Abstract
Propagation of signals from the T cell antigen receptor (TCR) involves a number of adaptor molecules. SH2 domain–containing protein 76 (SLP-76) interacts with the guanine nucleotide exchange factor Vav to activate the nuclear factor of activated cells (NF-AT), and its expression is required for normal T cell development. We report the cloning and characterization of a novel Grb2-like adaptor molecule designated as Grb2-related protein of the lymphoid system (GrpL). Expression of GrpL is restricted to hematopoietic tissues, and it is distinguished from Grb2 by having a proline-rich region. GrpL can be coimmunoprecipitated with SLP-76 but not with Sos1 or Sos2 from Jurkat cell lysates. In contrast, Grb2 can be coimmunoprecipitated with Sos1 and Sos2 but not with SLP-76. Moreover, tyrosine-phosphorylated LAT/pp36/38 in detergent lysates prepared from anti-CD3 stimulated T cells associated with Grb2 but not GrpL. These data reveal the presence of distinct complexes involving GrpL and Grb2 in T cells. A functional role of the GrpL–SLP-76 complex is suggested by the ability of GrpL to act alone or in concert with SLP-76 to augment NF-AT activation in Jurkat T cells.
Keywords: GrpL, SLP-76, Grb2, nuclear factor of activated cells, cell activation
Ligation of the T cell antigen receptor (TCR) complex activates protein tyrosine kinases (PTKs)1 including the Src family PTKs Fyn and Lck to phosphorylate immunoreceptor tyrosine-based motifs (ITAMs) within associating CD3 chains. This leads to the recruitment and activation of the Syk family kinase ZAP-70 and possibly other proteins such as Shc to phosphorylated CD3ζ. These activated PTKs in turn phosphorylate a variety of proteins, and thereby couple TCR ligation to a number of intracellular signaling cascades culminating in T cell activation. Principal downstream substrates/effectors for TCR-activated PTKs have been identified. They include phospholipase C-γ1 (1–3), the 85-kD subunit of the phosphatidylinositol 3-kinase (4), the src homology (SH)2 domain–containing leukocyte protein of 76 kD (SLP-76) (5, 6), the proto-oncogene product Cbl (5, 7), the Vav guanine nucleotide exchange factor (GEF) of the Rho/Rac/CDC42 family of GTP-binding proteins (8, 9), and the recently cloned and characterized linker for activation of T cells (LAT/pp36/38) (10–14). Functional interactions between these phosphorylated effector molecules are believed to be necessary for the operation of major pathways required for T cell maturation including (a) intracellular free Ca2+ release, (b) activation of the nuclear factor of activated cells (NF-AT), and (c) mitogen-activated protein kinase (MAPK) activation. These events ultimately converge to induce gene expression, proliferation, or differentiation of T cells (15–17).
SLP-76 is an essential adaptor molecule in T cells. When it is tyrosine phosphorylated, probably by ZAP-70, SLP-76 may become a ligand for Vav (9, 18, 19). In Jurkat cells, the tyrosine-phosphorylated SLP-76–Vav complex augments TCR-induced activation of NF-AT, and hence expression of IL-2 (9, 18–21). The in vivo function of SLP-76 in T lineage cells is further illustrated by recent studies of SLP-76– deficient mice. Mice deficient in SLP-76 expression show profound defects in T cell development, and contain no CD4/CD8 double positive thymocytes, resulting in a complete loss of peripheral T cells (22, 23).
One connection between the TCR and SLP-76 may be provided by Grb2 (24). The SH2 domain of Grb2 binds to both Shc and LAT/pp36/38, allowing Grb2 to be localized to the plasma membrane where most of the TCR-activated PTKs are (13, 14, 25). The SH3 domain(s) of Grb2 bind to Sos1 and Sos2 (GEF of the Ras family of GTP-binding protein) (26), SLP-76 (21, 27), and Cbl (7, 28, 29). The recruitment of Sos to the plasma membrane by Grb2 initiates the MAPK pathway, whereas the recruitment of SLP-76 (30), Vav (8, 9), and Cbl (7, 31, 32) to the plasma membrane probably facilitates their phosphorylation by TCR-activated PTK.
The studies above suggest that Grb2 may function to link multiple downstream effector molecules, e.g., SLP-76, to LAT and possibly to the vicinity of the activated TCR complex. However, more recent studies have revealed the presence of an additional Grb2-like adaptor molecule, Grap, which has the SH3-SH2-SH3 organization as Grb2 (33, 34). Interestingly, expression of Grap appears to be relatively restricted to hematopoietic tissues (33, 34). Besides binding to c-kit, the Bcr-Abl oncoprotein, and the erythropoietin receptor, Grap also interacts with LAT/pp36/38, Sos, Shc, and phospholipase C-γ1 in TCR-activated Jurkat cells (33, 34). Exactly how Grb2 and Grap coordinate their functions during TCR signaling remains to be elucidated.
In this paper, we report the molecular cloning and characterization of another linker protein, which shares a number of structural properties with Grb2 and Grap, yet differs from them by having a proline-rich region. Since transcripts encoding this linker protein appear to be preferentially expressed in lymphoid cells, we have designated this protein GrpL, for Grb2-related protein of the lymphoid system. GrpL associates with different effector molecules than does Grb2 in T cells and it regulates TCR-induced NF-AT activation.
Materials and Methods
Cells and Antibodies.
Various cell lines were maintained in standard culture media supplemented with l-glutamine, nonessential amino acids, sodium pyruvate, penicillin/streptomycin, and fetal bovine serum. A rabbit anti–human (h)GrpL serum was generated in this study against a His-Tag fusion protein containing the SH2 domain of hGrpL as described below. Antiphosphotyrosine 4G10 and anti-LAT/pp36/38 were purchased from Upstate Biotechnology, Inc. Anti–SLP-76, anti-Grb2, anti–SH2 domain–containing protein tyrosine phosphatase 2 (SHP-2), anti-Sos1, and anti-Sos2 were purchased from Santa Cruz Biotechnology. 64.1 (anti-CD3) was provided by Dr. John Hansen (Fred Hutchinson Cancer Research Center, Seattle, WA), C305 (anti-TCR) was provided by Dr. G. Koretzky (University of Iowa, Iowa City, IA), and OKT3 (anti-CD3) was obtained from the American Type Culture Collection.
Isolation of the Full-Length cDNA Clones Encoding Human and Murine GrpL.
A full-length cDNA encoding hGrpL was identified through random sequencing of clones from a library prepared from the acute myelogenous leukemia cell line KG-1a. Overlapping partial cDNA clones encoding murine (m)GrpL were isolated by low stringency reverse transcriptase (RT)-PCR using multiple hGrpL-specific primer sets and murine splenic total RNA as the template. RT-PCR at low annealing temperature was performed according to the Titan™ One Tube RT-PCR System protocol (Boehringer Mannheim Inc.). PCR products were resolved on 1.5% agarose gels. cDNA bands corresponding in sizes to the estimated lengths of hGrpL were excised from the gel and eluded using the QIAEX II Gel Extraction Kit (QIAGEN Inc.) according to the suppliers protocol. Isolated cDNAs were then ligated into a T/A cloning vector (Invitrogen) and transformed into competent E. coli (Invitrogen). Recombinant clones containing mGrpL inserts were selected by low stringency hybridization to a hGrpL cDNA probe and then sequenced. This allowed for the identification of primer sets specific for mGrpL for the isolation of cDNA clones encoding both the 5′ and 3′ ends of mGrpL by rapid amplification of cDNA ends. The gene specific primers chosen were: 5′ cDNA end, 5′-CTG AGC TTT CGC ACT GGA GAC ATT CTG; 3′ cDNA end, 5′-GCA TCA GCG TGG CGT TCA CCT CAC TTC C.
The Marathon-Ready™ cDNA kit (Clontech Laboratory Inc.) was used for RACE PCR and performed as recommended by the supplier. The resulting PCR products were cloned into T/A cloning vectors (Invitrogen) and sequenced.
Plasmid Constructs.
To generate the GrpL SH2 His-Tag fusion protein, PCR was used to amplify a piece of cDNA encoding the SH2 domains of GrpL using the full-length GrpL cDNA as template and the following primer pairs: forward primer, 5′-GCG GGG ATC CCC CAG TTT CCC AAA TGG TTT CAC; reverse primer, 5′-GCG GGG AAT TCC TCT GTC TCT AAG GAA GAT CTG. Underlined nucleotides represent BamHI and EcoRI sites in the forward and reverse primers, respectively. PCR products were cut with these enzymes and ligated into BamHI/EcoRI cut pET-28c(+) (Novagen, Inc.). To generate the GST fusion construct, the insert from the His-Tag fusion construct was cut out by BamHI/EcoRI and subcloned into BamHI/EcoRI cut pGEX-5X-2 (Amersham Pharmacia Biotech). The fusion protein constructs were then transformed into E. coli strain BL21(DE3) and induced with IPTG for protein production.
To generate the Myc-His tagged hGrpL expression plasmid the coding region of hGrpL was amplified using the following primer pairs: forward primer, 5′-CCT GAA GCT CGA GTC GGG TCA TGG GTG CCA CGT A; reverse primer, 5′-GCG GAA TTC GGA GGA TGG AAG CTG TTG CCA AGT TT. Underlined nucleotides represent XhoI and EcoRI sites in the forward and reverse primers, respectively. The PCR product was cloned into the pcDNA3.1(+)/Myc-His A vector (Invitrogen), in-frame with the c-Myc epitope and poly-His tags. The hGrpLMyc-His cassette was then excised with BamHI and PmeI and subcloned into the pEF expression vector (provided by Dr. G. Koretzky) at the BamHI and filled-in XbaI sites (pEF-GrpL). The pEF–SLP-76 and the NF-AT luciferase reporter (NF-AT Luc) constructs were also gifts from Dr. G. Koretzky.
Northern Blot Analysis and RT-PCR.
To detect GrpL message in cell lines, ∼30 μg of RNA were resolved on 1.2% morpholine propanesulfonic acid (MOPS)/formaldehyde agarose gels, transferred onto GeneScreen Plus membranes (NEN™ Life Science Products, Boston, MA), and fixed by UV cross-linking. Multiple tissue Northern blots were obtained from Clontech. Northern blots were prehybridized and hybridized to either the full-length GrpL cDNA or β-actin probes labeled with α-[32P]dCTP (NEN™ Life Science Products) according to the manufacturer's instructions. RT-PCR was performed using the Titan™ One Tube RT-PCR System (Boehringer Mannheim). The reaction was carried out using 1 μg of total RNA and 20 nM forward and reverse primers. Primers used were: forward primer, 5′-TGG AAG CTG TTG CCA AGT TTG ATT TCA C; reverse primer, 5′-CTT CTC GGG TTC TGT CTC TAA GGA AG.
This primer set amplifies a 452-bp fragment containing the NH2-terminal SH3 and the SH2 domains from cells expressing GrpL message. The amount of total RNA used was monitored by amplification of a 1-kb fragment encoding G3DPH using the following primer set: forward primer, 5′-TGA AGG TCG GAG TCA ACG GAT TTG GT; reverse primer, 5′-CAT GTG GGC CAT GAG GTC CAC CAC. Cycling parameters PCR were programmed according to the instructions provided with the Titan™ system (Boehringer Mannheim).
T Cell Stimulation, Immuoprecipitation, and Immunoblot Analysis.
Jurkat T cells were pelleted and resuspended at 5–10 × 106/ml in medium. Cells were allowed to equilibrate at 37°C for 15 min and then stimulated with either 5 or 10 μg/ml of anti-CD3 or 2.5 mM of H2O2 plus 250 μM of sodium orthovanadate (referred to as pervanadate). Either class-matched antibodies or medium only was used as control stimulus. Stimulation was terminated by dilution of cell suspensions into >10 vol of ice-cold PBS. Cells were pelleted and lysed in a buffer containing 150 mM NaCl, 50 mM Tris, pH 8.0, 5 mM EDTA, 0.5% NP-40, protease inhibitors (2 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 μg/ml pepstatin), and phosphatase inhibitors (10 mM NaF, 1 mM Na3VO4, and 5 mM Na4P2O7). Lysates were clarified by centrifugation before immunoprecipitation. Immunoprecipitation, SDS-PAGE, and Western blot analysis were conducted as described (35). Binding of primary antibodies to blots was detected with horseradish peroxidase–conjugated secondary antibodies or streptavidin (Jackson ImmunoResearch Labs.) and an enhanced chemiluminescence kit (Amersham Pharmacia Biotech).
T Cell Transfection and Luciferase Assays.
Jurkat cells in log-phase were washed with serum-free RPMI 1640 once and electroporated in triplicate with a total of 60 μg of plasmid at 240 V, 960 μF using a Gene Pulser (Bio-Rad) at 107 cells/400 μl serum-free RPMI 1640. In general, 20 μg of NF-AT Luc were cotransfected with either 20 μg of pEF, pEF–SLP-76, pEF-GrpL, or pEF–SLP-76 plus pEF-GrpL. Sonicated salmon sperm DNA (Sigma Chemical Co.) was used to maintain the total DNA at 60 μg whenever necessary. After electroporation, cells were incubated in fully supplemented RPMI 1640 for 14–18 h at 37°C, 5% CO2. Cells were then transferred into 96-well plates at 2–5 × 105 cells/ well in a total of 100 μl medium. Cells were stimulated in triplicate with 1:500 dilution of C305 anti-CD3 for 6–8 h at 37°C. Control cells were maintained in culture medium. After stimulation, cells were lysed in 50–100 μl Reporter Lysis buffer (Promega) for 15 min at room temperature. Luciferase activity was quantified by adding 10 μl of the lysate to 50 μl of luciferase assay substrate (Promega) and immediately measured with a GenProbe Leader™ 1 luminometer (Wallac Inc.). To normalize for variations in transfection efficiency, luciferase activities in different transfection conditions were expressed as a percentage of maximal NF-AT responses determined by treating cells with 50 ng/ml PMA and 1 μM ionomycin as previously described (18, 20).
Results
Identification of GrpL, a New Member of the Grb Family of Adaptor Proteins.
Through random sequencing of cDNA clones in a library prepared from the acute myelogenous leukemia cell line KG-1a, we identified a cDNA encoding a 330-amino acid polypeptide with a predicted molecular weight of ∼37 kD. The linear amino acid sequence of this polypeptide organizes into an NH2-terminal SH3 domain followed by an SH2 domain, a proline-rich region, and finally a COOH-terminal SH3 domain (Fig. 1 A). A comparison to other known proteins available in current sequence data banks revealed that this protein is most similar to the adaptor proteins, Grb2 and Grap (Fig. 1 B and Table I). Based on this similarity, we have designated this molecule, GrpL for Grb2-related protein of the lymphoid system (see below). Using RT-PCR and a combination of 5′- and 3′-RACE (Materials and Methods), we also isolated the full-length cDNA encoding the murine homologue of GrpL. Human and murine GrpL are highly conserved proteins sharing >90% identity in the SH3 and SH2 domains (Table I). The least conserved area is the proline-rich region with ∼74% identity (Table I). Amino acid sequence alignment between hGrpL, Grb2, and Grap revealed that hGrpL is 38–56% identical to Grb2 and Grap in the SH3 and SH2 domains (Fig. 1 B, Table I). This conservation is slightly less than that between Grb2 and Grap (49–68%; reference 33). The biggest difference between GrpL and Grb2 or Grap is the presence of a proline-rich region between the SH2 and COOH-terminal SH3 domain. This proline-rich region could potentially be bound by SH3 domain–containing proteins found in other adaptor molecules (36, 37). Taken together, the amino acid composition of GrpL suggests that it may interact with different proteins in lymphocytes when compared with Grb2 or Grap.
Figure 1.
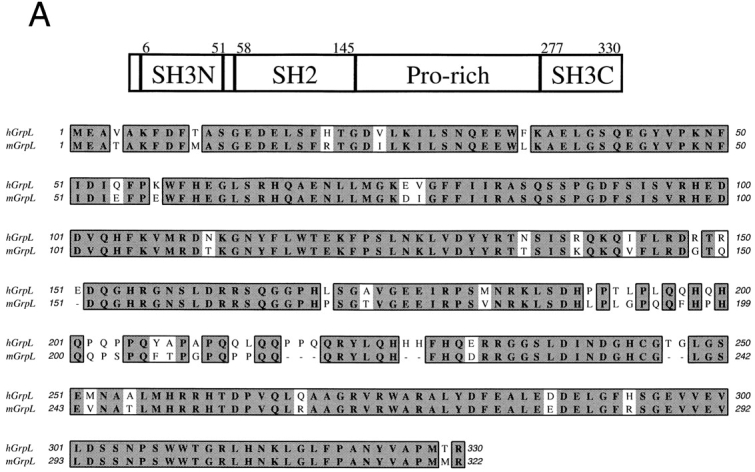
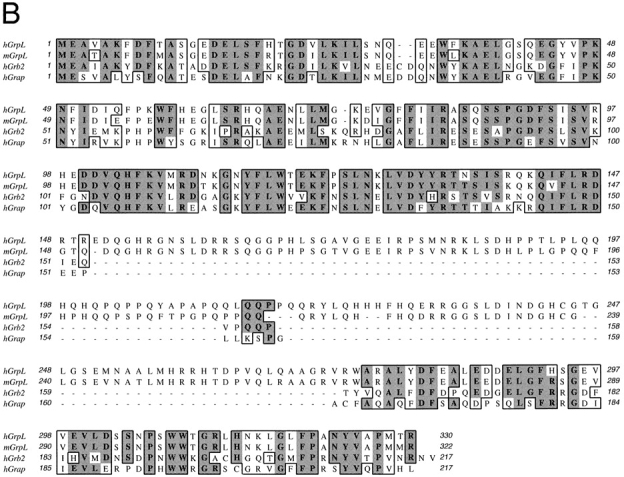
Structural organization of GrpL and amino acid homology comparison with other Grb2-like proteins. A schematic for the domain organization of GrpL is depicted in the upper part of A. Numbers on top mark amino acid residues at the boundaries of the different domains. The lower part of A shows the amino acid sequence comparison between mouse and human GrpL and B shows the amino acid sequence comparison among mGrpL, hGrpL, Grb2, and Grap. Boxed residues represent homologous regions. Amino acids in bold and shade denote identical residues, while unshaded amino acids denote conserved substitutions. The nucleotide sequences for hGrpL and mGrpL are available from EMBL/ GenBank/DDBJ under accession numbers AF129476 and AF129477.
Table I.
Amino Acid Sequence Comparison among hGrpL, mGrpL, Grb2, and Grap
| SH3N | SH2 | Proline-rich interdomain | SH3C | |||||
|---|---|---|---|---|---|---|---|---|
| hGrpL/mGrpL | 91 | 93 | 74 | 94 | ||||
| hGrpL/Grb2 | 52 | 56 | <5 | 50 | ||||
| hGrpL/Grap | 50 | 56 | <5 | 38 |
Numbers represent the percentage of identity between the different domains of the two proteins listed on the lefthand column.
Transcripts of GrpL Are Selectively Expressed in Hematopoietic Tissues and in B and T Lymphocytes.
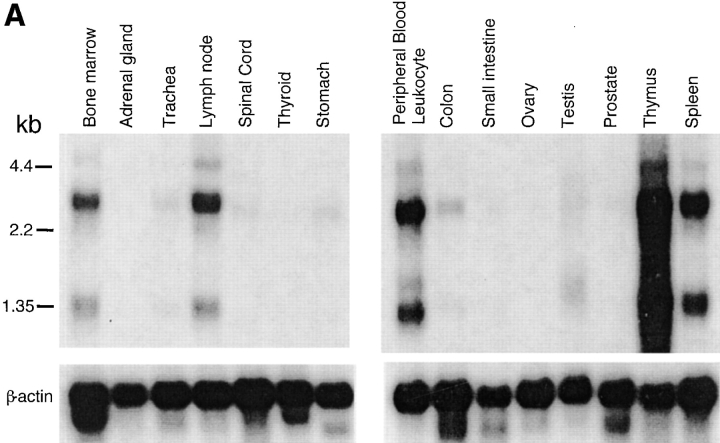
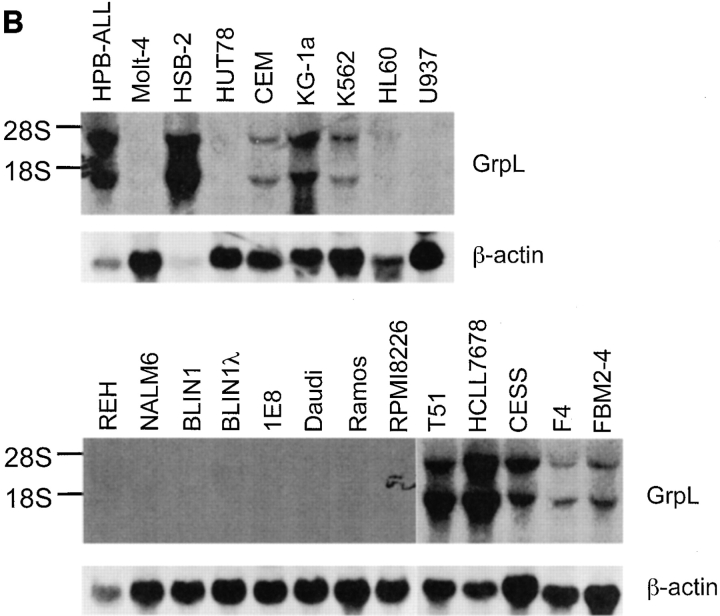
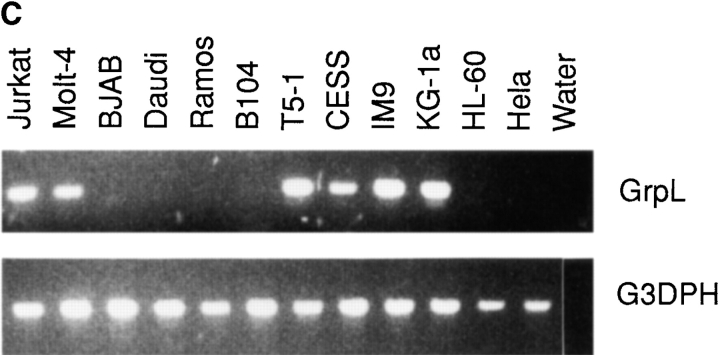
The transcripts for Grb2 are expressed ubiquitously in mammalian species (38), whereas Grap mRNA is expressed at considerably higher levels in lymphoid tissues than in other tissues (33, 34). Northern blot analysis of mRNA isolated from different human tissues demonstrated that the transcripts for GrpL are relatively restricted to hematopoietic tissues. Thus, transcripts of 1.4 kb and 3–3.5 kb were detected in lymph node, bone marrow, spleen, thymus, and peripheral blood lymphocytes, as well as weakly in testis, but not in stomach, thyroid, spinal cord, trachea, adrenal gland, prostate, ovary, or small intestine (Fig. 2 A). Probing total RNA from a panel of cell lines by Northern blot analysis and RT-PCR further confirmed the restricted expression pattern of GrpL (Fig. 2, B and C). GrpL transcripts were detected in the myeloid and erythroid progenitor cell lines KG-1a and K562, respectively, but not in the myeloid cell line HL60. In T cells, GrpL is expressed in the Jurkat, Molt-4, HSB-2, CEM, and HPB-ALL lines, but not in the HuT78 line. Interestingly, of all the B cell lines examined, GrpL transcripts were only found in EBV-transformed lymphoblastoid cell lines including T5-1, HCLL7678, CESS, F4, FBM2-4, and IM9. Other B cell lines, including the Burkitt's lymphomas (Daudi, BJAB, and Ramos), the pre-B cell lines (REH, NALM-6, and BLIN1), the sIgM+ early B cell lines (BLIN1 and 1E8), and the myeloma cell line RPMI8226, did not express any GrpL transcripts. Results from the above experiments show that GrpL expression is highly regulated and restricted to subsets within hematopoietic lineages.
Figure 2.
Expression of GrpL transcripts. Multiple tissue blots (Clontech) containing 2 μg per lane of polyA+ RNA (A) or blots containing 30 μg per lane of total RNA isolated from various hematopoietic cell lines (B) were probed with full-length GrpL. Blots were stripped and reprobed with β-actin to monitor RNA loading. (C) RT-PCR using conditions and an hGrpL primer as described in Materials and Methods was conducted on a subset of cell lines in B and additional cell lines to confirm and extend results obtained by Northern blot analysis. A primer set amplifying G3DPH was used to monitor RNA loading.
GrpL Associates with SLP-76 in T Cells.
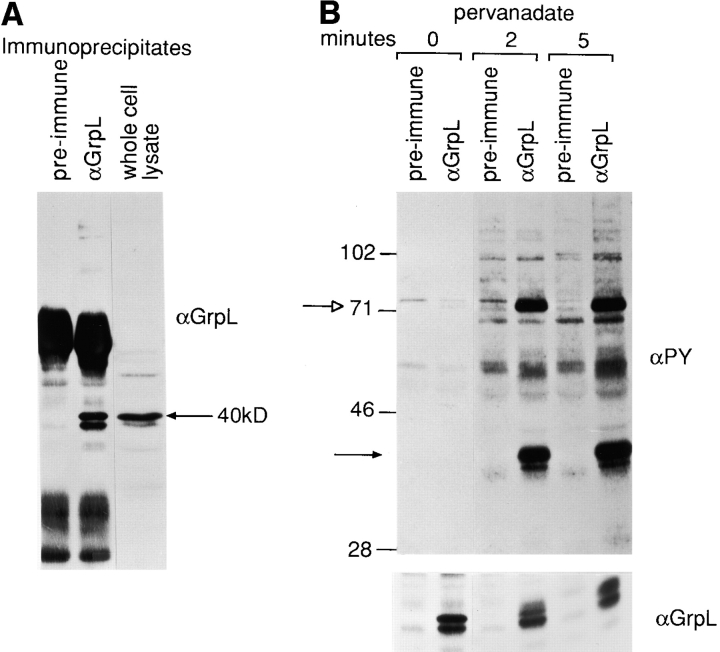
To characterize the GrpL protein and the proteins with which it interacts, we generated a rabbit hetero-antiserum specific for GrpL by immunizing with a His-Tag GrpLSH2 fusion protein. This antiserum can specifically immunoprecipitate from Jurkat T cells two proteins ∼38 and 40 kD in size (Fig. 3 A). Both proteins were detectable by Western blot analysis of either GrpL immunoprecipitates (IPs) or whole cell lysates. This antiserum does not cross-react with Grb2, which runs at a lower molecular weight on gels (Fig. 3 A and see Fig. 5), and by all criteria it appears to be specific for GrpL.
Figure 3.
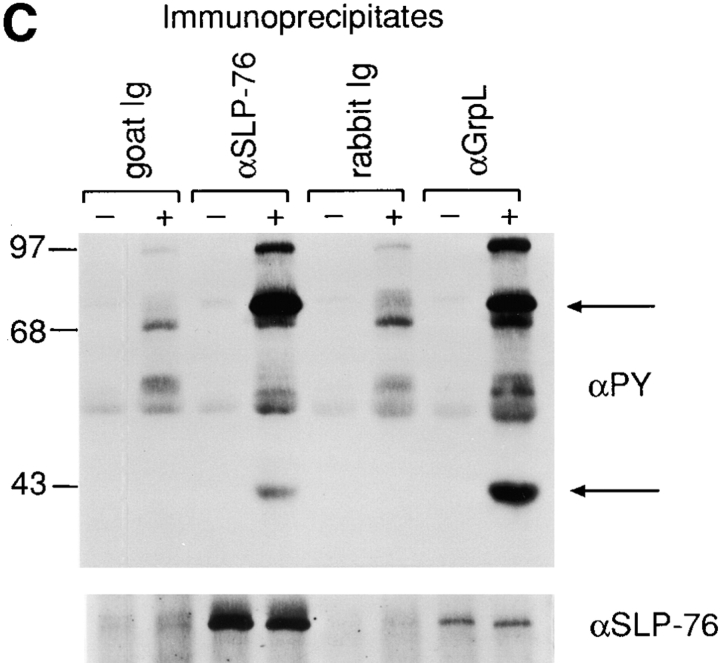
Biochemical characterization of GrpL. (A) Jurkat cell lysates were immunoprecipitated with an antiserum against GrpL generated by immunizing rabbits with the GST/GrpLSH2 fusion protein or preimmune serum. IPs were resolved along with whole cell lysate on reducing SDS-PAGE and immunoblotted with the same anti-GrpL serum. (B) Jurkat cells were stimulated for either 2 or 5 min with pervanadate as described in Materials and Methods. NP-40 lysates were immunoprecipitated with anti-GrpL or preimmune serum and immunoblotted with the antiphosphotyrosine antibody 4G10 (αPY). The amounts of GrpL in different lanes were monitored by immunoblotting with anti-GrpL (αGrpL). The open arrow indicates a predominant tyrosine-phosphorylated protein coimmunoprecipitating with GrpL, and the closed arrow indicates the mobility of GrpL. (C) NP-40 lysates prepared from unstimulated (−) or pervanadate-stimulated (+) Jurkat cells were immunoprecipitated with anti–SLP-76 (αSLP-76), anti-GrpL (αGrpL), or normal goat or rabbit control Ig. IPs were resolved on SDS-PAGE and immunoblotted with either antiphosphotyrosine or anti–SLP-76. The arrows indicate phosphoproteins migrating at the sizes of SLP-76 and GrpL.
Figure 5.
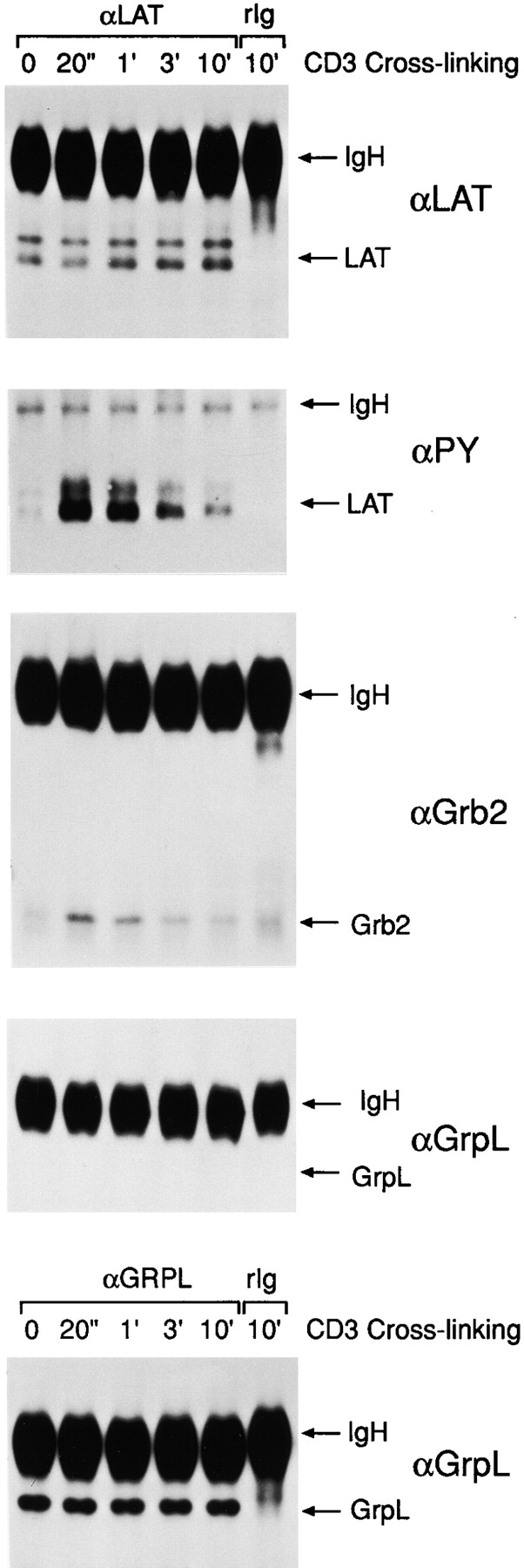
GrpL does not interact with LAT/pp36/38. NP-40 lysates were prepared from anti-CD3–stimulated Jurkat cells and were immunoprecipitated with anti-LAT/pp36/38. Normal rabbit Ig (rIg) was used as the negative control. The anti-LAT/ pp36/38 IPs were resolved by reducing SDS-PAGE on replicate gels and immunoblotted with anti-LAT/pp36/38 (αLAT/ pp36/38), antiphosphotyrosine (4G10) (αPY), anti-Grb2 (αGrb2), and anti-GrpL (αGrpL). Expression of GrpL in these cells during stimulation were monitored by immunoblotting the anti-GrpL IPs with anti-GrpL (αGrpL).
To detect proteins potentially capable of interacting with GrpL, we pharmacologically stimulated Jurkat cells with pervanadate, immunoprecipitated GrpL, and then Western blotted with either antiphosphotyrosine or anti-GrpL sera (Fig. 3 B). 2–5 min after stimulation, a major tyrosine phosphorylated protein 40 kD in size and a minor 38-kD phosphoprotein were detectable in anti-GrpL but not control serum IPs. This doublet most likely corresponds to GrpL itself, which migrates to the same position on gels (Fig. 3, A and B); it is possible that these bands represent differentially phosphorylated forms of GrpL. Compared with pervanadate, TCR ligation induced only weak tyrosine phosphorylation of GrpL (data not shown).
In addition to the 38–40-kD doublet, a second major tyrosine phosphorylated protein ∼68–70 kD in size was also detected in the GrpL IP after pervanadate treatment (Fig. 3, B and C). In T cells, substrates for protein tyrosine kinases in this size range include SLP-76, SHP-2, Syk, and ZAP-70. Indeed, in the GrpL IP obtained from both unstimulated and pervanadate-stimulated Jurkat cells, we were able to detected SLP-76 (Fig. 3 C) but not ZAP-70 (data not shown). However, the amount of SLP-76 associating with GrpL remained relatively unchanged after stimulation. Moreover, the SLP-76 molecule coimmunoprecipitated with GrpL before stimulation showed an almost undetectable level of tyrosine phosphorylation (Fig. 3 C). These results are consistent with the idea that GrpL can interact with SLP-76 independently of tyrosine phosphorylation as has been suggested for Grb2 and SLP-76 (21, 27). The increased signal of the 68–70-kD molecule coimmunoprecipitated with GrpL after stimulation may be a consequence of increased tyrosine phosphorylation of SLP-76 and/or the recruitment of additional tyrosine phosphorylated proteins of the same size to GrpL. In the SLP-76 IP obtained from pervanadate-stimulated Jurkat cells, we could also detect tyrosine-phosphorylated protein at 38– 40, 50–55, and 110 kD (Fig. 3 C).
GrpL and Grb2 Interact with Different Molecules.
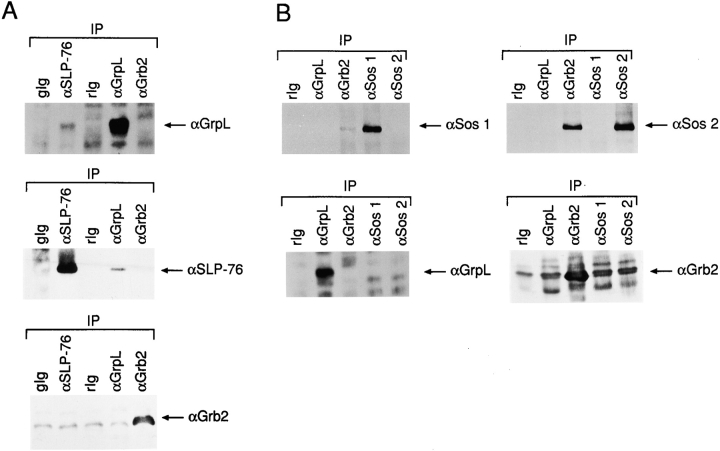
To examine the possibility of distinct functional roles played by different Grb2-like molecules in T cells, we asked whether GrpL and Grb2 interact with different proteins. We immunoprecipitated either GrpL or Grb2 from Jurkat cells and probed for associating proteins. SLP-76 was detected in GrpL IPs (Fig. 4 A), as also shown in Fig. 3 C. However, we could not detect any SLP-76 associating with Grb2. Consistent with this was the presence of GrpL but not Grb2 in the SLP-76 IPs (Fig. 4 A). On the other hand, we were able to detect Sos1 and Sos2, GEFs of Ras, in Grb2 IPs (Fig. 4 B), as has been reported in other cell systems. Interestingly, no detectable Sos1 or Sos2 was found to be coimmunoprecipitated with GrpL (Fig. 4 B). These results suggest that in Jurkat T cells, GrpL preferentially complexes with SLP-76, whereas Grb2 preferentially complexes with Sos1 or Sos2.
Figure 4.
GrpL and Grb2 interact with different effector molecules. (A) NP-40 lysates prepared from Jurkat cells were immunoprecipitated with anti–SLP-76 (αSLP-76), anti-GrpL (αGrpL), anti-Grb2 (αGrb2), normal goat Ig (gIg), or normal rabbit Ig (rIg). IPs were resolved on reducing SDS-PAGE and immunoblotted with antisera as indicated on the left. (B) NP-40 lysates prepared from Jurkat cells were immunoprecipitated with anti– SLP-76 (αSLP-76) anti-GrpL (αGrpL), anti-Sos1 (αSos1), anti-Sos2 (αSos2), or normal rabbit Ig (rIg). IPs were resolved on reducing SDS-PAGE and immunoblotted with antisera as indicated on the right.
Since both Grb2 (10–14) and Grap (33, 34) can associate with tyrosine-phosphorylated LAT/pp36/38 upon TCR ligation, we asked whether GrpL could also associate with tyrosine-phosphorylated LAT/pp36/38. Jurkat cells were stimulated by anti-CD3, and LAT/pp36/38 was immunoprecipitated from NP-40 Jurkat cell lysates at various time points after stimulation. As reported before (24), tyrosine-phosphorylation of LAT/pp36/38 increased dramatically after TCR ligation, and started to decline after 3 min of stimulation. This was correlated with the appearance of Grb2 in the LAT/pp36/38 IPs (Fig. 5). However, although the levels of GrpL proteins in NP-40 Jurkat cell lysates remained unchanged throughout stimulation, we consistently did not detect any GrpL associating with LAT/pp36/38 in these lysates (Fig. 5).
GrpL Regulates NF-AT Activation in T Cells.
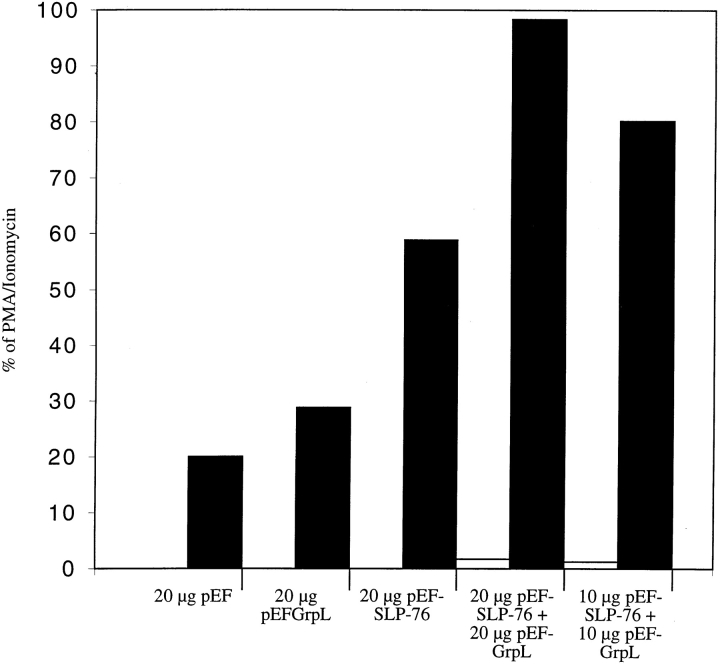
Since SLP-76 has been shown to regulate NF-AT activation in T cells (18, 20, 39), we investigated whether GrpL could play a role in this pathway. NF-AT activation was monitored by the expression of a NF-AT luciferase reporter construct in Jurkat cells. As described before, overexpression of SLP-76 by cotransfection of a SLP-76 expression vector augmented the anti-CD3–induced NF-AT activation (18, 20, 39). Overexpression of GrpL alone consistently augmented anti-CD3– induced NF-AT activation (Fig. 6). Moreover, an additive effect between GrpL and SLP-76 on NF-AT activation was observed when both were overexpressed in Jurkat cells.
Figure 6.
GrpL regulates NF-AT activation in T cells. Jurkat T cells were transfected in triplicate with 20 μg of the NF-AT Luc and empty vector (pEF), pEF-GrpL, pEF–SLP-76, or a combination of pEF-GrpL and pEF–SLP-76 at concentrations indicated in the figure. Cells were stimulated with the anti-TCR C305 (black bars) or left unstimulated in medium (white bars). Luciferase activities were then determined on each transfected cell population. The maximal NF-AT responses for the various transfection conditions were determined by treatment with PMA plus ionomycin, and did not differ significantly. The effects of overexpressing GrpL and/or SLP-76 are shown as percentage of the maximal NF-AT responses for each transfection conditions. The data are representative of four independent experiments.
The SH2 Domain of GrpL Recognizes Tyrosine-phosphorylated Protein Tyrosine Phosphatase SHP-2 In Vitro.
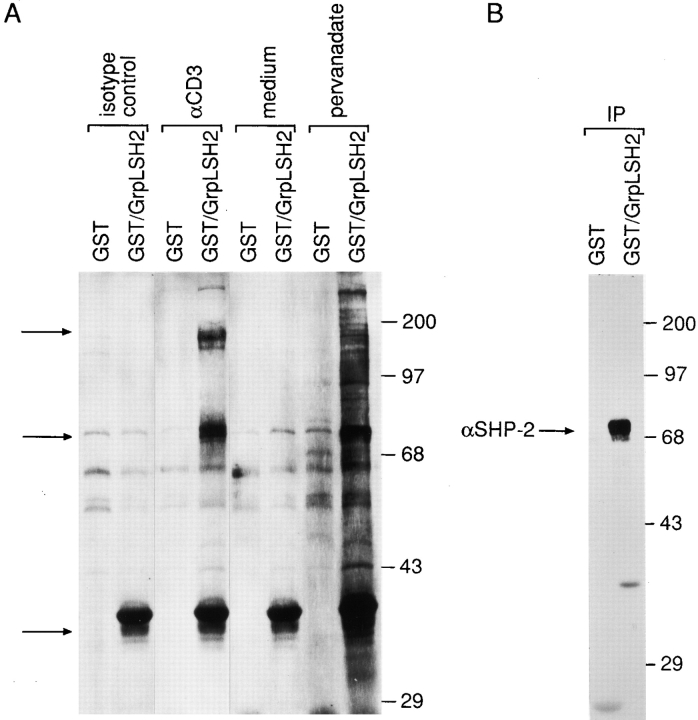
Since various SH2 domains can interact with a wide spectrum of tyrosine-phosphorylated targets, we tested to which tyrosine-phosphorylated protein(s) the SH2 domain of GrpL could bind. When the GST/GrpLSH2 fusion protein was used to probe pervanadate-stimulated Jurkat cells, tyrosine-phosphorylated proteins that migrated at 36, 69, and 71 kD could be readily detected (Fig. 7 A). In some experiments, we also detected 95- and 130-kD tyrosine-phosphorylated proteins. The GST/GrpLSH2 fusion protein also precipitated a similar pattern from lysates prepared from anti-CD3–stimulated Jurkat cells (Fig. 7 A). By analogy to Grb2 (24), these proteins could be LAT, ZAP-70, SHP-2, Vav, and perhaps c-Cbl. As shown in Fig. 7 B, we were able to confirm that SHP-2 was among the phosphoproteins precipitated by the GST/GrpLSH2 fusion protein from pervanadate-activated Jurkat cells. Although tyrosine-phosphorylated SLP-76 also migrates at ∼70 kD, we consistently did not detect any SLP-76 precipitated by GST/GrpSH2.
Figure 7.
The SH2 domain of GrpL interacts with SHP-2. (A) Jurkat T cells were stimulated with 10 μg/ml of anti-CD3 (αCD3) or pervanadate for 5 min. Control cells were treated with either isotype-matched control IgG or medium alone. NP-40 lysates were precipitated with either GST or GST/GrpLSH2, resolved on reducing SDS-PAGE, and immunoblotted with the antiphosphotyrosine antibody 4G10. Arrows indicates major tyrosine-phosphorylated proteins detected in the GST/ GrpLSH2 precipitates. (B) Replicate precipitates obtained from pervanadate-stimulated Jurkat cells were immunoblotted with anti–SLP-76.
Discussion
Critical functions are played by a variety of adaptor molecules in linking receptor proximal signal transduction events such as activation of receptor-associated kinases to downstream effector molecules. In this report, we describe the identification and characterization of a new member of the Grb family of adaptor proteins. GrpL shares similar structural organization as Grb2 and Grap. A distinguishing feature of GrpL not found in Grb2 or Grap is the presence of a 100-amino acid stretch connecting its SH2 and SH3C domains. Although this region is the least conserved between mGrpL and hGrpL, it does contain multiple proline and glutamine residues at conserved positions (Fig. 1 and Table I). Thus, two PxxP motifs are found in both hGrpL and mGrpL. In hGrpL, they are P191TLP and P202QPP; in mGrpL, they are P190LGP and P210QPP. These polyproline motifs conform to the consensus sequence recognized by different SH3 domains (36, 37). Accordingly, it is likely that the proline residues in GrpL do constitute a proline-rich domain capable of interacting with as yet to be defined SH3-containing proteins.
In contrast to the broadly expression Grb2, GrpL expression is restricted to hematopoietic cells (Fig. 2). In this regard, GrpL is similar to Grap that transcripts for both are found at much higher levels in hematopoietic tissues (33, 34). Although Grap is preferentially expressed in blood cells, little is known about its expression in different stages of T and B cell maturation or in different lymphocyte subsets. GrpL transcripts are expressed at lower levels in the bone marrow than in the secondary lymphoid organs. Importantly, the highest levels of GrpL transcripts were detected in the thymus (Fig. 2 A), suggesting that GrpL may play a crucial role in regulating early T cell development. Northern blot analysis on cell lines also demonstrates that GrpL transcripts are not expressed in all lymphocytes (Fig. 2, B and C), suggesting that their expression may be modulated during lymphocyte ontogeny by specific signals. To test this possibility, we fractionated normal B cells into subsets and measured levels of GrpL transcripts. A clear difference was observed in GrpL transcript expression in naive but not in germinal center B cells (Solow, S.A., A.J. Marshall, T.J. Yun, M.K. Ewings, and E.A. Clark, manuscript in preparation). To elucidate the signals involved in switching the expression of GrpL on and off will provide further insights into the functions of GrpL and its associating proteins at different stages of lymphocyte development.
We have identified one of the GrpL-interacting proteins to be SLP-76 (Figs. 3 and 4). Interaction between GrpL and SLP-76 appears to be constitutive, since it could be detected in both stimulated and unstimulated T cells (Figs. 3 and 4 A). Accordingly, this interaction may not require the tyrosine phosphorylation of either SLP-76 or GrpL. In support of this, results presented in Fig. 3 C show that the amount of SLP-76 coimmunoprecipitating with GrpL remained relatively constant, although SLP-76 became heavily tyrosine-phosphorylated after pervanadate treatment. Hence, our data argue against the possibility that the SH2 domain of GrpL mediates its interaction with SLP-76. Grb2 has been reported to be an adaptor molecule capable of associating with SLP-76 (24). In these studies, GST fusion proteins containing Grb2 or Grb2 SH3 domains recognized SLP-76 from lysates of resting or activated T cells independent of its tyrosine-phosphorylation (21, 27). Using deletional mutants of SLP-76, the Grb2 binding site was subsequently mapped to the central proline-rich region of SLP-76 from amino acid residues 224–244 (21). These results implicate the SH3 domain of Grb2 to be the primary mediator for interaction. Based on the similar structure between GrpL and Grb2 and the phosphorylation-independent interaction between GrpL and SLP-76, it is likely that the SH3 domain(s) of GrpL also mediates its interaction with SLP-76. Experiments are currently in progress to examine this possibility.
Several recent studies have provided strong evidence for an essential function served by SLP-76 in T lineage cells. Biochemical experiments show that SLP-76 can interact with both positive and negative regulators. TCR ligation induces ZAP-70 to phosphorylate SLP-76 at three NH2-terminal tyrosine residues (20, 30, 39). Once phosphorylated on its NH2-terminal tyrosine residues, in particular Y113 and Y128, SLP-76 can be bound by the SH2 domain of Vav (18, 19). Overexpression of SLP-76 augment IL-2 promoter activity (21), and SLP-76 also acts synergistically with Vav to augment IL-2 promoter activity (18). Moreover, the biological activity of SLP-76 is absolutely dependent on an intact proline-rich region, and hence, an interaction with adaptor proteins containing SH3 domain(s) (21). On the other hand, the COOH-terminal SH2 domain of SLP-76 binds to a 130-kD tyrosine-phosphorylated protein (40, 41). This protein has been cloned, designated to be SLAP-130/Fyb, and may be involved in downregulating the activating function of SLP-76 on the IL-2 promoter (41). The most compelling evidence to argue for an indispensable in vivo function of SLP-76 in T lineage cell is provided by the recent gene targeting experiments. SLP-76− /− mice are characterized by an early arrest in T cell development. CD4+/CD8+, CD4+, and CD8+ thymocyte populations are completely absent from thymuses of SLP-76 knockout mice, resulting in a corresponding lack of peripheral T cells (22, 23). In the SLP-76− /− mice, TCR-β chain genes appear to undergo normal rearrangement and exogenous anti-CD3 fails to drive the generation of double positive cells. Therefore, it is proposed that the absence of SLP-76 may render the pre-TCR complex fail to deliver maturational signals to developing T cells (22, 23).
Our observation that GrpL associates with SLP-76 in vivo suggests GrpL plays a pivotal role in TCR-mediated signal transduction cascades. Studies from different laboratories have convincingly demonstrated binding of recombinant Grb2 or Grb2 SH3 domains to SLP-76 both in cell lysates and on Western blots (9, 21, 27), but the presence of SLP-76 in Grb2 IPs and vice versa have not been reported thus far. Our data argue that SLP-76 is more likely to associate with GrpL than Grb2 under physiological conditions in T cells (Fig. 4). This GrpL/SLP-76 association may regulate signaling pathway(s) involving SLP-76 in T cells. One downstream target for SLP-76 is NF-AT. SLP-76 and Vav can synergistically enhance anti-CD3–induced NF-AT activation in T cells (18). Results in Fig. 6 demonstrate that overexpression of GrpL alone is sufficient to enhance the TCR-induced NF-AT. More importantly, GrpL cooperated with SLP-76 in this pathway as suggested by the additive effect upon overexpression of both GrpL and SLP-76. Given the functions of SLP-76 in regulating T cell development and gene expression through its interaction with the GEF Vav, we propose that GrpL may also modulate the guanine nucleotide exchange activity of Vav via its association with SLP-76 to regulate T cell development and functions. By contrast, Grb2 may function principally via the Sos1 and Sos2 GEFs.
Another feature that further distinguishes GrpL from Grb2 is the lack of association between GrpL and tyrosine-phosphorylated LAT/pp36/38 in NP-40 lysates prepared from TCR-activated T cells (Fig. 5). Detergent-insoluble membrane subdomains known as glycolipid-enriched microdomains or detergent-insoluble rafts concentrated with lipid-linked signaling proteins have been proposed to be focal areas from which receptor-mediated signaling originates (42–44). Recently, components of the activation TCR complex, including tyrosine-phosphorylated LAT, have been localized to such detergent-insoluble rafts, and disruption of these rafts inhibits signaling via the TCR (45, 46). Since GrpL appears to be an integral part of the TCR-mediated NF-AT activation pathway (Figs. 4–6), it is possible that GrpL may also be localized to rafts upon TCR ligation. This may allow GrpL to interact with upstream molecules activated by the TCR, including tyrosine-phosphorylated LAT in rafts. Experiments are now in progress to examine this possibility.
The in vitro binding of GrpLSH2 domain to tyrosine-phosphorylated SHP-2 suggests that SHP-2 may potentially interact with GrpL (Fig. 6). However, we have not yet detected an in vivo association between GrpL and SHP-2 in activated T cells (data not shown). SHP-2 possesses the dual functions of either potentiating or inhibiting signal transduction depending on the receptor system involved. For example, SHP-2 has been shown to be required for coupling receptors to the activation of the MAPK pathway (47, 48). In T cells, SHP-2 interacts with CTLA-4, which is believed to be the mechanism whereby CTLA-4 downregulates T cell activation, possibly via dephosphorylation of Shc (49). Nevertheless, a recent report suggests that SHP-2 may also be a positive regulator of TCR-mediated signaling. Thus, a multimeric complex involving Grb2, LAT/pp36/38, SHP-2, and a 110-kD tyrosine-phosphorylated protein has been reported in T cells (50). Interestingly, expression of a SHP-2 mutant devoid of phosphatase activity inhibits the TCR-induced MAPK pathway activation. Similar to Grb2, the SH2 domain of GrpL mediates binding to SHP-2. This binding is dependent on T cell activation and tyrosine phosphorylation of SHP-2 (Fig. 7). Hence, after TCR ligation, the interaction between Grb2 and SHP-2 may facilitate the redistribution of the GrpL– SLP-76 complex to the plasma membrane. Consequently, SLP-76 would become available for phosphorylation by ZAP-70 to generate the Vav binding sites.
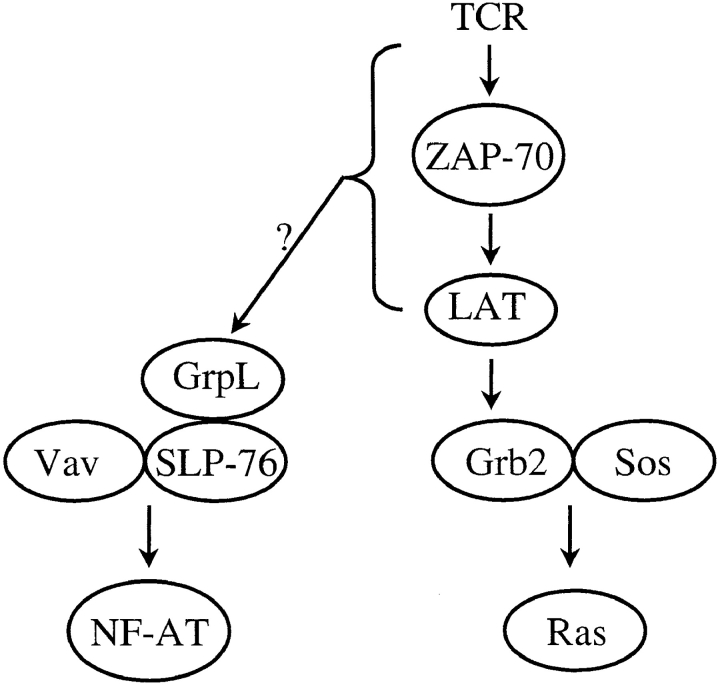
Taken together, our data support a model in which distinct complexes of Grb2/Sos1/Sos2 and GrpL–SLP-76 may be responsible for connecting the TCR to the Ras- and Rho/Rac/CDC42-mediated effector pathways, respectively (Fig. 8). Thus, Grb2 is the primary adaptor molecule responsible for the recruitment of the GEFs Sos1 and Sos2 for Ras (26) to LAT/pp36/38. The lymphoid-restricted, Grb2-like adaptor protein Grap can also serve a similar function by interacting with Sos and Ras (33, 34). Recruitment of Ras to the inner plasma membrane activates distal MAPKs including Erk1 and Erk2 (26). On the other hand, Vav, a GEF for the Rho/Rac/CDC42 family of GTP-binding proteins expressed in lymphoid cells (51, 52), binds to tyrosine-phosphorylated SLP-76 via its SH2 domain (9), and regulates NF-AT activation (18) and cytoskeletal reorganization in T cells (53). Since GrpL associates with SLP-76 and regulates NF-AT activation (Figs. 3–6), one testable possibility is that a trimolecular interaction among SLP-76, Vav, and GrpL may be required for optimal NF-AT activation and reorganization of cytoskeleton during T cell activation.
Figure 8.
A model for how GrpL may function in TCR- mediated signal transduction. Results from our study demonstrate that GrpL and Grb2 may differentially associate with SLP-76, LAT/pp36/p38, and Sos1/2, respectively. The preferential interaction between GrpL and SLP-76 suggests that GrpL may play a role in the regulation NF-AT pathway during TCR-mediated T cell activation.
During the course of this study, two other groups independently identified the same gene and protein as we describe herein, which they have designated as Mona (54) and Gads (55, 56). In vitro data suggest that in nonlymphoid cells, GrpL (Mona/Gads) may interact with either the M-CSF receptor, c-fms (54), or with c-kit (55). In agreement with our results, Liu et al. (56) independently found that GrpL can bind to SLP-76 and together with SLP-76 can promote the activation of NF-AT in T cells. We have recently identified another adaptor protein, BAM-32, which unlike GrpL (Fig. 6 and reference 56), SLP-76 (18–21), or the B cell adaptor BLNK (57), inhibits the activation NF-AT (Marshall, A.J., H. Niiro, T.J. Yun, and E.A. Clark, manuscript in preparation). Thus, antigen receptor–induced activation of NF-AT in lymphocytes is carefully regulated by adaptor proteins.
Acknowledgments
We thank Dr. Gary Koretzky for providing us with the pEF, pEF–SLP-76, and NF-AT Luc expression vectors and the C305 anti-TCR mAb. We also thank G. Lynn Law for access to the luminometer, and Drs. Nancy Boerth, Jennifer Westendorf, and Michelle Tallquist for suggestions on reporter gene assays.
This work was supported by National Institutes of Health grants GM42508, GM37905, and DE00859, and by fellowships from the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation (P.M. Chaudhary) and the Medical Research Council of Canada (A.J. Marshall).
Abbreviations used in this paper
- GEF
guanine nucleotide exchange factor
- GrpL
Grb2-related protein of the lymphoid system
- IP
immunoprecipitate
- MAPK
mitogen-activated protein kinase
- NF-AT
nuclear factor of activated T cells
- PTK
protein tyrosine kinase
- SH
src homology
- SHP
SH2 domain–containing protein tyrosine phosphatase
- SLP-76
SH2 domain–containing leukocyte protein of 76 kD
Footnotes
C.-L. Law's present address is Xcyte Therapies Inc., 2203 Airport Way South, Suite 300, Seattle, WA 98134.
C.-L. Law, M.K. Ewings, and P.M. Chaudhary all contributed equally to this study.
References
- 1.Park DJ, Rho HW, Rhee SG. CD3 stimulation causes phosphorylation of phospholipase C-gamma 1 on serine and tyrosine residues in a human T-cell line. Proc Natl Acad Sci USA. 1991;88:5453–5456. doi: 10.1073/pnas.88.12.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Secrist JP, Karnitz L, Abraham RT. T-cell antigen receptor ligation induces tyrosine phosphorylation of phospholipase C-gamma 1. J Biol Chem. 1991;266:12135–12139. [PubMed] [Google Scholar]
- 3.Weiss A, Koretzky G, Schatzman RC, Kadlecek T. Functional activation of the T-cell antigen receptor induces tyrosine phosphorylation of phospholipase C-gamma 1. Proc Natl Acad Sci USA. 1991;88:5484–5488. doi: 10.1073/pnas.88.13.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukazawa T, Reedquist KA, Panchamoorthy G, Soltoff S, Trub T, Druker B, Cantley L, Shoelson SE, Band H. T cell activation-dependent association between the p85 subunit of the phosphatidylinositol 3-kinase and Grb2/phospholipase C-gamma 1-binding phosphotyrosyl protein pp36/38. J Biol Chem. 1995;270:20177–20182. doi: 10.1074/jbc.270.34.20177. [DOI] [PubMed] [Google Scholar]
- 5.Motto DG, Ross SE, Jackman JK, Sun Q, Olson AL, Findell PR, Koretzky GA. In vivo association of Grb2 with pp116, a substrate of the T cell antigen receptor-activated protein tyrosine kinase. J Biol Chem. 1994;269:21608–21613. [PubMed] [Google Scholar]
- 6.Jackman JK, Motto DG, Sun Q, Tanemoto M, Turck CW, Peltz GA, Koretzky GA, Findell PR. Molecular cloning of SLP-76, a 76-kDa tyrosine phosphoprotein associated with Grb2 in T cells. J Biol Chem. 1995;270:7029–7032. doi: 10.1074/jbc.270.13.7029. [DOI] [PubMed] [Google Scholar]
- 7.Fukazawa T, Reedquist KA, Trub T, Soltoff S, Panchamoorthy G, Druker B, Cantley L, Shoelson SE, Band H. The SH3 domain-binding T cell tyrosyl phosphoprotein p120. Demonstration of its identity with the c-cbl protooncogene product and in vivo complexes with Fyn, Grb2, and phosphatidylinositol 3-kinase. J Biol Chem. 1995;270:19141–19150. doi: 10.1074/jbc.270.32.19141. [DOI] [PubMed] [Google Scholar]
- 8.Margolis B, Hu P, Katzav S, Li W, Oliver JM, Ullrich A, Weiss A, Schlessinger J. Tyrosine phosphorylation of vav proto-oncogene product containing SH2 domain and transcription factor motifs. Nature. 1992;356:71–74. doi: 10.1038/356071a0. [DOI] [PubMed] [Google Scholar]
- 9.Tuosto L, Michel F, Acuto O. p95vav associates with tyrosine-phosphorylated SLP-76 in antigen-stimulated T cells. J Exp Med. 1996;184:1161–1166. doi: 10.1084/jem.184.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilliland LK, Schieven GL, Norris NA, Kanner SB, Aruffo A, Ledbetter JA. Lymphocyte lineage- restricted tyrosine-phosphorylated proteins that bind PLC gamma 1 SH2 domains. J Biol Chem. 1992;267:13610–13616. [PubMed] [Google Scholar]
- 11.Buday L, Egan SE, Rodriguez V-P, Cantrell DA, Downward J. A complex of Grb2 adaptor protein, Sos exchange factor, and a 36-kDa membrane-bound tyrosine phosphoprotein is implicated in ras activation in T cells. J Biol Chem. 1994;269:9019–9023. [PubMed] [Google Scholar]
- 12.Sieh M, Batzer A, Schlessinger J, Weiss A. GRB2 and phospholipase C-gamma 1 associate with a 36- to 38-kilodalton phosphotyrosine protein after T-cell receptor stimulation. Mol Cell Biol. 1994;14:4435–4442. doi: 10.1128/mcb.14.7.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W, Sloan L-J, Kitchen J, Trible RP, Samelson LE. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 1998;92:83–92. doi: 10.1016/s0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
- 14.Weber JR, Ørstavik S, Torgersen KM, Danbolt NC, Berg SF, Ryan JC, Taskén K, Imboden JB, Vaage JT. Molecular cloning of the cDNA encoding pp36, a tyrosine-phosphorylated adaptor protein selectively expressed by T cells and natural killer cells. J Exp Med. 1998;187:1157–1161. doi: 10.1084/jem.187.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss A, Littman DR. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 16.Chan AC, Shaw AS. Regulation of antigen receptor signal transduction by protein tyrosine kinases. Curr Opin Immunol. 1996;8:394–401. doi: 10.1016/s0952-7915(96)80130-0. [DOI] [PubMed] [Google Scholar]
- 17.Wange RL, Samelson LE. Complex complexes: signaling at the TCR. Immunity. 1996;5:197–205. doi: 10.1016/s1074-7613(00)80315-5. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Motto DG, Koretzky GA, Weiss A. Vav and SLP-76 interact and functionally cooperate in IL-2 gene activation. Immunity. 1996;4:593–602. doi: 10.1016/s1074-7613(00)80485-9. [DOI] [PubMed] [Google Scholar]
- 19.Raab M, da Silva AJ, Findell PR, Rudd CE. Regulation of Vav-SLP-76 binding by ZAP-70 and its relevance to TCR zeta/CD3 induction of interleukin-2. Immunity. 1997;6:155–164. doi: 10.1016/s1074-7613(00)80422-7. [DOI] [PubMed] [Google Scholar]
- 20.Fang N, Motto DG, Ross SE, Koretzky GA. Tyrosines 113, 128, and 145 of SLP-76 are required for optimal augmentation of NFAT promoter activity. J Immunol. 1996;157:3769–3773. [PubMed] [Google Scholar]
- 21.Motto DG, Ross SE, Wu J, Hendricks T-LR, Koretzky GA. Implication of the GRB2-associated phosphoprotein SLP-76 in T cell receptor-mediated interleukin 2 production. J Exp Med. 1996;183:1937–1943. doi: 10.1084/jem.183.4.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clements JL, Yang B, Ross-Barta SE, Eliason SL, Hrstka RF, Williamson RA, Koretzky GA. Requirement for the leukocyte-specific adapter SLP-76 for normal T cell development. Science. 1998;281:416–419. doi: 10.1126/science.281.5375.416. [DOI] [PubMed] [Google Scholar]
- 23.Pivniouk V, Tsitsikov E, Swinton P, Rathbun G, Alt FW, Geha RS. Impaired viability and profound block in thymocyte development in mice lacking the adaptor protein SLP-76. Cell. 1998;94:229–238. doi: 10.1016/s0092-8674(00)81422-1. [DOI] [PubMed] [Google Scholar]
- 24.Koretzky GA. The role of Grb2-associated proteins in T-cell activation. Immunol Today. 1997;18:401–406. doi: 10.1016/s0167-5699(97)01088-8. [DOI] [PubMed] [Google Scholar]
- 25.Osman N, Lucas SC, Turner H, Cantrell D. A comparison of the interaction of Shc and the tyrosine kinase ZAP-70 with the T cell antigen receptor zeta chain tyrosine-based activation motif. J Biol Chem. 1995;270:13981–13986. doi: 10.1074/jbc.270.23.13981. [DOI] [PubMed] [Google Scholar]
- 26.Vojtek AB, Der JC. Increasing complexity of the Ras signaling pathway. J Biol Chem. 1998;273:19925–19928. doi: 10.1074/jbc.273.32.19925. [DOI] [PubMed] [Google Scholar]
- 27.Reif K, Buday L, Downward J, Cantrell DA. SH3 domains of the adapter molecule Grb2 complex with two proteins in T cells: the guanine nucleotide exchange protein Sos and a 75-kDa protein that is a substrate for T cell antigen receptor-activated tyrosine kinases. J Biol Chem. 1994;269:14081–14087. [PubMed] [Google Scholar]
- 28.Meisner H, Conway BR, Hartley D, Czech MP. Interactions of Cbl with Grb2 and phosphatidylinositol 3′-kinase in activated Jurkat cells. Mol Cell Biol. 1995;15:3571–3578. doi: 10.1128/mcb.15.7.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donovan JA, Ota Y, Langdon WY, Samelson LE. Regulation of the association of p120cbl with Grb2 in Jurkat T cells. J Biol Chem. 1996;271:26369–26374. doi: 10.1074/jbc.271.42.26369. [DOI] [PubMed] [Google Scholar]
- 30.Wardenburg JB, Fu C, Jackman JK, Flotow H, Wilkinson SE, Williams DH, Johnson R, Kong G, Chan AC, Findell PR. Phosphorylation of SLP-76 by the ZAP-70 protein-tyrosine kinase is required for T-cell receptor function. J Biol Chem. 1996;271:19641–19644. doi: 10.1074/jbc.271.33.19641. [DOI] [PubMed] [Google Scholar]
- 31.Buday L, Khwaja A, Sipeki S, Farago A, Downward J. Interactions of Cbl with two adapter proteins, Grb2 and Crk, upon T cell activation. J Biol Chem. 1996;271:6159–6163. doi: 10.1074/jbc.271.11.6159. [DOI] [PubMed] [Google Scholar]
- 32.Reedquist KA, Fukazawa T, Panchamoorthy G, Langdon WY, Shoelson SE, Druker BJ, Band H. Stimulation through the T cell receptor induces Cbl association with Crk proteins and the guanine nucleotide exchange protein C3G. J Biol Chem. 1996;271:8435–8442. doi: 10.1074/jbc.271.14.8435. [DOI] [PubMed] [Google Scholar]
- 33.Feng GS, Ouyang YB, Hu DP, Shi ZQ, Gentz R, Ni J. Grap is a novel SH3-SH2-SH3 adaptor protein that couples tyrosine kinases to the Ras pathway. J Biol Chem. 1996;271:12129–12132. doi: 10.1074/jbc.271.21.12129. [DOI] [PubMed] [Google Scholar]
- 34.Trub T, Frantz JD, Miyazaki M, Band H, Shoelson SE. The role of a lymphoid-restricted, Grb2-like SH3-SH2-SH3 protein in T cell receptor signaling. J Biol Chem. 1997;272:894–902. doi: 10.1074/jbc.272.2.894. [DOI] [PubMed] [Google Scholar]
- 35.Leprince C, Draves KE, Ledbetter JA, Torres RM, Clark EA. Characterization of molecular components associated with surface immunoglobulin M in human B lymphocytes: presence of tyrosine and serine/threonine protein kinases. Eur J Immunol. 1992;22:2093–2099. doi: 10.1002/eji.1830220820. [DOI] [PubMed] [Google Scholar]
- 36.Mayer BJ, Eck MJ. SH3 domains. Minding your p's and q's. Curr Biol. 1995;5:364–367. doi: 10.1016/s0960-9822(95)00073-x. [DOI] [PubMed] [Google Scholar]
- 37.Kuriyan J, Cowburn D. Modular peptide recognition domains in eukaryotic signaling. Annu Rev Biophys Biomol Struct. 1997;26:259–298. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- 38.Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, Ullrich A, Skolnik EY, Bar S-D, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 39.Musci MA, Motto DG, Ross SE, Fang N, Koretzky GA. Three domains of SLP-76 are required for its optimal function in a T cell line. J Immunol. 1997;159:1639–1647. [PubMed] [Google Scholar]
- 40.da Silva AJ, Li Z, de Vera C, Canto E, Findell P, Rudd CE. Cloning of a novel T-cell protein FYB that binds FYN and SH2-domain-containing leukocyte protein 76 and modulates interleukin 2 production. Proc Natl Acad Sci USA. 1997;94:7493–7498. doi: 10.1073/pnas.94.14.7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musci MA, Hendricks T-LR, Motto DG, Paskind M, Kamens J, Turck CW, Koretzky GA. Molecular cloning of SLAP-130, an SLP-76-associated substrate of the T cell antigen receptor-stimulated protein tyrosine kinases. J Biol Chem. 1997;272:11674–11677. doi: 10.1074/jbc.272.18.11674. [DOI] [PubMed] [Google Scholar]
- 42.Fra AM, Williamson E, Simons K, Parton RG. De novo formation of caveolae in lymphocytes by expression of VIP21-caveolin. Proc Natl Acad Sci USA. 1995;92:8655–8659. doi: 10.1073/pnas.92.19.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown DA, London E. Structure of detergent- resistant membrane domains: does phase separation occur in biological membranes? . Biochem Biophys Res Commun. 1997;240:1–7. doi: 10.1006/bbrc.1997.7575. [DOI] [PubMed] [Google Scholar]
- 44.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W, Trible RP, Samelson LE. LAT palmitoylation: its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation. Immunity. 1998;9:239–246. doi: 10.1016/s1074-7613(00)80606-8. [DOI] [PubMed] [Google Scholar]
- 46.Xavier R, Brennan T, Li Q, McCormack C, Seed B. Membrane compartmentation is required for efficient T cell activation. Immunity. 1998;8:723–732. doi: 10.1016/s1074-7613(00)80577-4. [DOI] [PubMed] [Google Scholar]
- 47.Xiao S, Rose DW, Sasaoka T, Maegawa H, Burke TR, Jr, Roller PP, Shoelson SE, Olefsky JM. Syp (SH-PTP2) is a positive mediator of growth factor-stimulated mitogenic signal transduction. J Biol Chem. 1994;269:21244–21248. [PubMed] [Google Scholar]
- 48.Zhao Z, Tan Z, Wright JH, Diltz CD, Shen SH, Krebs EG, Fischer EH. Altered expression of protein-tyrosine phosphatase 2C in 293 cells affects protein tyrosine phosphorylation and mitogen-activated protein kinase activation. J Biol Chem. 1995;270:11765–11769. doi: 10.1074/jbc.270.20.11765. [DOI] [PubMed] [Google Scholar]
- 49.Marengere LE, Waterhouse P, Duncan GS, Mittrucker HW, Feng GS, Mak TW. Regulation of T cell receptor signaling by tyrosine phosphatase SYP association with CTLA-4. Science. 1996;272:1170–1173. doi: 10.1126/science.272.5265.1170. [DOI] [PubMed] [Google Scholar]
- 50.Frearson JA, Alexander DR. The phosphotyrosine phosphatase SHP-2 participates in a multimeric signaling complex and regulates T cell receptor (TCR) coupling to the Ras/mitogen-activated protein kinase (MAPK) pathway in Jurkat T cells. J Exp Med. 1998;187:1417–1426. doi: 10.1084/jem.187.9.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crespo P, Schuebel KE, Ostrom AA, Gutkind JS, Bustelo XR. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature. 1997;385:169–172. doi: 10.1038/385169a0. [DOI] [PubMed] [Google Scholar]
- 52.Reif K, Cantrell DA. Networking Rho family GTPases in lymphocytes. Immunity. 1998;8:395–401. doi: 10.1016/s1074-7613(00)80545-2. [DOI] [PubMed] [Google Scholar]
- 53.Wardenburg JB, Pappu R, Bu J-Y, Mayer B, Chernoff J, Straus D, Chan AC. Regulation of PAK activation and the T cell cytoskeleton by the linker protein SLP-76. Immunity. 1998;9:607–616. doi: 10.1016/s1074-7613(00)80658-5. [DOI] [PubMed] [Google Scholar]
- 54.Bourette RP, Arnaud S, Myles GM, Blanchet J-P, Rohrschneider LR, Mouchiroud G. Mona, a novel hematopoietic-specific adaptor interacting with the macrophage colony-stimulating factor receptor, is implicated in monocyte/macrophage development. EMBO (Eur Mol Biol Organ) J. 1998;17:7273–7281. doi: 10.1093/emboj/17.24.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu SK, McGlade CJ. Gads is a novel SH2 and SH3 domain-containing adaptor protein that binds to tyrosine-phosphorylated Shc. Oncogene. 1998;17:3073–3082. doi: 10.1038/sj.onc.1202337. [DOI] [PubMed] [Google Scholar]
- 56.Liu SK, Fang N, Koretzky GA, McGlade CJ. The hematopoietic-specific adaptor protein Gads functions in T-cell signaling via interactions with the SLP-76 and LAT adaptors. Curr Biol. 1999;9:67–75. doi: 10.1016/s0960-9822(99)80017-7. [DOI] [PubMed] [Google Scholar]
- 57.Fu C, Turck CW, Kurosaki T, Chan AC. BLNK: a central linker protein in B cell activation. Immunity. 1998;9:93–103. doi: 10.1016/s1074-7613(00)80591-9. [DOI] [PubMed] [Google Scholar]