Abstract
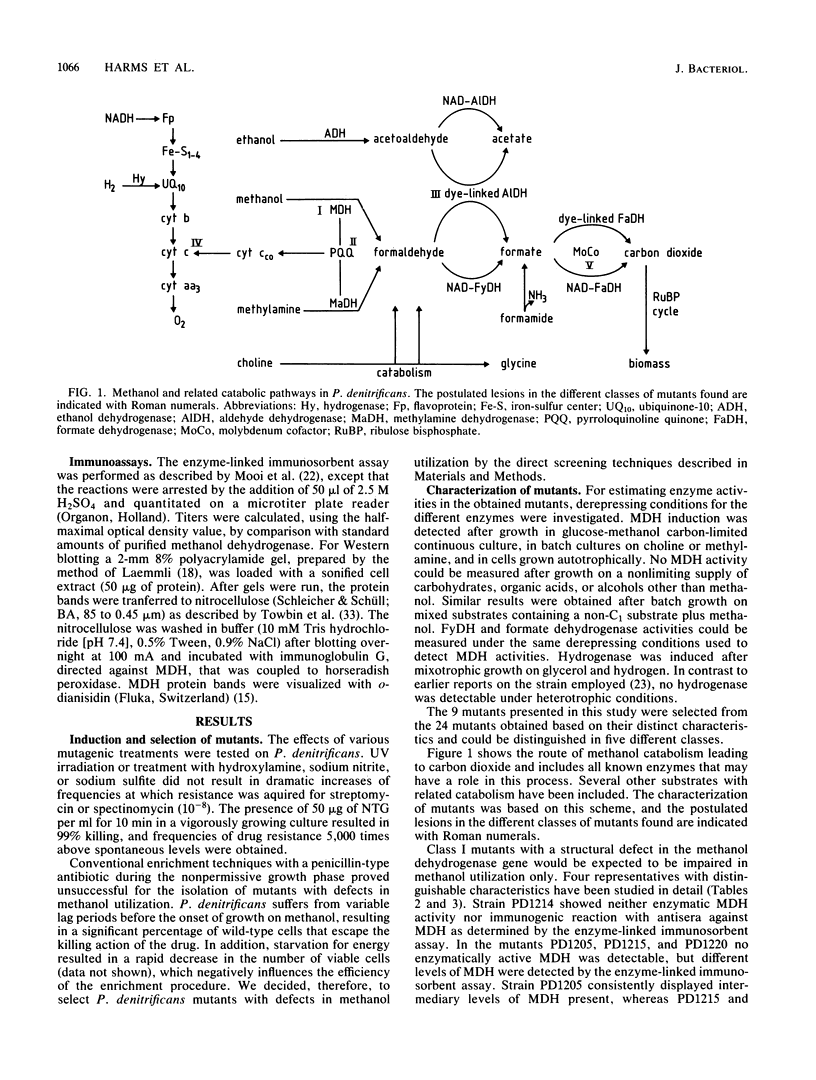
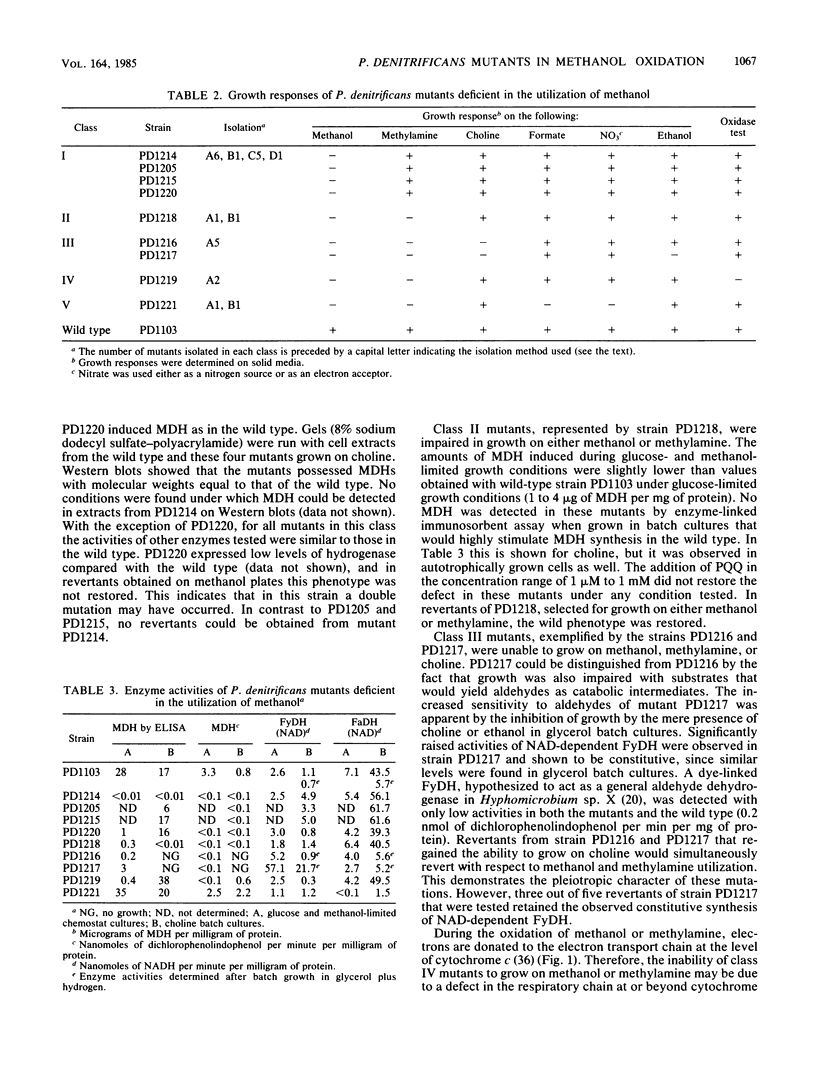
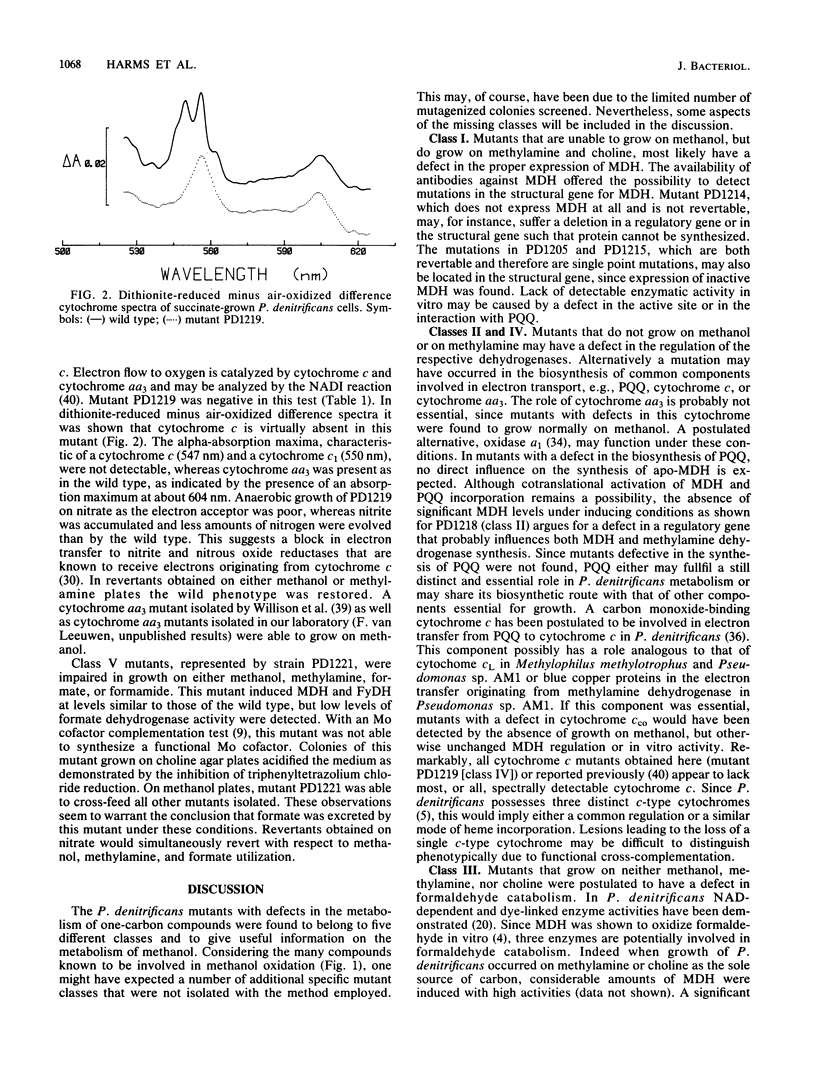
Mutants deficient in the metabolism of one-carbon compounds have been obtained by treating Paracoccus denitrificans with the mutagen N-methyl-N'-nitro-N-nitrosoguanidine. Mutants were selected without enrichment procedures by newly developed plate screening tests. The obtained mutants were characterized by their growth responses, cytochrome composition, enzyme activities, and immunogenic reaction with antisera against methanol dehydrogenase. By these criteria five mutant classes could be distinguished. Class I mutants are involved in the expression of methanol dehydrogenase. Three mutants of this class have a defect in the structural gene. A double mutant was found with defects in the expression of both methanol dehydrogenase and hydrogenase. Class II mutants have a defect in a regulatory gene involved in the regulation of both methanol dehydrogenase and methylamine dehydrogenase. Class III mutants are deficient in formaldehyde metabolism. A defect may exist in the expression of a second non-NAD-linked formaldehyde dehydrogenase which was postulated to be involved in C1 metabolism. Class IV mutants are deficient in cytochrome c. Mutants of class V have a defect in synthesis of the molybdenum cofactor essential for the function of formate dehydrogenase.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen L. N., Hanson R. S. Construction of broad-host-range cosmid cloning vectors: identification of genes necessary for growth of Methylobacterium organophilum on methanol. J Bacteriol. 1985 Mar;161(3):955–962. doi: 10.1128/jb.161.3.955-962.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. 2. The methanol-oxidizing enzyme of Pseudomonas sp. M 27. Biochem J. 1964 Sep;92(3):614–621. doi: 10.1042/bj0920614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamforth C. W., Quayle J. R. Aerobic and anaerobic growth of Paracoccus denitrificans on methanol. Arch Microbiol. 1978 Oct 4;119(1):91–97. doi: 10.1007/BF00407934. [DOI] [PubMed] [Google Scholar]
- Berry E. A., Trumpower B. L. Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c-aa3 complexes. J Biol Chem. 1985 Feb 25;260(4):2458–2467. [PubMed] [Google Scholar]
- Bochner B. R., Savageau M. A. Generalized indicator plate for genetic, metabolic, and taxonomic studies with microorganisms. Appl Environ Microbiol. 1977 Feb;33(2):434–444. doi: 10.1128/aem.33.2.434-444.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke K. A., Calder K., Lascelles J. Effects of molybdenum and tungsten on induction of nitrate reductase and formate dehydrogenase in wild type and mutant Paracoccus denitrificans. Arch Microbiol. 1980 Jun;126(2):155–159. doi: 10.1007/BF00511221. [DOI] [PubMed] [Google Scholar]
- Claassen V. P., Oltmann L. F., Bus S., v 't Riet J., Stouthamer A. H. The influence of growth conditions on the synthesis of molybdenum cofactor in Proteins mirabilis. Arch Microbiol. 1981 Sep;130(1):44–49. doi: 10.1007/BF00527070. [DOI] [PubMed] [Google Scholar]
- Cox R. B., Quayle J. R. The autotrophic growth of Micrococcus denitrificans on Methanol. Biochem J. 1975 Sep;150(3):569–571. doi: 10.1042/bj1500569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan P. M., Anthony C., Drabble W. T. Microbial metabolism of C 1 and C 2 compounds. The involvement of glycollate in the metabolism of ethanol and of acetate by Pseudomonas AM1. Biochem J. 1972 Jun;128(1):99–106. doi: 10.1042/bj1280099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich B., Heine E., Finck A., Friedrich C. G. Nickel requirement for active hydrogenase formation in Alcaligenes eutrophus. J Bacteriol. 1981 Mar;145(3):1144–1149. doi: 10.1128/jb.145.3.1144-1149.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton G. L., Nunn D. N., Lidstrom M. E. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp. strain AM1, a facultative methylotroph. J Bacteriol. 1984 Nov;160(2):718–723. doi: 10.1128/jb.160.2.718-723.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier F., Bonewald R. The use of plasmid R1162 and derivatives for gene cloning in the methanol-utilizing Pseudomonas AM1. Mol Gen Genet. 1980;178(2):375–380. doi: 10.1007/BF00270487. [DOI] [PubMed] [Google Scholar]
- Hawkes R., Niday E., Gordon J. A dot-immunobinding assay for monoclonal and other antibodies. Anal Biochem. 1982 Jan 1;119(1):142–147. doi: 10.1016/0003-2697(82)90677-7. [DOI] [PubMed] [Google Scholar]
- Heptinstall J., Quayle J. R. Pathways leading to and from serine during growth of Pseudomonas AM1 on C1 compounds or succinate. Biochem J. 1970 Apr;117(3):563–572. doi: 10.1042/bj1170563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. A., Quayle J. R. Microbial growth on C-1 compounds. 6. Oxidation of methanol, formaldehyde and formate by methanol-grown Pseudomonas AM-1. Biochem J. 1964 Nov;93(2):281–290. doi: 10.1042/bj0930281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mooi F. R., de Graaf F. K., van Embden J. D. Cloning, mapping and expression of the genetic determinant that encodes for the K88ab antigen. Nucleic Acids Res. 1979 Mar;6(3):849–865. doi: 10.1093/nar/6.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nokhal T. H., Schlegel H. G. The regulation of hydrogenase formation as a differentiating character of strains of Paracoccus denitrificans. Antonie Van Leeuwenhoek. 1980;46(2):143–155. doi: 10.1007/BF00444069. [DOI] [PubMed] [Google Scholar]
- O'Connor M. L. Extension of the model concerning linkage of genes coding for C-1 related functions in Methylobacterium organophilum. Appl Environ Microbiol. 1981 Feb;41(2):437–441. doi: 10.1128/aem.41.2.437-441.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M., Wopat A., Hanson R. S. Genetic transformation in Methylobacterium organophilum. J Gen Microbiol. 1977 Jan;98(1):265–272. doi: 10.1099/00221287-98-1-265. [DOI] [PubMed] [Google Scholar]
- PECK H. D., Jr, GEST H. A new procedure for assay of bacterial hydrogenases. J Bacteriol. 1956 Jan;71(1):70–80. doi: 10.1128/jb.71.1.70-80.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouthamer A. H., Boogerd F. C., van Verseveld H. W. The bioenergetics of denitrification. Antonie Van Leeuwenhoek. 1982;48(6):545–553. doi: 10.1007/BF00399540. [DOI] [PubMed] [Google Scholar]
- Van Verseveld H. W., Stouthamer A. H. Electron-transport chain and coupled oxidative phosphorylation in methanol-grown Paracoccus denitrificans. Arch Microbiol. 1978 Jul;118(1):13–20. doi: 10.1007/BF00406068. [DOI] [PubMed] [Google Scholar]
- Windass J. D., Worsey M. J., Pioli E. M., Pioli D., Barth P. T., Atherton K. T., Dart E. C., Byrom D., Powell K., Senior P. J. Improved conversion of methanol to single-cell protein by Methylophilus methylotrophus. Nature. 1980 Oct 2;287(5781):396–401. doi: 10.1038/287396a0. [DOI] [PubMed] [Google Scholar]
- van Verseveld H. W., Krab K., Stouthamer A. H. Proton pump coupled to cytochrome c oxidase in Paracoccus denitrificans. Biochim Biophys Acta. 1981 May 13;635(3):525–534. doi: 10.1016/0005-2728(81)90111-0. [DOI] [PubMed] [Google Scholar]


