Abstract
CD4+ T cells play a critical role in generating and maintaining immune responses against pathogens and alloantigens, and evidence suggests an important role for them in antitumor immunity as well. Although major histocompatibility complex class II–restricted human CD4+ T cells with specific antitumor reactivities have been described, no standard method exists for cloning the recognized tumor-associated antigen (Ag). In this study, biochemical protein purification methods were used in conjunction with novel mass spectrometry sequencing techniques and molecular cloning to isolate a unique melanoma Ag recognized by a CD4+ tumor-infiltrating lymphocyte (TIL) line. The HLA-DRβ1*0101–restricted Ag was determined to be a mutated glycolytic enzyme, triosephosphate isomerase (TPI). A C to T mutation identified by cDNA sequencing caused a Thr to Ile conversion in TPI, which could be detected in a tryptic digest of tumor-derived TPI by mass spectrometry. The Thr to Ile conversion created a neoepitope whose T cell stimulatory activity was enhanced at least 5 logs compared with the wild-type peptide. Analysis of T cell recognition of serially truncated peptides suggested that the mutated amino acid residue was a T cell receptor contact. Defining human tumor Ag recognized by T helper cells may provide important clues to designing more effective immunotherapies for cancer.
Keywords: melanoma, antigen, CD4+ T cells, HLA-DR1, triosephosphate isomerase
During the past decade, the field of human tumor immunology has evolved from describing interactions between lymphocytes and tumor cells on a cellular level, to dissecting mechanisms of tumor-specific immune recognition on a molecular and biochemical level. Attention has been focused predominantly on tumor-specific CD8+ T cells, since they exert important antitumor effector functions in many in vivo murine tumor models. Methods have been developed to identify MHC class I–restricted tumor-associated Ag recognized by human CD8+ T cells, through molecular cloning in stable or transient expression systems (1, 2), or through direct mass spectrometric sequencing of peptides eluted from tumor-derived MHC–peptide complexes (3). The availability of these technologies has facilitated a tremendous expansion of our knowledge of commonly expressed as well as mutated tumor-associated Ags recognized by MHC class I–restricted CD8+ T cells derived from cancer patients (for a review, see reference 4). Such knowledge has in turn led to the development of experimental Ag-specific immunotherapies, which have yielded encouraging clinical results (5). However, the partial and transient nature of many of the observed clinical responses indicates a need to develop approaches that recruit additional immunologic effector components.
CD4+ T cells play a central role in generating and maintaining immune responses, through interactions with CD8+ T cells and B cells, and in their capacity as memory cells. There is ample evidence from murine tumor models to suggest an important role for Th cells in regulating CTL-, macrophage-, eosinophil-, and antibody-dependent pathways of antitumor immunity (for a review, see reference 6). In humans, tumor-specific CD4+ T cells have been demonstrated in patients with many different types of cancers (6, 7), and we are just beginning to explore the nature of the specific Ags which are recognized. However, no standard method exists to facilitate this area of study. The molecular methods used successfully in class I systems are not directly applicable, due to the complexity of the class II– restricted Ag processing pathway with its specialized peptide loading component and requirement for accessory molecules. Likewise, the peptide elution strategy is also not readily applied, owing to the necessity for massive quantities of tumor tissue (>1011 cells) for study. We have previously identified tyrosinase as an Ag recognized by melanoma-specific CD4+ T cells from one patient (8, 9); however, this was done in the context of screening candidate Ags already known to be recognized by melanoma-specific CD8+ T cells. Thus, there is a critical need to develop new techniques for isolating and characterizing MHC class II– restricted tumor Ags. Here, we describe the identification of an HLA-DR1–restricted mutated triosephosphate isomerase (TPI)1 epitope recognized by melanoma-specific CD4+ T cells, using a strategy that combines biochemical purification with mass spectrometric sequencing and molecular cloning. This approach uses the exogenous route of Ag processing characteristic of class II systems, for pulsing components of subcellular compartments as well as sequentially purified tumor-derived protein fractions onto APCs for T cell recognition (8, 10, 11).
Materials and Methods
Cell Cultures.
T cell, B cell, and melanoma cell lines were initiated from specimens derived from patient 1558, a 48-yr-old Caucasian male who had a primary cutaneous melanoma lesion resected from the right shoulder 13 yr before the development of metastatic disease in multiple organ systems. Tumor-infiltrating lymphocytes (TIL) were cultured from a subcutaneous metastasis resected from patient 1558 according to methods described (12), and were maintained in RPMI 1640 with 10% human AB serum and recombinant IL-2 (600 IU/ml; Chiron) for up to 120 d without Ag restimulation. Under these conditions, CD4+ T cells selectively proliferated, reaching >97% after 4 wk of culture, and manifested specific recognition of autologous tumor cells which constitutively expressed MHC class II molecules. Melanoma cultures and EBV-transformed B cell lines were established in our laboratory and maintained in RPMI 1640 with 10% FCS (13). To grow large quantities of tumor cells for protein purification, the 1558-mel line, which was initiated from the same metastatic lesion used to generate TIL 1558, was maintained in 10-chamber cell factories (Nunc International). The monocytic leukemia cell line THP-1 (American Type Culture Collection) was grown in RPMI 1640 with 10% FCS; before use as APCs, these cells were cultured in the presence of IFN-γ 100 U/ml for 48–96 h to upregulate expression of MHC class II molecules (14).
HLA Typing.
HLA serotypes and DNA genotypes of fresh PBLs and cell lines were determined by the National Institutes of Health HLA Laboratory, as described (10). The class II genotype of patient 1558 was found to be HLA-DRβ1*0101, 1101; DQβ1*0301, 0501; DRβ3*0202.
Assessment of T Cell Responses to Tumor Cells, Tumor Lysates, Protein Fractions, and Peptides.
To assess recognition of whole melanoma cells, CD4+ TIL 1558 were cultured in flat-bottomed microtiter plates at 2 × 105 cells/well, in the presence of irradiated tumor cells (12,000 rad) at 105/well for 20–24 h (15). To assess T cell recognition of tumor lysates or fractionated tumor- derived proteins, EBV-B cells or IFN-γ–treated THP-1 cells were used as APCs. APCs cultured at 1.5 × 105 cells/well were pulsed for 16–24 h with freeze–thaw lysates of tumor cells (1–2 × 105 cell equivalents/well) or fractionated proteins derived from tumor cells (2 × 105 to 8 × 107 cell equivalents/well), after which T cells were added for 20 h (10). Peptide recognition was tested by pulsing APCs with peptides for 16–24 h, after which T cells were added to the assay for 20 h (9). Culture supernatants were then harvested, and GM-CSF secretion by CD4+ T cells was measured using a commercially available ELISA kit (R&D Systems; detection limit 8 pg/ml). In some experiments, anti-MHC mAbs were used to inhibit specific Ag recognition by T cells (10). Antibodies included L243 (against HLA-DR; IgG2a), IVA12 (HLA-DR, DP, ?DQ; IgG1), Genox 3.53 and G2b.2 (HLA-DQw1, which includes the DQβ1*0501 genotype; IgG1 and IgG2b, respectively), IVD12 (HLA-DQw3; IgG1), and W6/ 32 (HLA-A, B, C; IgG2a) (all purified from American Type Culture Collection hybridoma supernatants).
Preparation of Cell-free Extracts.
A total of 1.2 × 109 adherent 1558-mel cells were harvested from cell factories with trypsin/ EDTA, and cell pellets were washed twice in ice-cold PBS and stored dry at −70°C. Cell homogenization was achieved by suspending the cells in ice-cold sucrose buffer (150 mM sucrose, 2.5 mM dithiothreitol, 4 mM 4-[2-aminoethyl]-benzenesulfonyl fluoride hydrochloride) at a concentration of 108 cells/ml and disrupting them in a Wheaton Potter-Elvehjem tissue grinder. All successive purification steps were carried out on ice or at 4°C. Whole nuclei were separated from cytoplasm and membranes by centrifugation at 800 g for 10 min. Salts and protease inhibitors were subsequently added to minimize protein degradation in the supernatant. The mixture was adjusted to contain 50 mM Tris-HCl, 150 mM NaCl, 10% glycerol, 1 mM benzamidine, and 2.5 mM Na-EDTA, pH 7.5. Leupeptin, pepstatin, antipain, aprotinin, and chymostatin were added at 25 μg/ml homogenate. The cytosolic fraction was recovered by a high-speed centrifugation step (31,500 g for 60 min). The pellets which were recovered constituted the membrane fraction. The supernatant (cytosol) underwent addition of polyethyleneimine (PEI) 0.08% and centrifugation to precipitate nucleic acids, followed by a three-step fractionated ammonium sulfate precipitation saturating to 28, 52, and 75% with (NH4)2SO4. These fractions were dissolved in buffer A (25 mM Tris-HCl, pH 8.0, 25 mM NaCl, 2 mM dithiothreitol, 1 mM benzamidine, 1 mM Na-EDTA) and stored at −70°C if not used directly for further purification. A portion of each subcellular fraction was dialyzed against PBS for testing in bioassays.
Chromatographic Purifications.
The cytosolic fraction saturated to 75% with (NH4)2SO4 was desalted on Sephadex G25 M columns with buffer A (described above) and loaded onto an anion exchange chromatography column (HiTrap Q, 5 ml). Protein was eluted with an increasing NaCl gradient in buffer A (0.25 mM to 1 M NaCl). Portions of the eluted protein fractions were concentrated, equilibrated in PBS on Microcon-30 membrane units (Amicon, Inc.), and tested for T cell recognition. Bioactive fractions were then pooled and concentrated on a Centriprep-30 membrane unit (Amicon, Inc.) to ∼2 mg protein/ml, and loaded onto a Superdex 75 HiLoad 16/60 gel filtration column. After testing individual eluted fractions for bioactivity, active fractions were pooled and concentrated to ∼1 mg protein/ml. For hydrophobic interaction chromatography, the solution was adjusted to 1.5 M (NH4)2SO4 in buffer A by overnight dialysis at 4°C. A decreasing 1.5–0 M (NH4)2SO4 gradient in buffer A was performed on a Resource Phe hydrophobic interaction column to yield an active protein fraction with sufficient purity for sequence analysis. All chromatography columns were purchased from Amersham Pharmacia Biotech and applied on the AKTAexplorer liquid chromatography system (Amersham Pharmacia Biotech). Individual eluate fractions were analyzed by SDS-PAGE on 4–20% acrylamide Tris-glycine precast gels (Novex) stained with Coomassie blue (Novex) or silver stain (Bio-Rad Laboratories).
Nano-HPLC Microelectrospray Ionization Mass Spectrometry and Database Searching.
A bioactive purified protein fraction eluted from the hydrophobic interaction column was equilibrated in 50 mM ammonium carbonate, pH 7.8, using Microcon-30 membrane concentrators, for sequence analysis. 5 μg of protein was digested with 0.5 μg of modified trypsin (Promega) in a 50-μl vol at room temperature for 19 h. The digest was frozen at –35°C until analyzed. An aliquot containing ∼25 ng of digested protein was analyzed by nano-HPLC microelectrospray ionization (μESI) mass spectrometry (MS). All mass spectrometric analyses were performed on an LCQ ion trap mass spectrometer (Finnigan). The ESI head supplied with the instrument was removed and replaced with a μESI source. The ESI voltage (1.5–1.8 kV) was applied via a stainless steel union placed on the HPLC pump side of the reversed phase column. The μESI tips were made from 360-μm outer diameter (OD), 75-μm inner diameter (ID) fused silica capillary (Polymicro Technologies) pulled to ∼20-μm OD, 5-μm ID using a P-2000 laser puller (Sutter Co.). The μESI tips were connected directly to a nano-HPLC column using 350-μm ID Teflon tubing (Zues). Nano-HPLC columns were prepared by packing ∼10 cm of 5-μm C18 material (YMC Corp.) inside 360-μm OD, 75-μm ID fused silica capillary. Solvents A and B for HPLC were 0.1 M acetic acid in water and 0.1 M acetic acid in 70% acetonitrile. The HPLC gradient increased from 1 to 35% acetonitrile in 11 min. Variable flow chromatography (Settlage, R.E., R.E. Christian, J. Shabanowitz, and D.F. Hunt, manuscript submitted for publication; patent pending), technology in which the mass spectrometer data system controls the rate of sample introduction, was used for the analysis of the protein digest. Flow rate for mobile phase into the mass spectrometer was set at ∼200 nl/min at the beginning of the gradient, switched to <50 nl/min shortly before peptides began to elute, and returned to 200 nl/min once peptides were no longer detected. LCQ software programs used in the above experiment included those for data-dependent data acquisition, dynamic exclusion, and isotope exclusion. For data-dependent analysis, the instrument was programmed to continuously cycle through six scan events. Scan event 1 was an MS scan from 300 to 2,000 m/z. Scan events 2–6 were MS/MS scans on the five most abundant ions detected in scan event 1. The dynamic exclusion settings were as follows: 6 amu = exclusion width; 2 min = exclusion duration; 30 s = preexclusion duration. The isotope exclusion window was set to 4 amu. MS/MS spectra were analyzed using the SEQUEST (16, 17) search program, a database search algorithm capable of searching thousands of MS/MS spectra consecutively and automatically. SEQUEST compares experimental MS/MS spectra with theoretical MS/MS spectra of peptides derived from proteins in protein databases. Peptides from the protein databases are scored and ranked based on the similarity between the experimental and theoretical spectra. SEQUEST settings were as follows: parent ion mass tolerance = 2.0 amu; fragment ion mass tolerance = 0.4 amu; enzyme = trypsin; group scans = 40 scans; group scan parent tolerance = 0.5 amu; and no amino acid modifications.
Expression Cloning of TPI.
Total RNA was prepared from 1558-mel and 1558-EBV B cells using the Trizol reagent (GIBCO BRL), and mRNA was subsequently prepared from 1558-mel (PolyATract mRNA Isolation Systems; Promega). First-strand cDNA was synthesized (Superscript Preamplification System; GIBCO BRL) followed by PCR amplification of TPI (sequence data available from EMBL/GenBank/DDBJ under accession no. M10036). The 5′ oligonucleotide primer, 5′-AAAGGATCCTCGGCTCGGCCATGGCGCCCTCCAGGAAGTT-3′, contained an engineered BamHI restriction site at the 5′ end (initiation codon in bold type). The 3′ primer, 5′-AAAGGTACCTTA CTTGTCATCGTCATCCTTGTAGTC TTGTTTGG-CATTGATGAT-3′, contained an engineered KpnI site as well as a sequence encoding the 8-mer FLAG epitope DYKDDDDK allowing for mAb-mediated detection of TPI on Western blots (anti-FLAG M2 mAb; Sigma) (FLAG nucleotide sequence underlined, stop codon in bold type). Oligonucleotide primers were synthesized using an ABI 392 DNA/RNA synthesizer (Applied Biosystems). The resulting PCR fragment was ligated into the expression vector pCR3.1 (Eukaryotic TA Cloning Kit; Invitrogen), and ligation products were used to transform DH5-α Escherichia coli (GIBCO BRL). Bacterial colonies harboring recombinant plasmids with correct sequence orientation were detected by PCR. Plasmids were purified from bacterial clones using the QIAprep Spin Miniprep Kit (QIAGEN, Inc.). DNA sequencing of TPI inserts was performed with T7 forward and pCR3.1 reverse primers, or with the TPI sequence-specific primers described above, using the Big Dye Terminator Cycle Sequencing Kit (Perkin-Elmer/ABI). Sequences were determined with an ABI Prism 310 Genetic Analyzer (Perkin-Elmer). To test CD4+ T cell recognition of TPI, plasmids were transfected transiently into COS-7 cells (gift of Warren Leonard, National Institutes of Health) for 72 h with Lipofectamine Plus (GIBCO BRL), after which cell lysates were prepared with repeated freeze–thaw cycles for pulsing onto APCs.
Peptide Synthesis.
Peptides were synthesized and analyzed for sequence and purity as described (9).
Results
HLA-DR1–restricted CD4+ T Cell Recognition of a Unique Autologous Tumor Ag.
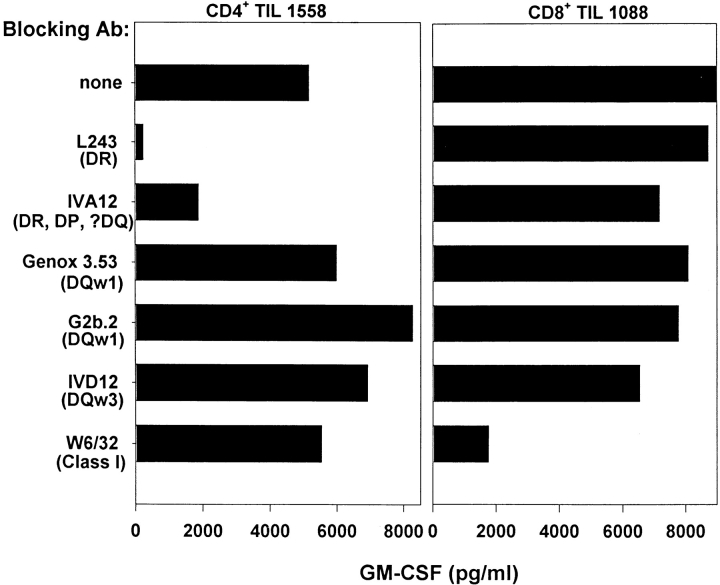
Most melanoma TIL cultures propagated in IL-2 under standard culture conditions will become predominantly CD8+ after 3–4 wk (18); however, TIL 1558 were >97% CD4+ by 26 d. When stimulated with MHC class II+ whole autologous tumor cells, either fresh or cultured, these TIL specifically secreted GM-CSF, IFN-γ, and TNF-α, but not IL-2 or IL-4. As shown in Fig. 1, TIL recognition of cultured 1558-mel, as measured by GM-CSF secretion, was inhibited by mAb directed against HLA-DR but not HLA-DQ or MHC class I molecules. TIL 1558 also recognized autologous EBV-B cells pulsed with lysates of 1558-mel, and mAb blocking experiments similar to that shown in Fig. 1 confirmed HLA-DR restriction. By using allogeneic B cell lines with known HLA types to process and present tumor lysate, it was determined that the operational restriction element was HLA-DRβ1*0101 (data not shown). The monocytic leukemia cell line THP-1, which expresses the DRβ1*0101 molecule, proved to have enhanced processing capabilities for the 1558-mel Ag relative to EBV-B cell lines, and so was used as the APCs for tumor lysates and protein fractions in all subsequent experiments.
Figure 1.
Melanoma-specific CD4+ TIL 1558 are HLA-DR restricted. TIL 1558 were coincubated with whole autologous 1558-mel cells for 20 h, in the absence or presence of anti-MHC mAb, and specific Ag recognition was measured by GM-CSF secretion. As a control for mAb blocking, the MART-1–specific, HLA-A2–restricted CD8+ TIL 1088 line was cocultured with its autologous 1088-mel. CD4+ TIL 1558 were inhibited by mAbs directed against HLA-DR (L243) and a monomorphic class II determinant (IVA12), whereas the CD8+ TIL were inhibited only by an anti–class I mAb (W6/32).
Although CD4+ TIL 1558 recognized autologous tumor cells, they failed to react against autologous normal cells, including cultured fibroblasts, EBV-B cells, and fresh PBLs. To assess whether the melanoma Ag recognized by TIL 1558 was commonly expressed among other human tumors, lysates were prepared from 45 different tumors for processing by THP-1 cells and presentation to TIL. CD4+ TIL 1558 were not stimulated by any lysates other than those derived from fresh or cultured autologous melanoma cells, including 19 allogeneic cultured melanomas, 7 fresh melanomas, 5 colon carcinomas, 2 breast carcinomas, 3 prostate carcinomas, 1 pancreatic carcinoma, 4 sarcomas, and 4 EBV-B cell lines (data not shown). These findings suggested that the Ag recognized by TIL 1558 was in fact a neoantigen resulting from a genetic mutation unique to the autologous melanoma.
Identification of the Specific Tumor Ag Recognized by CD4+ TIL 1558.
The ability of TIL 1558 to recognize the 1558-mel Ag when processed by APCs through the exogenous MHC class II pathway afforded an opportunity to apply a protein purification strategy for Ag identification. Through this approach, melanoma cells could be reduced to subcellular compartments and then to sequentially purified protein fractions, which could be assayed for bioactivity by feeding them to APCs for T cell recognition. Importantly, since the recognized protein Ag would be processed into smaller peptide fragments by APCs, it would not be necessary to preserve tertiary protein structure during the purification procedure.
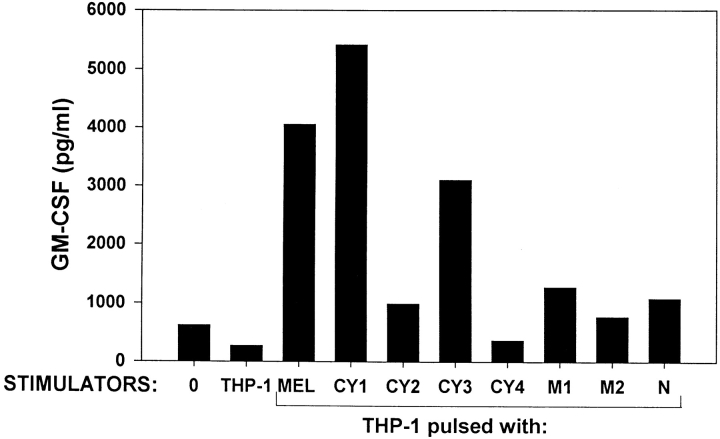
Melanoma cells disrupted by douncing in the presence of protease inhibitors were separated into nuclear, membrane, and cytosolic fractions by differential centrifugation. The cytosol was further fractionated by a stepwise (NH4)2SO4 precipitation, and each fraction was pulsed onto APCs for T cell recognition. As shown in Fig. 2, the cytosol provided a much stronger stimulus for melanoma-specific T cells than the membrane or nuclear fractions, and most of the cytosolic activity precipitated at 75% (NH4)2SO4 saturation. This finding was reproduced in two similar experiments. Titration experiments demonstrated that the 75% (NH4)2SO4 precipitate recovered from the cytosol was 5–25 times more stimulatory than the other fractions tested, although the protein content of the various fractions differed only by up to twofold (not shown). None of the 1558-mel fractions were capable of stimulating CD4+ TIL from another patient, and a cytosolic fraction prepared in a similar manner from a renal cell carcinoma line failed to stimulate TIL 1558. Thus, TIL 1558 seemed to recognize a cytosolic (soluble) protein unique to 1558-mel.
Figure 2.
CD4+ TIL 1558 recognize a cytosolic Ag. THP-1 cells (1.5 × 105 cells/well) were used as APCs for an unfractionated freeze–thaw lysate of 1558-mel, or for various subcellular fractions (2 × 105 cells equivalents/well) in a microtiter assay. CY1, unfractionated cytosol; CY2, cytosol precipitated at 50% (NH4)2SO4 saturation; CY3, cytosol precipitated at 75% (NH4)2SO4; CY4, proteins solubilized in 0.5 M KCl from a polyethyleneimine (PEI) precipitate of CY1; M1, membrane-bound proteins solubilized in 0.5 M KCl; M2, membrane proteins solubilized in 2% CHAPS detergent; N, nuclear fraction. T cell reactivity against protein-pulsed APCs was measured by GM-CSF secretion.
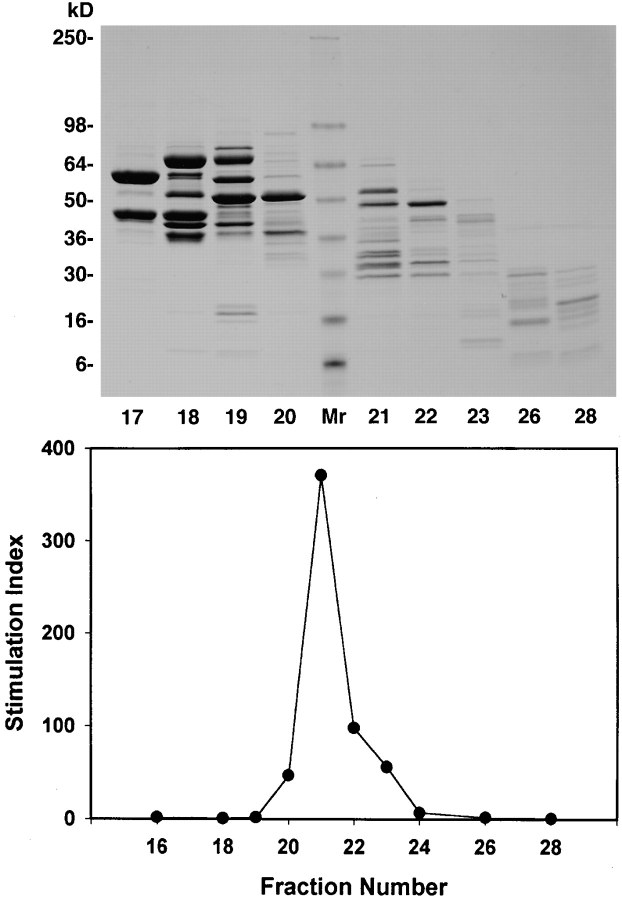
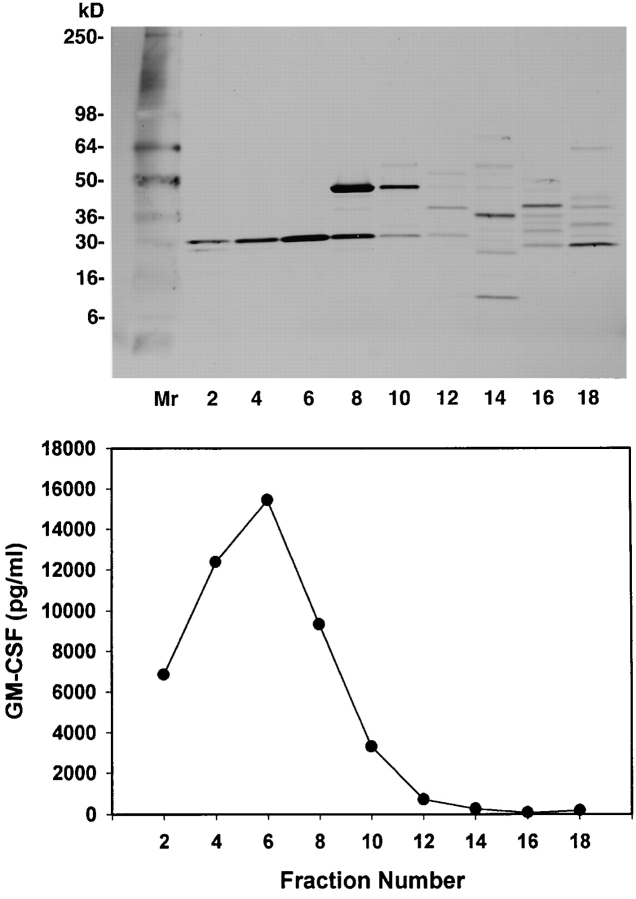
To isolate the 1558-mel Ag, the active cytosolic fraction was purified through a series of chromatographic separations. First, proteins that precipitated at 75% (NH4)2SO4 saturation were dissolved in buffer and applied to an anion exchange column. When individual fractions eluted from the column were pulsed onto APCs for T cell recognition, it was found that the bioactivity remained in the column run-through, and therefore the protein of interest had failed to bind to the column. However, SDS-PAGE analysis of all of the eluate fractions indicated that, although the bioactive run-through contained a complex mixture of proteins, many other proteins which lacked bioactivity had bound and separated adequately on the column (not shown); thus, a significant purification of the specific tumor Ag had been achieved. Next, the bioactive material recovered after anion exchange chromatography was separated according to molecular size on a gel filtration column. The results of testing individual fractions for T cell recognition are shown in Fig. 3, demonstrating a peak of bioactivity in eluate fractions 20–23, with GM-CSF secretion by TIL in response to fraction 21 reaching 371-fold above background secretion (29,300 compared with 79 pg/ml). Although a significant purification was achieved at this step, SDS-PAGE indicated that the bioactive fractions still contained a complex mixture of proteins. The final chromatographic purification was performed by combining the bioactive gel filtration fractions and applying them to a hydrophobic interaction column. Fig. 4 shows that this procedure yielded two highly bioactive fractions, numbers 4 and 6. Each appeared to contain a single protein band of ∼30 kD on SDS-PAGE.
Figure 3.
Purification of the 1558-mel Ag by size exclusion chromatography. Gel filtration chromatography was used, after anion exchange chromatography, to separate proteins derived from the 1558-mel cytosol according to size. SDS-PAGE of the eluted protein fractions is shown (gel stained with Coomassie blue), as well as the results of the corresponding bioassay. T cell recognition of APCs pulsed with individual protein fractions (4 × 107 cell equivalents/well) is shown as stimulation index, the fold specific secretion of GM-CSF above background secretion (T cells plus unpulsed APCs).
Figure 4.
Purification of the 1558-mel Ag by hydrophobic interaction chromatography. Individual eluate fractions (5 × 107 cell equivalents/ well) were pulsed onto APCs for T cell recognition. SDS-PAGE (silver stain) shows that bioactive fractions 4 and 6 appear to contain a single protein band of ∼30 kD. The protein in fraction 6 was sequenced via mass spectrometric analysis, identifying it as TPI. This was confirmed by NH2-terminal Edman degradation sequencing of fraction 4.
Fraction 6, which was eluted from the hydrophobic interaction column, was subjected to tryptic cleavage followed by nano-HPLC μESI ion trap MS. Using data- dependent data acquisition, dynamic exclusion, isotope exclusion, and SEQUEST database searching, 19 peptide sequences were obtained, representing a total of 187 amino acids, and all were identified as nonmutated human TPI. In addition, eluate fraction 4 was subjected to NH2-terminal sequencing with Edman degradation, yielding an 11–amino acid sequence corresponding to residues 2–12 of TPI. No other proteins were identified in these fractions. The complete TPI sequence (GenBank accession no. M10036) contains 249 amino acids with a calculated molecular mass of 26,700 daltons, corresponding to the observed mass of the 1558-mel Ag on SDS-PAGE.
Characterization of the Mutated TPI Epitope Recognized by CD4+ TIL 1558.
To confirm that TPI was indeed the tumor-specific Ag recognized by TIL 1558, cDNA encoding TPI was amplified from mRNA derived from 1558-mel, the cell line which stimulated CD4+ TIL, as well as from the autologous B cell line 1558-EBV, which did not stimulate TIL. TPI was cloned into a eukaryotic expression vector, and recombinant plasmids derived from individual bacterial colonies were transiently transfected into COS-7 cells. COS transfectants expressing TPI protein were then pulsed onto APCs as freeze–thaw lysates, for T cell recognition.
Table I shows the results of testing individual TPI clones derived from 1558-mel for T cell recognition. TPI encoded by three clones (designated 1B, 5B, and 11G) stimulated T cells, as manifested by GM-CSF secretion, while three other TPI clones were not stimulatory, nor was a β-actin clone used as a negative control. Western blotting with the anti-FLAG mAb, directed against a specific 8-mer sequence fused to the COOH terminus of each of these proteins, confirmed that all recombinant proteins were expressed in COS-7 cells at comparable levels (not shown). DNA sequencing demonstrated that the three TPI clones encoding a product recognized by T cells shared a missense mutation at nucleotide position 450, consisting of a C to T substitution. The effect of this mutation was to convert the wild-type threonine (ACT) at amino acid position 28 to an isoleucine (ATT). TPI clones 4B, 12A, and 12F, also derived from 1558-mel, did not contain this mutation and were not recognized by T cells. As an incidental finding, all six TPI clones derived from 1558-mel shared two silent nucleotide substitutions, at positions 652 (A to G) and 856 (C to G), presumed to represent DNA polymorphisms. Thus, the six TPI clones obtained from 1558-mel represented two alleles, one wild-type, the other containing a unique mutation. The nucleotide substitution at position 450 was not identified in six clones derived from 1558-EBV, suggesting that this was indeed a somatic mutation which had occurred in the tumor, and not an allelic polymorph.
Table I.
Recognition of Mutated TPI by CD4+ TIL 1558
| Stimulatorcell lysate |
Transfected gene |
cDNA sequence,nucleotide 450 |
GM-CSF | |||
|---|---|---|---|---|---|---|
| pg/ml | ||||||
| 1558-mel | None | T, C | 6,315 | |||
| 1558-EBV | None | C | <8 | |||
| COS-7 | TPI-1B | T | 3,485 | |||
| COS-7 | TPI-5B | T | 8,310 | |||
| COS-7 | TPI-11G | T | 6,340 | |||
| COS-7 | TPI-4B | C | 8 | |||
| COS-7 | TPI-12A | C | 11 | |||
| COS-7 | TPI-12F | C | 11 | |||
| COS-7 | β-actin | n.a. | <8 |
COS-7 cells were transfected for 4 d with the indicated genes. Cell lysates were then incubated with THP-1 monocytic leukemia cells for 20 h, after which TIL were added for 20 h. GM-CSF secretion by TIL was measured by ELISA. n.a., not applicable.
To confirm that the mutated TPI protein predicted by DNA sequencing was in fact present in 1558-mel cells, a novel mass spectrometric sequence analysis computer program was applied, which generated ∼5,000 mutant TPI sequences that differed from the wild-type sequence by a single amino acid. The MS/MS data from the TPI digest was then searched against the 5,000 mutant TPI sequences using SEQUEST programmed with the same settings as described in Materials and Methods. This experiment detected a new tryptic peptide corresponding to TPI residues 20–33, in which Thr at position 28 was substituted with either Ile or Leu. No other mutant peptides were detected in this analysis.
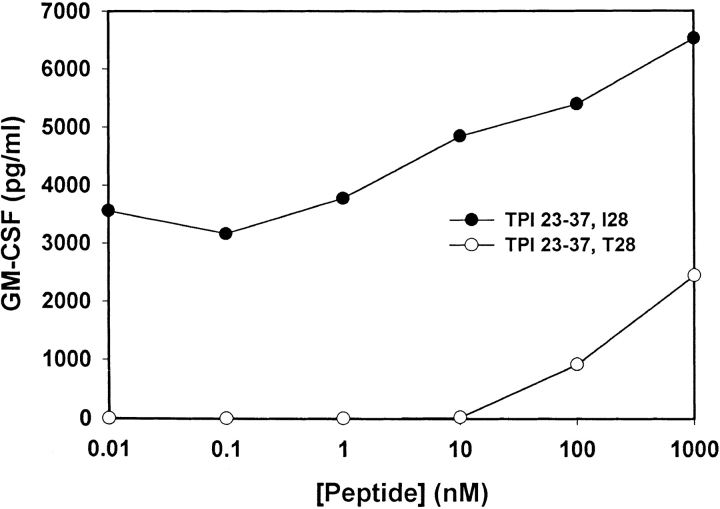
To further investigate the effect of the TPI mutation on T cell reactivity, peptides encompassing the site of mutation were synthesized and assessed for recognition by TIL 1558. A wild-type 15-mer peptide, TPI 23–37, was synthesized with the sequence GELIGTLNAAKVPAD, as well as a mutant peptide containing Ile instead of Thr at position 28 (TPImut). As shown in Fig. 5, T cell recognition of the mutated peptide was enhanced at least 5 logs compared with the wild-type. Also of note, subnanomolar concentrations of the mutant peptide were recognized consistently, reminiscent of the extremely sensitive CD4+ T cell responses typically observed against foreign microbial epitopes. This is in contrast to the CD4+ T cell recognition of nonmutated melanoma-associated tyrosinase epitopes described previously (9), which required peptide concentrations of 10 μM or more for half-maximal stimulation.
Figure 5.
Enhanced recognition of a mutated TPI peptide by CD4+ TIL 1558. 15-mer peptides containing a wild-type (open circles) or mutated (filled circles) sequence were pulsed onto THP-1 cells for recognition by T cells. T cell secretion of GM-CSF in response to peptide stimulation was measured by ELISA. Background secretion of GM-CSF by TIL cultured with THP-1 cells in the absence of peptide was undetectable (<8 pg/ml).
Finally, we wished to determine the immunologic function of the TPI mutation; that is, whether the mutated amino acid constituted an MHC-binding residue or a TCR contact. A substantial amount of information is available concerning the binding motif of peptides complexed to HLA-DRβ1*0101, which is the restriction element for TPImut 23–37 as demonstrated by mAb blocking studies and peptide presentation by allogeneic HLA-compatible APCs (not shown). The crystal structure of this HLA molecule complexed to a high-affinity peptide, influenza HA 306– 318, reveals that the peptide binding site has one large hydrophobic pocket accommodating the P1 peptide anchor residue, which is the major determinant of peptide binding (19). The crystal structure of HLA-DRβ1*0101 also demonstrates shallower, less hydrophobic pockets accommodating the P4, P6, P7, and P9 peptide anchor residues. Inspection of the sequences of peptides naturally binding to this MHC allele (20), as well as monitoring the effects of substituted analogues and truncated peptides on MHC binding (21), have confirmed the overriding importance of the hydrophobic P1 anchor and lesser roles for the other four anchor residues. In the case of the TPImut 23–37 peptide, potential P1 anchors include the hydrophobic residues Leu 25 and Ile 26, and the mutation itself, Ile 28. A series of truncated peptides were tested for T cell recognition, and as shown in Table II, Leu 25 was unimportant whereas Ile 26 was required for stimulating CD4+ TIL 1558. If Ile 26 were indeed the P1 anchor, then Val 34 would be the P9 anchor, and serial truncations at the COOH terminus of TPImut 23–37 confirmed that peptides lacking the Val 34 residue were not recognized. These data identify the 9–amino acid MHC binding core of the mutated TPI epitope as extending from Ile 26 to Val 34, and suggest that the site of mutation, Ile 28, is a TCR contact, as it resides in the P3 position. For peptides binding to HLA-DRβ1*0101, the role of the P3 residue as a TCR contact has been shown by crystal structure as well as by studies of substituted synthetic analogues which competitively inhibit TCR engagement (22, 23).
Table II.
Determining the PI Anchor Residue for TPImut 23–37
| TPImut residues | Sequence | GM-CSF | ||
|---|---|---|---|---|
| pg/ml | ||||
| 23–37 | G E L I G I L N A A K V P A D | 25,390 | ||
| 24–37 | E L I G I L N A A K V P A D | 28,460 | ||
| 25–37 | L I G I L N A A K V P A D | 11,750 | ||
| 26–38 | I G I L N A A K V P A D T | 7,210 | ||
| 27–39 | G I L N A A K V P A D T E | 38 | ||
| 28–40 | I L N A A K V P A D T E V | 22 | ||
| 29–41 | L N A A K V P A D T E V V | 34 | ||
| 20–34 | Q S L G E L I G I L N A A K V | 5,960 | ||
| 20–33 | Q S L G E L I G I L N A A K | 35 | ||
| 20–32 | Q S L G E L I G I L N A A | 25 | ||
| 20–31 | Q S L G E L I G I L N A | 24 | ||
THP-1 cells pretreated with IFN-γ were pulsed overnight with the indicated peptides at 100 nM, then T cells were added for 20 h. GM-CSF secretion by T cells was measured by ELISA. Background secretion of T cells plus THP-1 without peptide = 28 pg/ml.
Discussion
This study describes the isolation and characterization of a mutated MHC class II–restricted human tumor Ag through a multimodality approach that combines biochemical fractionation, mass spectrometric sequencing, and molecular cloning. Monach et al. have used a similar strategy to identify a class II–restricted murine tumor Ag (11); to our knowledge, this report is the first successful human application. This approach to Ag identification is uniquely suited to class II systems, in which Ag pulsed exogenously onto APCs can access the endosomal processing compartment (24). Since the end-point bioassay measures the presence of a processed peptide rather than an intact protein, the biochemical purification procedures need not preserve native tertiary structure. This represents a convenient advantage relative to traditional applications of similar protein purification schemes, such as those used in enzymology. Another innovation is the use of MS to sequence the purified protein. The combination of HPLC and ion trap MS has made it possible to sequence peptides at the 20–50 amol level. This technique is particularly well suited for sequencing proteins in complex mixtures and for characterizing posttranslational modifications. These features render mass spectrometric sequencing eminently suited for situations in which limited quantities of material are available, and for subtractive approaches comparing the constituents of two or more partially purified protein fractions with known bioactivities.
The protein containing the mutation recognized by CD4+ TIL 1558, TPI (EC 5.3.1.1), is an enzyme that catalyzes the interconversion of dihydroxyacetone phosphate and glyceraldehyde 3-phosphate, a critical step for generating energy in the glycolytic pathway. Its sequence is highly conserved among species, with ∼50% amino acid homology between E. coli and humans, and human TPI is considered to have evolved into a “perfect enzyme,” with one of the highest catalytic rates known (25). TPI is found among the dozen or so predominant proteins in two-dimensional gel databases generated from human tumors (26–28), which is not surprising in view of its energy-generating function as well as its link to nucleotide synthesis via the pentose phosphate shunt. Studies of thousands of individuals have failed to detect allelic variants of TPI (29). The several described mutant forms of the enzyme are all linked to human deficiency diseases; mutations at nucleotide position 450, associated with 1558-mel, have not been reported. However, there is evidence for a causal relationship between UV irradiation, an agent implicated in generating melanomas in vivo, and C to T nucleotide substitutions such as occurred in 1558-mel (30). Indeed, C to T mutations have been shown to generate MHC class I–restricted neoantigens recognized by CD8+ T cells from two melanoma patients. In those two instances, functional mutations in cyclin-dependent kinase 4 (31) and β-catenin (32) may have conferred an oncogenic advantage on the tumor cells, which could have counterbalanced or outweighed the negative effect of disclosing the tumor to immune recognition. The crystal structure of TPI indicates that Thr 28, the site of the 1558-mel mutation, is not located at an active catalytic site (33); in fact, deliberate attempts to enhance the function of TPI through site-directed mutagenesis of catalytic site residues have not been successful (34). However, one might speculate that the nonconservative Thr28Ile mutation could impact on the tertiary structure of the enzyme and hence its catalytic function, and the potential role of this mutation in enhancing TPI function will be explored in future studies.
By immunohistochemical analysis, the subcutaneous melanoma metastasis from which TIL 1558 were derived expressed the shared immunogenic melanoma-associated proteins tyrosinase, gp100, and melanoma antigen recognized by T cells (MART)-1 (35). Nevertheless, CD4+ TIL 1558 failed to react against these commonly expressed, nonmutated Ags, instead responding to a unique neoantigen. These TIL are representative of the majority of MHC class II–restricted melanoma-specific CD4+ T cells analyzed from nine patients, among which two recognized the commonly expressed nonmutated tyrosinase Ag, while the rest reacted against unique tumor-specific Ag (8, 10, 36, and our unpublished data). The apparent dichotomy between melanoma Ag recognized by CD4+ versus CD8+ T cells (unique versus shared Ag) may reflect events occurring during T cell development, with selective deletion of CD4+ but not CD8+ T cells reactive with lineage-specific “self” Ag expressed in melanomas. Since melanocytes do not normally reside in the thymus during development, their Ags would be represented there by migrating APCs which had ingested and processed these Ags, preferentially leading to presentation on MHC class II molecules and deletion of class II–restricted Th cells. Thus, while it is important to identify shared tumor-associated Ags recognized by both CD4+ and CD8+ T cells for devising more effective immunotherapies, it is also necessary to define optimally potent Ags which can mediate tumor rejection. These may not be one and the same, and immunotherapies incorporating unique neoantigens may be required to realize the full potential of this treatment modality.
Acknowledgments
The authors are grateful to Phillipa Marrack for insights into T cell development, John Wunderlich for providing TIL 1558, Paul Wingfield and Pat Spinella for Edman sequencing, Maria Parkhurst and Ellen Fitzgerald for peptide synthesis, Yong Li for DNA sequencing, and Paul Robbins for helpful discussions. We also thank Drew Pardoll for ideas and critical reading of the manuscript.
Footnotes
Abbreviations used in this paper: ID, inner diameter; MART, melanoma antigen recognized by T cells; μESI, microelectrospray ionization; MS, mass spectrometry; OD, outer diameter; TIL, tumor-infiltrating lymphocyte(s); TPI, triosephosphate isomerase.
References
- 1.Van Der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van Den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 2.Brichard V, Van Pel A, Wölfel T, Wölfel C, De Plaen E, Lethé B, Coulie P, Boon T. The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1993;178:489–495. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox AL, Skipper J, Chen Y, Henderson RA, Darrow TL, Shabanowitz J, Engelhard VH, Hunt DF, Slingluff CL., Jr Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science. 1994;264:716–719. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 4.Robbins PF, Kawakami Y. Human tumor antigens recognized by T cells. Curr Opin Immunol. 1996;8:628–636. doi: 10.1016/s0952-7915(96)80078-1. [DOI] [PubMed] [Google Scholar]
- 5.Rosenberg SA, Yang JC, Schwartzentruber DJ, Hwu P, Marincola FM, Topalian SL, Restifo NP, Dudley ME, Schwarz SL, Spiess PJ, et al. Immunologic and therapeutic evaluation of a synthetic peptide vaccine for the treatment of patients with metastatic melanoma. Nat Med. 1998;4:321–327. doi: 10.1038/nm0398-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardoll DM, Topalian SL. The role of CD4+T cell responses in antitumor immunity. Curr Opin Immunol. 1998;10:588–594. doi: 10.1016/s0952-7915(98)80228-8. [DOI] [PubMed] [Google Scholar]
- 7.Topalian SL. MHC class II restricted tumor antigens and the role of CD4+T cells in cancer immunotherapy. Curr Opin Immunol. 1994;6:741–745. doi: 10.1016/0952-7915(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 8.Topalian SL, Rivoltini L, Mancini M, Markus NR, Robbins PF, Kawakami Y, Rosenberg SA. Human CD4+ T cells specifically recognize a shared melanoma-associated antigen encoded by the tyrosinase gene. Proc Natl Acad Sci USA. 1994;91:9461–9465. doi: 10.1073/pnas.91.20.9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Topalian SL, Gonzales MI, Parkhurst M, Li UF, Southwood S, Sette A, Rosenberg SA, Robbins PF. Melanoma-specific CD4+T cells recognize nonmutated HLA-DR–restricted tyrosinase epitopes. J Exp Med. 1996;183:1965–1971. doi: 10.1084/jem.183.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Topalian SL, Rivoltini L, Mancini M, Ng J, Hartzman RJ, Rosenberg SA. Melanoma-specific CD4+T lymphocytes recognize human melanoma antigens processed and presented by Epstein-Barr virus-transformed B cells. Int J Cancer. 1994;58:69–79. doi: 10.1002/ijc.2910580113. [DOI] [PubMed] [Google Scholar]
- 11.Monach PA, Meredith SC, Siegel CT, Schreiber H. A unique tumor antigen produced by a single amino acid substitution. Immunity. 1995;2:45–59. doi: 10.1016/1074-7613(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 12.Topalian SL, Muul LM, Solomon D, Rosenberg SA. Expansion of human tumor infiltrating lymphocytes for use in immunotherapy trials. J Immunol Methods. 1987;102:127–141. doi: 10.1016/s0022-1759(87)80018-2. [DOI] [PubMed] [Google Scholar]
- 13.Topalian SL, Solomon D, Rosenberg SA. Tumor-specific cytolysis by lymphocytes infiltrating human melanomas. J Immunol. 1989;142:3714–3725. [PubMed] [Google Scholar]
- 14.Yunis JJ, Band H, Bonneville F, Yunis EJ. Differential expression of MHC class II antigens in myelomonocytic leukemia cell lines. Blood. 1989;73:931–937. [PubMed] [Google Scholar]
- 15.Schwartzentruber DJ, Topalian SL, Mancini M, Rosenberg SA. Specific release of granulocyte-macrophage colony-stimulating factor, tumor necrosis factor-α, and IFN-γ by human tumor-infiltrating lymphocytes after autologous tumor stimulation. J Immunol. 1991;146:3674–3681. [PubMed] [Google Scholar]
- 16.Eng J, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 17.Yates JR, III, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in a protein database. Anal Chem. 1995;8:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 18.Schwartzentruber DJ, Hom SS, Dadmarz R, White DE, Yannelli JR, Steinberg SM, Rosenberg SA, Topalian SL. In vitro predictors of therapeutic response in melanoma patients receiving tumor-infiltrating lymphocytes and interleukin-2. J Clin Oncol. 1994;12:1475–1483. doi: 10.1200/JCO.1994.12.7.1475. [DOI] [PubMed] [Google Scholar]
- 19.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 20.Rammensee H-G, Friede T, Stevanović S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 21.Hill CM, Liu A, Marshall KW, Mayer J, Jorgensen B, Yuan B, Cubbon RM, Nichols EA, Wicker LS, Rothbard JB. Exploration of requirements for peptide binding to HLA DRB1*0101 and DRB1*0401. J Immunol. 1994;152:2890–2898. [PubMed] [Google Scholar]
- 22.De Magistris MT, Alexander J, Coggeshall M, Altman A, Gaeta FCA, Grey HM, Sette A. Antigen analog-major histocompatibility complexes act as antagonists of the T cell receptor. Cell. 1992;68:625–634. doi: 10.1016/0092-8674(92)90139-4. [DOI] [PubMed] [Google Scholar]
- 23.Alexander J, Snoke K, Ruppert J, Sidney J, Wall M, Southwood S, Oseroff C, Arrhenius T, Gaeta FCA, Colón SM, Grey HM, Sette A. Functional consequences of engagement of the T cell receptor by low affinity ligands. J Immunol. 1993;150:1–7. [PubMed] [Google Scholar]
- 24.Yewdell JW, Bennink JR. The binary logic of antigen processing and presentation to T cells. Cell. 1990;62:203–206. doi: 10.1016/0092-8674(90)90356-j. [DOI] [PubMed] [Google Scholar]
- 25.Knowles JR, Albery WJ. Perfection in enzyme catalysis: the energetics of triosephosphate isomerase. Accounts of Chemical Research. 1977;10:105–111. [Google Scholar]
- 26.Ji H, Reid GE, Moritz RL, Eddes JS, Burgess AW, Simpson RJ. A two-dimensional gel database of human colon carcinoma proteins. Electrophoresis. 1997;18:605–613. doi: 10.1002/elps.1150180344. [DOI] [PubMed] [Google Scholar]
- 27.Clauser KR, Hall SC, Smith DM, Webb JW, Andrews LE, Tran HM, Epstein LB, Burlingame AL. Rapid mass spectrometric peptide sequencing and mass matching for characterization of human melanoma proteins isolated by two-dimensional PAGE. Proc Natl Acad Sci USA. 1995;92:5072–5076. doi: 10.1073/pnas.92.11.5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen RK, Ji H, Eddes JS, Moritz RL, Reid GE, Simpson RJ, Dorow DS. Two-dimensional electrophoretic analysis of human breast carcinoma proteins: mapping of proteins that bind to the SH3 domain of mixed lineage kinase MLK2. Electrophoresis. 1997;18:588–598. doi: 10.1002/elps.1150180342. [DOI] [PubMed] [Google Scholar]
- 29.Perry BA, Mohrenweiser HW. Human triosephosphate isomerase: substitution of Arg for Gly at position 122 in a thermolabile electromorph variant, TPI-Manchester. Hum Genet. 1992;88:634–638. doi: 10.1007/BF02265287. [DOI] [PubMed] [Google Scholar]
- 30.Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, Halperin AJ, Pontén J. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci USA. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wölfel T, Hauer M, Schneider J, Serrano M, Wölfel C, Klehmann-Hieb E, De Plaen E, Hankeln T, Meyer zum Büschenfelde K-H, Beach D. A p16INK4a-insensitive DCK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 32.Robbins PF, El-Gamil M, Li YF, Kawakami Y, Loftus K, Appella E, Rosenberg SA. A mutated β-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183:1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mande SC, Mainfroid V, Kalk KH, Goraj K, Martial JA, Hol WGJ. Crystal structure of recombinant human triosephosphate isomerase at 2.8 Å resolution. Triosephosphate isomerase-related human genetic disorders and comparison with the trypanosomal enzyme. Protein Sci. 1994;3:810–821. doi: 10.1002/pro.5560030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blacklow SC, Liu KD, Knowles JR. Stepwise improvements in catalytic effectiveness: independence and interdependence in combinations of point mutations of a sluggish triosephosphate isomerase. Biochemistry. 1991;30:8470–8476. doi: 10.1021/bi00098a026. [DOI] [PubMed] [Google Scholar]
- 35.Cormier JN, Abati A, Fetsch P, Hijazi YM, Rosenberg SA, Marincola FM, Topalian SL. Comparative analysis of the in vivo expression of tyrosinase, MART-1/ Melan-A, and gp100 in metastatic melanoma lesions: implications for immunotherapy. J Immunother. 1998;21:27–31. doi: 10.1097/00002371-199801000-00003. [DOI] [PubMed] [Google Scholar]
- 36.Markus NR, Rosenberg SA, Topalian SL. Analysis of cytokine secretion by melanoma-specific CD4+T lymphocytes. J Interferon Cytokine Res. 1995;15:739–746. doi: 10.1089/jir.1995.15.739. [DOI] [PubMed] [Google Scholar]