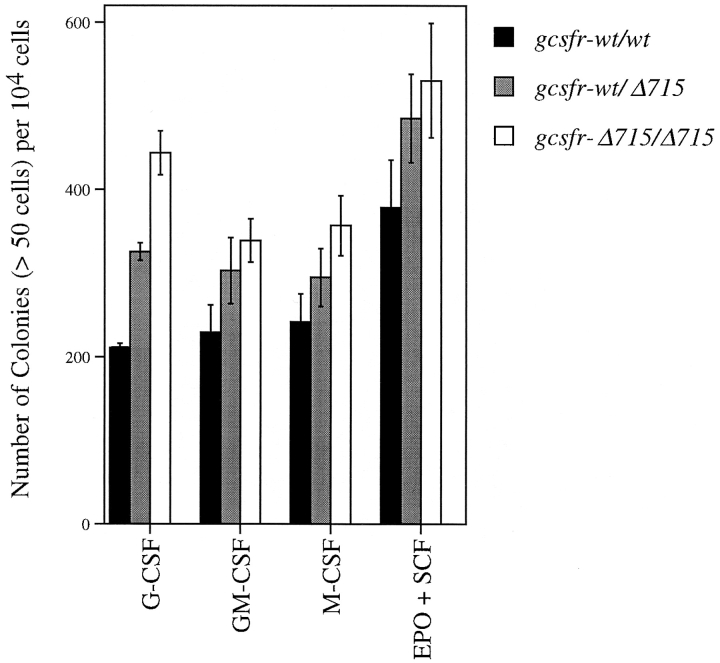
We have previously shown that gcsfr-Δ715 mice are neutropenic (46). To determine at what stage of myeloid development the effect of the Δ715 mutation becomes phenotypically overt, we performed in vitro colony assays in the presence of specific growth factors. Depending on the factor(s) added, these assays provide a quantitative measure of the number of neutrophilic precursors (G-CSF), granulocyte-macrophage precursors (GM-CSF), macrophage precursors (M-CSF), and erythroid precursors (EPO plus SCF). Data from a representative experiment are shown in Fig. 1. The frequency of G-CSF–responsive CFCs was increased by a factor 2.0 ± 0.6 (mean ± SD of four independent experiments) in homozygous Δ715 mice, and 1.8 ± 0.9 in heterozygous mice compared with wild-type littermates. Although the frequencies of GM-CSF, M-CSF, and EPO plus SCF responsive progenitors were also consistently higher in mutant mice, the difference between mutant and wild-type mice was less pronounced than that in the granulocytic lineage.
Figure 1.
Selective expansion of the G-CFC compartment in BM of gcsfr-Δ715 mice. BM cells of two mice of each genotype were pooled, enriched for progenitor cells, and plated in triplicate in methylcellulose-containing media supplemented with various growth factors. Hemopoietic colonies containing 50 cells or more were scored after 7–8 d. Data are mean ± SD.
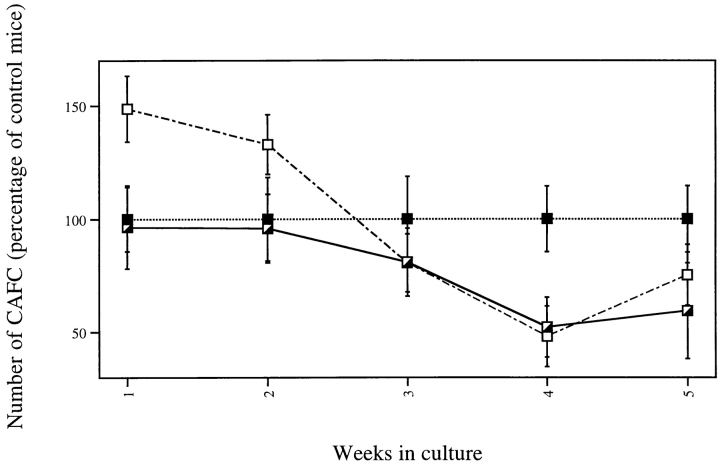
We next evaluated the effect of the mutation on more primitive hemopoietic cells in the CAFC assay. From the CAFC numbers per femur of gcsfr-wt/wt, wt/Δ715, and Δ715/Δ715 mice (Table I), we calculated the percentage of CAFCs relative to the CAFC numbers of wild-type mice (Fig. 2). Although CAFCs at week 1–2 (a measure of stem cells with short-term repopulating ability) were relatively more abundant in the gcsfr-Δ715/Δ715 mice, CAFCs at week 4–5 (primitive stem cells with long-term repopulating ability) were present in lower numbers than in control mice. Therefore, it appears that the absence of the COOH-terminal domain of the G-CSF–R leads to a selective expansion of a myeloid progenitor compartment, to some extent at the expense of primitive stem cells.
Table I.
Numbers of Progenitor Cells and Primitive Stem Cells Estimated in the CAFC Assay
| No. of CAFCs/femur | ||||||
|---|---|---|---|---|---|---|
| gcsfr-wt/wt | gcsfr-wt/Δ715 | gcsfr-Δ715/Δ715 | ||||
| Experiment 1 | ||||||
| Week 1 | 78,400 | 75,700 | 116,600 | |||
| 2 | 51,400 | 49,300 | 68,300 | |||
| 3 | 30,700 | 24,900 | 24,800 | |||
| 4 | 15,300 | 7,970 | 7,330 | |||
| 5 | 3,560 | 2,110 | 2,680 | |||
| Experiment 2 | ||||||
| Week 1 | 59,800 | 67,400 | 75,400 | |||
| 2 | 12,200 | 13,900 | 25,700 | |||
| 3 | 7,310 | 9,880 | 10,300 | |||
| 4 | 3,640 | 2,070 | 2,370 | |||
| 5 | 1,330 | 650 | 650 | |||
BM cells of two mice (one femur per mouse) were pooled and plated in a limiting dilution in 96-well plates on FBMD-1 stromal feeders. CAFCs were scored on weeks 1–5. Early appearing cobblestone areas (week 1–2) represent clones from the relatively mature stem/progenitor cell subset, whereas the most primitive hemopoietic stem cells, i.e., those with long-term repopulating abilities in vivo, are assessed at week 4–5. Data of two independent experiments are given.
Figure 2.
Numbers of progenitor cells and primitive hemopoietic stem cells in gcsfr-Δ715 mice measured by CAFC assays. BM cells were plated under a layer of stromal cells. Cobblestone areas were counted weekly and expressed as the number of CAFCs per femur relative to the CAFC number per femur of gcsfr-wt/wt mice. Data are mean ± SD of two independent experiments.
gcsfr-Δ715 Mice Develop Sustained Neutrophilia in Response to Long-term G-CSF Treatment.
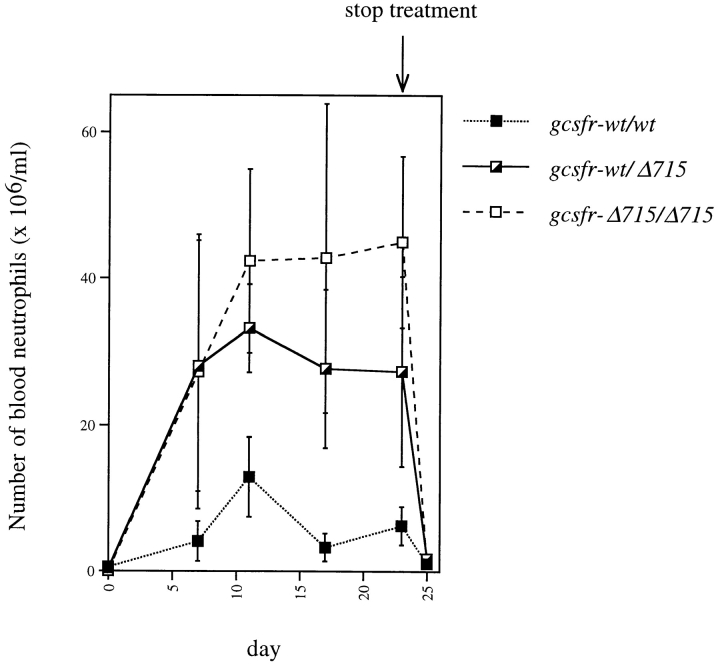
Previously, we showed that gcsfr-wt/Δ715 and gcsfr-Δ715/Δ715 mice injected with G-CSF for 1 wk have elevated blood neutrophil counts on days 6 and 7 compared with control littermates (46). To establish whether this effect is transient or sustained, gcsfr-wt/wt, gcsfr-wt/Δ715, and gcsfr-Δ715/Δ715 mice were injected daily with G-CSF (250 μg/kg) for 22 d, and blood neutrophil numbers were monitored. Wild-type mice responded to G-CSF treatment with a 10-fold increase in their blood neutrophil levels, mice heterozygous for the Δ715 mutation with a 65-fold increase, and gcsfr-Δ715/Δ715 mice with a 250-fold increase (Fig. 3). The average blood neutrophil count on day 23 was 45 × 106/ml in Δ715/Δ715 mice (n = 4), 27 × 106/ml in wt/Δ715 mice, and 6 × 106/ml in wt/wt mice. Thus, it is clear that G-CSF treatment leads to a sustained neutrophilia in gcsfr-Δ715 mice. Red blood cell and platelet counts were not influenced by the mutation, suggesting that the gcsfr-Δ715 mutation selectively enhances the proliferation of myeloid progenitors.
Figure 3.
Sustained neutrophilia in gcsfr-Δ715 mice induced by in vivo G-CSF treatment. Mice were injected subcutaneously with G-CSF (250 μg/ kg/d) for 22 d. Blood was collected and analyzed as described in Materials and Methods. Data are means ± SD of four animals for each genotype.
Kinetics of G-CSF–induced Activation of STAT Complexes in gcsfr-Δ715 Mice.
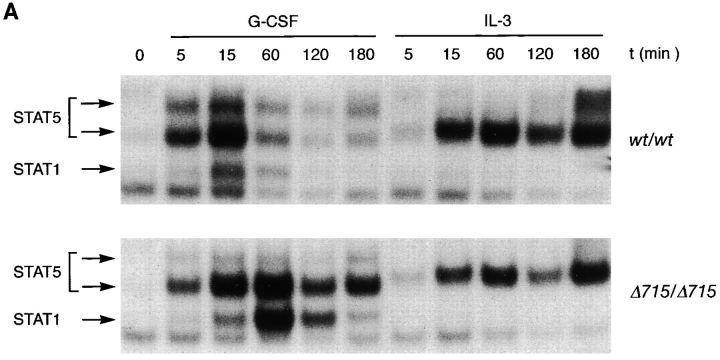
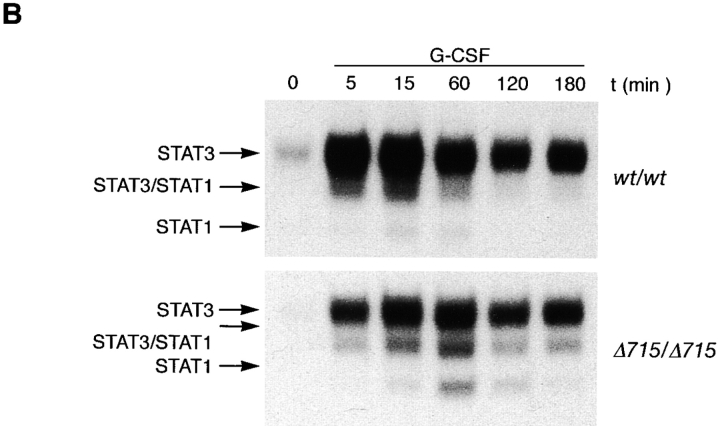
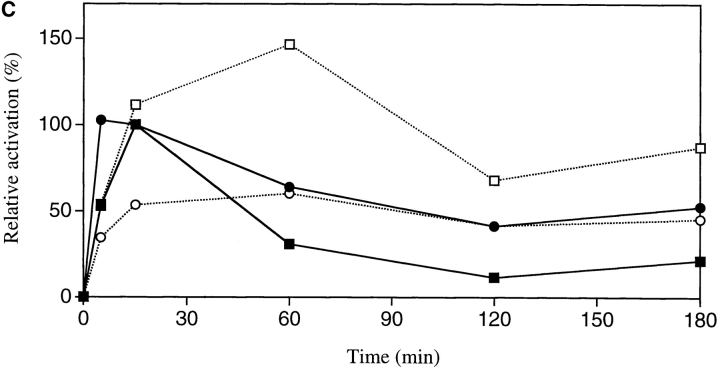
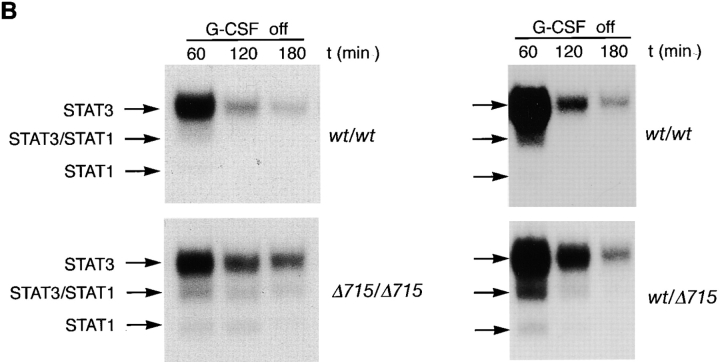
Because there is increasing evidence that STAT proteins play an important role in the control of proliferation and differentiation of hematopoietic cells, we analyzed both STAT5 and STAT3 activation in BM cells of gcsfr-wt/wt, wt/Δ715, and Δ715/Δ715 mice. At early time points (up to 15 min), G-CSF–induced activation of STAT5 was largely unaffected while STAT3 activation was reduced in Δ715/Δ715 BM cells (Fig. 4, A and B). However, at later time points of stimulation (60 and 120 min), when STAT5 activation in wt/wt cells declined, activation of STAT5 in Δ715/Δ715 cells persisted, whereas STAT3 levels became comparable in both. Quantification of the signals (Fig. 4 C) shows that after 1–3 h of stimulation, STAT5 activation in BM cells of Δ715 mice is four- to fivefold the STAT5 activation in wild-type BM cells, whereas STAT3 activation after 1–3 h of stimulation is similar. BM cells from heterozygous mice showed an intermediate response (data not shown). IL-3–induced activation of STAT complexes was equivalent in all genotypes.
Figure 4.
Sustained activation of STAT complexes by Δ715 G-CSF–R. Electrophoretic mobility shift assay of nuclear extracts prepared from BM cells, stimulated with G-CSF or IL-3 for the indicated times. (A) Time course of STAT1 and STAT5 activation using β-casein probe. Supershifts with antibodies against STAT1 and STAT5 indicated that the lower band contained STAT1 and the upper two bands contained STAT5 (reference 29). (B) Time course of STAT1 and STAT3 activation using m67 probe. Supershifts showed that the lower two bands contained STAT1, whereas the upper two bands contained STAT3 (reference 29). (C) Quantitative analysis of time course of STAT-binding kinetics, setting STAT5 activation of wild-type receptors at 15 min to 100%.
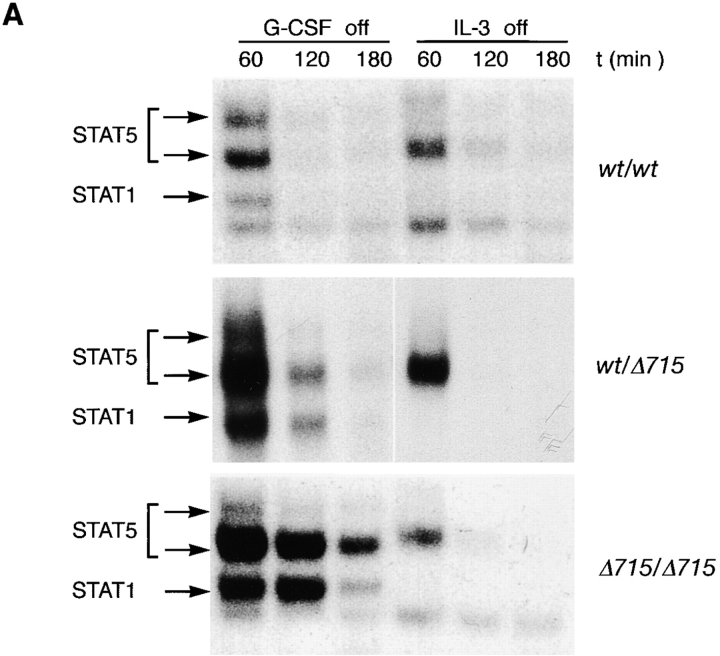
We also studied STAT activation after 10 min of stimulation followed by an extensive wash to remove G-CSF. Under these conditions, prolonged activation of STAT5, STAT3, and STAT1 was seen in Δ715/Δ715 cells and to a somewhat lesser extent in wt/Δ715 cells (Fig. 5, A and B). These findings suggest that the truncated receptors which bound ligand during the first 10 min of stimulation were responsible for the prolonged STAT activation, and that this was not due to stimulation of new receptors recruited to the cell surface. Because wild-type and truncated receptors show equivalent K d (41), this suggests that receptor deactivation is altered in truncated receptors.
Figure 5.
Prolonged activation of STAT complexes in gcsfr-Δ715 mice after removal of G-CSF. BM cells were stimulated with G-CSF or IL-3 for 10 min, washed extensively, and incubated in RPMI/10% FCS. Nuclear extracts were prepared at the indicated times and assayed as described above using STAT1- and STAT5-binding β-casein probe (A) or STAT1- and STAT3-binding m67 probe (B). Left and right panels represent independent experiments.
Ligand-mediated Internalization of Truncated G-CSF–R Is Delayed.
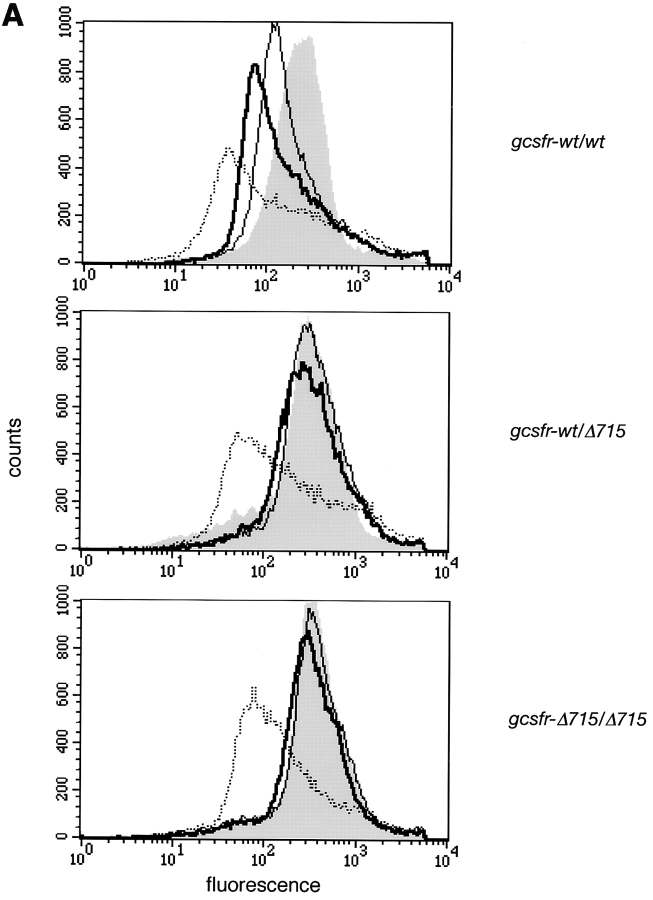
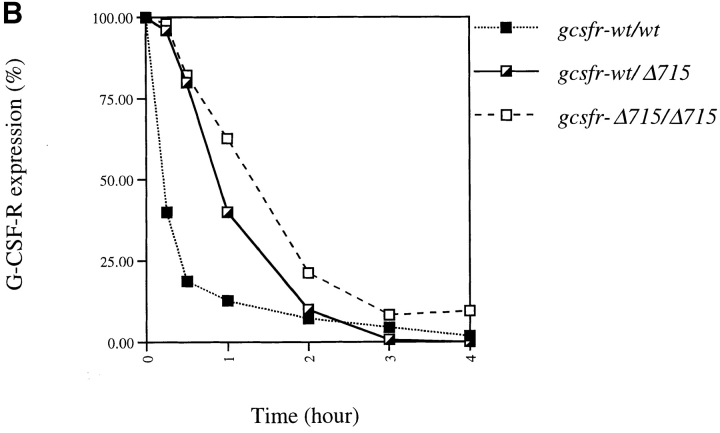
The COOH-terminal region of G-CSF–R contains a dileucine motif (STQPLL) that has been implicated in ligand-mediated receptor endocytosis (52, 53). Due to the truncation, Δ715 G-CSF–Rs lack this motif. When exposed to G-CSF, within 30 min, surface expression of wild-type receptors had decreased to 20% of the initial expression, whereas 80% of the mutant receptors were still present on the cell surface (Fig. 6, A and B). Thus, upon exposure to G-CSF, truncated G-CSF–Rs reside longer on the cell surface, which may contribute to their ability to induce sustained STAT activation.
Figure 6.
Comparison of internalization kinetics of full-length and truncated G-CSF–R. BM cells were incubated with biotinylated G-CSF at 37°C for the indicated periods and then stained with SA-PE. (A) Shaded histogram, t = 0; solid line, t = 15 min; bold line, t = 30 min; dotted line, t = 4 h. (B) Peak fluorescence standardized for t = 0 as a function of time.
Discussion
Before the therapeutic use of G-CSF for SCN patients, incidental cases terminating in AML were reported (54). After the introduction of G-CSF treatment of SCN patients, mortality due to severe opportunistic bacterial infections diminished, but the incidence of secondary AML gradually increased. The frequency of leukemic progression is currently estimated at 7.5–10% (55). After the discovery of mutations in the gene encoding the G-CSF–R in several SCN patients, a correlation was observed between the acquisition of such mutations and leukemic progression of the disease (37, 42). The consequences of these mutations for the signaling function of G-CSF–R have initially been investigated in myeloid cell lines. These studies revealed that truncated receptors transduce strong proliferative signals, but are defective in maturation signaling (40, 41).
To gain further insight in the contribution of GCSFR mutations to the pathogenesis of SCN and AML, we recently generated an in vivo model by introducing an SCN-derived nonsense mutation (gcsfr-Δ715) in mice (46). Both wt/Δ715 and Δ715/Δ715 mice have significantly reduced numbers of blood neutrophils compared with their wt/wt littermates, suggesting that the G-CSF–R COOH-terminal domain contributes to neutrophil development in vivo. This phenotype has remained consistent and independent of genetic background in the mice generated so far. In contrast, other investigators have reported that blood neutrophil numbers in mice with a similar mutation in the gcsfr gene were not significantly altered (56). However, a complication of their model is that the expression of the mutant allele was significantly higher than the wild-type allele. This might be due to the fact that the PGK-neo cassette used for selection of embryonic stem cells within the gcsfr gene had not been removed before the generation of animals. This increased expression of truncated G-CSF–R may compensate for the mutation, and could explain the absence of neutropenia.
In gcsfr-Δ715 mice, the numbers of CAFCs at week 5 appear slightly reduced in BM, whereas the numbers of G-CFCs are selectively expanded. As yet, we cannot explain this relative paucity of primitive stem cells in Δ715 mice. One possibility is that expression of truncated G-CSF–Rs on these cells would lead to an increase in cell cycling and commitment. However, whether primitive hemopoietic stem cells express G-CSF–R is still controversial (57, 58). The fact that the numbers of IL-3–, M-CSF–, and GM-CSF–responsive CFCs are also increased in gcsfr-Δ715 mice suggests that the GM-CFC compartment is already somewhat expanded. Despite the increased G-CFC compartment, the number of circulating neutrophils is significantly reduced in gcsfr-Δ715 mice (46). This implies that truncated receptors do impair the development to blood neutrophil, but at what stage between the G-CFC and mature neutrophil this defect becomes manifest remains unclear.
The gcsfr-Δ715 mice showed a sustained neutrophilia in response to G-CSF administration. The fact that this hyperproliferation persisted after prolonged exposure to G-CSF is compatible with the notion that the rate of proliferation of the early myeloid progenitor compartment is increased in these mice. Increased proliferation due to G-CSF–R truncation has also been observed in cell lines (40, 41). However, immortalized cell lines contain multiple genetic aberrations that influence cell cycle regulation. We have shown here in primary cells that the gcsfr-Δ715 mutation itself is sufficient for an in vivo hyperproliferative response to G-CSF. This supports the idea that acquisition of this type of mutation represents a step in leukemic transformation. In SCN patients, expansion of progenitor cells with acquired G-CSF–R mutations is observed. Thus, it is conceivable that under continuous G-CSF administration, such as that received by SCN patients, progenitor cells that acquire a GCSFR mutation would have a proliferative advantage leading to their in vivo expansion.
STAT proteins are involved in control of multiple cellular processes regulated by cytokines and growth factors, including cell differentiation, proliferation, and survival. Since the BM from gcsfr-wt/wt, gcsfr-wt/Δ715, and gcsfr-Δ715/Δ715 mice appears similar in composition, also within the neutrophilic lineage (46), we could directly compare G-CSF–induced activation of STAT proteins in the BM of these mice. BM cells from gcsfr-Δ715 mice displayed normal levels of STAT5, but decreased levels of STAT3 activation. The latter is most likely due to the absence of specific STAT3-activating mechanism(s) (26, 29). In view of the putative role of STAT3 in G-CSF–induced differentiation of neutrophilic progenitors (32), the decreased STAT3 activation could, at least partially, explain the defective maturation to blood neutrophil which leads to basal neutropenia in gcsfr-wt/Δ715 and gcsfr-Δ715/Δ715 mice. Importantly, truncated G-CSF–Rs induced prolonged activation of STAT complexes and showed slower internalization rates. However, the relative levels of G-CSF– induced activation of STAT5 versus STAT3 in gcsfr-Δ715 BM were consistently higher than in gcsfr-wt BM. This may shift the proliferation/differentiation balance within the neutrophilic progenitor cells. There is increasing evidence that oncogenesis is associated with abnormal STAT signaling (59). For example, constitutive STAT activation has been reported in cells transformed by Src, Abl, and other oncoproteins (60–64). In human tumors, e.g., breast carcinoma cells, lymphomas, and leukemias, STATs are also frequently activated (63, 65–67). In view of these findings, our observation that the kinetics of STAT activation by truncated G-CSF–Rs are altered further corroborates the idea that GCSFR mutations truncating the COOH terminus of the receptor are a step in leukemogenesis in SCN patients.
Acknowledgments
We are grateful to Karola van Rooyen for graphical assistance and Bernd Groner for antibodies.
Abbreviations used in this paper
- AML
acute myeloid leukemia
- BM
bone marrow
- CAFC
cobblestone area–forming cell
- CFC
colony-forming cell
- EPO
erythropoietin
- SCF
stem cell factor
- SCN
severe congenital neutropenia
- STAT
signal transducer and activator of transcription
Footnotes
This work was financed by a grant from the Netherlands Organization for Scientific Research NWO (to M.H.A. Hermans, C. Antonissen, and I.P. Touw), a European Molecular Biology Organization Long-term Fellowship (to A.C. Ward), and grants from the Dutch Cancer Society (to I.P. Touw).
References
- 1.Welte K, Boxer LA. Severe chronic neutropenia: pathophysiology and therapy. Semin Hematol. 1997;34:267–278. [PubMed] [Google Scholar]
- 2.Kostmann, R. 1956. Infantile genetic agranulocytosis. Acta Paediatr. Scand. 45:1 (Suppl. 105). [PubMed]
- 3.Komiyama A, Yamazaki M, Yoda S, Saitoh H, Morosawa H, Akabane T. Morphologic and functional heterogeneity of chronic neutropenia of childhood with normal neutrophil colony formation in vitro. Am J Hematol. 1981;11:175–182. doi: 10.1002/ajh.2830110209. [DOI] [PubMed] [Google Scholar]
- 4.L'Esperance P, Brunning R, Deinard AS, Park BY, Biggar WD, Good RA. Congenital neutropenia: impaired maturation with diminished stem-cell input. Birth Defects Orig Artic Ser. 1975;11:59–65. [PubMed] [Google Scholar]
- 5.Parmley RT, Ogawa M, Darby CP, Jr, Spicer SS. Congenital neutropenia: neutrophil proliferation with abnormal maturation. Blood. 1975;46:723–734. [PubMed] [Google Scholar]
- 6.Lieschke GJ, Grail D, Hodgson G, Metcalf D, Stanley E, Cheers C, Fowler KJ, Basu S, Zhan YF, Dunn AR. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 1994;84:1737–1746. [PubMed] [Google Scholar]
- 7.Liu F, Wu HY, Wesselschmidt R, Kornaga T, Link DC. Impaired production and increased apoptosis of neutrophils in granulocyte colony-stimulating factor receptor-deficient mice. Immunity. 1996;5:491–501. doi: 10.1016/s1074-7613(00)80504-x. [DOI] [PubMed] [Google Scholar]
- 8.Glasser L, Duncan BR, Corrigan JJ., Jr Measurement of serum granulocyte colony-stimulating factor in a patient with congenital agranulocytosis (Kostmann's syndrome) Am J Dis Child. 1991;145:925–928. doi: 10.1001/archpedi.1991.02160080103029. [DOI] [PubMed] [Google Scholar]
- 9.Mizuno Y, Hara T, Nagata M, Omori F, Shimoda K, Okamura S, Niho Y, Ueda K. Serum granulocyte colony-stimulating factor levels in chronic neutropenia of infancy. Pediatr Hematol Oncol. 1990;7:377–381. doi: 10.3109/08880019009033415. [DOI] [PubMed] [Google Scholar]
- 10.Mempel K, Pietsch T, Menzel T, Zeidler C, Welte K. Increased serum levels of granulocyte colony-stimulating factor in patients with severe congenital neutropenia. Blood. 1991;77:1919–1922. [PubMed] [Google Scholar]
- 11.Pietsch T, Buhrer C, Mempel K, Menzel T, Steffens U, Schrader C, Santos F, Zeidler C, Welte K. Blood mononuclear cells from patients with severe congenital neutropenia are capable of producing granulocyte colony-stimulating factor. Blood. 1991;77:1234–1237. [PubMed] [Google Scholar]
- 12.Guba SC, Sartor CA, Hutchinson R, Boxer LA, Emerson SG. Granulocyte colony-stimulating factor (G-CSF) production and G-CSF receptor structure in patients with congenital neutropenia. Blood. 1994;83:1486–1492. [PubMed] [Google Scholar]
- 13.Welte K, Zeidler C, Reiter A, Muller W, Odenwald E, Souza L, Riehm H. Differential effects of granulocyte-macrophage colony-stimulating factor and granulocyte colony-stimulating factor in children with severe congenital neutropenia. Blood. 1990;75:1056–1063. [PubMed] [Google Scholar]
- 14.Fukunaga R, Ishizaka-Ikeda E, Pan CX, Seto Y, Nagata S. Functional domains of the granulocyte colony-stimulating factor receptor. EMBO (Eur Mol Biol Organ) J. 1991;10:2855–2865. doi: 10.1002/j.1460-2075.1991.tb07835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demetri GD, Griffin JD. Granulocyte colony-stimulating factor and its receptor. Blood. 1991;78:2791–2808. [PubMed] [Google Scholar]
- 16.Fukunaga R, Ishizaka-Ikeda E, Seto Y, Nagata S. Expression cloning of a receptor for murine granulocyte colony-stimulating factor. Cell. 1990;61:341–350. doi: 10.1016/0092-8674(90)90814-u. [DOI] [PubMed] [Google Scholar]
- 17.Nicholson SE, Oates AC, Harpur AG, Ziemiecki A, Wilks AF, Layton JE. Tyrosine kinase JAK1 is associated with the granulocyte-colony-stimulating factor receptor and both become tyrosine-phosphorylated after receptor activation. Proc Natl Acad Sci USA. 1994;91:2985–2988. doi: 10.1073/pnas.91.8.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong F, van Paassen M, van Buitenen C, Hoefsloot LH, Lowenberg B, Touw IP. A point mutation in the granulocyte colony-stimulating factor receptor (G-CSF-R) gene in a case of acute myeloid leukemia results in the overexpression of a novel G-CSF-R isoform. Blood. 1995;85:902–911. [PubMed] [Google Scholar]
- 19.Tian SS, Tapley P, Sincich C, Stein RB, Rosen J, Lamb P. Multiple signaling pathways induced by granulocyte colony-stimulating factor involving activation of JAKs, STAT5, and/or STAT3 are required for regulation of three distinct classes of immediate early genes. Blood. 1996;88:4435–4444. [PubMed] [Google Scholar]
- 20.Nicholson SE, Novak U, Ziegler SF, Layton JE. Distinct regions of the granulocyte colony-stimulating factor receptor are required for tyrosine phosphorylation of the signaling molecules JAK2, Stat3, and p42, p44MAPK. Blood. 1995;86:3698–3704. [PubMed] [Google Scholar]
- 21.Corey SJ, Burkhardt AL, Bolen JB, Geahlen RL, Tkatch LS, Tweardy DJ. Granulocyte colony-stimulating factor receptor signaling involves the formation of a three-component complex with Lyn and Syk protein- tyrosine kinases. Proc Natl Acad Sci USA. 1994;91:4683–4687. doi: 10.1073/pnas.91.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corey SJ, Dombrosky-Ferlan PM, Zuo S, Krohn E, Donnenberg AD, Zorich P, Romero G, Takata M, Kurosaki T. Requirement of Src kinase Lyn for induction of DNA synthesis by granulocyte colony-stimulating factor. J Biol Chem. 1998;273:3230–3235. doi: 10.1074/jbc.273.6.3230. [DOI] [PubMed] [Google Scholar]
- 23.Ward AC, Monkhouse JL, Csar XF, Touw IP, Bello PA. The Src-like tyrosine kinase Hck is activated by granulocyte colony-stimulating factor (G-CSF) and docks to the activated G-CSF receptor. Biochem Biophys Res Commun. 1998;251:117–123. doi: 10.1006/bbrc.1998.9441. [DOI] [PubMed] [Google Scholar]
- 24.Ihle JN, Nosaka T, Thierfelder W, Quelle FW, Shimoda K. Jaks and Stats in cytokine signaling. Stem Cells. 1997;1:105–111. doi: 10.1002/stem.5530150814. [DOI] [PubMed] [Google Scholar]
- 25.Ihle JN, Kerr IM. Jaks and Stats in signaling by the cytokine receptor superfamily. Trends Genet. 1995;11:69–74. doi: 10.1016/s0168-9525(00)89000-9. [DOI] [PubMed] [Google Scholar]
- 26.de Koning JP, Dong F, Smith L, Schelen AM, Barge RM, van der Plas DC, Hoefsloot LH, Lowenberg B, Touw IP. The membrane-distal cytoplasmic region of human granulocyte colony-stimulating factor receptor is required for STAT3 but not STAT1 homodimer formation. Blood. 1996;87:1335–1342. [PubMed] [Google Scholar]
- 27.Shimoda K, Feng J, Murakami H, Nagata S, Watling D, Rogers NC, Stark GR, Kerr IM, Ihle JN. Jak1 plays an essential role for receptor phosphorylation and Stat activation in response to granulocyte colony-stimulating factor. Blood. 1997;90:597–604. [PubMed] [Google Scholar]
- 28.Nicholson SE, Starr R, Novak U, Hilton DJ, Layton JE. Tyrosine residues in the granulocyte colony-stimulating factor (G-CSF) receptor mediate G-CSF-induced differentiation of murine myeloid leukemic (M1) cells. J Biol Chem. 1996;271:26947–26953. doi: 10.1074/jbc.271.43.26947. [DOI] [PubMed] [Google Scholar]
- 29.Ward AC, Hermans MHA, Smith L, van Aesch YM, Schelen AM, Antonissen C, Touw IP. Tyrosine-dependent and -independent mechanisms of STAT3 activation by the human granulocyte colony-stimulating factor (G-CSF) receptor are differentially utilized depending on G-CSF concentration. Blood. 1999;93:113–124. [PubMed] [Google Scholar]
- 30.Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO (Eur Mol Biol Organ) J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 31.Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, Akira S. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc Natl Acad Sci USA. 1996;93:3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimozaki K, Nakajima K, Hirano T, Nagata S. Involvement of STAT3 in the granulocyte colony-stimulating factor-induced differentiation of myeloid cells. J Biol Chem. 1997;272:25184–25189. doi: 10.1074/jbc.272.40.25184. [DOI] [PubMed] [Google Scholar]
- 33.Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle JN. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 34.Onishi M, Nosaka T, Misawa K, Mui AL, Gorman D, McMahon M, Miyajima A, Kitamura T. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol Cell Biol. 1998;18:3871–3879. doi: 10.1128/mcb.18.7.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong F, Hoefsloot LH, Schelen AM, Broeders CA, Meijer Y, Veerman AJ, Touw IP, Lowenberg B. Identification of a nonsense mutation in the granulocyte-colony-stimulating factor receptor in severe congenital neutropenia. Proc Natl Acad Sci USA. 1994;91:4480–4484. doi: 10.1073/pnas.91.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong F, Dale DC, Bonilla MA, Freedman M, Fasth A, Neijens HJ, Palmblad J, Briars GL, Carlsson G, Veerman AJ, et al. Mutations in the granulocyte colony-stimulating factor receptor gene in patients with severe congenital neutropenia. Leukemia. 1997;11:120–125. doi: 10.1038/sj.leu.2400537. [DOI] [PubMed] [Google Scholar]
- 37.Dong F, Brynes RK, Tidow N, Welte K, Lowenberg B, Touw IP. Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N Engl J Med. 1995;333:487–493. doi: 10.1056/NEJM199508243330804. [DOI] [PubMed] [Google Scholar]
- 38.Bernard T, Gale RE, Evans JP, Linch DC. Mutations of the granulocyte-colony stimulating factor receptor in patients with severe congenital neutropenia are not required for transformation to acute myeloid leukaemia and may be a bystander phenomenon. Br J Haematol. 1998;101:141–149. doi: 10.1046/j.1365-2141.1998.00652.x. [DOI] [PubMed] [Google Scholar]
- 39.Tidow N, Pilz C, Kasper B, Welte K. Frequency of point mutations in the gene for the G-CSF receptor in patients with chronic neutropenia undergoing G-CSF therapy. Stem Cells. 1997;1:113–119. doi: 10.1002/stem.5530150815. [DOI] [PubMed] [Google Scholar]
- 40.Fukunaga R, Ishizaka-Ikeda E, Nagata S. Growth and differentiation signals mediated by different regions in the cytoplasmic domain of granulocyte colony-stimulating factor receptor. Cell. 1993;74:1079–1087. doi: 10.1016/0092-8674(93)90729-a. [DOI] [PubMed] [Google Scholar]
- 41.Dong F, van Buitenen C, Pouwels K, Hoefsloot LH, Lowenberg B, Touw IP. Distinct cytoplasmic regions of the human granulocyte colony-stimulating factor receptor involved in induction of proliferation and maturation. Mol Cell Biol. 1993;13:7774–7781. doi: 10.1128/mcb.13.12.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welte K, Touw IP. G-CSF receptor mutations in patients with severe chronic neutropenia: a step in leukemogenesis? . Blood. 1997;90:433a. . (Abstr.) [Google Scholar]
- 43.Bernard T, Gale RE, Linch DC. Analysis of granulocyte colony-stimulating factor receptor isoforms, polymorphisms and mutations in normal haemopoietic cells and acute myeloid leukaemia blasts. Br J Haematol. 1996;93:527–533. doi: 10.1046/j.1365-2141.1996.d01-1696.x. [DOI] [PubMed] [Google Scholar]
- 44.Carapeti M, Soede-Bobok A, Hochhaus A, Sill H, Touw IP, Goldman JM, Cross NC. Rarity of dominant-negative mutations of the G-CSF receptor in patients with blast crisis of chronic myeloid leukemia or de novo acute leukemia. Leukemia. 1997;11:1005–1008. doi: 10.1038/sj.leu.2400697. [DOI] [PubMed] [Google Scholar]
- 45.Shibata S, Asano Y, Yokoyama T, Shimoda K, Nakashima H, Okamura S, Niho Y. Analysis of the granulocyte colony-stimulating factor receptor gene structure using PCR-SSCP in myeloid leukemia and myelodysplastic syndrome. Eur J Haematol. 1998;60:197–201. doi: 10.1111/j.1600-0609.1998.tb01022.x. [DOI] [PubMed] [Google Scholar]
- 46.Hermans MH, Ward AC, Antonissen C, Karis A, Lowenberg B, Touw IP. Perturbed granulopoiesis in mice with a targeted mutation in the granulocyte colony-stimulating factor receptor gene associated with severe chronic neutropenia. Blood. 1998;92:32–39. [PubMed] [Google Scholar]
- 47.Ploemacher RE, van der Sluijs JP, Voerman JS, Brons NH. An in vitro limiting-dilution assay of long-term repopulating hematopoietic stem cells in the mouse. Blood. 1989;74:2755–2763. [PubMed] [Google Scholar]
- 48.Aardal NP, Laerum OD. Circadian variations in mouse bone marrow. Exp Hematol. 1983;11:792–801. [PubMed] [Google Scholar]
- 49.Lowell CA, Fumagalli L, Berton G. Deficiency of Src family kinases p59/61hck and p58c-fgr results in defective adhesion-dependent neutrophil functions. J Cell Biol. 1996;133:895–910. doi: 10.1083/jcb.133.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner BJ, Hayes TE, Hoban CJ, Cochran BH. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO (Eur Mol Biol Organ) J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang D, Stravopodis D, Teglund S, Kitazawa J, Ihle JN. Naturally occurring dominant negative variants of Stat5. Mol Cell Biol. 1996;16:6141–6148. doi: 10.1128/mcb.16.11.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dittrich E, Haft CR, Muys L, Heinrich PC, Graeve L. A di-leucine motif and an upstream serine in the interleukin-6 (IL-6) signal transducer gp130 mediate ligand-induced endocytosis and down-regulation of the IL-6 receptor. J Biol Chem. 1996;271:5487–5494. doi: 10.1074/jbc.271.10.5487. [DOI] [PubMed] [Google Scholar]
- 53.Hunter M, Avalos B. A dileucine motif in the G-CSFR may mediate the dominant negative phenotype in SCN/AML. Blood. 1998;90:433a. . (Abstr.) [Google Scholar]
- 54.Gillio AP, Gabrilove JL. Cytokine treatment of inherited bone marrow failure syndromes. Blood. 1993;81:1669–1674. [PubMed] [Google Scholar]
- 55.Freedman MH. Safety of long-term administration of granulocyte colony-stimulating factor for severe chronic neutropenia. Curr Opin Hematol. 1997;4:217–224. doi: 10.1097/00062752-199704030-00011. [DOI] [PubMed] [Google Scholar]
- 56.McLemore ML, Poursine-Laurent J, Link DC. Increased granulocyte colony-stimulating factor responsiveness but normal resting granulopoiesis in mice carrying a targeted granulocyte colony-stimulating factor receptor mutation derived from a patient with severe congenital neutropenia. J Clin Invest. 1998;102:483–492. doi: 10.1172/JCI3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orlic D, Anderson S, Biesecker LG, Sorrentino BP, Bodine DM. Pluripotent hematopoietic stem cells contain high levels of mRNA for c-kit, GATA-2, p45 NF-E2, and c-myb and low levels or no mRNA for c-fms and the receptors for granulocyte colony-stimulating factor and interleukins 5 and 7. Proc Natl Acad Sci USA. 1995;92:4601–4605. doi: 10.1073/pnas.92.10.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKinstry WJ, Li CL, Rasko JE, Nicola NA, Johnson GR, Metcalf D. Cytokine receptor expression on hematopoietic stem and progenitor cells. Blood. 1997;89:65–71. [PubMed] [Google Scholar]
- 59.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot RP, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao X, Tay A, Guy GR, Tan YH. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol Cell Biol. 1996;16:1595–1603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Danial NN, Pernis A, Rothman PB. Jak-STAT signaling induced by the v-abl oncogene. Science. 1995;269:1875–1877. doi: 10.1126/science.7569929. [DOI] [PubMed] [Google Scholar]
- 62.Yu CL, Meyer DJ, Campbell GS, Larner AC, Carter-Su C, Schwartz J, Jove R. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 63.Garcia R, Yu CL, Hudnall A, Catlett R, Nelson KL, Smithgall T, Fujita DJ, Ethier SP, Jove R. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differ. 1997;8:1267–1276. [PubMed] [Google Scholar]
- 64.Zong C, Yan R, August A, Darnell JE, Jr, Hanafusa H. Unique signal transduction of Eyk: constitutive stimulation of the JAK-STAT pathway by an oncogenic receptor-type tyrosine kinase. EMBO (Eur Mol Biol Organ) J. 1996;15:4515–4525. [PMC free article] [PubMed] [Google Scholar]
- 65.Chai SK, Nichols GL, Rothman P. Constitutive activation of JAKs and STATs in BCR-Abl-expressing cell lines and peripheral blood cells derived from leukemic patients. J Immunol. 1997;159:4720–4728. [PubMed] [Google Scholar]
- 66.Watson CJ, Miller WR. Elevated levels of members of the STAT family of transcription factors in breast carcinoma nuclear extracts. Br J Cancer. 1995;71:840–844. doi: 10.1038/bjc.1995.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sartor CI, Dziubinski ML, Yu CL, Jove R, Ethier SP. Role of epidermal growth factor receptor and STAT-3 activation in autonomous proliferation of SUM-102PT human breast cancer cells. Cancer Res. 1997;57:978–987. [PubMed] [Google Scholar]