Abstract
Gene expression patterns can provide vital clues to the pathogenesis of neoplastic diseases. We investigated the expression of 950 genes in Hodgkin's disease (HD) by analyzing differential mRNA expression using microarrays. In two independent microarray experiments, the HD-derived cell lines L428 and KMH2 were compared with an Epstein-Barr virus (EBV)-immortalized lymphoblastoid B cell line, LCL-GK. Interleukin (IL)-13 and IL-5 were found to be highly expressed in the HD-derived cell lines. Examination of IL-13 and IL-5 expression by Northern blot analysis and enzyme-linked immunosorbent assay confirmed these results and revealed the expression of IL-13 in a third HD-derived cell line, HDLM2. Control LCL and EBV-negative non-Hodgkin lymphoma–derived cell lines did not express IL-13. In situ hybridization of lymph node tissue from HD patients showed that elevated levels of IL-13 were specifically expressed by Hodgkin/Reed-Sternberg (H/RS) tumor cells. Treatment of a HD-derived cell line with a neutralizing antibody to IL-13 resulted in a dose-dependent inhibition of H/RS cell proliferation. These data suggest that H/RS cells produce IL-13 and that IL-13 plays an important role in the stimulation of H/RS cell growth, possibly by an autocrine mechanism. Modulation of the IL-13 signaling pathway may be a logical objective for future therapeutic strategies.
Keywords: lymphoma, cytokines, cDNA microarray, gene expression, proliferation
Hodgkin's disease (HD)1 is unique among human lymphoma in that the tumor cells, known as Hodgkin/ Reed-Sternberg (H/RS) cells, are exceedingly rare. Investigation of H/RS cells has been impeded by their low frequency (0.1–1%) within the lymphoma tissue (1–4). PCR-based assays of DNA from single H/RS cells have revealed rearranged immunoglobulin genes and somatic mutations, suggesting that H/RS cells are clonal and may be derived from germinal center B cells (5, 6). However, the precise pathogenesis of HD remains to be determined.
Much evidence has accumulated that suggests that the proliferation and survival of HD-derived cells is due to cytokine signaling. It is well established that the unique histology and eosinophilia of HD tissues, as well as secondary symptoms in the patient such as fever, weight loss, and night sweats, are induced by a pathological pattern of cytokine secretion (1, 2). However, it has not been possible to consistently demonstrate the overexpression of a particular cytokine or group of cytokines in all HD-derived cell lines and primary H/RS tumor cells. Thus, the identity of particular cytokines playing a role in the pathogenesis of HD remains rather controversial.
The tool of microarray analysis has recently been developed (7–9) as a very efficient means of studying the differential expression of many genes simultaneously. Gene expression in two different samples is compared using competitive hybridization of two probes labeled with different fluorescent dyes. We have used microarrays containing 950 cDNA segments from genes relevant to inflammation and neoplasia to investigate gene expression patterns in HD-derived cell lines. Microarray expression patterns in the aneuploid, clonal cell lines L428 and KMH2 (2), which were cultured from Hodgkin tissues and are regarded as HD cell lines, were compared with microarray expression patterns in a control EBV-infected lymphoblastoid B cell line LCL-GK, which was derived from a healthy individual. Of the 950 genes on the microarray, IL-5 and IL-13 were the most strongly differentially expressed in both HD-derived cell lines, but only IL-13 was consistently overexpressed in a third HD cell line and primary H/RS tumor cells examined. Moreover, functional analysis revealed a role for IL-13 in stimulating the proliferation of H/RS cells.
Our study strongly suggests that IL-13 is a crucial factor in the pathogenesis of HD, and demonstrates the utility of cDNA microarray analysis of gene expression patterns in unraveling the complexities underlying neoplastic diseases like HD.
Materials and Methods
Cell Culture.
The cell lines were grown in IMDM medium supplemented with penicillin, streptomycin, and 10% FCS. The cell lines HDLM2 and LCL-HO were purchased from the German Collection of Microorganisms and Cell Cultures. The cell line LCL-GK was a gift from Dr. M. Dosch (Hospital for Sick Children, Toronto, Ontario, Canada), and the EBV-negative non-Hodgkin lymphoma (NHL)-derived cell lines Ly4, Ly7 and Ly13.2 were provided by Dr. H. Messner (Ontario Cancer Institute/Princess Margaret Hospital, Toronto, Ontario, Canada).
Microarray Experiments.
Polyadenylated [poly(A)+] mRNA was prepared from total RNA on Oligotex-dT resin (Qiagen). Each mRNA sample was converted into fluorescent-labeled cDNA probes using the microarray (GEM) probe labeling kit (Synteni). The microarray used in these experiments contained 950 human genes involved in inflammation and neoplasia (Synteni). Two microarrays were independently probed with 200 ng of a 1:1 mixture of fluorescein-labeled cDNA from HD-derived L428 cells and control LCL-GK cells on Microarray 1 (M1) or from HD-derived KMH2 cells and LCL-GK cells on microarray 2 (M2), respectively. Known concentrations of mRNA synthesized from inter–open reading frame regions from Saccharomyces cerevisiae were used as quantitation standards. The hybridization was performed as previously described (7).
Northern Blot Analysis.
Total RNA was isolated using Trizol reagent (GIBCO BRL). 15 μg total RNA was separated on a 1% formaldehyde agarose gel. Samples were run in 0.02 M MOPS (3-[N-morpholino]propanesulfonic acid), pH 7.0, 8 mM sodium acetate, 1 mM EDTA. The gel was blotted overnight in 20× SSC on a Hybond N+ nylon membrane (Amersham Pharmacia Biotech) and cross-linked using UV irradiation. Membranes were hybridized at 65°C to [α-32P]dATP-labeled (Mutiprime DNA Labeling System, Amersham Pharmacia Biotech) cDNA fragments specific for the human Notch2, NF-IL3, urokinase, IL-13, and human β-actin genes. Membranes were washed first in 0.1% SDS/2× SSC (30 min, 65°C) and then in 0.1% SDS/0.2× SSC (30 min, 65°C).
Cytokine Production.
Supernatants of cell cultures (105 cells/ml) were recovered 48 h after medium exchange and assayed for IL-5, IL-13, and GM-CSF production by specific ELISA using Quantikine-kits (R&D Systems).
Fixation of Biopsied Material.
Freshly biopsied lymph nodes were fixed in 10% formalin for 24 h before overnight processing and paraffin embedding.
In Situ Hybridization.
For in situ hybridization, paraffin sections were mounted and fixed according to standard protocols. The IL-13 probe used was a 388-bp human cDNA probe synthesized using reverse transcriptase PCR and IL-13–specific primers as follows: 5′ GTTGACCACGGTCATTGCTCTCACT and 3′ TTCAGTTGAACCGTCCCTCGCGAA. The cDNA was cloned into the pCRII vector (TA Cloning Kit; Invitrogen). Sense and antisense probes were synthesized from the linearized vector with SP6 or T7 polymerase, labeled with [33P]UTP, and processed as previously described (10). The sections were counterstained with toluidine blue using a standard protocol.
Immunohistochemistry.
Immunohistochemistry was performed using mAbs against CD30 (Dako) and CD15 (Becton Dickinson). Formalin-fixed paraffin sections were digested with pepsin and incubated (1 h) with the primary antibody. The slides were washed in PBS and incubated with biotinylated rabbit anti– mouse IgG (20 min) and with streptavidin–biotin complex labeled with horseradish peroxidase (20 min) after a second washing step (Ultra Streptavidin Detection System; Signet). The enzyme reaction was developed with AEC (3-amino-9-ethyl carbazole) and the slides were counterstained with hematoxylin.
Analysis of Cell Proliferation after Neutralization of IL-13.
HD-derived L428, KMH2, and HDLM2 cells and LCL control cells (2 × 104/well) were cultured in 96-well flat-bottomed plates for 24, 48, or 72 h in the presence of anti–IL-13, anti–IL-5, or isotype control antibodies (PharMingen) at 5, 10, 20, 30, 50, 100, or 150 μg/ml. Cells were treated either with 0.5, 1, 5, 10, 50, 100, and 200 ng/ml IL-13 (R&D Systems) or with these doses of IL-13 combined with 20 μg/ml anti–IL-13. [3H]thymidine (1 μCi/ well) was added to each well for an 8-h incubation. The cells were harvested on filters and the incorporation of [3H]thymidine into cellular DNA was measured as previously described (11).
Results and Discussion
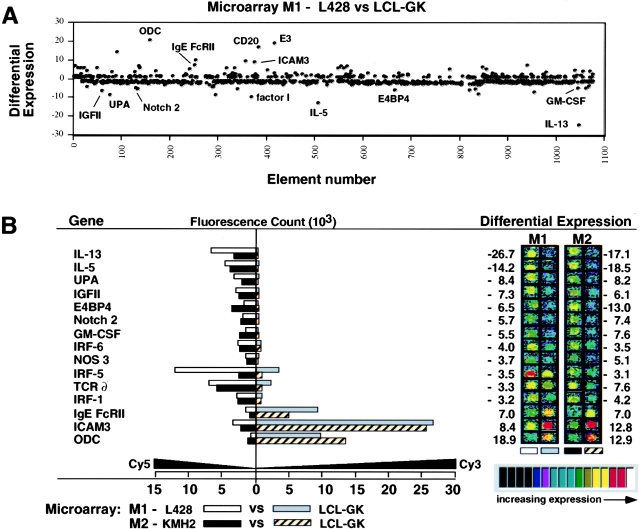
Of 950 genes displayed on the microchip, the genes showing a greater than threefold difference in expression in both HD-derived cell lines were IL-13; IL-5; ornithine decarboxylase (ODC); ICAM-3; urokinase plasminogen activator (UPA); IgE Fc receptor II; insulin-like growth factor (IGF) II; NF-IL3/E4BP4; Notch2; GM-CSF; interferon regulatory factor (IRF)-1, IRF-5, and IRF-6; nitric oxide synthase (NOS) 3; and the TCR δ chain (Fig. 1 B). IL-13 expression in HD-derived L428 and KMH2 cells was increased compared with that in control LCL-GK cells by 26.7- and 17.1-fold, respectively, and IL-5 expression was increased by 14.2- and 18.5-fold (Fig. 1 B).
Figure 1.
Differential expression of 950 genes investigated by microarray hybridization. (A) Genes showing the greatest differential expression are indicated. The vertical axis shows differential expression calculated as the ratio of the values of the two channels recording Fluorescence1 signal (Cy3) and Fluorescence2 signal (Cy5). A positive number indicates that Cy3 (channel one) is greater than Cy5 (channel two), whereas a negative number indicates that Cy5 is greater than Cy3. (B) Genes showing differences in expression that were greater than threefold on two microarrays (M1, M2). Fluorescent scans are represented in pseudocolor and correspond to hybridization intensities of genes. False colors ranging from black (at the low end) through blue, green, yellow, and red to white (at the high end) indicate the intensity of the signal. E3, retinoic acid-inducible E3 protein; IGFII, insulin-like growth factor II; IRF, interferon regulatory factor; NOS 3, nitric oxide synthase 3; ODC, ornithine decarboxylase; UPA, urokinase plasminogen activator.
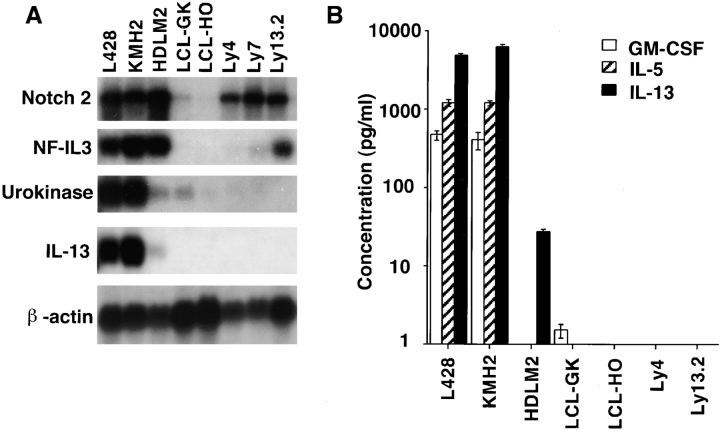
To evaluate the significance of these results, we used Northern blots and ELISA to compare the expression of IL-13, IL-5, GM-CSF, NF-IL3, Notch2, and urokinase in the HD-derived EBV-negative cell lines L428, KMH2, and HDLM2 (2) with that in the lymphoblastoid EBV- infected B cell lines LCL-GK and LCL-HO, and in three EBV-negative NHL-derived cell lines with either B cell (Ly4, Ly7) or T cell (Ly13.2) phenotype. Notch2, urokinase, and NF-IL3 were upregulated in LCL or NHL cell lines as well as in the HD-derived cell lines, but the overexpression of IL-13, IL-5, and GM-CSF was restricted to the HD-derived cell lines (Fig. 2). Although IL-5 and GM-CSF expression and secretion could be demonstrated only in L428 and KMH2 cells, mRNA expression and secretion of IL-13 could be detected in all three HD-derived cell lines. HDLM2 cells secreted a moderate amount of IL-13 (27 pg/ml), whereas L428 and KMH2 cells demonstrated intense IL-13 (4,800 and 6,100 pg/ml, respectively) secretion (Fig. 2 B).
Figure 2.
Differential expression of selected genes in HD-derived cell lines and control LCL and NHL-derived cell lines. (A) Northern blot analysis of total RNA from the EBV-negative, HD-derived cell lines L428, KMH2, and HDLM2; the EBV-immortalized lymphoblastoid B cell lines LCL-GK and LCL-HO; and the EBV-negative NHL-derived cell lines Ly4, Ly7, and Ly13.2. (B) Immunoassays to detect human IL-13, IL-5, and GM-CSF produced by the cell lines in A.
To confirm that IL-13 was expressed by the H/RS tumor cells, in situ hybridization using sense and antisense RNA probes for IL-13 was performed on lymph node biopsies from four untreated patients newly diagnosed with classical nodular sclerosis HD. The diagnosis was based on the nodular sclerosis architecture of the lymph nodes, the presence of H/RS cells in the appropriate context, and the immunohistological detection of CD30 and CD15 expression by cells with H/RS morphology. In all four patients, significant numbers of cells expressing IL-13 were detected (Fig. 3 A). Virtually all IL-13–expressing cells displayed morphological features of H/RS cells such as large size, prominent nucleoli, and multinuclear and/or lacunar cell morphology. Also, some IL-13–expressing cells were mitotic (Fig. 3 B). To test whether cells with a positive signal for expression of IL-13 express the HD markers CD30 and CD15, serial sections from two patients were cut and stained by immunohistochemistry for CD30 or CD15, or used for in situ hybridization with an IL-13 antisense probe. On these serial sections, it was possible to correlate a high proportion of IL-13 mRNA–containing cells with the presence of CD30 and CD15 expression by the same tumor cell (Fig. 3, C–E). A benign reactive lymph node hybridized in parallel with the IL-13 probe as well as a lymph node from a HD patient hybridized with an IL-13 sense probe did not yield any signal (data not shown), illustrating the specificity of IL-13 expression by H/RS cells in lymph nodes from HD patients.
Figure 3.

IL-13 expression in the lymph node of a patient with Hodgkin's disease. Paraffin sections of the diagnostic formalin-fixed cervical lymph node from a 53-yr-old male patient with Hodgkin's disease of the nodular sclerosis subtype, clinical stage IIA. (A, B, and D) In situ hybridization for IL-13, black silver grains denote IL-13 expression. (A) Multinuclear Reed-Sternberg and Hodgkin cells express IL-13. (B) Mitotic H/RS cell with a positive IL-13 signal. (C–E) Serial sections were stained for CD30 (C) or CD15 (E) or hybridized with an IL-13 antisense probe (D). The photographs are taken from the same area of three consecutive serial sections and illustrate the expression of CD30, IL-13, and CD15 by the same cell. Original magnification: A, ×400; B, ×1,000; C–E, ×200.
To investigate the effects of IL-13 and IL-5 on the proliferation of H/RS cells, L428, KMH2, and HDLM2 cells were incubated with medium alone, medium containing neutralizing antibodies to IL-13 or IL-5, or isotype controls. Proliferation was measured by determining [3H]thymidine incorporation at 24, 48, or 72 h after treatment. The viability of cultured cell lines after treatment with anti–IL-13 neutralizing antibodies was examined by 7-aminoactinomycin D (7-AAD) staining; no significant differences between HD-derived cells and controls were detected (data not shown). Neutralizing antibodies against IL-13 and IL-5 had no effect on the proliferation of control LCL-HO cells (Fig. 4, A and B). However, after 72 h of treatment with 20 μg/ml anti–IL-13 neutralizing antibody, the proliferation of HDLM2 cells (which secrete moderate levels of IL-13) was suppressed to 27% of that of untreated control cells (Fig. 4 A). Treatment of L428 and KMH2 cells with ≤150 μg/ml anti–IL-13 antibody had no effect on the proliferation of the cell lines (data not shown), perhaps because of their vigorous secretion of IL-13 (Fig. 2). No significant changes in proliferation were observed in control groups of HDLM2 cells treated with an anti–IL-5 neutralizing antibody (Fig. 4 B) or isotype control antibodies (Fig. 4 C). A combination of anti–IL-13 and anti–IL-5 antibodies did not inhibit proliferation to any greater extent than anti–IL-13 alone (data not shown). Treatment with increasing concentrations of anti–IL-13 antibody demonstrated that the effect on the proliferation of HD-derived cells was dose dependent (Fig. 4 D). Furthermore, the antiproliferative effect of anti–IL-13 on HDLM2 cells could be overcome by the addition of exogenous IL-13 (Fig. 4 E). Treatment of HDLM2 cells with exogenous IL-13 alone did not result in an increase in proliferation over that of untreated cells (Fig. 4 F), suggesting that the cells produce saturating levels of IL-13 sufficient to support maximal proliferation.
Figure 4.
Effect of IL-13 treatment on the proliferation of HD-derived cells. (A–C) KMH2, HDLM2, and control LCL-HO cells (105 cells/ml) were treated with either 20 μg/ml of anti– IL-13 (A); anti–IL-5 neutralizing antibodies (B); or the matching IgGκ1 and IgGκ2 isotype controls (C). Proliferation was measured at 24 and 72 h by [3H]thymidine incorporation. The y-axis shows the proliferation of the treated culture as a percentage of the untreated control, calculated as the mean of triplicate samples. (D–F) Relative proliferation (percentage of the [3H]thymidine incorporated by the untreated control) of HDLM2 cells 48 h after treatment with various doses of anti–IL-13 antibody or isotype control (D); a combination of 20 μg/ml anti–IL-13 with various doses of IL-13 (E); or various doses of IL-13 alone (F). For the control, 11,031 cpm was taken to represent 100% proliferation.
The systemic symptoms of HD and the unique reactive cellular background exhibited in HD biopsies suggest that cytokines are involved in HD pathogenesis. The elevated expression of various cytokines has been reported in HD biopsies and several HD-derived cell lines (1, 2). Overexpression of IL-5 in H/RS cells has been demonstrated previously by in situ hybridization, but only in HD patients exhibiting eosinophilia (12, 13). To date, no cytokine has been consistently reported as being overexpressed in HD-derived cell lines or in primary H/RS cells.
Of 950 genes tested, we found that only IL-13 was consistently and specifically overexpressed in H/RS tumor cells. IL-13 is a T cell–derived cytokine with immunomodulatory and antiinflammatory properties (14). The biological effects of IL-13 on B cells, macrophages, and monocytes are very similar to those of IL-4, probably because the IL-4 and IL-13 receptors share a common α chain. In B cells, IL-13 promotes proliferation, differentiation, and Ig heavy chain class switching to IgE and IgG4 (15). Proliferation results from a signaling pathway in which the engagement of the IL-13 receptor activates JAK1, which in turn activates STAT6 (16). Our results suggest that a similar mechanism may operate in the B cell–like H/RS cells, since neutralization of IL-13 dramatically inhibited the proliferation of HD-derived HDLM2 cells (Fig. 4 A). Indeed, in the microarray experiments, mRNA for the IL-13 receptor was found to be expressed in both of the HD-derived cell lines tested (data not shown). The secretion of IL-13 by H/RS cells, the expression of mRNA for the IL-13 receptor, and the specific effect of the neutralizing anti–IL-13 antibody suggest that an autocrine mechanism may control the growth of H/RS tumor cells.
Evidence for a role for IL-13 in the etiology of HD is indirect but consistent. IgE is elevated in HD tissues and serum samples from HD patients (13, 17), and IL-13 is known to promote Ig class switching to IgE. IL-13–deficient mice exhibit lower basal levels of serum IgE (18). Furthermore, studies of IL-4–deficient, IL-13–transgenic mice have demonstrated that IL-13 can promote class switching to IgE independently of IL-4 (19), emphasizing that IL-4 and IL-13 have distinct roles in regulating B cell functions. Our results have clearly demonstrated that HD-derived cell lines and H/RS tumor cells express elevated levels of IL-13. The elevation of IgE in H/RS cells and in the serum of HD patients therefore could be explained if IL-13 secreted by H/RS cells affects both the H/RS cells themselves and the bystander B cells.
Other aspects of the HD phenotype may also be attributable to the effects of IL-13. A recent study of IL-13–deficient mice has shown that cultures of Th2 cells from these animals produce significantly reduced levels of IL-4, IL-5, and IL-10 compared with the results from the wild-type animals, suggesting an important role for IL-13 as a regulator of Th2 cell commitment (18). If IL-13 is also important for promoting the differentiation of Th2 cells in humans, it could explain why H/RS cells (which secrete IL-13) are surrounded by Th2 cells in HD biopsies. In addition, because fibroblasts express the IL-13 receptor and can be activated by IL-13 (20), the secretion of IL-13 by H/RS cells may underlie the pathogenesis of the fibrosis observed in nodular sclerosis HD.
Our microarray hybridization also showed that the expression of NF-IL3 and Notch2 was upregulated by more than threefold in HD-derived cell lines (Fig. 1). Northern blot analysis revealed NF-IL3 and Notch2 expression also in NHL-derived cell lines (Fig. 2 A). The basic leucine zipper transcription factor NF-IL3 acts downstream of IL-3 and has been shown to prevent apoptosis after forced expression in an IL-3–dependent pro-B cell line (21). The relevance of NF-IL3 in HD is unknown. Notch2 is a transmembrane receptor that has been shown to be involved in cell fate decisions (22). The human Notch2 gene has been mapped to chromosome 1 at position 1p13-p11, which is a region of translocations associated with neoplasia (23). Chromosome 1 has also been linked to the HD phenotype, since structural rearrangements of chromosome 1 are frequently observed in HD (24). Interestingly, the gene for CD30, which was first identified in the HD-derived cell line L428 (25) and is considered a marker for the disease, is also located on chromosome 1 at 1p36 (26). CD30 is highly expressed in lymphoblastoid cells and H/RS cells and has been shown to promote a Th2 phenotype (27). The possible roles of NF-IL3 and Notch2 and their relationship to IL-13 and the CD30 antigen in the pathogenesis of HD lymphoma require further investigation.
In conclusion, our data combined with recent reports on the function of IL-13 indicate that IL-13 may be a critical cytokine in the etiology of HD. We have shown both that IL-13 is secreted by HD tumor cells and that IL-13 specifically promotes the proliferation of these cells. Our results are consistent with a role for IL-13 in the pathogenesis of the fibrosis, increased IgE production, and bias towards Th2 cells, all of which are typical of classical HD.
Acknowledgments
The authors thank E. Tong and N. Jamal for technical support; M. Dosch and H. Messner for the LCL and NHL-derived cell lines; S. Daefler, V. Stambolic, J. Penninger, D. Siderovski, M. Minden, and H. Messner for valuable critical comments; and M. Saunders for scientific editing.
Abbreviations used in this paper
- HD
Hodgkin's disease
- H/RS
Hodgkin/ Reed-Sternberg
- NHL
non-Hodgkin lymphoma
Footnotes
U. Kapp's current address is University Medical Center, Department of Hematology/Oncology, Hugstetter Strasse 55, 79106 Freiburg, Germany.
References
- 1.Drexler HG. Recent results on the biology of Hodgkin and Reed-Sternberg cells. I. Biopsy material. Leuk Lymphoma. 1992;8:283–313. doi: 10.3109/10428199209051008. [DOI] [PubMed] [Google Scholar]
- 2.Drexler HG. Recent results on the biology of Hodgkin and Reed-Sternberg cells. II. Continuous cell lines. Leuk Lymphoma. 1993;9:1–25. doi: 10.3109/10428199309148499. [DOI] [PubMed] [Google Scholar]
- 3.Kadin ME. Pathology of Hodgkin's disease. Curr Opin Oncol. 1994;6:456–463. doi: 10.1097/00001622-199409000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Cossman J, Messineo C, Bagg A. Reed-Sternberg cell: survival in a hostile sea. Lab Invest. 1998;78:229–235. [PubMed] [Google Scholar]
- 5.Kanzler H, Küppers R, Hansmann ML, Rajewsky K. Hodgkin and Reed-Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med. 1996;184:1495–1505. doi: 10.1084/jem.184.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Küppers R, Rajewski K. The origin of Hodgkin and Reed/Sternberg cells in Hodgkin's disease. Annu Rev Immunol. 1998;16:471–493. doi: 10.1146/annurev.immunol.16.1.471. [DOI] [PubMed] [Google Scholar]
- 7.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 8.Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shalon D, Smith SJ, Brown PO. A DNA microarray system analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 10.Hui CC, Joyner AL. A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenetic deletion of the Gli3 gene. Nat Genet. 1993;3:241–246. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- 11.Shahinian A, Pfeffer K, Lee KP, Kundig TM, Kishihara K, Wakeham A, Kawai K, Ohashi PS, Thompson CB, Mak TW. Differential T cell costimulatory requirements in CD28-deficient mice. Science. 1993;261:609–612. doi: 10.1126/science.7688139. [DOI] [PubMed] [Google Scholar]
- 12.Samoszuk M, Nansen L. Detection of interleukin-5 messenger RNS in Reed-Sternberg cells of Hodgkin's disease with eosinophilia. Blood. 1990;75:13–16. [PubMed] [Google Scholar]
- 13.Samoszuk M. IgE in Reed-Sternberg cells of Hodgkin's disease with eosinophilia. Blood. 1992;79:1518–1522. [PubMed] [Google Scholar]
- 14.Minty A, Chalon P, Deroque J-M, Dumont X, Guillemont J-C, Kaghad M, Labit C, Leplatois P, Liauzun P, Miloux B, et al. Interleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature. 1993;362:248–250. doi: 10.1038/362248a0. [DOI] [PubMed] [Google Scholar]
- 15.Zurawski G, deVries JE. Interleukin 13, an interleukin 4-like cytokine that acts on monocytes and B cells, but not on T cells. Immunol Today. 1994;15:19–26. doi: 10.1016/0167-5699(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 16.Lin JX, Migone TS, Tsang M, Friedmann M, Weatherbee JA, Zhou L, Yamauchi A, Bloom ET, Mietz J, John S, et al. The role of shared receptor motifs and common stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity. 1995;2:331–339. doi: 10.1016/1074-7613(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 17.Thomas MR, Steinberg P, Votaw ML, Bayne NK. IgE levels in Hodgkin's disease. Ann Allergy. 1976;37:416–419. [PubMed] [Google Scholar]
- 18.McKenzie GJ, Emson CL, Bell SE, Anderson S, Fallon P, Zurawski G, Murray R, Grencis R, McKenzie AN. Impaired development of Th2 cells in IL-13-deficient mice. Immunity. 1998;9:423–432. doi: 10.1016/s1074-7613(00)80625-1. [DOI] [PubMed] [Google Scholar]
- 19.Emson CL, Bell SE, Jones A, Wisden W, McKenzie AN. Interleukin (IL)-4–independent induction of immunoglobulin (Ig)E, and pertubation of T cell development in transgenic mice expressing IL-13. J Exp Med. 1998;188:399–404. doi: 10.1084/jem.188.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doucet C, Brouty-Boye D, Pottin-Clemenceau C, Canconica GW, Jasmin C, Azzarone B. Interleukin (IL) 4 and IL-13 act on human lung fibroblasts. Implication in asthma. J Clin Invest. 1998;101:2129–2139. doi: 10.1172/JCI741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikushima S, Inukai T, Inaba T, Nimer SD, Cleveland JL, Look AT. Pivotal role for the NFIL3/ E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc Natl Acad Sci USA. 1997;94:2609–2614. doi: 10.1073/pnas.94.6.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robey E. Notch in vertebrates. Curr Opin Genet Dev. 1997;7:551–557. doi: 10.1016/s0959-437x(97)80085-8. [DOI] [PubMed] [Google Scholar]
- 23.Larsson C, Lardelli M, White I, Lendahl U. The human Notch1, 2, and 3 genes are located at chromosome positions 9q34, 1p13-p11, and 19p13.2-p13.1 in regions of neoplasia-associated translocation. Genomics. 1994;24:253–258. doi: 10.1006/geno.1994.1613. [DOI] [PubMed] [Google Scholar]
- 24.Thangavelu M, Le Beau MM. Chromosomal abnormalities in Hodgkin's disease. Hematol Oncol Clin N Am. 1989;3:221–236. [PubMed] [Google Scholar]
- 25.Schwab U, Stein H, Gerdes J, Lemke H, Kirchner H, Schaadt M, Diehl V. Production of a monoclonal antibody specific for Hodgkin and Reed-Sternberg cells of Hodgkin's disease and a subset of normal lymphoid cells. Nature. 1982;299:65–67. doi: 10.1038/299065a0. [DOI] [PubMed] [Google Scholar]
- 26.Fonatsch C, Latza U, Durkop H, Rieder H, Stein H. Assignment of the human CD30 (Ki-1) gene to 1p36. Genomics. 1992;14:825–826. doi: 10.1016/s0888-7543(05)80203-4. [DOI] [PubMed] [Google Scholar]
- 27.Del Prete G, De Carli M, D'Elios MM, Daniel KC, Almerigogna F, Alderson M, Smith CA, Thomas E, Romagnani S. CD30-mediated signaling promotes the development of human T helper type 2–like T cells. J Exp Med. 1995;182:1655–1661. doi: 10.1084/jem.182.6.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]