Abstract
Bacterial lipopolysaccharide (LPS) provokes a vigorous, generalized proinflammatory state in the infected host. Genetic regulation of this response has been localized to the Lps locus on mouse chromosome 4, through study of the C3H/HeJ and C57BL/10ScCr inbred strains. Both C3H/HeJ and C57BL/10ScCr mice are homozygous for a mutant Lps allele (Lpsd/d) that confers hyporesponsiveness to LPS challenge, and therefore exhibit natural tolerance to its lethal effects. Genetic and physical mapping of 1,345 backcross progeny segregating this mutant phenotype confined Lps to a 0.9-cM interval spanning 1.7 Mb. Three transcription units were identified within the candidate interval, including Toll-like receptor 4 (Tlr4), part of a protein family with members that have been implicated in LPS-induced cell signaling. C3H/HeJ mice have a point mutation within the coding region of the Tlr4 gene, resulting in a nonconservative substitution of a highly conserved proline by histidine at codon 712, whereas C57BL/ 10ScCr mice exhibit a deletion of Tlr4. Identification of distinct mutations involving the same gene at the Lps locus in two different hyporesponsive inbred mouse strains strongly supports the hypothesis that altered Tlr4 function is responsible for endotoxin tolerance.
Keywords: lipopolysaccharide, inflammation, positional cloning, Salmonella, mice/ inbred C3H
Serious infection caused by Gram-negative bacteria is a problem of increasing magnitude, especially among the elderly and immunocompromised populations (1). Virulence and pathogenic potential among this group of organisms are attributed mainly to the presence of LPS, an abundant glycolipid in the outer bacterial membrane (2). The proinflammatory and immunostimulatory properties of LPS are due to the inner conserved lipid A component of LPS; purified lipid A administered in vivo is capable of eliciting a generalized inflammatory response now described as the systemic inflammatory response syndrome (3, 4). To defend against serious infection by Gram-negative bacteria, eukaryotes have developed innate and exquisitely sensitive immunologic surveillance mechanisms that can detect minute quantities of bacterial LPS in vivo. Recognition of LPS by various cell types, including lymphocytes and macrophages, leads to pleiotropic cellular activation and enhanced microbicidal capacities, all of which are aimed at elimination of the inciting pathogen.
Mouse models have been used to characterize the host response to purified LPS for approximately 50 years. Most inbred strains of laboratory mice exhibit reproducible susceptibility to the proinflammatory and toxic properties of LPS. Remarkably, the C3H/HeJ strain exhibits natural tolerance to otherwise lethal doses of LPS, as well as an altered inflammatory response to experimental challenge (5–7). Several cell types derived from the C3H/HeJ strain are hyporesponsive to LPS in vitro. Macrophages fail to develop an activation phenotype or die when exposed to high concentrations of LPS; B lymphocytes do not respond to the mitogenic, adjuvant, or immunogenic properties of LPS, and fibroblasts fail to undergo metabolic activation upon LPS exposure. One consequence of altered endotoxin responsiveness is that C3H/HeJ mice are extremely susceptible to overwhelming infection with virulent Gram-negative bacteria, including Salmonella typhimurium, a natural pathogen of both mice and humans (8).
C3H/HeJ mice, along with the C3H/HeN substrain, were both derived from the progenitor strain, C3H/He, in 1947. Neither C3H/He nor C3H/HeN mice have the LPS tolerance phenotype characteristic of C3H/HeJ. Study of LPS responsiveness among several sublines derived from C3H/HeJ led to the conclusion that the mutant phenotype arose through spontaneous mutation between 1960 and 1968 (9). Initial genetic analysis established that this phenotype is under the control of a single locus, named Lps, existing in two allelic forms, Lpsn (responsive) and Lpsd (hyporesponsive) (10). Using the BXH recombinant inbred strain panel, the Lps locus was assigned to chromosome 4 by its linkage to Mup1 (11) and is distinct from the IFN-α gene cluster (Ifa [12]). Further genetic mapping using two backcross panels positioned Lps between Mup1 and Ps with the following order and map distance: Mup1-(6±2)-Lps- (13±7)-Ps (13). A congenic BALB/c mouse strain containing a 5.5-cM segment of chromosome 4 surrounding the mutant Lps locus has been developed and exhibits LPS hyporesponsiveness in vitro and in vivo (14).
Tolerance to the stimulatory properties of LPS has also been observed in the C57BL/10ScCr inbred strain (15). As with C3H/HeJ, the mechanism responsible for LPS tolerance among C57BL/10ScCr mice is unknown. The C57BL/ 10Sn progenitor strain from which C57BL/10ScCr was derived is LPS responsive, indicating that this mutant phenotype was acquired after 1953. Complementation was not observed among F1 hybrid mice derived from C3H/HeJ and C57BL/10ScCr parents, indicating that both strains have a mutant allele at the Lps locus (15).
To identify the nature of LPS hyporesponsiveness in C3H/HeJ mice by positional cloning, we previously localized Lps to a 1.1-cM interval through high-resolution genetic mapping of mid-chromosome 4 (16). Through additional genetic mapping, the candidate interval for Lps has been narrowed to 0.9 cM between two microsatellite markers, D4Nds9 and D4Mit178, and we have assembled a 1.7-Mb DNA contig spanning this interval. Four unique transcription units were identified within the cloned region, including the gene encoding Toll-like receptor 4 (Tlr4)1 (17). Tlr4 is an exceptionally strong candidate for the Lps mutation based on its chromosomal location, the presence of two independent mutant alleles, and the demonstrated role of members of the TLR family in innate immune recognition of infectious pathogens (18, 19).
Materials and Methods
Fine Genetic Mapping.
Procedures for construction of a high-resolution genetic map at the Lps locus have been published (16). Using publicly available genetic mapping databases (http://www. informatics.jax.org/maptools.html), one additional informative microsatellite (D4Nds9) derived from a flow-sorted 4:6 Robertsonian chromosome library was identified and typed by PCR using flanking oligonucleotides, as described previously (20). An anonymous microdissected clone from the same flow-sorted library (D4Rck137; gift of Dr. Jeffrey Friedman, Rockefeller University, New York) was typed by RFLP using TaqI, as described (21).
Identification of Novel Simple Sequence Length Repeats.
DNA samples (5 μg) from 10 BAC, 2 P1, and 58 cosmid clones derived from YAC 89A3 were digested with AluI or a combination of HaeIII, AvaII, and HincII using conditions provided by the supplier (New England Biolabs). Individual DNA digests were purified using standard protocols (22), ligated into EcoRV site of pBluescript II KS+ (Stratagene), and transformed to Escherichia coli strain XL1-MRF′. Nylon filter replicates (Hybond N; Amersham Life Science) were hybridized in Church's solution at 60°C and probed with poly dAdC:dTdG (Pharmacia Biotech) labeled to high specific activity with [α-32P]dATP by the random-primer method (23). Filters were washed for 30 min at 65°C in 0.5× SSC and 0.1% SDS and exposed for 4–16 h at −70°C using Kodak XAR film and an intensification screen. Plasmid DNA inserts from positive colonies were sequenced by the dideoxy chain termination method (Pharmacia Biotech) to derive flanking primer sequences for D4Mcg5 to D4Mcg60. Oligonucleotide primer pairs for simple sequence length repeat (SSLP) markers D4Mcg61–D4Mcg86 were identified directly from the sequence of randomly cloned sheared BAC DNA.
Genomic Library Screening and Contig Assembly.
Mouse genomic clones corresponding to microsatellites (D4Nds9, D4Mit25, D4Mit178) and the microdissected probe D4Rck137 were identified from a YAC library (24; Research Genetics) by PCR screening of DNA pools. The primer pair for D4Rck137 was derived from its nucleotide sequence and amplified a unique 122-bp fragment of the original probe. Genomic clones from a mouse BAC library (25; Research Genetics) were identified by PCR using microsatellites D4Nds9, D4Mit25, and D4Mcg6. P1 clones corresponding to D4Rck137 and D4Mit178 were obtained from Genome Systems. Conditions for PCR were essentially as described for genetic mapping except that 1–3 μl of each DNA pool was used as a template. Products were separated on a 7% polyacrylamide gel containing 7 M urea and exposed for 1–7 d at room temperature using Kodak BioMax film. Nucleotide sequencing of the ends of BAC clone inserts was performed using the Thermosequenase radiolabeled terminator cycle sequencing kit (Amersham Life Science). YAC ends were cloned using bubble or inverse PCR methods (26). Additional BAC clones were identified through library screening with oligonucleotide primer pairs derived from nonrepetitive insert-end sequences and SSLPs obtained from BAC and cosmid clones. Assembly of the YAC and BAC/P1 contig spanning the D4Nds9–D4Mit178 interval was done by sequence-tagged site (STS) content and restriction mapping using rare-cutter enzymes and pulsed field gel electrophoresis (PFGE), as described previously (27).
PFGE Analysis.
Genomic DNA from yeast clones containing YACs was prepared in agarose blocks for PFGE analysis (28). Agarose block slices (20 μl) were run undigested or after digestion with NotI, MluI, BssHII, or SacII restriction enzymes (New England Biolabs) using conditions suggested by the supplier. DNA was separated by electrophoresis in a 1% agarose gel (SeaKem GTG; FMC Bioproducts) containing 0.5× TBE (Tris-boric acid-EDTA) using a contour-clamped homogenous electrophoretic field (CHEF DRII; BioRad). Electrophoresis was performed at 130 V for 24 h at 14°C with 15–60-s pulse times to resolve fragments of 50–2,000 kb. DNA of Lambda concatemers and of Saccharomyces cerevisiae chromosomes were used as size standards. Southern blots of these gels were prepared, and hybridization was performed using [α-32P]dATP-radiolabeled single-copy probes from the region, total mouse genomic DNA, and pBR322 sequences specific for each arm of the pYAC4 vector.
Subcloning of DNA Inserts from YAC Clones.
Agar-embedded genomic DNA from individual yeast clones carrying YAC 89A3 was partially digested with Sau3AI to produce an average fragment size of 30–50 kb and ligated into the BamHI cloning site of the cosmid vector, SuperCosI (Stratagene). The ligation mixture was packaged in vitro (Gigapack II Gold; Stratagene) and used to infect E. coli XL1-MRF′, which was subsequently plated on LB agar containing 100 μg/ml ampicillin. Replicate filters (Hybond N; Amersham Life Science) were made for each plate and hybridized with [α-32P]dATP-radiolabeled total mouse genomic DNA to identify cosmids containing mouse inserts. Positive colonies were selected and expanded in culture, and cosmid DNA was purified by standard protocols (22). 71 cosmid subclones were ordered relative to the originating YAC 89A3 and the BAC contig by restriction mapping with EcoRI, and by hybridization of total genomic DNA as described previously (29) and specific probes derived from the Lps candidate region.
cDNA Selection.
Direct cDNA selection was performed on BAC clones or cosmids according to the method of Rommens et al. (30). 6–10 μg of BAC or cosmid DNA was electrophoresed in 0.9% agarose gel (SeaKem GTG) and immobilized on a nylon membrane (Hybond N) by Southern transfer. Two rounds of hybridization were conducted in Church's solution at 60°C for 72 h using 1 μg each of mouse cDNA prepared from mouse spleen, hypothalamus, heart, kidney, testis, and skin polyA+ RNA. Selected cDNA was PCR-amplified with the oligonucleotide (CUA)4GCGGAATTCTCGAGATCT and cloned by using the uracil DNA glycosylase cloning system (GIBCO BRL). Automated sequencing of cDNA was performed using an ABI 373 apparatus (Applied Biosystems). Sequences were compared with nucleotide, expressed sequence tag (EST), and protein databases using the BLAST server (http://www.ncbi.nih.nlm.gov/cgi-bin/BLAST/); repetitive elements were identified using RepeatMasker.
Sheared BAC Library Construction.
Individual BAC clones comprising the minimal tiling path across the Lps region were cultured overnight in 500–1,000 ml of LB broth with 12.5 μg/ml chloramphenicol, and plasmid DNA was purified using Midi or Maxi Prep kits (QIAGEN). Random shearing was performed by passing argon through an AeroMist nebulization chamber (Cis-Us, Inc.) containing 50–100 μg BAC DNA in 1.5 ml of shearing buffer (35 mM Tris-HCl, pH 8.0, 15 mM MgCl2, 25% glycerol) for 5–15 min. DNA ends were repaired with T4 polynucleotide kinase and T7 DNA polymerase using conditions recommended by the supplier (New England Biolabs). Samples were electrophoresed in 1.5% low-melting-point agarose gel in 1× TAE (Tris-acetic acid-EDTA), and fragments of 1.5–3.0 kb in size were excised and purified using β-agarase. Ligation of phosphorylated linkers (oligo 1, 5′-CTCTAAAG-3′, and oligo 2, 5′-CTTTAGAGCACA-3′, were mixed in a 1:1 ratio and annealed by heating to 95°C for 10 min followed by cooling to room temperature) in 100-fold molar excess was done overnight at 16°C. Linkers were removed by centrifugation for 5 min at 6,000 rpm through a 40-bp MWCO column (Millipore) followed by electrophoresis in 1.5% low-melting-point agarose. Linkered DNA was purified with β-agarase, ligated into the BstxI site of pmin, and transformed to DH10β competent cells (GIBCO BRL). Automated sequencing of 960 subclones was performed for each sheared BAC; nucleotide sequences were compared with nucleotide, protein, and EST databases using BLAST.
Northern and Southern Blotting.
Mouse multiple tissue polyA+ Northern blots (Clontech) were hybridized with [α-32P]dATP-radiolabeled probes obtained by direct selection (1d8), cDNA library screening (3f3), or reverse transcription (RT)-PCR (Tlr4). Hybridization was done at 42°C in a mixture of 50% formamide, 5× SSC, 1% SDS, 10% dextran sulfate, 1× Denhardt's solution, 0.02 M Tris (pH 7.5), and denatured herring sperm DNA (200 μg/ml) for 16 h with final washing at 65°C for 30 min in 0.2× SSC, 0.1% SDS. Autoradiography was performed with Kodak XAR film at −70°C and an intensifying screen for 16 h. Southern blots of genomic DNA (5 μg) from C57BL/6J, C3H/HeJ, DBA/2J, and C57BL/10ScCr mouse strains that had been digested with either HindIII or BamHI as described (16) were probed with [α-32P]dATP radiolabeled 0.65-, 1.2-, and 2.9-kb probes corresponding to nucleotides 1–649, 243–1443, and 1–2943 of the Tlr4 coding sequence using the same conditions as for Northern blots; the final wash was performed with 0.5× SSC and 0.1% SDS.
cDNA Library Screening.
Approximately 106 clones of a λgt11 pre-B cell line 70/Z3 cDNA library were plated and screened with the 3f3 probe isolated by cDNA selection following standard techniques (22). Positive phages were plaque purified, and phage DNA was isolated according to standard procedures. The inserts of positive clones were released by EcoRI digestion, subcloned into the EcoRI site of pBluescript II (KS+) vector (Stratagene), and PCR cycle-sequenced (Amersham Life Science). Overlapping sequences were aligned to obtain the complete open reading frame (ORF) of 3f3.
RT-PCR.
Total spleen RNA from C57BL/6J, C3H/HeJ, and C57BL/10ScCr mouse strains was extracted using Trizol (GIBCO BRL) and used to isolate strain-specific cDNA clones for Tlr4. First-strand cDNA synthesis was carried out using 2 μg of total RNA, 100 ng of random hexamers, and 200 U of Moloney murine leukemia virus reverse transcriptase (GIBCO BRL). The reaction conditions for cDNA synthesis were as described previously (29). Tlr4 cDNA clones were obtained by PCR amplification of three overlapping fragments covering the entire coding region of the mRNA. Fragment A was 710 bp in length and was amplified using primers 5′-TCTGCCCTGCCACCATTTAC-3′ and 5′-ACATGTCTAAAGAGAGATTGAC-3′; fragment B was 1,190 bp in length and was amplified using primers 5′-GCTGGATTTATCCAGGTG-3′ and 5′-GAAAGAATTGCCAGCCATTTT-3′; and fragment C was 1,681 bp in length and was amplified using primers 5′-AGCTGCAGCACCTGGATTT-3′ and 5′-AAATTGGAATGAAGACCTCTCA-3′. Amplified products were directly sequenced using the Thermosequenase radiolabeled terminator cycle sequencing kit.
Single-strand Conformational Polymorphism Analysis.
To detect single nucleotide changes in alleles of different inbred mouse strains, radiolabeled PCR products were denatured and electrophoresed on 6% polyacrylamide gels containing 5% glycerol at 30 W for 4–6 h at room temperature. The nucleotide sequences of polymorphisms detected by single-strand conformational polymorphism (SSCP) analysis were determined with the Thermosequenase radiolabeled terminator cycle sequencing kit.
Identification of ESTs Mapping to the Lps Interval.
40 ESTs from the Human Transcript Map (http://www4.ncbi.nih.nlm.gov/science96/) corresponding to map positions D9S160 and D9S258 were obtained from the IMAGE consortium (31) via Research Genetics. Clone inserts were isolated according to the suppliers' protocols and individually hybridized to dot blots of BAC DNA from the Lps candidate region after [α-32P]dATP radiolabeling. Hybridization was performed for 16 h at 55°C in 6× SSC, 5× Denhardt's solution, 0.5% SDS, and denatured herring sperm DNA (200 μg/ml); final washing was performed at 55°C for 15–30 min in 0.5× SSC, 0.1% SDS. Autoradiography was performed using Kodak XAR film with an intensifying screen for 4–16 h at −70°C.
Rapid Amplification of cDNA Ends.
5′- and 3′-rapid amplification of cDNA ends (RACE) were performed with Marathon-ready mouse spleen cDNA (0.5 ng) using the Advantage cDNA PCR kit (Clontech). Various gene-specific primers derived from our Tlr4 sequence were used in conjunction with the AP1 and AP2 primers to perform primary and secondary nested PCR. After electrophoresis in 1.5% low-melting-point agarose gel, amplification products were excised and purified using β-agarase (New England Biolabs). Cycle sequencing was performed with gene-specific primers using the Thermosequenase radiolabeled terminator cycle sequencing kit (Amersham Life Science).
Results
Lps Maps to a 0.9-cM Interval Spanning 1.7 Mb.
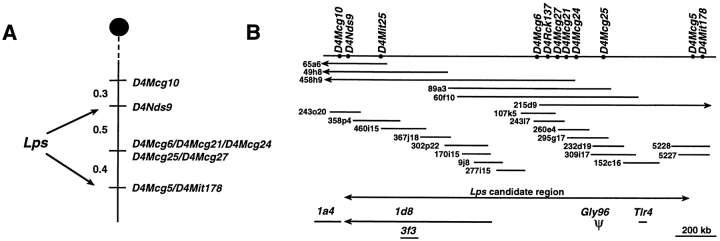
Previously, we reported the construction of a high-resolution linkage map encompassing the Lps locus, using 597 (DBA/ 2J × C3H/HeJ)F1 × C3H/HeJ and 748 (C57BL/6J × C3H/HeJ)F1 C3H/HeJ backcross progeny segregating the recessive mutant (Lpsd/d) phenotype and a preexisting interspecific backcross panel of 259 (Mus spretus × C57BL/ 6J)F1 × C57BL/6J (13). LPS hyporesponsiveness, identified by an absent splenocyte mitogenic response after in vitro stimulation, as well as susceptibility to in vivo infection with S. typhimurium, was compared with allelic inheritance of SSLP markers originating from mid-chromosome 4. Both phenotypes were concordantly inherited and localized within a 1.1-cM interval to which no candidate genes had previously been mapped. To refine the location of Lps, additional SSLPs obtained from public databases (http://www.informatics.jax.org/) as well as 7 informative microsatellites among 86 novel (D4Mcg) markers derived from our cloned DNA contig were mapped in recombinant progeny (Table I). D4Mcg markers were isolated either by [CA]n screening of small-insert libraries constructed from P1 clones (5226, 5227), BAC clones (243o20, 358p4, 302p22), and 58 cosmids derived from YAC 89A3, or via sequence analysis of clones obtained by cDNA selection and randomly cloned sheared BAC libraries. Five of these SSLPs did not recombine with Lps among 748 (D4Mcg6, D4Mcg21) or 1,345 (D4Mcg24, D4Mcg25, D4Mcg27) meioses tested. D4Mcg10 mapped 0.3 cM centromeric to D4Nds9, and D4Mcg5 cosegregated with the distal flanking marker D4Mit178, confining the genetic interval containing Lps to 0.9 cM (Fig. 1 A). Most of the other novel D4Mcg markers were informative in the interspecific backcross panel (data not shown) and useful in assembly of the DNA contig spanning this interval.
Table I.
New Polymorphic Markers Mapped to the Lps Region
| Locus | Probe | Repeat unit* | Primers 5′ → 3′ | Allele size | Polymorphism‡ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| bp | ||||||||||
| P1-5227 | D4Mcg5 | [CT]23 [GT]23 | F: GGGAGGATGTAAAGCTTGAT | 141 | H>B=D>S | |||||
| R: CCAACATCCTCTTACCTCCA | ||||||||||
| P1-5226 | D4Mcg6 | [GT]24 | F: GCATAGGAACAGCTTCTGTGT | 122 | H=D>B | |||||
| R: TTCTATAAATAAGTACCT | ||||||||||
| Bac243o20 | D4Mcg10 | [CA]4TA[CA]25 | F: TCCTCAGTCACACTAGATTG | 120 | B=D>H>S | |||||
| R: TAGCCTGTGGTGTGATTAGTT | ||||||||||
| Yac89a3 | D4Mcg21 | [GT]19N22[AT]11 | F: CAGGGAGGCATATTCATA | 168 | H=D<B<S | |||||
| R: TCTAATTGCCTTTCAATC | ||||||||||
| Yac89a3 | D4Mcg24 | [GT]16 | F: GGTACCTCTATTAGGTGAATC | 113 | S>H>B=D | |||||
| R: ACAGTCTGACCTAGGTAGAGG | ||||||||||
| Yac89a3 | D4Mcg25 | [CA]24 | F: CCATGCTGGGAATCTCACAAC | 164 | D<H=B<S | |||||
| R: CTATTCTAGAGCCAGTCATTTC | ||||||||||
| Yac89a3 | D4Mcg27 | [GT]25 | F: CTTTCATTAGACAGATTATAT | 140 | H>B=D>S | |||||
| R: CAGACAGTATGAACTCCTG |
The base composition of each simple sequence repeat is indicated. A, adenine; C, cytosine; G, guanine; T, thymidine. Numeric subscripts denote the length of each dinucleotide repeat shown in brackets. N22 = 22-bp nonrepetitive sequence.
Relative allele sizes among inbred mouse strains as generated by PCR using flanking forward (F) and reverse (R) primers. B, C57BL/6J; D, DBA/ 2J; H, C3H/HeJ; S, M. spretus.
Figure 1.
Schematic representation of the Lps candidate region. (A) Genetic map of the Lps interval. A genetic map of mouse chromosome 4 was generated from haplotype analysis of 1,345 backcross progeny segregating the Lps phenotype. The Lps locus was mapped between D4Nds9 and D4Mit178. Genetic distances are given to the left in cM. (B) Physical map of the Lps region. Solid bar, genomic DNA. The order of markers was established based on the YAC, BAC, and P1 contig. Unique clone addresses are indicated. Arrow, the candidate Lps region. There are six unresolved recombination events between the proximal marker D4Nds9 and D4Mcg6, and five unmapped crossovers between D4Mcg25 and D4Mcg5. The position of the four transcription units and the pseudogene identified within the candidate region is shown immediately below the cloned contig. The direction of the transcription is shown for clone 1d8.
A 1.7-Mb genomic DNA contig encompassing the Lps locus was constructed using flanking (D4Nds9, D4Mit178) and internal (D4Mit25, D4Rck137) markers as entry probes for a bidirectional chromosome walk within YAC, BAC, and P1 libraries. A total of 31 YAC, 29 BAC, and 5 P1 clones that together form a robust contig across the candidate interval for Lps were sequentially identified through repeated isolation of novel internal or end-insert markers from previously identified clones, followed by rescreening of 1 or more libraries. All clones were ordered by sequence-tagged site (STS) content mapping and sized by PFGE; only those used for generation of novel probes or transcription unit identification are shown (Fig. 1 B). The integrity of clones was confirmed by mapping of informative novel probes in the interspecific backcross panel and/or by Southern hybridization to DNA derived from the contig.
Tlr4 and Two Novel Genes Map within the Lps Candidate Region.
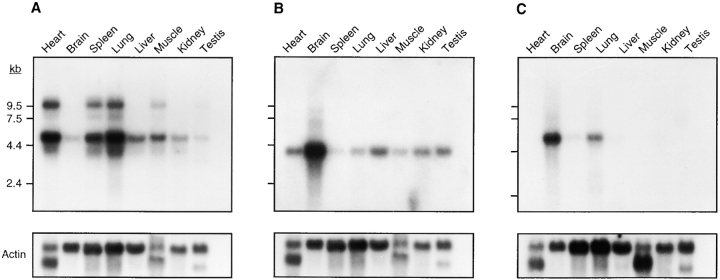
Direct cDNA selection, sequence analysis of randomly cloned sheared BAC DNA from our contig, and comparative mapping of ESTs and genes that had been assigned to regions of human chromosome 9 syntenic with mouse chromosome 4 were used for transcript identification. Three transcription units corresponding to the DNA contig spanning the Lps locus were initially identified by direct cDNA selection. Two rounds of hybridization with expanded cDNA from heart, hypothalamus, skin, testis, kidney, and spleen were performed on 13 BAC clones and 5 cosmid clones, yielding a total of 516 selected cDNA. 95 repeat-free mouse DNA fragments were identified by partial sequence analysis; 35 of these specifically hybridized to clones of the DNA contig at 12 distinct positions. Unique transcription units corresponding to three cDNA clones (1a4, 1d8, 3f3) were identified through a combination of Northern blot analysis and database comparisons (Fig. 2). Clone 1a4 exhibited significant sequence homology to human pregnancy-associated plasma protein A (PAPPA; reference 32), and mapped to BAC 243o20, the most proximal clone in the contig. Human PAPPA had previously been mapped to chromosome 9q33.1, a region known to be homologous to a portion of mouse chromosome 4 proximal to Lps. Northern blot analysis with clone 3f3 revealed a single ∼3.7-kb transcript that was most highly expressed in the mouse brain, and visible in all other tissues. Using 3f3 to probe a pre-B cell cDNA library, a set of overlapping clones mapping specifically to BAC 460i15 was isolated. Sequence analysis revealed that the transcription unit corresponding to 3f3 has an ORF of 1,959 bp encoding 653 amino acids. At the nucleotide level, this transcript is 85% homologous to a human zinc finger protein (sequence data available from EMBL/GenBank/DDBJ under accession no. U18543) known to interact with the activation domain of lentiviral Tat proteins (33). Northern blot analysis with 1d8 revealed a single ∼6-kb transcript most strongly expressed in the brain, with lower expression levels in the lung, spleen, and liver, and no detectable message in other tissues. This clone shares 68% nucleotide homology to mouse astrotactin (EMBL/GenBank/DDBJ accession no. U48797), a ligand for neuron-glia binding during neuronal migration (34). Southern hybridization of additional probes (human EST WI-14828 corresponding to human IMAGE clone 46391 [31] and mouse partial cDNA clones) corresponding to different regions of the same transcription unit demonstrated that the genomic DNA encoding this astrotactin homologue spans ∼750 kb of the proximal half of the BAC contig (Fig. 1 B). A fourth transcription unit was identified by comparative mapping. The human Toll-like receptor 4 gene (TLR4; sequence data available from EMBL/GenBank/DDBJ under accession no. U93091) maps to chromosome 9q32-33 and is a member of an emerging family of molecules implicated in innate immune recognition (17). PCR primers derived from the human and rat sequences (EMBL/GenBank/DDBJ accession no. AF057025) were used to localize mouse Tlr4 to BAC 152c16. Northern blot analysis using a 1.2-kb partial mouse Tlr4 cDNA probe revealed expression of an ∼5-kb transcript in all tissues, corresponding to the length of the cDNA we have cloned. An ∼9-kb transcript was also detected in heart, spleen, lung, muscle, and testis. The transcription unit corresponding to Pappa was excluded as a candidate Lps gene because it mapped outside the Lps candidate region defined by D4Nds9 and D4Mit178. Although the three genes corresponding to 1d8, 3f3, and Tlr4 are candidate Lps genes based on their expression in reticuloendothelial organs and location within the Lps candidate region, we chose to focus our analysis on Tlr4 because of its homology to a gene family including Drosophila Toll and TLR2, both of which are components of the innate immune response and capable of mediating LPS-induced cell signaling (18, 19).
Figure 2.
Expression analysis of three transcription units identified within the Lps candidate region. Mouse multiple-tissue Northern blots of polyA+ RNA (Clontech) were hybridized with (A) Tlr4; (B) clone 3f3; and (C) clone 1d8. β-actin was used as control. Exposure times were all 24 h and reflect the relative abundance of each transcript. RNA size markers (in kb) are shown on the left side of each autoradiogram.
The Amino Acid Sequence of Human and Mouse Tlr4 Is Highly Conserved.
Tlr4 cDNA fragments were isolated by RT-PCR using primers derived from the human TLR4 gene (EMBL/GenBank/DDBJ accession no. U93091) and a rat EST (accession no. AF057025) sequence that shows strong homology with the human TLR4 gene. We determined the sequence of a 3,864 bp cDNA containing one 2,505-bp ORF predicted to encode a protein of 835 amino acids (Fig. 3). Our sequence, combined with estimation of the lengths of the 5′ and 3′ untranslated regions (UTRs) by RACE PCR (data not shown), corresponds to the smaller transcript observed by Northern blot analysis. The nature of the larger mouse transcript identified by our Tlr4 probe will require further investigation. In humans, two transcripts of ∼5.5 and ∼4.4 kb are present in all tissues, with predominant expression in the spleen and peripheral blood leukocytes. The observed size difference of the two human TLR4 transcripts was explained by differential splicing of the 3′ UTR (18).
Figure 3.

Predicted 835 amino acid sequence of the mouse Tlr4 gene. Amino acid positions are numbered from the start of the 2.5-kb ORF. The protein sequences of mouse Tlr4 (top) and human TLR4 (bottom; EMBL/GenBank /DDBJ accession no. U93091) are aligned. Amino acid identity is shown between the two sequences; a + represents similarity between the corresponding pair of amino acid residues. The predicted transmembrane domain is underlined. The position of the proline to histidine substitution at position 712 associated with LPS hyporesponsiveness is highlighted in bold. These sequence data are available from EMBL/ GenBank/DDBJ under accession no. AF110133.
The predicted amino acid sequence of mouse Tlr4 was aligned with the corresponding human protein (Fig. 3). The mouse Tlr4 sequence is homologous (67% identical, 79% similar at the amino acid level) over its entire length to human TLR4, a type I transmembrane protein with an extracellular leucine-rich repeat (LRR) domain and a cytoplasmic Toll IL-1 receptor (TIR) homology signaling domain (18). Several imperfect LRRs were identified by sequence and pattern analysis of the extracellular segment of mouse Tlr4 using the program PRINTS (http://www.biochem. ucl.ac.uk/bsm/dbbrowser/PRINTS/PRINTS.html). LRRs are found in a variety of protein families with widely different functions and are thought to be involved in mediating protein–protein interactions (35). Amino acid conservation between the mouse and human TLR4 is stronger in the cytoplasmic signaling domain compared with the extracellular portion of the protein (89 vs. 68%). The intracytoplasmic segment corresponding to TIR signaling domain is a core module shared by TLR genes, IL-1R, mammalian MyD88 factors, and plant disease resistance proteins (36). Mouse Tlr4 exhibits a lower degree of homology (40–46% similarity) with other members of the Toll family, including human TLR1, TLR2, TLR3, TLR5, and RP105.
Endotoxin-hyporesponsive Mice Have Unique, Distinct Mutations in Tlr4.
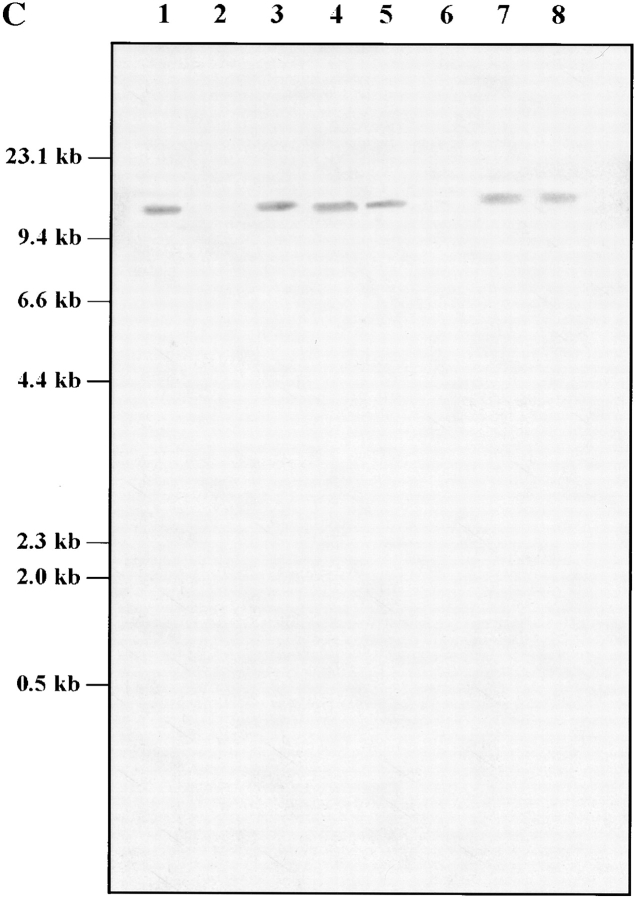
Three pairs of oligonucleotide primers were used to analyze mRNA transcripts from C3H/HeJ (Lpsd/d) and C57BL/6J (Lpsn/n) mouse strains by RT-PCR for the presence of sequence alterations in the coding portion of the Tlr4 gene. The only significant mutation detected was a C to A transversion resulting in a single nonconservative amino acid substitution (proline712 to histidine712) within the cytoplasmic domain of Tlr4 of the LPS-tolerant C3H/ HeJ strain (Fig. 4 A). To compare the segregation of Tlr4 with the Lps phenotype, PCR primers (5′-GTGCCCCGCTTTCACCTCTG-3′ and 5′-TCTAGACACTACCACAATAA-3′) flanking the region predicted to contain the C3H/HeJ mutation were used to amplify genomic DNA from our two panels of recombinant animals. No recombinants were detected among 1,345 meioses, as the C to A transversion showed perfect cosegregation with the Lps phenotype. Since the Pro712 to His712 missense mutation identified in C3H/HeJ Tlr4 does not suggest an obvious loss of function, the corresponding region was PCR-amplified using genomic DNA from 46 classical and wild-derived mouse strains and analyzed for nucleotide changes by SSCP and sequencing. Of three different migration patterns detected by SSCP, one was unique to C3H/HeJ mice, whereas the second and third patterns were present in 12 and 34 mouse strains, respectively (Table II). Sequence analysis confirmed that the unique C3H/HeJ SSCP pattern is caused by the C to A transversion at codon 712 (CCT to CCA, Pro712 to His712) and that the two other SSCP patterns resulted from a silent mutation (T to C transition) at codon Asn719 (Table II, and Fig. 4 A). Tlr4 cDNA from the C57BL/10ScCr (Lpsd/d) mouse strain was not amplified with any of these primer pairs (Fig. 4 B). Therefore, five additional primer pairs covering the entire coding region and the 3′ UTR of the mouse Tlr4 cDNA were used to amplify RT RNA from the spleens of normal and mutant mice. PCR amplification products of the anticipated size were generated with all sets of primers using RT RNA from normal C57BL/6J and LPS-hyporesponsive C3H/HeJ strains, whereas no PCR products were obtained using C57BL/ 10ScCr spleen RT RNA (Fig. 4 B). A control Gapdh RT-PCR product was generated in all mouse species examined. Southern blot analysis of genomic DNA from normal (C57BL/6J and DBA/2J) and LPS-hyporesponsive (C3H/ HeJ and C57BL/10ScCr) mice revealed the absence of hybridizing bands with a Tlr4 cDNA probe in the C57BL/ 10ScCr strain, suggesting that the absence of Tlr4 mRNA is due to a major chromosomal rearrangement (Fig. 4 C).
Figure 4.
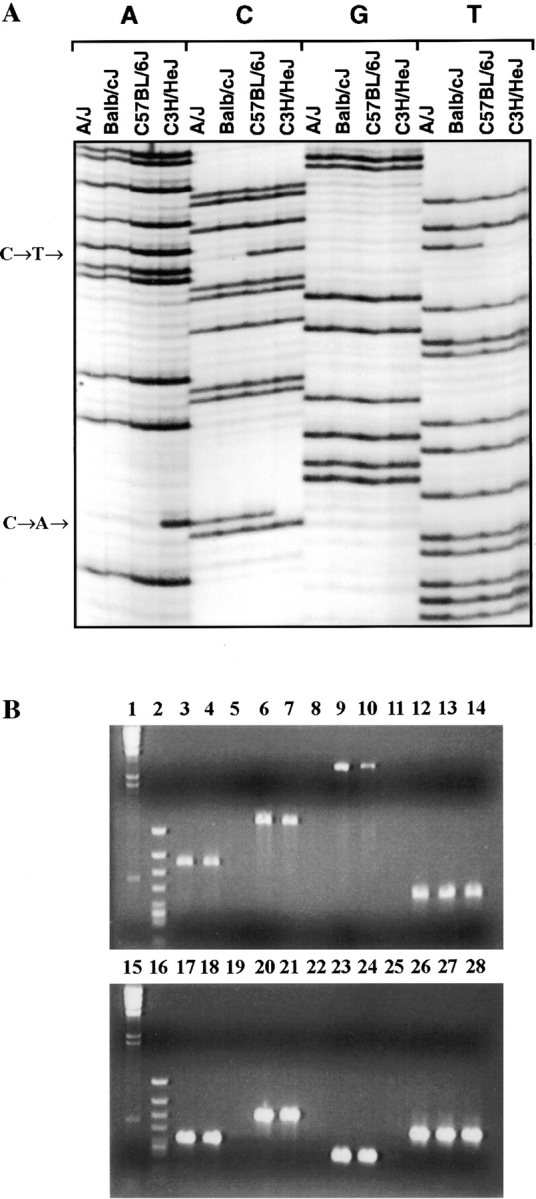
Tlr4 mutation analysis in C3H/HeJ and C57BL/10ScCr. (A) C3H/HeJ Lps mutation. Sequence analysis of Tlr4 shows the missense mutation, a C to A transversion (Pro712His). This change is present only in C3H/HeJ mice. A silent mutation (C to T substitution, Asn719) is also present in several inbred strains of mice, including A/J and BALB/cJ. (B and C) C57BL/10ScCr Lps mutation. (B) RT-PCR of spleen mRNA from C57BL/6J (lanes 3, 6, 9, 12, 17, 20, 23, and 26), C3H/HeJ (lanes 4, 7, 10, 13, 18, 21, 24, and 27), and C57BL/10ScCr (lanes 5, 8, 11, 14, 19, 22, 25, and 28) using six different combinations of primers. Molecular size markers λ DNA-HindIII digest (lanes 1 and 15) and φX-174 RF HincII digest (lanes 2 and 16) are shown. Primer positions are given in brackets according to the position of the first coding ATG. Lanes 3–5: fragment A (696 bp) was amplified with primers 5′-TCTGCGCTGCCACCAGTTAC-3′ (−69) and 5′-ACATGTCTAAAGAGAGATTGAC-3′ (627). Lanes 6–8: fragment B (1,200 bp) was amplified with primers 5′-GCTGGATTTATCCAGGTG-3′ (242) and 5′-GAAAGAATTGCCAGCCATTTT-3′ (1,442). Lanes 9 and 10: fragment C (2,700 bp) was amplified with primers 5′-GCTGGATTTATCCAGGTG-3′ (242) and 5′-AAATTGGAATGAAGACCTCTCA-3′ (2,942). Lanes 17–19: fragment D (409 bp) was amplified with primers 5′-TGACACCCTCCATAGACTTC-3′ (1,541) and 5′-GGTATATCAGAAATGCTACA-3′ (1,950). Lanes 20–22: fragment E (641 bp) was amplified with primers 5′-GTCCTTGAGAAGGTTGAGAAGTCCC-3′ (2,301) and 5′-AAATTGGAATGAAGACCTCTCA-3′ (2,942). Lanes 23–25: fragment F (299 bp) was amplified with primers 5′-GAAGGAAGTAGCACTGACACCTTC-3′ (2,643) and 5′-AAATTGGAATGAAGACCTCTCA-3′ (2,942). Lanes 12–14 and 26–28: Gapdh (459 bp) was amplified with primers 5′-TTTGTGATGGGTGTGAACCACGAG-3′ and 5′-GGAGACAACCTGGTCCTCAGTGTA-3′. A single Tlr4 product of the expected size is seen only in RT-PCR of spleen mRNA from C57BL/6J and C3H/HeJ, although Gapdh mRNA was amplified from all RT-PCR samples including C57BL/10ScCr. (C) Southern blot analysis of chromosomal rearrangement associated with the C57BL/10ScCr Lps allele. Southern blot of a Tlr4 partial cDNA (position 242–1442) hybridized to C57BL/6J (lanes 1 and 5), C57BL/10ScCr (lanes 2 and 6), C3H/HeJ (lanes 3 and 7), and DBA/2J (lanes 4 and 8) DNA digested with BamHI (1– 4) or HindIII (5–8). Size markers (in kb) are indicated to the left of the autoradiogram.
Table II.
Distribution of Nucleotide Sequence Variations in the Coding Portion of Mouse Tlr4
| Pro712 (CCT) | His712 (CAT) | |||
|---|---|---|---|---|
| Asn719 (AAT) | A/J, BALB/cJ, NZW/LacJ, PERA/Ei, PERA/Rk | |||
| PERC/Ei, RBA/Dn, RBB/Dn, Poschiavinos/Tirano, | ||||
| Poschiavinos/Zaleno, WMP/Pas, WSB/Ei | ||||
| 129/J, AKR/J, BUB/BnJ, C3H/HeN, C3H/HeOuJ, | ||||
| Asn719 (AAC) | C3H/HeSnJ, C3HeB/FeJ, C57BL/6J, C57BL/10SnJ, | C3H/HeJ | ||
| C57BR/cdJ, C57L/J, C58/J, CE/J, DBA/2J, LP/J, NZB/BINJ, | ||||
| P/J, PL/J, RIIIS/J, RF/J, NOD/LtJ, SPRET/Ei, CAST/Ei, | ||||
| CZECH II/Ei, MOLC/Rk, MOLD/Rk, MOLE/Rk, MOLF/Ei, | ||||
| MOLG/Dn, Mus caroli, SF/CamEi, SK/CamEi, SK/CamRk, | ||||
| SKIVE/Ei | ||||
Discussion
Using high-resolution genetic mapping techniques, we have restricted the position of the Lps gene to a 0.9-cM region of chromosome 4, flanked proximally by D4Nds9 and distally by D4Mit178. A 1.7-Mb cloned DNA contig spanning this interval was sequentially assembled using YAC, BAC, and P1 clones. Our data differ significantly from another recently published physical map encompassing the Lps locus (37). In this contig, a gap (estimated at 100 kb by fluorescence in situ hybridization) exists in the BAC contig between D4Nds9 and D4Mit178. Comparison of BAC clone addresses common to both maps suggests that this gap corresponds to the center of our contig, and is ∼950 kb in size. Finally, through cDNA selection and nucleotide sequencing of randomly cloned sheared BACs from our contig, we have identified three transcription units within the Lps candidate region, including Tlr4, and two novel genes.
We provide evidence that implicates mouse Tlr4 as a critical regulator of the innate host response to bacterial LPS. TLRs have clearly been identified as components of innate immunity against different classes of microbes in evolutionarily diverse organisms, including plants, invertebrates, and mammals (36). By using a mouse mutant that is naturally hyporesponsive to LPS, we have extended the role of TLRs, specifically Tlr4, to innate recognition and defense against Gram-negative bacteria in vivo. Several lines of evidence support the contention that the Tlr4 gene is a compelling candidate for the Lps mutation. First, Tlr4 maps within the genetic interval encompassing Lps as defined by 1,345 meioses derived from 2 independent backcrosses segregating the mutant phenotype. Second, throughout the entire Tlr4 ORF, a single missense mutation was uniquely observed in the mutant C3H/HeJ inbred strain among all inbred strains examined, and this mutation shows exact cosegregation with the mutant Lps phenotype. The resulting amino acid substitution involves a highly conserved proline within the cytoplasmic signaling domain, the portion of Tlr4 that exhibits the strongest degree of evolutionary conservation. The structural integrity of this domain appears to be essential for LPS-induced signaling by TLRs (19). The point mutation observed in the C3H/ HeJ strain is not predicted to alter transcription of Tlr4; however, the resulting substitution of a proline residue by a basic residue would be expected to alter the topology of the Tlr4 signaling domain and potentially disrupt protein– protein interactions with downstream molecules. Finally, corroborating evidence of a genomic deletion and absent Tlr4 transcription was observed in C57BL/10ScCr, the other endotoxin-tolerant mouse strain known to have a mutation at the Lps locus. C57BL/10ScCr mice would not be expected to produce Tlr4 protein, supporting the hypothesis that their mutant phenotype results from a loss of Tlr4 function. Independent studies done by another group have also recently led to the proposal of Tlr4 as a candidate gene for the Lps mutation (37).
Mammalian TLRs are part of a protein family with homologues in insects and plants, implying an ancient evolutionary origin (36). The first Toll receptor was identified during the study of embryonic development in Drosophila melanogaster (38). The role of this protein family in innate immune function was subsequently revealed in the adult fruit fly, as Toll mutants were unable to mount an effective antimicrobial response and succumbed to overwhelming fungal infection (39). Remarkable parallels have subsequently been identified between the Toll and IL-1R signaling pathways leading from the cell membrane to transcriptional activation within the nucleus. The cytoplasmic domains of Toll and the mammalian IL-1R (TIR domain) are homologous, and both activate signaling pathways leading to the expression of genes under the regulation of the nuclear factor (NF)-κB family of transcription factors. MyD88, an adaptor protein known to interact with IL-1R through its own TIR domain, was recently identified as a direct downstream element of the human Toll signaling pathway (36). Human Toll and IL-1R pathways share other downstream components, including the serine-threonine innate immunity kinase IRAK and TNFR-associated factor 6 (TRAF6), although the range of genes induced by these two receptors may not be identical. Additional components of these two signaling pathways are likely to be identified and will clarify their specific roles in regulation of innate immune response genes.
A family of TLRs (TLR1–5) whose genes are dispersed throughout the human genome has been identified through homology comparisons of protein architecture (17). Stable transfection of TLR2 into human embryonic kidney 293 cells induced cellular signaling, including activation of NF-κB in response to LPS and LPS binding protein (LBP), suggesting that recognition of LPS may be an attribute common to all TLRs (19). Our finding that a mutation in mouse Tlr4 creates LPS hyporesponsiveness is consistent with and supported by these in vitro observations. An endotoxin-hyporesponsive phenotype has been documented in humans (40); however, the hypothesis that TLR4 mediates LPS-induced signaling in vivo requires further investigation. Recent observations demonstrating that the innate immune system of Drosophila is able to discriminate between various classes of infecting microorganisms and thereby regulate the production of different, specific antimicrobial peptides argue against significant functional redundancy of the TLR family (41).
LPS is among the most well-characterized and intensively studied bacterial virulence factors. While not intrinsically toxic, LPS is a prototypical cell stimulator capable of provoking an intense and potentially lethal host inflammatory response. A great deal is known about the intracellular events that follow LPS stimulation, yet a critical gap exists in our understanding of the mechanism by which LPS at the cell surface induces host cell activation across the cytoplasmic membrane (42, 43). In the current model, LPS monomers are catalytically transferred by a lipid exchange molecule, LBP, to CD14, a major LPS receptor that lacks a transmembrane domain (44). A putative LPS transmembrane coreceptor is then responsible for initiation of cellular responses after interaction with the LPS–CD14 complex. Based on our assertion that mutation of a transmembrane molecule creates a state of LPS hyporesponsiveness, it is conceivable that Tlr4 is not only a strong candidate for Lps, but also represents a putative LPS coreceptor.
Identification of the precise mechanisms of LPS-induced cellular stimulation is of substantial significance for the understanding of bacterial pathogenesis and protective host defense. Dysregulation of the host response may lead to multiple system organ dysfunction and possible death. A molecular map of the pathways usurped by LPS will clearly be useful in future development of strategies to protect the host from the deleterious consequences of Gram-negative bacterial infection. The identification of Tlr4 as a strong candidate for the naturally occurring mouse mutation at the Lps locus is reminiscent of the use of Drosophila mutants in the original description of Toll, and emphasizes the tremendous degree of conservation of structure and function within the innate immune system of multicellular organisms. Further studies of the role of Tlr4 in host responses to LPS should pave the way for better understanding of the fundamental mechanisms of cellular activation.
Acknowledgments
We are grateful to Dr. Silvia Vidal and Dr. Kenneth Morgan, who made helpful suggestions during the course of this work. We thank Dr. J. Friedman for providing the D4Rck137 probe, Dr. D. Nagle and Lisa Dirocco for assistance with BAC subcloning, Waan-Jeng Nir for help with cDNA selection, and Line Laroche for technical assistance.
This work was supported by research grants to D. Malo from the Medical Research Council of Canada and to D. Malo and P. Gros from Millennium Pharmaceuticals Inc. D. Malo is a Scholar of Fonds de la Recherche en Santé du Québec, and P. Gros is an International Research Scholar of the Howard Hughes Medical Institute and a Senior Scientist of the Medical Research Council of Canada. S.T. Qureshi is a recipient of a Clinician Scientist Award from the Medical Research Council of Canada and a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
Abbreviations used in this paper
- EST
expressed sequence tag
- LRR
leucine-rich repeat
- NF
nuclear factor
- ORF
open reading frame
- PAPPA
pregnancy-associated plasma protein A
- PFGE
pulsed field gel electrophoresis
- RACE
rapid amplification of cDNA ends
- RT
reverse transcription
- SSCP
single-strand conformational polymorphism
- SSLP
simple sequence length repeat
- STS
sequence-tagged site
- TIR
Toll IL-1 receptor
- TLR
Toll-like receptor
- Tlr4
murine Toll-like receptor 4
- UTR
untranslated region
References
- 1.Increases in national hospital discharge survey rates for septicemia—United States, . –1987. 1990. MMWR Morb Mortal Wkly Rep. 1979;39:31–34. [PubMed] [Google Scholar]
- 2.Glauser MP, Heumann D, Baumgartner JD, Cohen J. Pathogenesis and potential strategies for prevention and treatment of septic shock: an update. Clin Infect Dis. 1994;18:S205–S216. doi: 10.1093/clinids/18.supplement_2.s205. [DOI] [PubMed] [Google Scholar]
- 3.Burrell R. Human responses to bacterial endotoxin. Circ Shock. 1994;43:137–153. [PubMed] [Google Scholar]
- 4.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RMH, Sibbald WJ. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 5.Sultzer BM. Genetic control of leukocyte responses to endotoxin. Nature. 1968;219:1253–1254. doi: 10.1038/2191253a0. [DOI] [PubMed] [Google Scholar]
- 6.Rosenstreich, D.L. 1985. Genetic control of the endotoxin response: C3H/HeJ mice. In Handbook of Endotoxin, Vol. 3. L.J. Berry, editor. Elsevier Science B.V., Amsterdam. 82–122.
- 7.Vogel, S.N. 1992. The Lps gene. Insights into the genetic and molecular basis of LPS responsiveness and macrophage differentiation. In Tumor Necrosis Factors: The Molecules and Their Emerging Role in Medicine. B. Beutler, editor. Raven Press, Ltd., New York. 485–513.
- 8.O'Brien AD, Rosenstreich DL, Scher I, Campbell GH, MacDermott RP, Formal SB. Genetic control of susceptibility to Salmonella typhimurium in mice: role of the Lpsgene. J Immunol. 1980;124:20–24. [PubMed] [Google Scholar]
- 9.Glode LM, Rosenstreich DL. Genetic control of B cell activation by bacterial lipopolysaccharide is mediated by multiple distinct genes or alleles. J Immunol. 1976;117:2061–2066. [PubMed] [Google Scholar]
- 10.Watson J, Riblet R. Genetic control of responses to bacterial lipopolysaccharides in mice. I. Evidence for a single gene that influences mitogenic and immunogenic responses to lipopolysaccharides. J Exp Med. 1974;140:1147–1161. doi: 10.1084/jem.140.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson J, Riblet R, Taylor B. The response of recombinant inbred strains of mice to bacterial lipopolysaccharides. J Immunol. 1977;118:2088–2093. [PubMed] [Google Scholar]
- 12.Fultz MJ, Vogel SN. The physical separation of Lps and Ifaloci in BXH recombinant inbred mice. J Immunol. 1989;143:3001–3006. [PubMed] [Google Scholar]
- 13.Watson J, Kelly K, Largen M, Taylor B. The genetic mapping of a defective LPS response gene in C3H/ HeJ mice. J Immunol. 1978;120:422–424. [PubMed] [Google Scholar]
- 14.Vogel SN, Wax JS, Perera PY, Padlan C, Potter M, Mock BA. Construction of a BALB/c congenic mouse, C.C3H-Lpsd, that expresses the Lpsd allele: analysis of chromosome 4 markers surrounding the Lpsgene. Infect Immun. 1994;62:4454–4459. doi: 10.1128/iai.62.10.4454-4459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coutinho A, Meo T. Genetic basis for unresponsiveness to lipopolysaccharide in C57BL/10Cr mice. Immunogenetics. 1978;7:17–24. doi: 10.1007/BF01843983. [DOI] [PubMed] [Google Scholar]
- 16.Qureshi ST, Larivière L, Sebastiani G, Clermont S, Skamene E, Gros P, Malo D. A high-resolution map in the chromosomal region surrounding the Lpslocus. Genomics. 1996;31:283–294. doi: 10.1006/geno.1996.0050. [DOI] [PubMed] [Google Scholar]
- 17.Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF. A family of human receptors structurally related to DrosophilaToll. Proc Natl Acad Sci USA. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the DrosophilaToll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 19.Yang R-B, Mark MR, Gray A, Huang A, Xie MH, Goddard A, Wood WI, Gurney AL, Godowski PJ. Toll-like receptor-2 mediates lipopolysaccharide-induced cell signalling. Nature. 1998;395:284–288. doi: 10.1038/26239. [DOI] [PubMed] [Google Scholar]
- 20.Cornall RJ, Friedman JM, Todd JA. Mouse microsatellites from a flow-sorted 4:6 Robertsonian chromosome. Mamm Genome. 1992;3:620–624. doi: 10.1007/BF00352478. [DOI] [PubMed] [Google Scholar]
- 21.Bahary N, McGraw DE, Shilling R, Friedman JM. Microdissection and microcloning of mid-chromosome 4: genetic mapping of 41 microdissection clones. Genomics. 1993;16:113–122. doi: 10.1006/geno.1993.1148. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 23.Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 24.Kusumi K, Smith JS, Segre JA, Koos DS, Lander ES. Construction of a large-insert yeast artificial chromosome library of the mouse. Mamm Genome. 1993;4:391–392. doi: 10.1007/BF00360591. [DOI] [PubMed] [Google Scholar]
- 25.Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachliri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coliusing an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dracopoli, N.C., J.L. Haines, B.R. Korf, D.T. Moir, C.C. Morton, C.E. Seidman, J.G. Seidman, and D.R. Smith. 1994–1997. Current Protocols in Human Genetics, Vol. 1. John Wiley & Sons, Inc., New York.
- 27.Malo D, Vidal S, Lieman JH, Ward DC, Gros P. Physical delineation of the minimal chromosomal segment encompassing the murine host resistance locus . Bcg Genomics. 1993;17:667–675. doi: 10.1006/geno.1993.1388. [DOI] [PubMed] [Google Scholar]
- 28.Carle GF, Olson MV. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alteration gel electrophoresis. Nucleic Acids Res. 1984;12:5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasite: isolation of a candidate for Bcg. . Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 30.Rommens, J.M., L. Mar, J. McArthur, L.-C. Tsui, and S.W. Scherer. 1994. Towards a transcriptional map of the q21-q22 region of chromosome 7. In Identification of Transcribed Sequences. U. Hochgeschwender and K. Gardiner, editors. Plenum Press, New York. 65–79.
- 31.Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 32.Silahtaroglu AN, Tumer Z, Kristensen T, Sottrup-Jensen L, Tommerup N. Assignment of the human gene for pregnancy-associated plasma protein A (PAPPA)to 9q33.1 by fluorescence in situ hybridization to mitotic and meiotic chromosomes. Cytogenet Cell Genet. 1993;62:214–216. doi: 10.1159/000133479. [DOI] [PubMed] [Google Scholar]
- 33.Fridell RA, Harding LS, Bogerd HP, Cullen BR. Identification of a novel human zinc finger protein that specifically interacts with the activation domain of lentiviral Tat proteins. Virology. 1995;209:347–357. doi: 10.1006/viro.1995.1266. [DOI] [PubMed] [Google Scholar]
- 34.Zheng C, Heintz N, Hatten M. CNS gene encoding astrotactin, which supports neuronal migration along glial fibres. Science. 1996;272:417–419. doi: 10.1126/science.272.5260.417. [DOI] [PubMed] [Google Scholar]
- 35.Kobe B, Deisenhofer J, Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats Nature. 1993;366:751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- 36.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadien A, Chen C, Ghosh S, Janeway CA., Jr MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol Cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 37.Poltorak A, Smirnova I, He X, Liu M-Y, Van Huffel C, Birdwell D, Alejos E, Silva M, Du X, Thompson P, et al. Genetic and physical mapping of the lpslocus: identification of toll-4 receptor as a candidate gene in the critical region. Blood Cells Mol Dis. 1998;15:340–355. doi: 10.1006/bcmd.1998.0201. [DOI] [PubMed] [Google Scholar]
- 38.Hashimoto D, Hudson KL, Anderson KV. The Toll gene of Drosophila, required for dorso-ventral embryonic polarity, appears to encode a transmembrane protein. Cell. 1988;52:269–279. doi: 10.1016/0092-8674(88)90516-8. [DOI] [PubMed] [Google Scholar]
- 39.Lemaitre B, Nicolas E, Michaut L, Reichhart J-M, Hoffman J. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophilaadults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 40.Kuhns DB, Long DA, Priel, Gallin JI. Endotoxin and IL-1 hyporesponsiveness in a patient with recurrent bacterial infections. J Immunol. 1997;158:3959–3964. [PubMed] [Google Scholar]
- 41.Lemaitre B, Reichhart J-M, Hoffman J. Drosophilahost defense: differential induction of antimicrobial peptides after infection by various classes of microorganisms. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raetz CRH, Ulevitch RJ, Wright SD, Sibley CH, Ding A, Nathan CF. Gram-negative endotoxin: an extraordinary lipid with profound effects on eukaryotic signal transduction. FASEB J. 1991;5:2652–2660. doi: 10.1096/fasebj.5.12.1916089. [DOI] [PubMed] [Google Scholar]
- 43.Ulevitch RJ, Tobias PS. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu Rev Immunol. 1995;13:437–457. doi: 10.1146/annurev.iy.13.040195.002253. [DOI] [PubMed] [Google Scholar]
- 44.Yu B, Wright SD. Catalytic properties of lipopolysaccharide (LPS) binding protein. J Biol Chem. 1996;271:4100–4105. doi: 10.1074/jbc.271.8.4100. [DOI] [PubMed] [Google Scholar]