Abstract
Molecular interactions with the extracellular domains of class I major histocompatibility complex proteins are major determinants of immune recognition that have been extensively studied both physically and biochemically. However, no immunological function has yet been placed on the transmembrane or cytoplasmic amino acid sequences of these proteins despite strict conservation of unique features within each class I major histocompatibility complex locus. Here we report that lysis by a subset of natural killer (NK) cells inhibited by target cell expression of human histocompatibility leukocyte antigen (HLA)-Cw6 or -Cw7 was not inhibited by expression of chimeric proteins consisting of the extracellular domains of HLA-C and the COOH-terminal portion of HLA-G. Assays using transfectants expressing a variety of HLA-Cw6 mutants identified the transmembrane sequence and, in particular, cysteine at position 309 as necessary for inhibition of 68% (25/37) of NK cell lines and 23% (33/145) of NK clones tested. Moreover, these NK clones inhibited by target cell expression of HLA-Cw6 and dependent upon the transmembrane sequence were found not to express or to only dimly express NK inhibitory receptors (NKIR1) that are EB6/HP3E4-positive. Furthermore, assays using monoclonal antibody blocking suggest that an NK receptor other than NKIR1 or CD94 is responsible for recognition dependent upon the transmembrane sequence of HLA-Cw6.
Keywords: NK cells, class I MHC protein, transmembrane sequence, NK receptors, cysteine
NK cells are lymphocytes that provide innate immunity by killing virally infected or tumor cells apparently independently of prior specific antigen stimulation (1). Reduced or absent cell surface expression of class I MHC proteins, which occurs, for example, in virally infected or tumor cells deficient in class I MHC heavy chain β2-microglobulin (β2m)1 or transporter associated with antigen processing (TAP) gene expression, is one factor that renders a cell susceptible to NK cell–mediated lysis. Thus, NK cells survey target cells for absence of self class I MHC proteins, the missing self hypothesis (2, 3), although absent or low class I MHC protein expression is not always necessary or sufficient for susceptibility to NK cell–mediated lysis (4).
NK cells express a wide variety of receptors for class I MHC proteins that are capable of inhibiting or augmenting NK cell cytotoxicity. In particular, 58-kD and 70-kD natural killer inhibitory receptors (NKIR) for class I MHC proteins, containing two and three Ig domains, respectively, inhibit NK cell cytotoxicity (5–7), whereas related 50-kD receptors associated with DAP12 (8) augment NK cell cytotoxicity (9). Other receptors from the Ig superfamily have also been identified (10–14). The C-type lectin complex CD94/NKG2A, or C, can also transduce inhibitory or costimulatory signals, respectively, in NK cells (15) upon recognition of HLA-E (16–18). At least some of these receptors can also regulate the activation of a subset of T cells (19, 20). The repertoire of NK receptors expressed can vary considerably among different donors (21, 22), and the mechanisms by which such a multitude of receptors interact in the regulation of NK cell cytotoxicity remain unclear (23–25). Whether only the extracellular portions of class I MHC proteins are sufficient to facilitate inhibition of NK cell cytotoxicity by initiating signaling through these receptors, or whether class I MHC proteins can play a more active role in NK cell inhibition, e.g., by directly signaling within target cells or by association with other target cell surface proteins, has been little studied.
NKIR enable NK cells to be inhibited only by interaction with particular class I MHC allotypes. In contrast, HLA-G, a class Ib (nonclassical) MHC protein expressed at the maternal–fetal interface (26), can be involved in inhibition of NK cells of diverse phenotypes (27). In part to search for the mechanism facilitating the ability of HLA-G to inhibit NK cell cytotoxicity, the strikingly short cytoplasmic tail and distinct transmembrane region of HLA-G compared with that of HLA-C was investigated. To this end, constructs were made encoding mutants of HLA-C or chimeric proteins made from extracellular portions of HLA-C with the cytoplasmic tail and/or transmembrane region of HLA-G and transfected into the B cell line 721.221, deficient in cell surface expression of class I MHC protein. Transfectants were then tested for their susceptibility to lysis by various NK cell lines and clones. These studies unexpectedly led to the delineation of a role for the transmembrane region in the inhibitory function of HLA-C with respect to a subset of human NK cells.
Materials and Methods
Plasmids Encoding Wild-Type, Chimeric, and Mutant Class I MHC Proteins and Cell Transfectants.
Plasmids containing wild-type genes encoding HLA-C and -G proteins were used as described (27, 28). Plasmids encoding chimeric HLA-Cw6 or HLA-Cw7 with the region encoding the conserved residue Arg 201 (near the beginning of the α3 domain) to the end of the mature protein replaced by that of HLA-G were also prepared as described (29). Proteins encoded by these plasmids were termed HLA-Cw6/ G-tail and HLA-Cw7/G-tail in the investigation of their relative turnover at the cell surface (29), but in the current report, they are renamed HLA-Cw6/G end and HLA-Cw7/G end to emphasize that these proteins contain part of the α3 domain and the complete transmembrane sequence as well as the cytoplasmic tail of HLA-G (Fig. 1).
Figure 1.
Alignment of amino acid sequences of HLA-Cw6, -Cw7, -G, -A2, and the chimera HLA-Cw6/TM G used in the current study. The location of the Bsu36I restriction site used to make the chimeras HLA-Cw6/G end and HLA-Cw7/G end is marked. Residues underlined were those mutated into stop codons in HLA-Cw6, and the cysteine at position 309 that was mutated into tryptophan is marked bold and underlined. The region of HLA-Cw6 altered to make HLA-Cw6/TM G is shaded.
Plasmid inserts encoding HLA-Cw6 truncated by the insertion of a stop codon were obtained by PCR from the plasmid encoding HLA-Cw6 in pcDNA3 and cloned back into pcDNA3 (Invitrogen Corp.) as KpnI–EcoRI fragments. The primers used for the PCR were 5′-GGGGTACCCCGCCGCCACCATGCGGGTCATGGCGCCCCGAACC-3′ for the 5′ end of HLA-Cw6, including an added KpnI restriction site and the Kozak sequence to enhance expression, coupled with each of the following for the 3′ end encoding the EcoRI restriction site and stop codon (underlined): 5′-CCGGAATTCTCATCCACCTGAGCTCTTCCT-3′ for HLA-Cw6/315 stop, 5′-CCGGAATTCTCACGCAGCCTGAGAGCAGCT-3′ for HLA-Cw6/325 stop, 5′-CCGGAATTCTCAGCCCTGGGCACTGTTGCT-3′ for HLA-Cw6/332 stop, and 5′-CCGGAATTCTCACTCATCAGAGCCCTGGGC-3′ for HLA-Cw6/335 stop.
To obtain the plasmid encoding HLA-Cw6 with cysteine at position 309 replaced by tryptophan, the residue in that position in HLA-A and HLA-G, two overlapping fragments were obtained by PCR using the plasmid encoding HLA-Cw6 in pcDNA3 as the template. The 5′ fragment was obtained using a T7 oligo (New England Biolabs, Inc.) as the 5′ primer together with 5′-GCTCTTCCTCCTCCACATCACAAC-3′, and the 3′ fragment was obtained using an SP6 oligo (New England Biolabs, Inc.) as the 3′ primer together with 5′-GTTGTGATGTGGAGGAGGAAGACC-3′. These two fragments were joined by PCR using SP6 and T7 oligos as primers and cloned into pcDNA3 as a KpnI–EcoRI fragment.
The plasmid encoding HLA-Cw6 in which the transmembrane sequence had been replaced by that of HLA-G was made by using three fragments obtained by PCR from the plasmids encoding HLA-Cw6 and HLA-G. The 5′ fragment encoding the cytoplasmic tail of HLA-Cw6 was obtained by PCR using SP6 as the 5′ primer together with 5′-GCTGCTGTGCTGTGGAGGAGGAAGAGCTCA-3′. The fragment encoding the transmembrane domain of HLA-G was obtained by PCR using 3′-TGAGCTCTTCCTCCTCCACAGCACAGCAGC-5′ at the 3′ end and 5′-CTGAGATGGGAGCCATCTTCCCTGCCCACC-5′ at the 5′ end. These two fragments were then joined together by PCR using the appropriate flanking primers, producing a fragment encoding the cytoplasmic tail of HLA-Cw6 attached to the transmembrane domain of HLA-G. The 3′ fragment encoding the extracellular portion of HLA-Cw6 was obtained by PCR with T7 as the 3′ primer together with 5′-GGTGGGCAGGGAAGATGGCTCCCATCTCAG-3′. This fragment was then joined to the fragment encoding the cytoplasmic tail of HLA-Cw6 and transmembrane domain of HLA-G by PCR using T7 and SP6 oligos as primers. This insert was then cloned as a KpnI– EcoRI fragment into pcDNA3. All primers were purchased from Life Technologies and all plasmid inserts were sequenced by the Core Facilities, Dana-Farber Cancer Institute (Boston, MA) using T7, SP6, and primers internal to the class I MHC–encoded region.
The human B cell line 721.221, deficient in cell surface expression of class I MHC proteins (30), was obtained from the American Type Culture Collection (ATCC). 721.221 cells were transfected with 100 μg of linearized plasmids by electroporation as described (28) and continually kept in the selection medium. Transfectants were sorted by flow cytometry to express equivalent levels of class I MHC protein detected by the conformation-specific mAb W6/32 (31). An exception, however, was that the cell surface expression of class I MHC protein in the HLA-G transfectant used was significantly higher than that of the other transfectants, as shown to be necessary for inhibition of NK cell cytotoxicity (27, 32).
Transfectants expressing the extracellular portions of HLA-C also expressed equivalent levels of the epitope for mAb L31 by flow cytometry. This mAb recognizes naturally occurring HLA-C heavy chains not associated with β2m (33). Transfectants were routinely stained with mAb W6/32 or G46-2.6 and analyzed by flow cytometry to ensure that equivalent cell surface expression of class I MHC proteins remained after time in culture. Transfectants also expressed equivalent levels of proteins previously found to be associated with class I MHC proteins (see Discussion), namely, CD20, CD45, CD53, CD81, CD82, HLA-DR, and IL-2R γ chain as determined by flow cytometry using appropriate antibodies. Expression of correctly sized heavy chains from HLA-Cw6, -Cw7, -G, -Cw6/G end, and -Cw7/G end associated with the light chain β2m was confirmed by immunoprecipitation from the transfectants with mAb BBM.1 that recognizes β2m (34) followed by SDS-PAGE analysis. cDNA made from mRNA extracted from these transfectants was used as a template for PCR of the class I MHC–encoding products, which were sequenced to further confirm the sequence of the appropriate MHC product in each transfectant.
Cytotoxicity Assays.
The cytolytic activity of NK lines and clones against various target cell lines and transfectants was assessed in 5-h 35S-release assays as described (28). Assays were performed in duplicate or triplicate, and data values differed by <10% (and on average, ∼5%) of the mean. In all presented cytotoxicity assays, the spontaneous release of 35S was <25% (and on average, ∼10%) of the maximal release.
NK Cell Lines and Clones and mAbs.
NK cell lines 986, I, 264, 268, and 532 were prepared from healthy donors as described (28). NK cell lines 15, 26, 53, 60, 69, 71, 86, and X1 were prepared from cells derived from healthy donors sorted to be CD56+CD3− by flow cytometry and plated at 30 cells/well (FACStarPLUS®; Becton Dickinson). These sorted cells were grown in X-Vivo 10 medium (Bio-Whittaker) supplemented with 0.1% PHA (Murex), 100 U/ml rIL-2 (Boehringer Mannheim), 10% lymphocult (Biotest), 5% human serum (Bio-Whittaker), 100 mM MEM sodium pyruvate (Life Technologies), 1% MEM nonessential amino acids (Life Technologies), 1% 2-ME (Life Technologies), and 50,000/well irradiated PBLs from two allogeneic donors and 10,000/well irradiated RPMI 8866. These sorted cells were restimulated with irradiated feeder cells once every 14 d. All NK lines and clones were periodically monitored to be CD3− and CD56+ by antibody staining. NK cell cytotoxicity assays were performed at least 4 d after restimulation with feeder cells or fresh rIL-2 and were performed using only cells that appeared healthy through a standard laboratory microscope.
mAb to NKIR1, EB6, and HP3E4 were purchased from Immunotech or were a gift from M. Lopez-Botet (Hospital Universitario de la Princesa, Madrid, Spain), respectively. mAb HP3E4 and EB6 both recognize p58 NKIR and can block NK cell recognition of HLA-C alleles belonging to group 1, including HLA-Cw6 and -Cw4, but can recognize serologically distinct epitopes (35–37). mAb recognizing other proteins were used as follows: HLA-C (L31; a gift from P. Giacomini and A.G. Siccardi, San Raffaele Scientific Institute, Milan, Italy), NKB1 (DX9; a gift from L. Lanier, DNAX), CD3 (SK7; Becton Dickinson), CD20 (2H7; PharMingen), CD45 (HI30; PharMingen), CD53 (HI29; PharMingen), CD56 (NCAM16.2; Becton Dickinson), CD81 (JS-81; PharMingen), CD82 (50F11; PharMingen), CD94 (HP3D9; PharMingen), HLA-A, B, C (G46-2.6; PharMingen and W6/32; ATCC), β2m (BBM.1; ATCC), HLA-DR (G46-6; PharMingen), IL-2R γ chain (38024.11; R & D Systems, Inc.). For mAb blocking of NKIR1 and CD94, TEPC183 (Sigma Chemical Co.) and GL183 (mAb to NKIR2; Coulter Immunology) were used as control IgM and IgG1 mAb, respectively.
Results
The COOH-terminal Portion of HLA-C Is a Determinant in Inhibition of NK Cell Cytotoxicity.
The Bsu36I restriction enzyme site in exon 4 of class I MHC genes was used to construct plasmids encoding chimeric proteins of HLA-Cw6 and -Cw7 attached to most of the α3, transmembrane, and cytoplasmic domains of HLA-G, termed HLA-Cw6/G end and HLA-Cw7/G end, respectively (Fig. 1). These chimeric proteins were expressed in the HLA-A,B,C–negative B cell line 721.221 and compared with wild-type HLA-Cw6 and -Cw7 proteins for their capacity to inhibit lysis by various NK cell lines (Fig. 2).
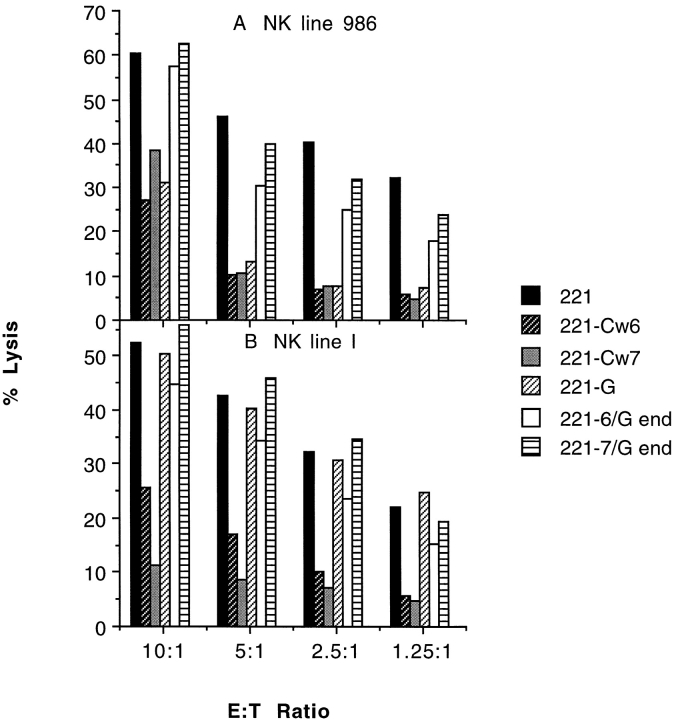
Figure 2.
The COOH-terminal portion of HLA-C is a determinant for inhibition of NK cell cytotoxicity. NK cell lines 986 (A) and I (B), selected by their susceptibility to inhibition by the expression of HLA-C proteins in 721.221 cells, were assayed for their capacity to be inhibited by expression of HLA-Cw6/G end and HLA-Cw7/G end at various E/T cell ratios. The data set shown is representative of three independent experiments using two or three independently transfected target cell lines for each class I MHC protein variant, except HLA-Cw7 and -G, for which only one transfectant was used.
NK lines 986 and I (and four other lines not shown), independently derived from healthy donors, efficiently lysed 721.221 cells at the E/T ratios shown but were inhibited by target cell expression of either HLA-Cw6 or -Cw7. NK line 986, but not line I, was also inhibited by target cell expression of HLA-G. However, neither of these lines (or the four other lines not shown) was inhibited by target cell expression of HLA-Cw6/G end or -Cw7/G end (Fig. 2).
The Cytoplasmic Tail of HLA-C Is Not a Determinant in Inhibition of NK Cell Cytotoxicity.
HLA-G has a short, 6–amino acid cytoplasmic tail compared with the 33–amino acid cytoplasmic tail of HLA-C. Thus, transfectants were made expressing mutants of HLA-Cw6 in which the cytoplasmic tail was truncated by the insertion of stop codons at the various positions marked in Fig. 1. NK lines 268 and 264, derived from healthy donors, with phenotypes of inhibition like lines 986 or I in Fig. 2, were also efficiently inhibited by expression of HLA-Cw6 truncated at positions Lys 315, Ser 325, Ser 332 (not shown), and Ser 335 (Fig. 3). Thus, the cytoplasmic tail after residue 314 can be removed from HLA-Cw6 with no effect on its capacity to inhibit NK cell cytotoxicity.
Figure 3.
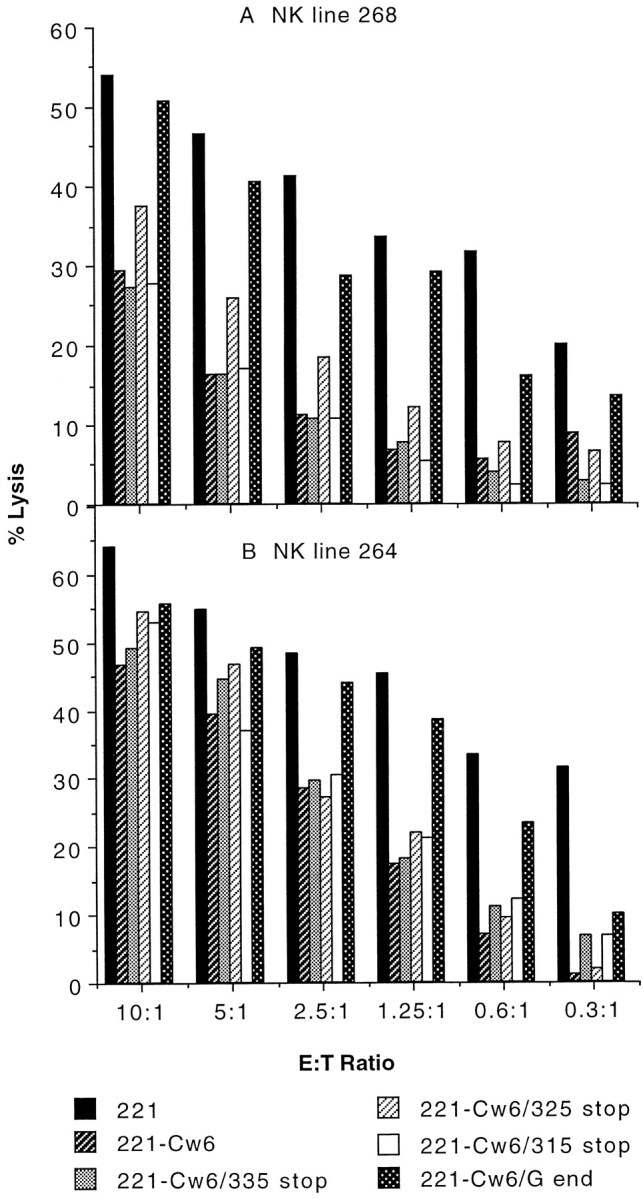
The cytoplasmic tail of HLA-Cw6 is not a determinant in inhibition of NK cell–mediated lysis. NK cell lines 268 (A) and 264 (B), selected by their susceptibility to inhibition by target cell expression of HLA-Cw6, were assayed for their capacity to be inhibited by target cell expression of HLA-Cw6 with truncations placed along various positions in the cytoplasmic tail. Assays were performed at various E/T cell ratios shown. Data sets shown are representative of three independent experiments for each NK line. Another independently made transfectant for HLA-Cw6/315 stop behaved similarly. A transfectant expressing HLA-Cw6 with a stop codon placed at position 332 behaved similarly to other truncated -Cw6 transfectants.
The Transmembrane Sequence and, in Particular, Cysteine at Position 309 of HLA-C Has a Critical Role in Inhibition of NK Cell Cytotoxicity.
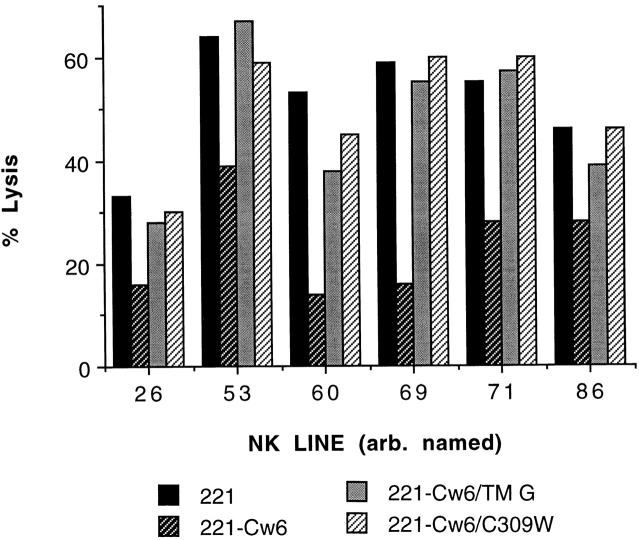
To further investigate the sequence of HLA-Cw6 between residues 201 (the Bsu36I cleavage site) and 315 that, when replaced by that of HLA-G, incapacitates the HLA-Cw6 inhibition of NK cell cytotoxicity, a transfectant expressing HLA-Cw6 with the transmembrane sequence of HLA-G (HLA-Cw6/TM G) was made. Similarly, as the most striking difference between the transmembrane sequences of HLA-Cw6 and HLA-G is at position 309, a transfectant expressing HLA-Cw6 with cysteine at position 309 mutated to tryptophan (HLA-Cw6/C309W), the corresponding residue present in HLA-A and -G, was also made. 25/37 (68%) NK lines were inhibited by target cell expression of HLA-Cw6 but efficiently lysed target cells expressing HLA-Cw6/TM G or HLA-Cw6/C309W (six representative lines shown, Fig. 4). Some of the other 12 NK lines inhibited by target cell expression of HLA-Cw6 partially lysed target cells expressing HLA-Cw6/TM G or -Cw6/C309W. Thus, the transmembrane sequence and, in particular, cysteine at position 309 are critical determinants of the capacity of HLA-Cw6 to inhibit the cytotoxicity of many NK cell lines.
Figure 4.
The transmembrane sequence, and particularly cysteine at position 309, controls the capacity of HLA-Cw6 to inhibit lysis by some NK cell lines. Various arbitrarily numbered NK lines selected by their susceptibility to inhibition by target cell expression of HLA-Cw6 were assayed for their capacity to be inhibited by target cell expression of HLA-Cw6 with the transmembrane sequence replaced by that of HLA-G (HLA-Cw6/TM G) or the cysteine at position 309 replaced by tryptophan (HLA-Cw6/C309W). Lysis was assayed at the E/T ratio of ∼3:1, and at least two independent experiments were performed for each NK line. Two other independently made transfectants of HLA-Cw6/TM G gave similar results. The transfectant expressing HLA-Cw6/G end behaved similarly to that expressing -Cw6/TM G or -Cw6/C309W with all NK lines tested (not shown).
The Subset of NK Clones that Stain Brightly with mAbs EB6 and HP3E4 Is Not Sensitive to the Transmembrane Sequence of HLA-Cw6.
To investigate which NK cells are sensitive to the transmembrane sequence of HLA-Cw6, many NK clones were examined. Three phenotypes of inhibition are illustrated in Fig. 5. NK clone 4 was efficiently inhibited by target cell expression of HLA-Cw6, -Cw6/TM G, and -Cw6/C309W (Fig. 5 A). NK clone 2 was efficiently inhibited by expression of HLA-Cw6 but not significantly inhibited by expression of HLA-Cw6/TM G or -Cw6/ C309W (Fig. 5 B). NK clone 82 was partially inhibited by target cell expression of HLA-Cw6 yet was unaffected by target cell expression of HLA-Cw6/TM G or -Cw6/C309W (Fig. 5 C). In total, 33/145 (23%) NK clones selected for their ability to be inhibited by target cell expression of HLA-Cw6 behaved similarly to NK clones 2 or 82 (Fig. 5, B and C), i.e., they were not inhibited by target cell expression of HLA-Cw6/TM G or -Cw6/C309W. 112/145 (77%) NK clones behaved similarly to NK clone 4 (Fig. 5 A) in that they could be efficiently or partially inhibited by target cell expression of HLA-Cw6, -Cw6/TM G, or -Cw6/C309W. One rare NK clone was even costimulated by target cell expression of HLA-Cw6 but not by expression of -Cw6/TM G or -Cw6/C309W (not shown). Also, many of the NK clones found to be inhibited by target cell expression of HLA-Cw7 efficiently lysed target cells expressing HLA-Cw7/G end (not shown), implying that the importance of the transmembrane sequence in NK cell inhibition is not restricted to HLA-Cw6.
Figure 5.
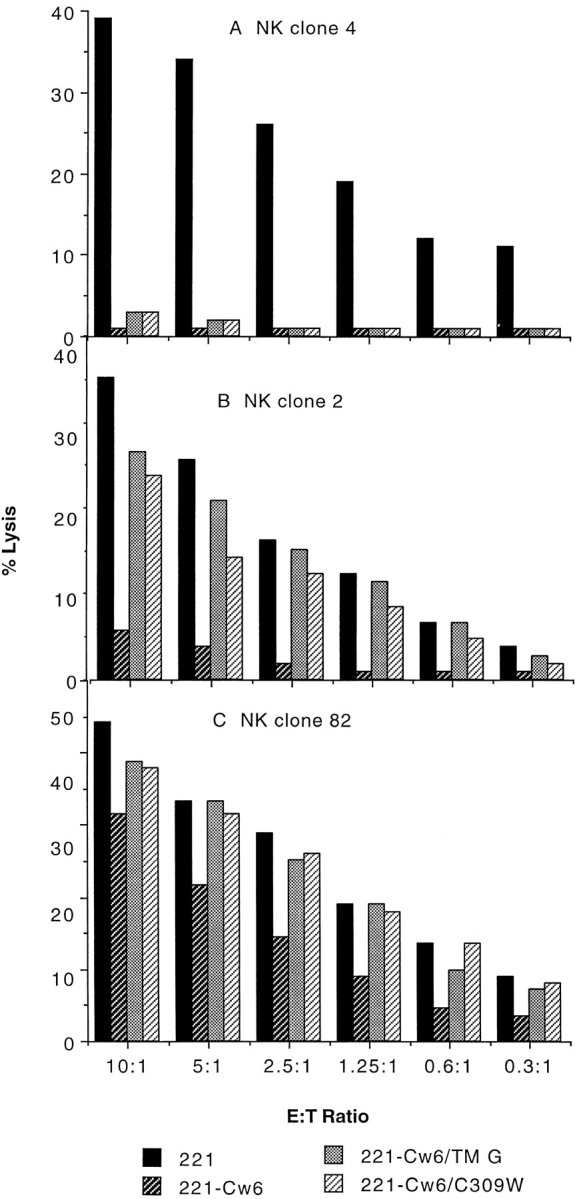
NK clones inhibited by target cell expression of HLA-Cw6 differ in their sensitivity to the transmembrane sequence of the protective class I MHC protein. NK clone 4 (A), NK clone 2 (B), and NK clone 82 (C), selected by their susceptibility to be inhibited by target cell expression of HLA-Cw6, were assayed for lysis of HLA-Cw6/TM G and -Cw6/C309W transfectants at the various E/T cell ratios shown. Data sets shown are representative of two independent experiments performed. Another independently made HLA-Cw6/TM G transfectant gave similar results (not shown). The transfectant expressing HLA-Cw6/G end behaved similarly to that expressing -Cw6/TM G or -Cw6/C309W (not shown).
Some of the NK clones that were inhibited by HLA-Cw6 were phenotyped by mAb staining. All of those tested were CD3− and CD56+, as used in the selection, and happened to be GL183− (mAb for NKIR2) and DX9− (mAb for NKB1). However, they varied considerably in their expression of NKIR1, being either EB6bright/HP3E4bright (clone 84), EB6dim/HP3E4dim (clone G), EB6−/HP3E4− (clone 57) (Fig. 6), or EB6−/HP3E4dim (clones 2 and 82; Table I). Strikingly, clones inhibited by target cell expression of HLA-Cw6, -Cw6/TM G, or -Cw6/C309W were bright for both mAbs EB6 and HP3E4, whereas those inhibited by target cell expression of HLA-Cw6 and not by target cell expression of -Cw6/TM G or -Cw6/C309W were negative or dim for EB6 or HP3E4 (Table I). Furthermore, only those clones bright for EB6 and HP3E4 were also able to be inhibited by target cell expression of HLA-Cw4, as expected by the specificity of the NKIR1 (data not shown).
Figure 6.
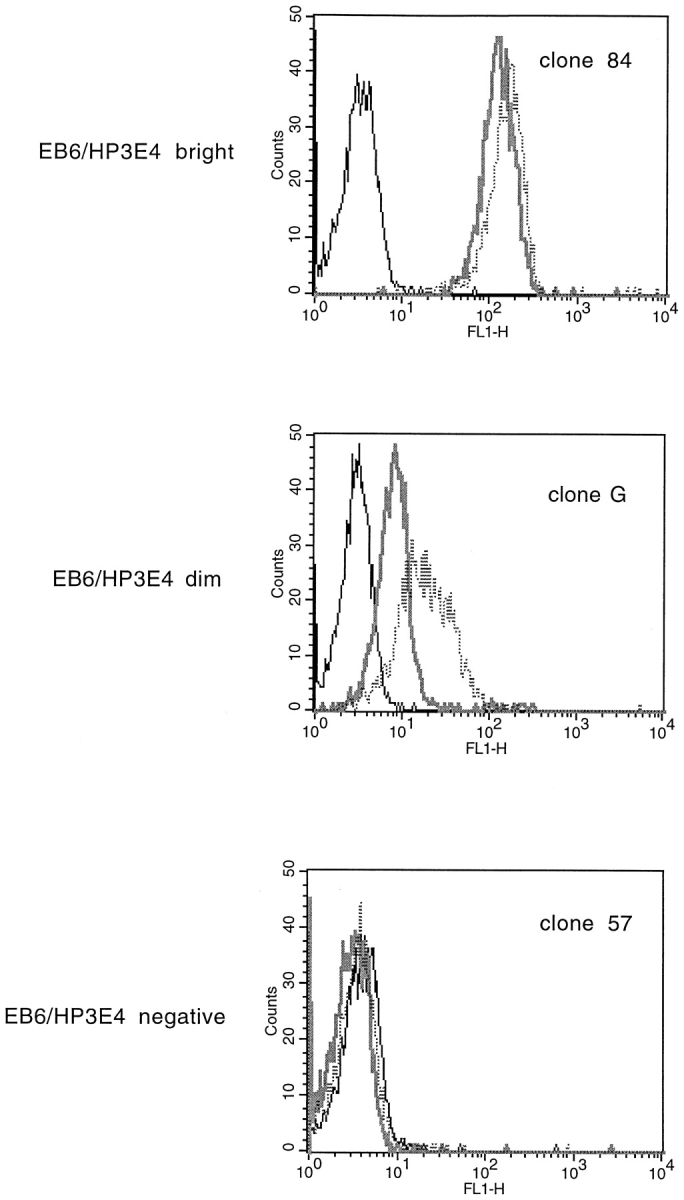
Staining NK clones with mAbs to NK receptors reveals distinct phenotypes between clones that are sensitive to the transmembrane sequence of HLA-Cw6 and those that are not. Histograms of fluorescence intensity of NK clones stained with EB6 (thick gray line), HP3E4 (dotted line), and no first antibody (thin black line), followed by FITC- labeled goat anti–mouse Ig secondary antibody.
Table I.
Relative Intensity of Staining NK Clones by Antibodies Listed with Corresponding Percent Lysis of 721.221 Cells and Transfectants at an E/T Cell Ratio of ∼3:1
| Percent lysis of each transfectant | Antibody staining of NK clone (arbitrary units) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NK clone | 221 | 221-Cw6 | 221-Cw6/C309W | No mAb | HP3E4 | EB6 | ||||||
| B | 56 | 5 | 10 | 2 | 100 | 175 | ||||||
| F | 40 | 0 | 4 | 3 | 81 | 135 | ||||||
| H | 57 | 2 | 13 | 3 | 86 | 87 | ||||||
| 18 | 26 | 11 | 4 | 3 | 67 | 184 | ||||||
| 35 | 27 | 11 | 2 | 3 | 67 | 170 | ||||||
| 95 | 58 | 6 | 8 | 4 | 111 | 125 | ||||||
| 84 | 45 | 1 | 5 | 3 | 165 | 124 | ||||||
| 4 | 39 | 1 | 3 | 2 | 115 | 593 | ||||||
| 2 | 37 | 6 | 28 | 2 | 29 | 4 | ||||||
| 82 | 32 | 16 | 29 | 2 | 48 | 6 | ||||||
| 57 | 57 | 23 | 46 | 4 | 4 | 4 | ||||||
| 59 | 59 | 11 | 45 | 4 | 4 | 3 | ||||||
| 64 | 22 | 6 | 21 | 3 | 3 | 3 | ||||||
| 67 | 57 | 25 | 50 | 4 | 4 | 4 | ||||||
| G | 42 | 25 | 36 | 3 | 19 | 8 | ||||||
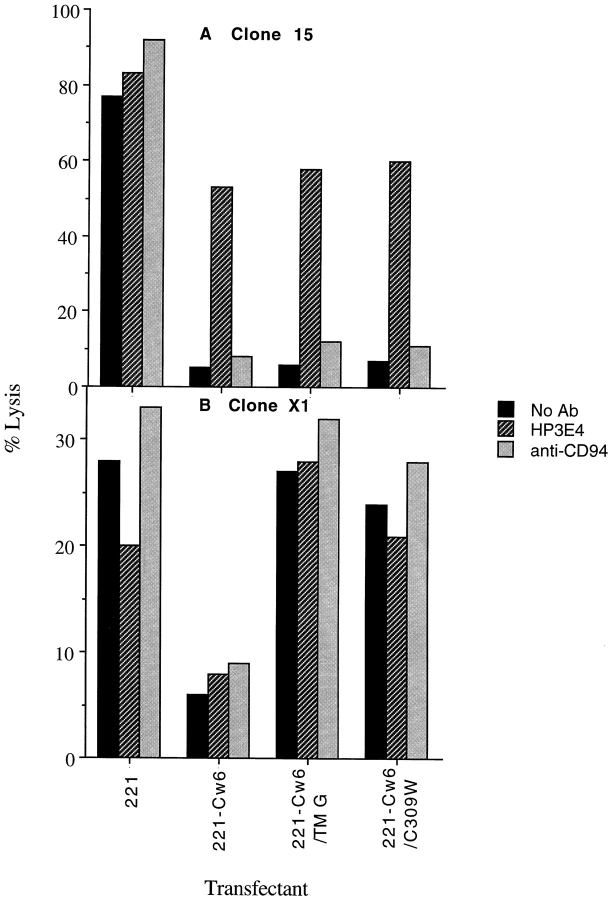
Reversal of NK Cell Inhibition by mAb to NK Receptors.
To directly determine the NK receptors that facilitate inhibition of clones by target cell expression of HLA-Cw6, the effect of blocking NKIR and CD94 with mAb HP3E4 and HP3D9, respectively, was assayed. Two representative clones are shown (Fig. 7). NK clone 15 was efficiently inhibited by target cell expression of either HLA-Cw6, -Cw6/TM G, or -Cw6/C309W, and this inhibition could be efficiently blocked by mAb HP3E4 to NKIR1 and not by mAb HP3D9 to CD94. In contrast, NK clone X1 efficiently lysed target cells expressing HLA-Cw6/TM G or -Cw6/C309W, and in this case, the inhibition mediated by expression of HLA-Cw6 was unaffected by mAb to either NKIR1 or CD94. This implies that an NK receptor other than NKIR1 or CD94 mediates inhibition dependent upon the transmembrane sequence of HLA-Cw6.
Figure 7.
NK clones inhibited by target cell expression of HLA-Cw6 differing in their sensitivity to the transmembrane sequence of the protective class I MHC protein also differ in their sensitivity to blocking by mAb HP3E4. NK clone 15 (A) and NK clone X1 (B), selected by their susceptibility to inhibition by target cell expression of HLA-Cw6, were assayed for lysis of HLA-Cw6/TM G and -Cw6/C309W transfectants at the E/T cell ratio of 3:1 in the presence and absence of mAb HP3E4 (anti-NKIR1), as ascites diluted 1:1,000, and 20 μg/ml anti-CD94 mAb. Isotype control mAb TEPC183 (IgM ascites control for HP3E4) and GL183 (IgG1 control for CD94) had no effect on the NK cell lysis of any of the target transfectants (not shown). Data sets shown are representative of two independent experiments performed. Several other NK lines and clones behaved similarly to those shown. Inhibition of a minor subset of NK lines and clones by target cell expression of HLA-Cw6 could be blocked by anti-CD94 mAb with no correlation to whether or not each particular NK line or clone could be inhibited by target cell expression of HLA-Cw6/TM G or -Cw6/C309W (not shown).
Discussion
The attachment of the COOH-terminal portion of HLA-G after conserved codon Arg 201 renders the α1 and α2 extracellular domains of HLA-Cw6 or -Cw7 ineffective at inhibition of 25/37 NK cell lines. Thus, cell surface expression of the extracellular domains of an appropriate HLA-C protein is not sufficient for inhibition of NK cell– mediated lysis. Unexpectedly, the transmembrane sequence, and particularly Cys 309, was found to be critical in facilitating HLA-Cw6–mediated inhibition of cytotoxicity of many NK cell lines.
68% (25/37) of NK lines, yet only a smaller subset of NK clones, i.e., 33/145 (23%), were sensitive to the transmembrane sequence of HLA-Cw6. This apparent discrepancy may be because (a) clones are not a proportionate representation of cells within a line, i.e., a selection occurs during the cloning procedure; (b) a selection occurs by the use of NK lines that were inhibited by HLA-Cw6, and the clones used were not necessarily obtained from lines that were inhibited by HLA-Cw6; or (c) a minority of cells within an NK cell line can dominate the overall cytotoxicity of the line through secretion of inhibitory or costimulating cytokines or another method of intercellular communication.
All EB6/HP3E4 NKIR1+ NK clones recognized HLA-Cw6 independently of the transmembrane sequence (Table I and Fig. 7 A). However, inhibition of EB6−/HP3E4− or EB6dim/HP3E4dim NK clones by HLA-Cw6 was clearly dependent on the transmembrane region and particularly on Cys 309. This inhibition is unlikely to function through the CD94/NKG2/HLA-E interaction, as there was no correlation between NK lines and clones that were sensitive to the transmembrane sequence of HLA-Cw6 and those for which inhibition could be reversed by anti-CD94 mAb (Fig. 7 B). Moreover, the same leader peptide that facilitates HLA-E expression (16–18) is encoded within each HLA-Cw6 construct. Thus, the NK receptor responsible for recognition dependent upon the transmembrane sequence of HLA-Cw6 is unknown. It may be a previously cloned receptor such as a member of the ILT/LIR family (10–12), or it could be an uncloned receptor such as that postulated to associate with NKIR via zinc (38). Allogeneic T cell lines that lyse 721.221 transfectants expressing HLA-Cw6 equally lysed target cells expressing -Cw6/TM G or -Cw6/ C309W (data not shown), implying that these receptors do not function on the bulk of T cells.
A glycosylphosphatidylinositol (GPI)-linked variant of the mouse class I MHC protein, H-2Dd, in H-2b tumor cells was sufficient to protect against rejection by NK cells after transplantation (39). Thus, replacement of the transmembrane and cytoplasmic domains of this class I MHC protein with GPI does not affect the capacity of the mouse class I MHC protein to protect against NK cell cytotoxicity. The apparent discrepancy between this finding and the current work may be accounted for in several ways: (a) the GPI linkage and subsequent association of cytoplasmic tyrosine kinases (40) facilitates signaling within the target cell that may converge with the natural transmembrane-mediated signal pathway; (b) the GPI linkage causes similar cell surface localization/clustering that the cysteine-containing transmembrane sequence facilitates; or (c) the repertoire of inhibitory ligands on mouse NK cells is different from that expressed on human NK cells, and mouse NK cell receptors are less or not at all sensitive to the transmembrane sequence of the inhibitory MHC protein.
Aspects of class I MHC protein expression that may be altered by mutation of the transmembrane sequence include: (a) oligomerization (41, 42); (b) association with another protein on the target cell surface (43–46); (c) expression of a protein associated in transport; (d) particular localization within cell membrane domains, perhaps facilitated by bound palmitic acid (47, 48); (e) conformational change, perhaps induced by association of particular lipids (49); or (f) capacity for internalization of protein from the cell surface (29). However, dimerized or oligomeric class I MHC protein at the cell surface was not detected by immunoprecipitating surface-biotinylated class I MHC protein from HLA-Cw6 or -Cw6/TM G transfectants, and no evidence was found of a second specific protein component in the immunoprecipitated material (data not shown). Moreover, confocal fluorescence microscopy failed to reveal any change in the surface or intracellular localization of HLA-Cw6 with an altered transmembrane sequence (data not shown). However, this does not rule out the possibility that the transmembrane sequence is critical in facilitating transient dimerization of HLA-Cw6 or association with another protein upon ligation of an NK receptor.
Cysteine at position 309 of class I MHC proteins is most likely located at the interface of the inner leaflet of the lipid bilayer and cytoplasm where the local environment is probably not dissimilar to the extremely viscous surfactant/ aqueous interface widely studied in model colloidal systems such as lipid vesicles and micelles (50, 51). The unique physical conditions of the interfacial environment may be critical to the molecular mechanism by which the cysteine facilitates some aspect of class I MHC protein presentation through noncovalent interactions. For example, the transmembrane sequence may control the lateral mobility of class I MHC protein within the cell membrane.
In summary, a critical physiological function for the transmembrane sequence of class I MHC proteins in its interaction with a subset of NK cells has been shown. A cysteine within this transmembrane sequence located at or near the interface of the inner leaflet of the lipid bilayer and cytoplasm is particularly important. This cysteine is present in HLA-B and -C proteins but not in HLA-A, -G, -E, -F or CD1a, b, c, or d (although CD1d has a cysteine located centrally within the transmembrane region). HLA-B and -C are major ligands for NKIR, and it is thus intriguing to wonder whether the presence of a cysteine at this location was conserved specifically to facilitate some aspect of NK cell recognition.
Acknowledgments
We thank those cited for gifts of cells and mAbs. D.M. Davis is grateful for a useful mix of critical discussion and welcome encouragement from Katie Nicholls, Clive Gerner, Hugh Reyburn, Laszlo Pazmany, Guido Guidotti, Mar Vales-Gomez, Brian Wilson, George Cohen, Pratap Malik, and Basya Rybalov.
This work was supported by a National Institutes of Health research grant (CA-47554), by a postdoctoral fellowship from The Irvington Institute for Immunological Research, New York, to D.M. Davis, and by the Cancer Research Fund of The Damon Runyon-Walter Winchell Foundation Fellowship grant No. DRG 1454 to O. Mandelboim.
Abbreviations used in this paper
- β2m
β2-microglobulin
- GPI
glycosylphosphatidylinositol
- NKIR
natural killer cell inhibitory receptor
Footnotes
Presented in abstract form at The British Society for Immunology 6th Annual Congress, Harrogate, England. 1–4 December 1998.
References
- 1.Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ljunggren H-G, Karre K. In search of the missing self: MHC molecules and NK recognition. Immunol Today. 1990;11:7–10. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 3.Karre K. How to recognise a foreign submarine. Immunol Rev. 1997;155:5–9. doi: 10.1111/j.1600-065x.1997.tb00935.x. [DOI] [PubMed] [Google Scholar]
- 4.Malnati MS, Lusso P, Ciccone E, Moretta A, Moretta L, Long EO. Recognition of virus-infected cells by natural killer cell clones is controlled by polymorphic target cell elements. J Exp Med. 1993;178:961–969. doi: 10.1084/jem.178.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colonna M, Samaridis J. Cloning of immunoglobulin superfamily members associated with HLA-C allele and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 6.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati MS, Vitale M, Bottino C, Moretta L, Moretta A, Long EO. Molecular clones of the p58 NK cell receptor reveal immunoglobulin related molecules with diversity in both the extra- and intra-cellular domains. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 7.D'Andrea A, Chang C, Franz-Bacon K, McClanahan T, Phillips JH, Lanier LL. Molecular cloning of NKB1: a natural killer cell receptor for HLA-B haplotype. J Immunol. 1995;155:2306–2310. [PubMed] [Google Scholar]
- 8.Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 9.Moretta A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R, Bottino C, Moretta L. Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J Exp Med. 1995;182:875–884. doi: 10.1084/jem.182.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colonna M, Navarro F, Bellon T, Liano M, Garcia P, Samaradis J, Angman L, Cella M, Lopez-Botet M. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu M-L. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 12.Borges L, Hsu M-L, Fanger N, Kubin M, Cosman D. A family of human lymphoid and myeloid Ig-like receptors, some of which bind to MHC class I molecules. J Immunol. 1997;159:5192–5196. [PubMed] [Google Scholar]
- 13.Selvakumar A, Steffans U, Dupont B. NK cell receptor gene of the KIR family with two IG domains but highest homology to KIR receptors with three IG domains. Tissue Antigens. 1996;48:285–295. doi: 10.1111/j.1399-0039.1996.tb02647.x. [DOI] [PubMed] [Google Scholar]
- 14.Cantoni C, Verdiani S, Falco M, Pessino A, Cilli M, Conte R, Pende D, Ponte M, Mikaelsson MS, Moretta L, et al. p49, a putative HLA class I-specific inhibitory NK receptor belonging to the immunoglobulin superfamily. Eur J Immunol. 1998;28:1980–1990. doi: 10.1002/(SICI)1521-4141(199806)28:06<1980::AID-IMMU1980>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Botet M, Perez-Villar JJ, Carretero M, Rodriguez A, Melero I, Bellon T, Liano M, Navarro F. Structure and function of the CD94 C-type lectin receptor complex involved in recognition of HLA class I molecules. Immunol Rev. 1997;155:165–174. doi: 10.1111/j.1600-065x.1997.tb00949.x. [DOI] [PubMed] [Google Scholar]
- 16.Borrego F, Ulbert M, Weiss EH, Coloigen JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA-E) complexed with HLA class I signal sequence–derived peptide by CD94/NKG2 confers protection from natural killer cell–mediated lysis. J Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Lanier LL, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B, and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 18.Lee N, Liano M, Carretero M, Ishitani A, Navarro F, Lopez-Botet M, Geraghty DE. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/ NKG2. Proc Natl Acad Sci USA. 1998;95:5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips JH, Gumperz JE, Parham P, Lanier LL. Superantigen dependent, cell mediated cytotoxicity inhibited by MHC class I receptors on T lymphocytes. Science. 1995;268:403–405. doi: 10.1126/science.7716542. [DOI] [PubMed] [Google Scholar]
- 20.Mandelboim O, Davis DM, Reyburn HT, Vales-Gomez M, Sheu EG, Pazmany L, Strominger JL. Enhancement of class II-restricted T cell responses by costimulatory NK receptors for class I MHC proteins. Science. 1996;274:2097–2100. doi: 10.1126/science.274.5295.2097. [DOI] [PubMed] [Google Scholar]
- 21.Valiante NM, Uhrberg M, Shilling HG, Lienart-Weidenbach K, Arnett KL, D'Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 22.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 23.Renard V, Cambiaggi A, Vely F, Blery M, Olcese L, Olivero S, Bouchet M, Vivier E. Transduction of cytotoxic signals in natural killer cells: a general model of fine tuning between activatory and inhibitory pathways in lymphocytes. Immunol Rev. 1997;155:205–221. doi: 10.1111/j.1600-065x.1997.tb00953.x. [DOI] [PubMed] [Google Scholar]
- 24.Lanier LL. NK cells: from no receptors to too many. Immunity. 1997;6:371–378. doi: 10.1016/s1074-7613(00)80280-0. [DOI] [PubMed] [Google Scholar]
- 25.Lanier LL. NK cell receptors. Adv Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 26.McMaster MT, Librach CL, Zhou Y, Lim K-H, Janatpour MJ, DeMars R, Kovats S, Damsky C, Fisher SJ. Human placental HLA-G expression is restricted to differentiated cytotrophoblasts. J Immunol. 1995;154:3771–3778. [PubMed] [Google Scholar]
- 27.Pazmany L, Mandelboim O, Vales-Gomez M, Davis DM, Reyburn HT, Strominger JL. Protection from natural killer cell-mediated lysis by HLA-G expression on target cells. Science. 1996;274:792–795. doi: 10.1126/science.274.5288.792. [DOI] [PubMed] [Google Scholar]
- 28.Mandelboim O, Reyburn HT, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, Strominger JL. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis DM, Reyburn HT, Pazmany L, Chiu I, Mandelboim O, Strominger JL. Impaired spontaneous endocytosis of HLA-G. Eur J Immunol. 1997;27:2714–2719. doi: 10.1002/eji.1830271035. [DOI] [PubMed] [Google Scholar]
- 30.Shimizu Y, DeMars RJ. Production of human cells expressing individual transferred HLA-A, -B, -C, genes using an HLA-A, -B, -C, null human cell line. J Immunol. 1989;142:3320–3328. [PubMed] [Google Scholar]
- 31.Parham P, Barnstable CJ, Bodmer WF. Use of monoclonal antibody (W6/32) in structural studies of HLA-A,B,C antigens. J Immunol. 1979;23:342–349. [PubMed] [Google Scholar]
- 32.Mandelboim O, Pazmany L, Davis DM, Vales-Gomez M, Reyburn HT, Rybalov B, Strominger JL. Multiple receptors for HLA-G on human natural killer cells. Proc Natl Acad Sci USA. 1997;94:14666–14670. doi: 10.1073/pnas.94.26.14666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giacomini P, Beretta A, Nicotra MR, Ciccarelli G, Martayan A, Cerboni C, Lopalco L, Bini D, Delfino L, Ferrara GB, et al. HLA-C heavy chains free of β2-microglobulin: distribution in normal tissues and neoplastic lesions of non-lymphoid origin and interferon-γ responsiveness. Tissue Antigens. 1997;50:555–566. doi: 10.1111/j.1399-0039.1997.tb02913.x. [DOI] [PubMed] [Google Scholar]
- 34.Brodsky FM, Bodmer WF, Parham P. Characterization of a monoclonal anti-beta 2-microglobulin antibody and its use in the genetic and biochemical analysis of major histocompatibility antigens. Eur J Immunol. 1979;9:536–545. doi: 10.1002/eji.1830090709. [DOI] [PubMed] [Google Scholar]
- 35.Lanier LL, Gumperz JE, Parham P, Melero I, Lopez-Botet M, Phillips JH. The NKB1 and HP3E4 NK cell receptors are structurally distinct glycoproteins and independently recognise polymorphic HLA-B and HLA-C molecules. J Immunol. 1995;154:3320–3327. [PubMed] [Google Scholar]
- 36.Melero I, Salmeron A, Balboa MA, Aramburu J, Lopez-Botet M. Tyrosine kinase-dependent activation of human NK cell functions upon stimulation through a 58-kDa surface antigen selectively expressed on discrete subsets of NK cell and T lymphocytes. J Immunol. 1994;152:1662–1673. [PubMed] [Google Scholar]
- 37.Moretta A, Bottino C, Pende D, Tripodi G, Tambussi G, Viale O, Orengo A, Barbaresi M, Merli A, Ciccone E, Moretta L. Identification of four subsets of human CD3−CD16+natural killer (NK) cells by expression of clonally distributed functional surface molecules: correlation between subset assignment of NK clones and ability to mediate specific alloantigen recognition. J Exp Med. 1990;172:1589–1598. doi: 10.1084/jem.172.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajagopalan S, Long EO. Zinc bound to the killer cell-inhibitory receptor modulates the negative signal in human NK cells. J Immunol. 1998;161:1299–1305. [PubMed] [Google Scholar]
- 39.Sentman CL, Olsson-Alheim MY, Lendahl U, Karre K. Influence of glycosylphosphatidylinositol-linked H-2Dd molecules on target cell protection and natural killer cell specificity in transgenic mice. Eur J Immunol. 1996;26:2127–2132. doi: 10.1002/eji.1830260925. [DOI] [PubMed] [Google Scholar]
- 40.Bohuslav J, Cinek T, Horejsi V. Large, detergent-resistant complexes containing murine antigens Thy-1 and Ly-6 and protein tyrosine kinase p56lck. Eur J Immunol. 1993;23:825–831. doi: 10.1002/eji.1830230409. [DOI] [PubMed] [Google Scholar]
- 41.Heldin C-H. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 42.Klemm JD, Schreiber SL, Crabtree GR. Dimerization as a regulatory mechanism in signal transduction. Annu Rev Immunol. 1998;16:569–592. doi: 10.1146/annurev.immunol.16.1.569. [DOI] [PubMed] [Google Scholar]
- 43.Szollosi J, Horejsi V, Bene L, Angelisova P, Damjanovich S. Supramolecular complexes of MHC class I, MHC class II, CD20 and tetraspan molecules (CD53, CD81, and CD82) at the surface of a B cell line JY. J Immunol. 1996;157:2939–2946. [PubMed] [Google Scholar]
- 44.Skov S. Intracellular signal transduction mediated by ligation of MHC class I molecules. Tissue Antigens. 1998;51:215–223. [PubMed] [Google Scholar]
- 45.Wagner N, Engel P, Vega M, Tedder TF. Ligation of MHC class I and class II molecules can lead to heterologous desensitization of signal transduction pathways that regulate homotypic adhesion in human lymphocytes. J Immunol. 1994;152:5275–5287. [PubMed] [Google Scholar]
- 46.Tscherning T, Claesson MH. Signal transduction via MHC class-I molecules in T cells. Scand J Immunol. 1994;39:117–121. doi: 10.1111/j.1365-3083.1994.tb03349.x. [DOI] [PubMed] [Google Scholar]
- 47.Kaufman JF, Krangel MS, Strominger JL. Cysteines in the transmembrane region of major histocompatibility antigens are fatty acylated via thioester bonds. J Biol Chem. 1984;259:7230–7238. [PubMed] [Google Scholar]
- 48.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 49.Dan N, Safran SA. Effect of lipid characteristics on the structure of transmembrane proteins. Biophys J. 1998;75:1410–1414. doi: 10.1016/S0006-3495(98)74059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hasegawa M, Sugimura T, Kuraishi K, Shindo Y, Kitahara A. Microviscosity in AOT reverse micellar core determined with a viscosity sensitive fluorescence probe. Chem Lett. 1992;7:1373–1380. [Google Scholar]
- 51.Davis DM, McLoskey D, Birch DJ, Gellert PR, Kittlety RS, Swart RM. The fluorescence and circular dichroism of proteins in reverse micelles: application to the photophysics of human serum albumin and N-acetyl-l-tryptophanamide. Biophys Chem. 1996;60:63–77. doi: 10.1016/0301-4622(96)00016-6. [DOI] [PubMed] [Google Scholar]