Abstract
The natural resistance associated macrophage protein (Nramp) gene family is composed of two members in mammals, Nramp1 and Nramp2. Nramp1 is expressed primarily in macrophages and mutations at this locus cause susceptibility to infectious diseases. Nramp2 has a much broader range of tissue expression and mutations at Nramp2 result in iron deficiency, indicating a role for Nramp2 in iron metabolism. To get further insight into the function and mechanism of action of Nramp proteins, we have generated isoform specific anti-Nramp1 and anti-Nramp2 antisera. Immunoblotting experiments indicate that Nramp2 is present in a number of cell types, including hemopoietic precursors, and is coexpressed with Nramp1 in primary macrophages and macrophage cell lines. Nramp2 is expressed as a 90–100-kD integral membrane protein extensively modified by glycosylation (>40% of molecular mass). Subcellular localization studies by immunofluorescence and confocal microscopy indicate distinct and nonoverlapping localization for Nramp1 and Nramp2. Nramp1 is expressed in the lysosomal compartment, whereas Nramp2 is not detectable in the lysosomes but is expressed primarily in recycling endosomes and also, to a lower extent, at the plasma membrane, colocalizing with transferrin. These findings suggest that Nramp2 plays a key role in the metabolism of transferrin-bound iron by transporting free Fe2+ across the endosomal membrane and into the cytoplasm.
Keywords: iron, anemia, transport, infection, macrophage
Naturally occurring (1) or experimentally induced (2) mutations at the Nramp1 (natural resistance associated macrophage protein 1)1 locus in vivo impair macrophage function and cause susceptibility to infection by intracellular pathogens such as Salmonella, Leishmania, and Mycobacterium in mice. In humans, polymorphic variants at Nramp1 are associated with increased susceptibility to tuberculosis and leprosy (3, 4). Studies in vitro in explanted cell populations have indicated that mutations at Nramp1 affect the ability of the macrophage to restrict the intracellular replication of antigenically unrelated microorganisms. We cloned the Nramp1 gene (5) and showed that its mRNA is expressed abundantly in macrophages (6) and in neutrophils (7) and is inducible in macrophages by exposure to cytokines and bacterial endotoxin (6). Predicted amino acid sequence analysis indicates that Nramp1 has many characteristics of an integral membrane transport protein including 12 putative transmembrane (TM) domains, several predicted N-linked glycosylation sites, and a sequence signature previously identified in a number of eukaryotic and prokaryotic transport proteins (5). In macrophages, direct biochemical studies have shown that Nramp1 is a membrane phosphoglycoprotein of apparent mass 90–110 kD (8), which is expressed in the Lamp1-positive lysosomal compartment (9). Moreover, studies in phagosomes containing either latex beads or intact bacteria have shown that upon phagocytosis, Nramp1 is recruited to the membrane of the phagosome, where it remains during its maturation to phagolysosome (9). These findings suggest that Nramp1 may affect resistance to infection by modulating the intravesicular milieu of the bacterial phagosome.
We have identified a second Nramp gene in mammals, Nramp2, which encodes a protein highly similar to Nramp1 (78% identity over the hydrophobic core) (10). As opposed to the phagocyte-specific expression of Nramp1, Nramp2 mRNA expression has been detected in most tissues and cell types analyzed (10–12). Recently, it was shown that the Nramp2 gene is mutated (G185R) in two animal models of iron deficiency, the mk mouse (13) and the Belgrade rat (14). The mk mouse displays deficiency in intestinal iron uptake and microcytic anemia (15, 16). The Belgrade rat also shows a defect in intestinal iron absorption (17). Moreover, studies in oocytes have shown that Nramp2 can transport a number of divalent cations such as Fe2+, Zn2+, and Mn2+ in a pH-dependent, electrogenic fashion associated with the symport of a single proton (12). In addition, transient overexpression of the wild type but not G185R Nramp2 in HEK293T cells results in a robust stimulation of cellular 55Fe uptake (15). Taken together, these results indicate that Nramp2 is the transferrin-independent system responsible for dietary iron absorption in the intestine. However, the ubiquitous expression of Nramp2 mRNA suggests that it may be involved in iron metabolism in other tissues as well. As opposed to Nramp1, where the cellular and subcellular localization of the protein have been established, the lack of isoform-specific, anti-Nramp2 antibodies has precluded the identification of the cell type and of the subcellular compartment expressing this protein. Such information is critical to elucidate the role of the Nramp2 protein in cellular iron metabolism. In particular, the demonstrated H+-driven, Fe2+ transport activity of Nramp2, as well as its expression in a wide variety of tissues, make it a likely candidate not only for transferrin-independent iron absorption in the intestine but also for the transferrin- dependent uptake of iron in peripheral tissues. It is well established that acidification of the endosomal compartment causes Fe3+ release from transferrin and that reductases then convert the Fe3+ to Fe2+, but the mechanism of transport of Fe2+ across the endosomal membrane has not yet been elucidated.
Materials and Methods
Immunogens.
For the production of isoform-specific polyclonal antisera directed against Nramp2, rabbits were immunized with fusion proteins containing glutathione S-transferase (GST) fused to a peptide segment derived from the amino terminal region of Nramp2 (residues 1–71; for amino acid numbering see reference 10). This peptide is in a region of the protein which is not conserved in other Nramp family members, including Nramp1 (18). The GST–Nramp2 fusion protein was constructed in the plasmid vector pGEX (Pharmacia) as follows: The Nramp2 sequence was amplified by PCR using oligonucleotides NF2 (5′-AAAGATCTATGGTGTTGGATCC-3′) and NR (5′-CTGAA TTCGAACGCCCAGAGT-3′) (nucleotides 1–268), and the full-length Nramp2 cDNA as template. The PCR product was digested with BglII and EcoRI and the fragment was subcloned into pGEX digested with BamHI and EcoRI to create the in-frame GST fusion protein. Overexpression of the Nramp2–GST fusion protein was carried out in large scale cultures of Escherichia coli and the protein was purified from bacterial lysates using glutathione–Sepharose 4B (Pharmacia) as previously described (19). Purified proteins were analyzed by 7.5% SDS-PAGE and excised from the gel after light staining with 0.05% Coomassie blue in ddH2O.
Production of Anti-Nramp2 Antibodies.
Polyclonal antibodies were produced in male New Zealand White rabbits as described previously (8). A system for affinity purification of the antibodies was devised using the same Nramp2 peptide fused to a second fusion partner, dihydrofolate reductase modified by the addition of eight consecutive histidine residues (his–DHFR). The fusion protein construct was made as described above, except the PCR product was digested with EcoRI and the resulting overhangs repaired using the Klenow fragment of DNA polymerase I (Pharmacia) before digestion with BglII. The digested PCR product was ligated into BglII- and SmaI-digested pQE40 plasmid vector (Qiagen). The in-frame his–DHFR–Nramp2 fusion protein construct was transformed into E. coli strain M15(pREP4) for expression (Qiagen). Purification was performed on Ni–NTA agarose according to experimental conditions suggested by the manufacturer (Qiagen). The polyclonal antiserum directed against the GST fusion protein was purified against the his–DHFR fusion protein by a preparative immunoblot procedure (20). The anti-Nramp1 polyclonal antiserum (8) was affinity purified against the corresponding Nramp1–GST fusion protein by the same protocol.
Cell Culture.
The mouse monocyte–macrophage cell lines RAW 264.7 and J774a, the mouse Sertoli cell line TM4, and the mouse kidney line mIMCD-3 were obtained from the American Type Culture Collection (ATCC). They were cultured in media and under conditions recommended by the ATCC. WEHI 3B (myelomonocyte), WEHI 231 (B lymphocyte), BI 141 (T lymphocyte), and 70Z/3 (pre-B cell) cells were cultured as described previously (21). Chinese hamster ovary (CHO) cells LR73 (22) were grown in α-MEM supplemented with 10% fetal calf serum, 2 mM L-glutamine, 50 U/ml penicillin and 50 μg/ml streptomycin. All media and media supplements were purchased from GIBCO BRL. Murine macrophages were obtained by peritoneal lavage, as previously described (8). We have previously described the production and characterization of RAW macrophages expressing a transfected wild-type Nramp1 fused to a c-myc epitope (9). The c-myc–tagged Nramp2 expression plasmid was constructed by excising the myc-tagged Nramp2 cDNA from plasmid pBluescript (23) using SpeI and EcoRV sites from the polylinker, followed by cloning into the mammalian expression plasmid pCB6 (24). For expression in CHO cells, the same insert was cloned into the expression vector pMT2 (25). CHO cells were transfected by the calcium phosphate coprecipitation method (26). RAW cells were transfected by electroporation as described previously (9). Clones of stable transfectants were selected in geneticin (G418, 1 mg crude/ml final; GIBCO BRL) for 10–14 d, picked and expanded individually, and tested for protein expression by immunofluorescence using the anti–c-myc tag monoclonal antibody 9E10 (Babco).
Immunoblotting and Immunoprecipitation.
Crude membrane fractions from the various cells were prepared as described previously (27). Protein concentration of the membrane fraction was determined by the Bradford assay (BioRad). Proteins were separated on SDS–polyacrylamide gels and transferred by electroblotting to nitrocellulose membranes. For experiments where the membrane was to be stripped and reprobed, a polyvinylidene fluoride membrane was used (Westran; Schleicher and Schuell) to reduce protein loss from the membrane during stripping. Equal loading and transfer of proteins was verified by staining the blots with Ponceau S (Sigma Chemical Co.). The blots were blocked in TBST (10 mM Tris/Cl, pH 8, 150 mM NaCl, 0.05% Tween 20, pH 8) plus 5% skim milk powder for 1 h at room temperature. Primary antibodies used were as follows: affinity purified rabbit anti– mouse Nramp2 (1:100 dilution); affinity purified rabbit anti– mouse Nramp1 (1:200); mouse monoclonal anti c-myc–epitope tag 9E10 (Babco; 1:100), rat anti–mouse transferrin receptor (Biosource International; 1:200), and rat anti–mouse Lamp1 (1: 200). Anti–rabbit, anti–rat, and anti–mouse secondary antibodies conjugated to horseradish peroxidase were used at 1:10,000 (Amersham). Chemiluminescence was used for detection of immune complexes on the immunoblot (ECL; Amersham). For immunoprecipitation, CHO cells and Nramp1 and Nramp2 CHO transfectants were metabolically labeled with [35S]methionine by incubating overnight in 100 μCi/ml of [35S]methionine (DuPont) in methionine-free DMEM (GIBCO BRL) containing 10% heat-inactivated, dialyzed fetal bovine serum, 2 mM L-glutamine, and 2 mM Hepes. Immunoprecipitation was performed exactly as described previously (8).
Immunofluorescence.
Cells were grown on glass coverslips and fixed with 4% paraformaldehyde in PBS for 30 min at 4°C. Immunofluorescence was performed as previously described (9) with the following modifications: incubation with the primary antibody was 1 h at 20°C for the anti–c-myc mouse monoclonal 9E10 (1:200; Babco), and the anti-Lamp1 rat monoclonal (1:200), or overnight at 4°C for the anti-Nramp2 antiserum (1:800) followed by anti–mouse, anti–rat, or anti–rabbit secondary antibodies conjugated to rhodamine (1:300) or FITC (1:200) (Jackson Immunochemicals). Immunofluorescence was analyzed with a Nikon microscope using the 100× oil immersion objective. Certain colocalization studies were carried out using a Zeiss laser confocal microscope with a 63× objective. Composites of confocal images were assembled and labeled using PhotoShop, Metamorph, and Freehand software. To label the lysosomal compartment, cells were incubated with 1 mg/ml lysine-fixable FITC–dextran (Molecular Probes) in growth medium for 4 h at 37°C in 5% CO2. After washing, cells were incubated an additional 30 min to chase the dextran from the early endosomal to the lysosomal compartments. For identification of the early and recycling endosomal compartment, cells were incubated in serum-free medium containing 50 μg/ml FITC–transferrin (Molecular Probes) for 30 min at 37°C in 5% CO2. Phagosomes were formed by incubating the cells with 3 μm latex beads (Sigma Chemical Co.) diluted 1:200 in complete culture medium for 15 min at 37°C in 5% CO2. After treatments to identify the specific subcellular compartments, cells were fixed in 4% paraformaldehyde and processed for immunofluorescence.
Glycosidase Treatments.
Endo–β-acetylglucosaminidase H (Endo H) and peptide N-glycosidase F (PNGase F) were obtained from New England Biolabs. Aliquots of membrane preparations from J774a and CHO cells were denatured before digestion in a buffer containing 0.5% SDS and 0.1 M β-mercaptoethanol for 2 min at 70°C. For Endo H digestion, samples were diluted twofold and incubated with 4,000 U of Endo H in 50 mM sodium citrate, pH 5.5, for 1 h at 37°C. The −Endo H controls were treated identically except an equivalent volume of ddH2O was added in place of Endo H. For PNGase F digestion, samples were diluted twofold and incubated in 50 mM sodium phosphate, pH 7.5, 1% NP-40, with 500 U of PNGase F for 1 h at 37°C. The −PNGase F controls were treated identically except an equivalent volume of ddH2O was added in place of PNGase F.
RNA Isolation and Hybridization Studies.
Total cellular RNA was isolated using guanidinium–HCl solubilization and differential ethanol precipitation (28). 20 μg of total cellular RNA were denatured in a formamide–formaldehyde mixture and loaded onto denaturing agarose gels containing 0.66 M formaldehyde. Blots were prehybridized in a solution containing 10% dextran sulfate, 1 M NaCl, 1% SDS, and heat-denatured salmon sperm DNA (200 μg/ml) at 65°C for 2–16 h. Hybridization was for 24 h at 65°C in the same buffer containing the radiolabeled probe (106 cpm/ml of hybridization buffer; specific activity 109 cpm/μg DNA). Blots were washed under conditions of increasing stringency up to 0.1× SSC and 0.1% SDS at 65°C and then exposed to Kodak XR film with two intensifying screens at −70°C for 18 h to 7 d at −80°C.
Phagosome Fractionation.
Phagosomes were isolated from J774a cells by a modification of a method described previously (9). 10 subconfluent 150 mm dishes of each cell line were fed with a 1:200 dilution of blue-dyed latex beads (0.8 μm; Sigma Chemical Co.) in culture medium for 1 h at 37°C in 5% CO2. The cells were then washed in PBS and harvested in the presence of protease inhibitors (1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin, and 100 μg/ml PMSF; all Boehringer Mannheim) and recovered by centrifugation (2,000 g, 5 min). The cell pellets were washed and resuspended in homogenization buffer (8.5% sucrose, 3 mM imidazole, pH 7.4) and homogenized by passage through a 22G needle until 90% of the cells were broken, as monitored by light microscopy. Nuclei and unbroken cells were pelleted and the supernatant loaded onto a sucrose step gradient as follows: the supernatant was brought up to 40% sucrose by addition of 62% sucrose and loaded on top of a 1-ml 62% sucrose cushion. Layers of 2 ml of 35, 25 and finally 10% sucrose (all sucrose solutions wt/wt in 3 mM imidazole, pH 7.4, plus protease inhibitors) were sequentially added to the top of the tube, and the gradients were centrifuged at 100,000 g for 1 h at 4°C (SW41; Beckman). Phagosomes were recovered from the 10–25% sucrose interface, washed with PBS containing protease inhibitors, and recovered by a final centrifugation at 40,000 g in an SW41 rotor at 4°C. The final pellets were resuspended in 2× Laemmli sample buffer. Phagosomes prepared by this protocol have been previously shown to be free of endoplasmic reticulum (endoplasmin, BiP, and calnexin) and Golgi apparatus (galactosyl transferase) contaminants (29).
Results
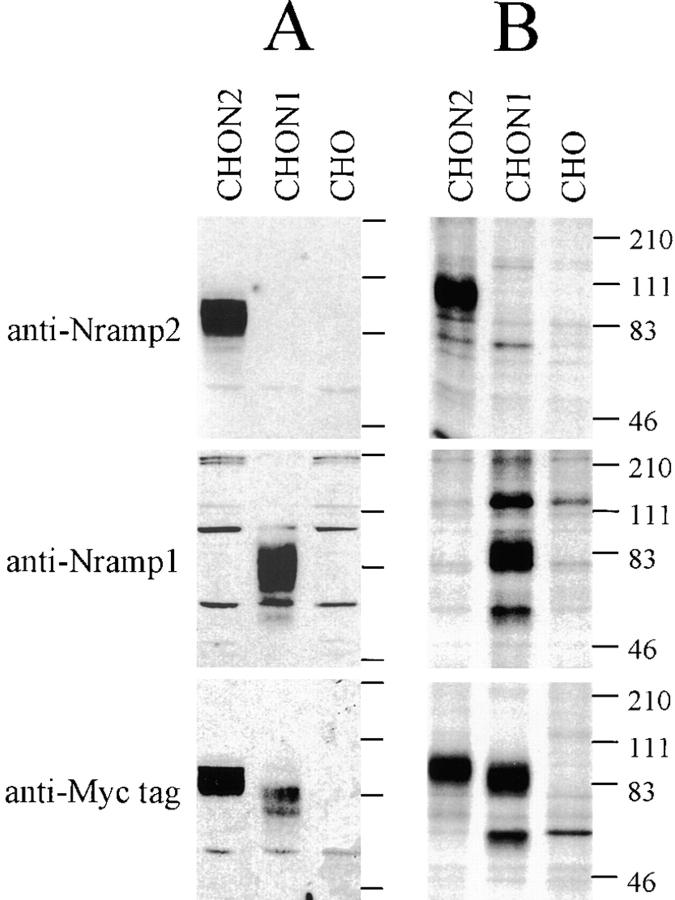
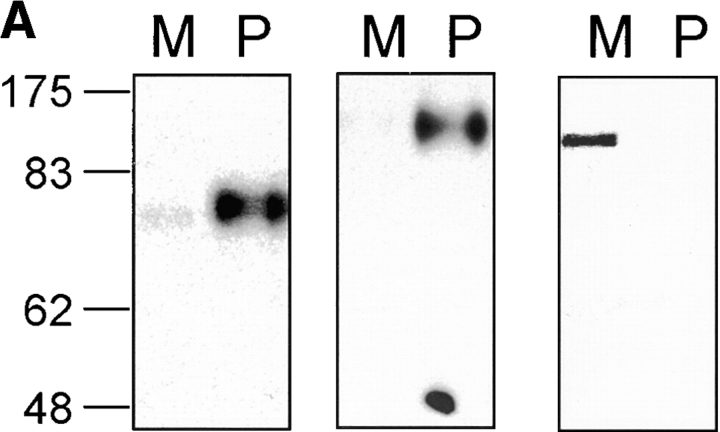
To generate an isoform-specific anti-Nramp2 antiserum, a protein segment derived from the amino terminus of Nramp2 was selected based on its predicted antigenicity and its sequence divergence from Nramp1 (28% identity). A GST–Nramp2 fusion protein containing the amino terminal Nramp2 segment was produced and used for immunization, and the anti-Nramp2 fraction was further isolated from the immune serum by affinity purification against a second immobilized Nramp2–DHFR fusion partner. The antiserum was tested for specificity by immunoblotting crude membrane fractions as well as by immunoprecipitation of [35S]methionine–labeled cell lysates from transfected CHO cell clones expressing either c-myc–tagged Nramp1 or c-myc–tagged Nramp2 proteins (Fig. 1). The anti–c-myc monoclonal antibody (9E10 [30]) specifically recognized a species of apparent molecular mass 90–100 kD in the Nramp2-transfected CHO cells, and a protein of 85–95 kD in the Nramp1-transfected cells that were absent from extracts of untransfected CHO controls (Fig. 1, A and B, bottom). These protein species migrated as broad bands in SDS–acrylamide gels. In membrane preparations from Nramp2-transfected CHO cells (CHON2), the affinity-purified anti-Nramp2 antiserum recognized a single protein species of apparent molecular mass 90–100 kD (Fig. 1 A, top). Immunoblotting analysis of the same set of membrane fractions with anti-Nramp1 antiserum revealed a single protein species of 85–95 kD in membranes from the Nramp1-transfected CHO cells (Fig. 1 A, middle). These immunoreactive bands were absent from untransfected CHO cells and from transfected cells expressing the other Nramp isoform. The electrophoretic mobility characteristics of the Nramp1 and Nramp2 detected by the respective polyclonal antisera were very similar to those of the species detected by the anti–c-myc antibody in the same cell extracts (Fig. 1 A, bottom). The reactivity and isoform specificity of the antibodies were confirmed by immunoprecipitation studies of [35S]methionine metabolically labeled cell extracts from CHO cells or from Nramp1 and Nramp2 CHO transfectants (Fig. 1 B). These data indicate that the anti-Nramp1 and anti-Nramp2 antisera produced and purified according to our protocol are isoform specific.
Figure 1.
After affinity purification, anti-Nramp1 and anti-Nramp2 antisera were tested for specificity and reactivity by immunoblotting against crude membrane fractions (A) as well as by immunoprecipitation of [35S]methionine–labeled cell lysates (B) from CHO cells or from the same cells transfected with a c-myc–tagged (tag sequence EQKLISEEDL) Nramp1 (CHON1) or a c-myc–tagged Nramp2 (CHON2). Protein extracts were separated by SDS-PAGE and either transferred to membranes (A) or exposed to x-ray films after immunoprecipitation (B). Antisera directed against Nramp2 (top), Nramp1 (center) or a commercially available mouse monoclonal anti–c-myc antibody (9E10; bottom) were used. The size of the molecular mass markers (in kD) is indicated to the right of the gels.
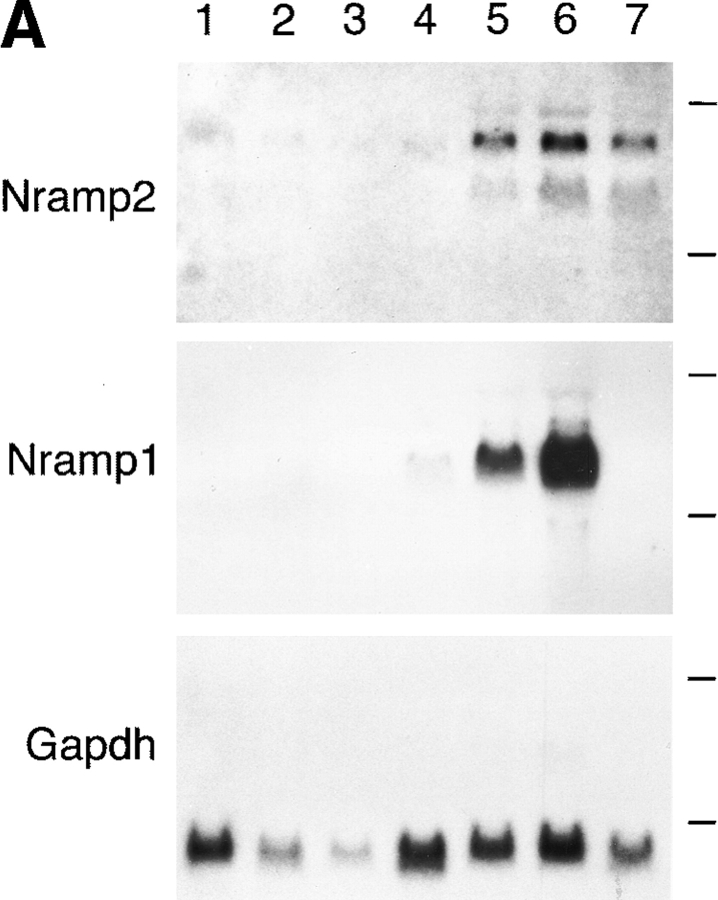
Northern blotting and in situ hybridization studies have shown that, as opposed to Nramp1, which is expressed almost exclusively in mononuclear phagocytes, Nramp2 mRNA is expressed in most tissues (10–12). We questioned whether the two Nramp proteins would display an overlapping or mutually exclusive expression pattern. Northern blot analysis of total cellular RNA from a panel of murine hematological cell lines revealed a readily detectable level of Nramp2 mRNA expression in the macrophage lines RAW 264.7 and J774a as well as in Friend virus– transformed erythroleukemia (MEL) cells (Fig. 2 A, top; exposure time 1 wk). A much lower level of expression of Nramp2 was found in other cell lines: WEHI 231 (B lymphocyte), WEHI 3B (myelomonocyte), 70/Z (pre-B lymphocyte), and BI 141 (T lymphocyte). In comparison, Nramp1 mRNA expression was restricted to the macrophage cell lines RAW264.7 and J774a and is expressed at levels ∼50-fold higher than that of Nramp2 (Fig. 2 A, middle; exposure time 24 h). Thus, Nramp1 and Nramp2 mRNAs are coexpressed in macrophages.
Figure 2.
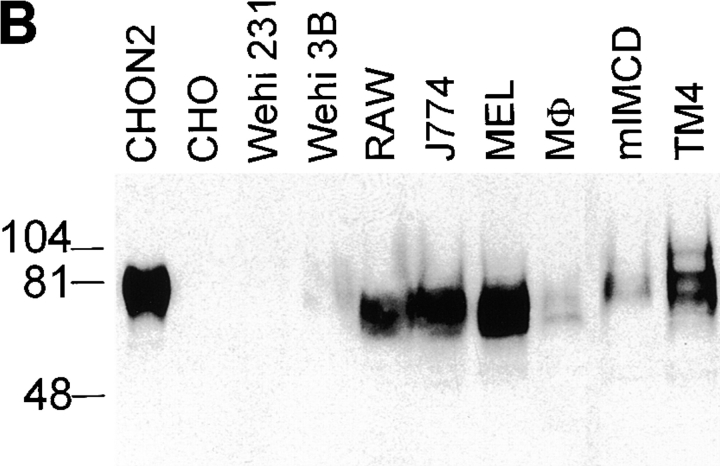
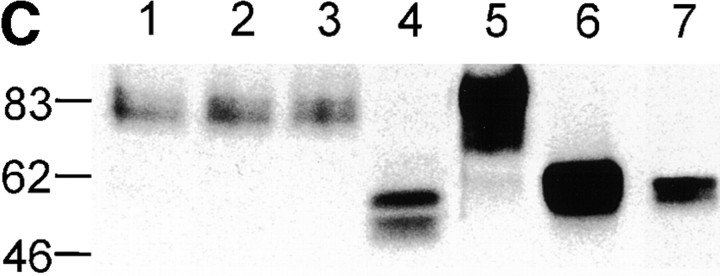
(A) Nramp2 mRNA expression in cultured cell lines. Northern blot analysis of total cellular RNA (20 μg) from mouse cell lines: BI 141 (T lymphocyte; lane 1), 70/Z (pre-B lymphocyte; lane 2), WEHI 231 (B lymphocyte; lane 3), WEHI 3B (myelomonocyte; lane 4), RAW 264.7 (macrophage; lane 5), J774a (macrophage; lane 6), and MEL (erythroleukemia; lane 7). The RNA was electrophoresed in a denaturing agarose gel, followed by transfer to a nylon membrane and hybridization to gene-specific cDNA segments from either Nramp2 (top), Nramp1 (center), or glyceraldehyde phosphate dehydrogenase (Gapdh, bottom). Exposure time was 7 d for Nramp2 and 1 d for Nramp1 and Gapdh. (B) Nramp2 protein expression in cultured cell lines. Crude membrane fractions were prepared from different cultured cell lines as well as from thioglycolate-induced primary mouse macrophages (Mø) and were separated by SDS-PAGE. Immunoblotting was performed using the anti-Nramp2 antiserum. (C) Glycosylation of Nramp2 protein. Membrane fractions from J774a cells (lanes 1–4) and Nramp2-transfected CHO cells (lanes 5–7) were treated with endoglycosidases followed by electrophoresis and immunoblotting with the anti-Nramp2 antiserum. Membrane fractions were either mock treated (lanes 1, 3, and 5) or incubated with Endo H (lane 2) or PNGaseF (lanes 4, 6, and 7). A lighter exposure of lane 6 is shown in lane 7.
To analyze Nramp2 protein expression in macrophages, membrane fractions were prepared from thioglycolate- induced primary mouse macrophages, from J774a and RAW 264.7 cultured macrophages. For comparison, membranes were also prepared from cell lines derived from tissues previously shown to express a high level of Nramp2 mRNA: the Sertoli cell line TM4 and the kidney inner medullary collecting duct line mIMCD-3 (17). Membranes were also prepared from control and Nramp2-transfected CHO cells, as well as from two cell lines expressing low levels of Nramp2 mRNA (Fig. 2 A, WEHI 231 and WEHI 3B). The membranes were analyzed by immunoblotting with the anti-Nramp2 antibody (Fig. 2, B and C). The antibody detected a major heterogeneous immunoreactive protein species of broad electrophoretic mobility with an apparent molecular mass of 80–90 kD in all cells tested, with the exception of WEHI 231 cells and untransfected CHO cells. The protein was most abundant in TM4, RAW 264.7, J774a and MEL cells. The protein was also detected in the membranes prepared from primary mouse macrophages and mIMCD-3 cells, although at a lower level. WEHI 3B membranes showed the lowest level of Nramp2 expression, whereas WEHI 231 and untransfected CHO cells were negative for Nramp2 expression. In the positive membrane samples, the electrophoretic mobility and heterogeneity of the immunoreactive species varied, possibly due to different posttranslational modification of the protein in these cell types. Thus, Nramp2 is expressed in a wide variety of tissues, including macrophages, and macrophages coexpress Nramp1 and Nramp2.
The apparent mass of endogenous Nramp2 estimated by SDS-PAGE is considerably greater than the 62.3 kD molecular mass predicted by the primary amino acid sequence of the cDNA. Together with the broadness of the immunoreactive band, this anomalous mobility suggests that Nramp2 may be posttranslationally modified by glycosylation. To test this hypothesis, membrane fractions from J774a cells and Nramp2-transfected CHO cells were treated with endoglycosidases followed by electrophoresis and immunoblotting. Nramp2 was resistant to digestion with Endo H (Fig. 2 C, lanes 1 and 2), which specifically cleaves high mannose and some hybrid N-linked oligosaccharides from glycoproteins. In contrast, PNGaseF, which hydrolyzes high mannose, hybrid, and complex oligosaccharides, converted the 82-kD Nramp2 species into smaller forms of approximate apparent molecular masses of 50–55 kD (Fig. 2 C, lane 4). PNGase treatment of membranes from the Nramp2 CHO transfectants also resulted in a shift of the apparent molecular mass of the protein from 85 to ∼56 kD (Fig. 2 C, lanes 6 and 7). Therefore, Nramp2 is posttranslationally modified extensively by complex N-linked glycosylation.
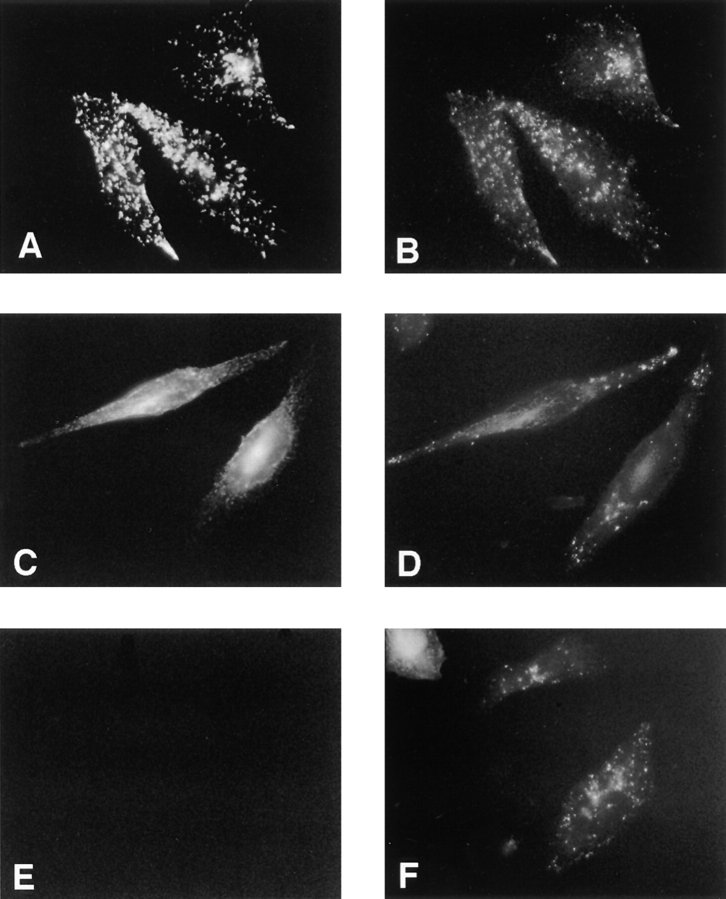
To gain insight into the subcellular localization of Nramp2, we first performed immunofluorescence studies on CHO and RAW 264.7 transfected cells, using an antibody directed against the c-myc epitope attached to the carboxy terminus of the transfected Nramp2 protein. The high levels of transfected protein in these cell lines facilitated nonambiguous localization of Nramp2, without possible limitation associated with low levels of expression of the endogenous protein. We have previously observed that the c-myc–tagged Nramp2 protein is functional in both CHO and RAW macrophage backgrounds and carries out active Fe2+ transport in these cells (Govoni, G., and P. Gros, unpublished results). We initially tested whether Nramp2 localizes to the late endosomal/lysosomal compartment, as found earlier for Nramp1 (9). To label the lysosomal compartment, cells were cultured in the presence of FITC–conjugated dextran, followed by a chase period of 30 min to remove the dextran from the early endosomal compartments, before fixation and immunostaining with the anti–c-myc antibody. In Nramp1-transfected CHO cells, there was clear colocalization of the anti–c-myc staining (Fig. 3 A) and the dextran-loaded late endosomal/lysosomal compartment (Fig. 3 B). Nramp1-stained vesicles negative for FITC–dextran were also detected. In contrast, in Nramp2-transfected CHO cells, anti–c-myc staining revealed an intracellular network of finer punctate vesicles distributed throughout the cytoplasm (Fig. 3 C). This staining does not appear to colocalize with the FITC–dextran (Fig. 3 D). Similar results were obtained in parallel experiments using the c-myc–Nramp1 and c-myc–Nramp2-transfected RAW cells (data not shown), indicating that Nramp2 is not expressed in the lysosomal compartment. Thus, Nramp1 and Nramp2 clearly appear to have distinct, nonoverlapping subcellular sites of expression.
Figure 3.
Subcellular localization of Nramp1 and Nramp2 in transfected CHO cells. Immunofluorescence was performed on CHO cells transfected with c-myc–tagged Nramp1 proteins (A and B), c-myc–tagged Nramp2 proteins (C and D), or with untransfected CHO controls (E and F). Immunofluorescence was with the anti–c-myc mouse monoclonal antibody 9E10 (A, C, and E) which detects the transfected Nramp1 and Nramp2 proteins. Lysosomes were labeled by preincubation of the cells with FITC–conjugated dextran (B, D, and F). Cells were then fixed and reacted with the anti–c-myc tag antibody 9E10 and a rhodamine-conjugated secondary antibody (A, C, and E), followed by examination by fluorescence microscopy and photography.
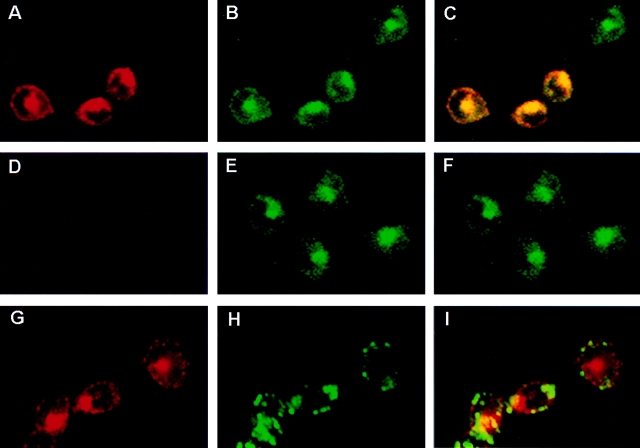
Since Nramp2 is implicated in cellular iron uptake, it appears logical that Nramp2 be present at the plasma membrane and/or in recycling endosomes. To label these compartments, CHO (Fig. 4) and RAW transfected cells (Fig. 5) were cultured in the presence of FITC–conjugated transferrin before fixation and immunostaining with the anti–c-myc antibody. Analysis by confocal microscopy indicated that, as expected, transferrin (green) stained both the plasma membrane (ring-like staining at the edge of the cells) and the recycling endosomes (subcellular punctate staining) (Fig. 4 B). A very similar and overlapping pattern was observed for Nramp2, as revealed by the anti–c-myc antibody (red, Fig. 4 A). Superimposition of the two images (Fig. 4 C) clearly identifies colocalization (yellow) of the two signals. Certain cells stained with FITC–transferrin but were negative for the c-myc staining, suggesting that although these cells are positive for the pSV2neo plasmid and are resistant to G418, they failed to express c-myc–tagged Nramp2. Such cells provide an internal control for the specificity of the anti–c-myc staining and for the overlapping staining with FITC–transferrin. Similarly, when untransfected CHO cells were identically processed and examined, the cells were not stained with the anti–c-myc antibody (Fig. 4 D), but displayed a normal pattern of staining with FITC–transferrin (Fig. 4 E). Finally, when the lysosomes of the Nramp2-transfected cells were stained with FITC–dextran (Fig. 4 H) and with the anti–c-myc antibody for Nramp2 (Fig. 4 G), no significant overlap between the two signals was detected (Fig. 4 I). A similar colocalization of Nramp2 and FITC–transferrin was noted in parallel experiments with RAW macrophages expressing c-myc– Nramp2 (Fig. 5). These results confirm that Nramp1 and Nramp2 have nonoverlapping, subcellular localization, and that Nramp2 colocalizes with transferrin in the early recycling endosomal compartment.
Figure 4.
Colocalization of Nramp2 and transferrin in early endosomes was determined by double immunofluorescence and confocal microscopy in either CHO control cells (D–F) or CHO cells transfected with a c-myc–tagged Nramp2 protein (A–C and G–I). Cells were cultured in the presence of FITC–conjugated transferrin (A–F) or FITC–conjugated dextran (G–I) before fixation and immunostaining with the anti–c-myc tag antibody and a rhodamine-conjugated secondary antibody (A, D, and G). The slides were then examined by confocal microscopy, and the FITC (green; B, E, and H) and rhodamine (red; A, D, and G) images were overlaid (yellow identifies colocalization; C, F, and I). In the Nramp2-transfected CHO cells, the Nramp2 staining (A and G) showed extensive overlap with the distribution of FITC–transferrin (A + B = C) but not with lysosomal FITC–dextran (G + H = I). FITC–transferrin staining was observed in the untransfected CHO cells (E), but the cells were negative for anti–c-myc staining (D).
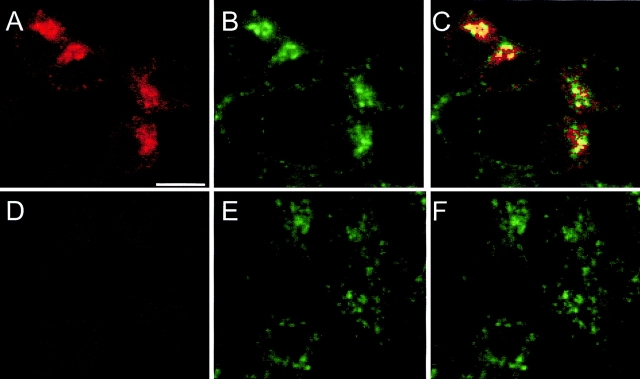
Figure 5.
Colocalization of Nramp2 and transferrin in early endosomes was determined by double immunofluorescence and confocal microscopy in RAW macrophages transfected with a c-myc–tagged Nramp2 (A–C) and in untransfected control cells (D–F). Cells were cultured in the presence of FITC–conjugated transferrin before fixation and immunostaining with the primary anti–c-myc tag antibody (9E10) and a secondary rhodamine-conjugated, anti–mouse antibody. The slides were then examined by confocal microscopy, and the FITC (green; B and E) and rhodamine (red; A and D) images were overlaid to identify colocalization (C and F). The image in C shows colocalization of Nramp2 and FITC–transferrin in several of the cells in the field.
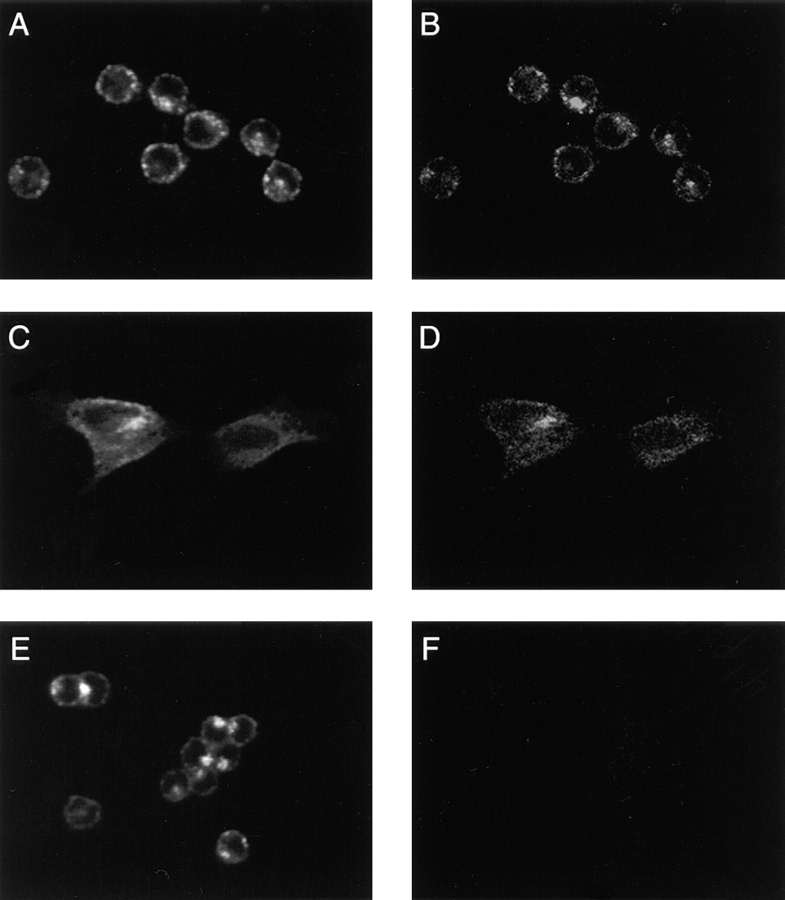
To confirm the endosomal localization of Nramp2 determined in transfected CHO and RAW cells, we performed immunofluorescence in nontransfected cell lines that tested positive (MEL, TM4) or negative (WEHI 231) for Nramp2 expression by immunoblotting (Fig. 2). Immunofluorescence was performed with the anti-Nramp2 polyclonal antiserum and FITC–transferrin, and results are shown in Fig. 6. In MEL (Fig. 6, A and B) and TM4 (C and D) cells, the endogenous Nramp2 protein (B and D) showed very similar staining pattern to that generated by FITC–transferrin (A and C), similar to that seen in transfected cells (Figs. 4 and 5). Finally, in agreement with the absence of Nramp2 expression in WEHI 231 cells noted by immunoblotting (Fig. 2 B), no Nramp2 staining was observed in WEHI231 cells (Fig. 6 F), although the endosomal compartment of these cells could readily be labeled by FITC–transferrin (Fig. 6 E). Together, these results verify data obtained in transfected CHO and RAW cells.
Figure 6.
Subcellular localization of endogenous Nramp2 in cell lines. Cell lines previously shown by immunoblotting to be either positive (MEL, TM4) or negative (WEHI 231) for Nramp2 expression were cultured in the presence of FITC–transferrin before fixation and immunostaining with the anti-Nramp2 antiserum and a rhodamine-conjugated secondary antibody. The slides were then examined by confocal microscopy, and FITC (A, C, and E; transferrin) and rhodamine (B, D, and H; Nramp2) images were obtained from the samples. The same settings for the confocal microscope were used for all samples examined. The images show very similar subcellular localization of Nramp2 and FITC–transferrin in MEL (A and B) and TM4 (C and D) cells, whereas WEHI (E and F) cells are negative for Nramp2.
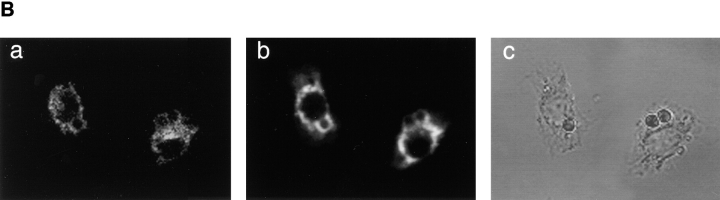
The localization of Nramp2 to the plasma membrane and endosomal network raised the possibility that like Nramp1, Nramp2 may become associated with phagosomal membranes after phagocytosis. Phagosomes are initially derived from the plasma membrane and are known to sequentially interact and acquire proteins from both early and late endosomes before their final fusion with lysosomes (29). We have shown that Nramp1 is acquired during the phagosomal maturation process, using the model system of latex bead–containing phagosomes (9). Latex bead–containing phagosomes are ideal for microscopic examination and can also be purified from cell homogenates by flotation on sucrose gradients. To determine whether Nramp2 can associate with the phagosomal membrane, J774a cells were fed latex beads for 1 h at 37°C, and the phagosomal fraction was isolated from cell homogenates by fractionation on sucrose gradient. Equal amounts of the purified phagosomal fraction and of a crude total membrane fraction were separated by SDS-PAGE, and the relative amount of endogenous Nramp2 in each sample was determined by immunoblotting. As shown in Fig. 7 A (left), Nramp2 was significantly enriched in purified phagosomes as compared to the crude membrane preparation. In these experiments, the Lamp1 protein (marker of the phagolysosome, center) and the transferrin receptor (marker of the plasma membrane, right) were used as controls. As expected, significant enrichment of Lamp1 was seen in the phagosomal fraction whereas the transferrin receptor was not enriched in phagosomes even though it is readily detectable in crude membrane fractions. These results suggest that Nramp2 becomes associated with the phagosome during its maturation to phagolysosome. Possible association of Nramp2 with latex beads phagosomes was further analyzed in J774a cells by immunofluorescence and confocal microscopy. J774a cells were fed latex beads and then fixed and processed by double immunofluorescence using anti-Nramp2 and anti-Lamp1 antibodies (Fig. 7 B). In J774a cells, the Nramp2 signal obtained with our antibody was weak, which limited the analysis. Nevertheless, in several of the sections analyzed a portion of the Nramp2 signal could clearly be seen at the periphery of the bead, suggesting association with the phagosome. However, this signal was much weaker than that obtained using the anti-Lamp1 antibody. Thus, results from immunoblotting and immunofluorescence suggest the possibility that a portion of the endosomal Nramp2 protein becomes associated with the phagosome during phagolysosome maturation.
Figure 7.
Nramp2 association with phagosomes. (A) Immunoblotting of latex bead–containing phagosomes isolated from J77a cells. Latex bead–containing phagosomes were purified from cell homogenates by subcellular fractionation on sucrose density gradients as described in Materials and Methods. Equal amounts of phagosomal proteins (P) and of a crude membrane protein extract prepared prior to phagocytosis (M) were separated by SDS-PAGE on a 7.5% gel. Proteins were transferred to nitrocellulose and the immunoblot was sequentially analyzed with anti-Nramp2 antiserum (left), anti-Lamp1 antibody (center), and anti-transferrin receptor antibody (right). The position of molecular mass markers (in kD) is indicated on the left side of the immunoblot. (B) Localization of the Nramp2 and Lamp1 proteins in J774a macrophages by immunofluorescence. J774a macrophages were allowed to phagocytose latex beads, then fixed and stained with the anti-Nramp2 antiserum and a rhodamine-conjugated secondary antibody (a), and the anti-Lamp1 antibody and a secondary antibody coupled to FITC (b). A phase contrast image of the cells shown in panels a and b is also included in panel c.
Discussion
A large body of biochemical data supports the proposal that Nramp2 functions as a transporter for several divalent cations, including Fe2+ (12–14, 23, 31). Nramp2 is mutated in the mk mouse and in the Belgrade rat (13, 14), with both animals exhibiting a severe microcytic hypochromic anemia and a severe defect in iron absorption by intestinal cells (15, 16). However, in vitro studies have shown that iron acquisition is also decreased in the peripheral cells and tissues of these animals (17, 32–36) and that the anemia cannot be corrected by direct iron injections (37), suggesting a second block of iron entry into peripheral tissues. Thus, physiological consequences of Nramp2 mutations in vivo strongly suggest that Nramp2 is not only involved in iron uptake at the level of the intestinal enterocyte but also participates in iron acquisition in other cell types as well. In peripheral tissues, cellular iron uptake is through the transferrin cycle (for review see reference 38). Diferric transferrin binds to the transferrin receptor and is internalized, and acidification of the internalized vesicles results in release of iron from transferrin followed by alkalinization of the vesicles and recycling of the receptor to the cell surface (39). Iron escapes the acidified endosomal compartment to reach the cytoplasm, where it can be captured by mitochondria for heme biosynthesis and incorporation into heme-containing proteins, stored in the cytoplasm in the form of ferritin, and/or used directly for synthesis of nonheme-containing proteins (e.g., ribonucleotide reductase). The mechanism by which iron is extruded from the acidified endosome to enter the cytoplasm is unknown and has been a matter of considerable debate.
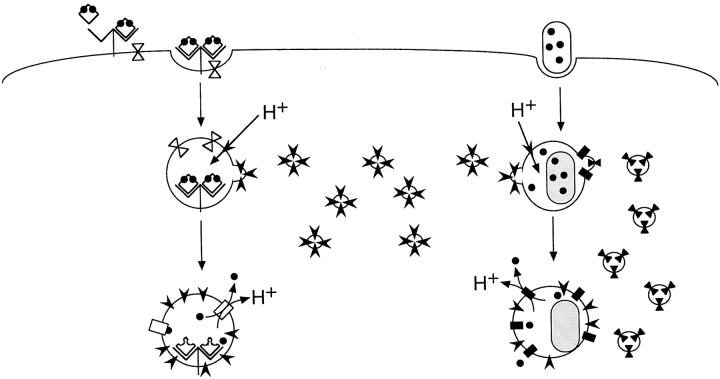
In the current study, we have raised isoform specific anti-Nramp2 antiserum and have used it to verify a number of structural and biochemical features of Nramp2 predicted from the primary amino acid sequence deduced from the cDNA. These analyses have shown that Nramp2 is an integral membrane protein which is extensively modified by N-linked glycosylation. As opposed to Nramp1, which is macrophage-specific, Nramp2 protein was found ubiquitously expressed in a majority of cell lines analyzed. The current study has also clearly established that Nramp2 and Nramp1 localize to distinct subcellular compartments. Whereas Nramp1 colocalizes with FITC–dextran in the lysosomal compartment, Nramp2 is not detectable in this compartment but rather shows clear colocalization with FITC–transferrin both at the plasma membrane and in recycling endosomes. The demonstration of Nramp2 expression in several peripheral tissues, the colocalization of Nramp2 and transferrin in plasma membrane and recycling endosomes, the iron transport properties of the Nramp2 protein, and the effect of Nramp2 mutations on iron metabolism in peripheral tissues are strong evidence that Nramp2 is responsible for transporting Fe2+ into the cytoplasm after acidification of the transferrin-positive endosome (Fig. 8). Interestingly, this acidification would simultaneously provide a gating mechanism for iron transport by Nramp2 and for release from transferrin (Fig. 8). Indeed, the pH dependence of iron transport by Nramp2 has been demonstrated in several systems, including Xenopus oocytes (12), transfected HEK293T cells (14), and CHO cells (Govoni, G., and P. Gros, unpublished results). Likewise, release of iron from transferrin and its subsequent release from endosomes is dependent on endosome acidification, which can be inhibited by bafilomycin (40) and concanamycin (41), specific inhibitors of the vacuolar H+–ATPase, but is insensitive to the Na+, K+–ATPase inhibitor ouabain (41, 42). This suggests a critical role for the vacuolar H+– ATPase in this process. Indeed, the association of vacuolar H+–ATPase with transferrin-positive endosomal vesicles has been demonstrated by immunohistochemical means in LLC–porcine kidney epithelial 1 cells (43).
Figure 8.
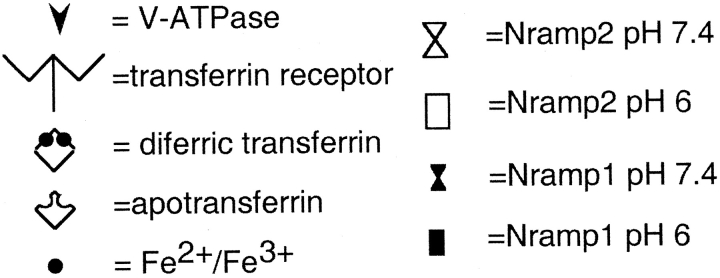
Schematic representation of Nramp 2 (left) and Nramp1 (right) function. Nramp2 is present at the plasma membrane and recycling endosomes but is not functional at neutral pH. When diferric transferrin binds to the transferrin receptor, the complex is internalized. After recruitment of the V–ATPase, the endosome acidifies, causing dissociation of iron from transferrin as well as activation of Nramp2 function. Iron can then be cotransported along with a proton across the endosomal membrane into the cytosol. In the case of Nramp1, bacteria that contain and require iron are taken up in phagosomes at the plasma membrane. These phagosomes acquire both Nramp1 and the V–ATPase. Acidification of the phagosome provides the driving force for the cotransport of iron and protons out of the phagosome in order to sequester iron away from the bacteria.
The demonstration that Nramp1 and Nramp2 are coexpressed in the same cell type with distinct subcellular localizations suggests possible functional parallels between Nramp1 and Nramp2 (Fig. 8). Both the yeast Smf1 and the mammalian Nramp2 proteins can transport Mn2+, with the latter also transporting Fe2+ and other divalent cations. Nramp2 and Smf1 share approximately 40% sequence identity within the conserved hydrophobic core (18). As the mammalian Nramp1 and Nramp2 proteins share almost 80% sequence identity within their hydrophobic cores, it is likely that Nramp1 is involved in the transport of divalent cations as well. The removal of such metabolically essential ions from the phagosomal space would provide an attractive explanation for the observed pleiotropic effect of Nramp1 mutations in vivo on the replicative potential of internalized microbes that inhabit the phagosomal space in macrophages. Additionally, the observation that a portion of Nramp2 associates with latex bead phagosomes in J774a cells suggests that Nramp2 may also play a role in depleting the phagosomal space of divalent cations necessary for microbial survival.
Despite considerable efforts, transport studies in CHO and RAW cells transfected and overexpressing Nramp1 protein have so far failed to demonstrate an Nramp1-mediated transport of either 54Mn or 55Fe transport in these cells (Govoni, G., and P. Gros, unpublished results). The distinct, nonoverlapping distribution of Nramp1 and Nramp2 reported here provides an explanation for this apparent lack of transport activity associated with Nramp1. While the two proteins may have the same transport potential, the observed targeting of Nramp2 to the plasma membrane and recycling endosome compartment would result in a net increase in cellular accumulation of extracellularly added, radiolabeled ligand under acidic pH transport assay conditions. On the other hand, the restricted expression of Nramp1 to the lysosomal compartment would not cause a similar increased cellular uptake of a ligand presented in the extracellular milieu, although it could act on such ligand if present in the phagolysosomal space. Therefore, it is tempting to speculate that both Nramp1 and Nramp2 have similar transport function but act at different, nonoverlapping, intracellular sites (Fig. 8). If Nramp1 and Nramp2 do indeed transport the same substrates, it is also tempting to speculate that vesicular acidification via the vacuolar H+– ATPase may provide a key common gating mechanism for the activation of both transporters, through fusion with vacuolar H+–ATPase-positive vesicles (Fig. 8). Possible similarities and differences in the mechanism of action and regulation of Nramp1 and Nramp2 in macrophages are currently being investigated.
Acknowledgments
The authors would like to thank Elia Abi-Jaoude and Dr. Danny Baranes (McGill University) for expert technical help and advice, Dr. W. Trimble (Hospital for Sick Children, Toronto) for advice on antibody purification, Dr. M. Desjardins for the gift of the anti-Lamp1 antibody, and Dr. A. Veillette for his gift of cell lines.
This work was supported by National Institutes of Health grant 1 R01 A1 35237-06 to P. Gros and a Medical Research Council of Canada (MRC) grant to S. Grinstein. P. Gros and S. Grinstein are International Scholars of the Howard Hughes Medical Institute and Career Scientists of the MRC.
Footnotes
Abbreviations used in this paper: CHO, Chinese hamster ovary; CHON, Nramp-transfected CHO cells; Endo H, Endo-β-acetylglucosaminidase H; GST, glutathione S-transferase; MEL, erythroleukemia; Nramp, natural resistance associated macrophage protein; TM, transmembrane.
References
- 1.Malo D, Vogan K, Vidal S, Hu J, Cellier M, Schurr E, Fuks A, Bumstead N, Morgan K, Gros P. Haplotype mapping and sequence analysis of the mouse Nramp gene predict susceptibility to infection with intracellular parasites. Genomics. 1994;23:51–61. doi: 10.1006/geno.1994.1458. [DOI] [PubMed] [Google Scholar]
- 2.Vidal S, Tremblay ML, Govoni G, Gauthier S, Sebastiani G, Malo D, Skamene E, Olivier M, Jothy S, Gros P. The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J Exp Med. 1995;182:655–666. doi: 10.1084/jem.182.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abel L, Sanchez FO, Oberti J, Thuc NV, Hoa LV, Lap VD, Skamene E, Lagrange PH, Schurr E. Susceptibility to leprosy is linked to the human NRAMP1 gene. J Infect Dis. 1998;177:133–145. doi: 10.1086/513830. [DOI] [PubMed] [Google Scholar]
- 4.Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, Hill AV. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N Engl J Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 5.Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 6.Govoni G, Gauthier S, Billia F, Iscove NN, Gros P. Cell-specific and inducible Nramp1 gene expression in mouse macrophages in vitro and in vivo. J Leukoc Biol. 1997;62:277–286. doi: 10.1002/jlb.62.2.277. [DOI] [PubMed] [Google Scholar]
- 7.Cellier M, Shustik C, Dalton W, Rich E, Hu J, Malo D, Schurr E, Gros P. Expression of the human NRAMP1 gene in professional primary phagocytes: studies in blood cells and in HL-60 promyelocytic leukemia. J Leukoc Biol. 1997;61:96–105. doi: 10.1002/jlb.61.1.96. [DOI] [PubMed] [Google Scholar]
- 8.Vidal SM, Pinner E, Lepage P, Gauthier S, Gros P. Natural resistance to intracellular infections: Nramp1 encodes a membrane phosphoglycoprotein absent in macrophages from susceptible (Nramp1 D169) mouse strains. J Immunol. 1996;157:3559–3568. [PubMed] [Google Scholar]
- 9.Gruenheid S, Pinner E, Desjardins M, Gros P. Natural resistance to infection with intracellular pathogens: the Nramp1 protein is recruited to the membrane of the phagosome. J Exp Med. 1997;185:717–730. doi: 10.1084/jem.185.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruenheid S, Cellier M, Vidal S, Gros P. Identification and characterization of a second mouse Nramp gene. Genomics. 1995;25:514–525. doi: 10.1016/0888-7543(95)80053-o. [DOI] [PubMed] [Google Scholar]
- 11.Moffett P, Dayo M, Reece M, McCormick MK, Pelletier J. Characterization of msim, a murine homologue of the Drosophila sim transcription factor. Genomics. 1996;35:144–155. doi: 10.1006/geno.1996.0333. [DOI] [PubMed] [Google Scholar]
- 12.Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 13.Fleming MD, Trenor CC, III, Su MA, Foernzler D, Beier DR, Dietrich WF, Andrews NC. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 14.Fleming MD, Romano MA, Su MA, Garrick LM, Garrick MD, Andrews NC. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proc Natl Acad Sci USA. 1998;95:1148–1153. doi: 10.1073/pnas.95.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell ES, Nash DJ, Bernstein SE, Kent EL, McFarland EC, Matthews SM, Norwood MS. Characterization and genetic studies of microcytic anemia in house mouse. Blood. 1970;35:838–850. [PubMed] [Google Scholar]
- 16.Edwards JA, Hoke JE. Defect of intestinal mucosal iron uptake in mice with hereditary microcytic anemia. Proc Soc Exp Biol Med. 1972;141:81–84. doi: 10.3181/00379727-141-36720. [DOI] [PubMed] [Google Scholar]
- 17.Farcich EA, Morgan EH. Diminished iron acquisition by cells and tissues of Belgrade laboratory rats. Am J Physiol. 1992;262:R220–224. doi: 10.1152/ajpregu.1992.262.2.R220. [DOI] [PubMed] [Google Scholar]
- 18.Cellier M, Prive G, Belouchi A, Kwan T, Rodrigues V, Chia W, Gros P. Nramp defines a family of membrane proteins. Proc Natl Acad Sci USA. 1995;92:10089–10093. doi: 10.1073/pnas.92.22.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ausubel, F., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1996. Current Protocols in Molecular Biology. Vol. 2. K. Janssen, editor. John Wiley and Sons Inc., New York. 16.7.1–16.7.5.
- 20.Smith DE, Fisher PA. Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophilaembryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol. 1984;99:20–28. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chow LM, Davidson D, Fournel M, Gosselin P, Lemieux S, Lyu MS, Kozak CA, Matis LA, Veillette A. Two distinct protein isoforms are encoded by ntk, a csk-related tyrosine protein kinase gene. Oncogene. 1994;9:3437–3448. [PubMed] [Google Scholar]
- 22.Pollard JW, Stanners CP. Characterization of cell lines showing growth control isolated from both the wild type and a leucyl-tRNA synthetase mutant of Chinese hamster ovary cells. J Cell Physiol. 1979;98:571–585. doi: 10.1002/jcp.1040980315. [DOI] [PubMed] [Google Scholar]
- 23.Pinner E, Gruenheid S, Raymond M, Gros P. Functional complementation of the yeast divalent cation transporter family SMF by NRAMP2, a member of the mammalian natural resistance-associated macrophage protein family. J Biol Chem. 1997;272:28933–28938. doi: 10.1074/jbc.272.46.28933. [DOI] [PubMed] [Google Scholar]
- 24.Canfield, V.A., and R. Levenson. 1993. Transmembrane organization of the Na,K-ATPase determined by epitope addition. Biochemistry. 32:13782–13786 (erratum published 33: 3142). [DOI] [PubMed]
- 25.Kaufman RJ, Davies MV, Pathak VK, Hershey JWB. The phosphorylation site of eucaryotic initiation factor 2 alters translational efficiency of specific mRNAs. Mol Cell Biol. 1989;9:946–958. doi: 10.1128/mcb.9.3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kast C, Canfield V, Levenson R, Gros P. Membrane topology of P-Glycoprotein as determined by epitope insertion: transmembrane organization of the N-terminal domain of mdr3. Biochemistry. 1995;34:4402–4411. doi: 10.1021/bi00013a032. [DOI] [PubMed] [Google Scholar]
- 27.Devault A, Gros P. Two members of the mouse mdr gene family confer multidrug resistance with overlapping but distinct drug specificities. Mol Cell Biol. 1990;10:1652–1663. doi: 10.1128/mcb.10.4.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 29.Desjardins M, Huber LA, Parton RG, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol. 1994;124:677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Supek F, Supekova L, Nelson H, Nelson N. A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc Natl Acad Sci USA. 1996;93:5105–5110. doi: 10.1073/pnas.93.10.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards JA, Hoke JE. Red cell iron uptake in hereditary microcytic anemia. Blood. 1975;46:381–388. [PubMed] [Google Scholar]
- 33.Edwards JA, Garrick LM, Hoke JE. Defective iron uptake and globin synthesis by erythroid cells in the anemia of the Belgrade laboratory rat. Blood. 1978;51:347–357. [PubMed] [Google Scholar]
- 34.Edwards JA, Sullivan AL, Hoke JE. Defective delivery of iron to the developing red cell of the Belgrade laboratory rat. Blood. 1980;55:645–648. [PubMed] [Google Scholar]
- 35.Farcich EA, Morgan EH. Uptake of transferrin-bound and nontransferrin-bound iron by reticulocytes from the Belgrade laboratory rat: comparison with Wistar rat transferrin and reticulocytes. Am J Hematol. 1992;39:9–14. doi: 10.1002/ajh.2830390104. [DOI] [PubMed] [Google Scholar]
- 36.Garrick MD, Gniecko K, Liu Y, Cohan DS, Garrick LM. Transferrin and the transferrin cycle in Belgrade rat reticulocytes. J Biol Chem. 1993;268:14867–14874. [PubMed] [Google Scholar]
- 37.Bannerman, R.M. 1976. Genetic defects of iron transport. Fed. Proc. 35. 11:2281–2285. [PubMed]
- 38.Ponka P, Beaumont C, Richardson DR. Function and regulation of transferrin and ferritin. Semin Hematol. 1998;35:35–54. [PubMed] [Google Scholar]
- 39.Dautry-Varsat A, Ciechanover A, Lodish HF. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc Natl Acad Sci USA. 1983;80:2258–2262. doi: 10.1073/pnas.80.8.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teter K, Chandy G, Quinones B, Pereyra K, Machen T, Moore HPH. Cellubrevin-targeted fluorescence uncovers heterogeneity in the recycling endosomes. J Biol Chem. 1998;273:19625–19633. doi: 10.1074/jbc.273.31.19625. [DOI] [PubMed] [Google Scholar]
- 41.Hackam DJ, Rotstein OD, Zhang W, Gruenheid S, Gros P, Grinstein S. Host resistance to intracellular infection: mutation of natural resistance–associated macrophage protein 1 (Nramp1) impairs phagosomal acidification. J Exp Med. 1998;188:351–364. doi: 10.1084/jem.188.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sipe DM, Jesurum A, Murphy RF. Absence of Na+,K(+)-ATPase regulation of endosomal acidification in K562 erythroleukemia cells. Analysis via inhibition of transferrin recycling by low temperatures. J Biol Chem. 1991;266:3469–3474. [PubMed] [Google Scholar]
- 43.Yurko MA, Gluck S. Production and characterization of a monoclonal antibody to vacuolar H+ATPase of renal epithelia. J Biol Chem. 1987;262:15770–15779. [PubMed] [Google Scholar]