Abstract
We have recently shown that expression of the enzyme indoleamine 2,3-dioxygenase (IDO) during murine pregnancy is required to prevent rejection of the allogeneic fetus by maternal T cells. In addition to their role in pregnancy, IDO-expressing cells are widely distributed in primary and secondary lymphoid organs. Here we show that monocytes that have differentiated under the influence of macrophage colony-stimulating factor acquire the ability to suppress T cell proliferation in vitro via rapid and selective degradation of tryptophan by IDO. IDO was induced in macrophages by a synergistic combination of the T cell–derived signals IFN-γ and CD40-ligand. Inhibition of IDO with the 1-methyl analogue of tryptophan prevented macrophage-mediated suppression. Purified T cells activated under tryptophan-deficient conditions were able to synthesize protein, enter the cell cycle, and progress normally through the initial stages of G1, including upregulation of IL-2 receptor and synthesis of IL-2. However, in the absence of tryptophan, cell cycle progression halted at a mid-G1 arrest point. Restoration of tryptophan to arrested cells was not sufficient to allow further cell cycle progression nor was costimulation via CD28. T cells could exit the arrested state only if a second round of T cell receptor signaling was provided in the presence of tryptophan. These data reveal a novel mechanism by which antigen-presenting cells can regulate T cell activation via tryptophan catabolism. We speculate that expression of IDO by certain antigen presenting cells in vivo allows them to suppress unwanted T cell responses.
Keywords: macrophage; indoleamine 2,3-dioxygenase; T cells; tryptophan; macrophage colony-stimulating factor
Certain macrophages (Møs)1 and possibly other subsets of APCs suppress T cell responses (1, 2). Immunosuppressive APCs have been hypothesized to play an important role in maintaining peripheral T cell tolerance. We have previously shown that Møs that differentiate in vitro under the influence of macrophage colony-stimulating factor (MCSF) acquire the ability to suppress T cell proliferation (3, 4). This attribute was not constitutively present but rather was invoked only in response to attempted T cell activation. Suppressor activity was restricted to specific Mø phenotypes (e.g., the phenotype produced by MCSF), with other phenotypes supporting normal T cell activation (3). Taken together, these characteristics suggested that the inhibitory properties of MCSF-derived Møs might reflect a physiologic system for regulating T cell activation. However, the mechanism of this inhibition was unknown.
In the course of our studies, we found that MCSF-derived Møs were capable of rapidly and selectively depleting the essential amino acid tryptophan from cocultures and that this depletion occurred only in response to attempted T cell activation. Møs are known to possess the inducible tryptophan-degrading enzyme indoleamine 2,3-dioxygenase (IDO), which catalyzes the initial and rate-limiting step in the metabolism of tryptophan along the kynurenine pathway (5–8). It has been postulated that the role of IDO is to inhibit proliferation of eukaryotic intracellular pathogens (9–13) or tumor cells (14) by depriving them of tryptophan. At the time of this study, however, no role had been proposed for IDO in regulating T cell responses. Recently, we have reported that IDO expression in placenta is critically involved in preventing rejection of the allogeneic fetus by maternal T cells (15). The current study tests the hypothesis that tryptophan depletion via IDO is the mechanism by which MCSF-derived Møs inhibit T cell activation in vitro and identifies a tryptophan-sensitive cell cycle arrest point during T cell activation.
Materials and Methods
Reagents.
Recombinant human MCSF was the gift of Genetics Institute, Cambridge, MA. Recombinant human IFN-γ was the gift of Genentech, South San Francisco, CA. Recombinant human CD40 ligand (CD40L) homotrimer was the gift of W. Fanslow, Immunex Corp., Seattle, WA. The IDO inhibitor 1-methyl-d,l-tryptophan (16) was purchased from Aldrich Chemical Co. 6-nitro-tryptophan (17) was synthesized by D. Boykin, Georgia State University, Atlanta, GA, using a modification of the method of Moriya et al. (18). Polyclonal antiserum against human IFN-γ was obtained from Biosource International. All other reagents were obtained from Sigma Chemical Co. unless otherwise specified.
Cell Isolation and Culture.
Human peripheral blood monocytes and lymphocytes were isolated from healthy volunteer donors by leukocytapheresis and counterflow centrifugal elutriation, following appropriate informed consent under a protocol approved by our Institutional Review Board. Monocytes (>95% purity by cell surface markers) were cultured in 96-well plates as previously described (4) using RPMI 1640 with 10% newborn calf serum (Hyclone) plus MCSF (200 U/ml).
T cell activation studies in cocultures were performed as previously described (4), using the above medium supplemented with an additional 5% FCS. In brief, Møs (5 × 104 cells/well) were allowed to differentiate for 4–6 d in MCSF, and then autologous lymphocytes (2 × 105 cells/well) were added along with mitogen. The mitogens used in this study were anti-CD3 mAb (100 ng/ml, clone OKT3; American Type Culture Collection) and staphylococcal enterotoxin B (5 μg/ml; Sigma Chemical Co.). Both gave equivalent results; the data shown are from anti-CD3 unless otherwise specified. T cell proliferation was assessed by standard thymidine incorporation assay as described (3). When T cell activation was studied without Møs, fresh autologous monocytes were added (1:4) as nonsuppressive accessory cells.
Conditioned medium from cocultures of T cells and Møs was prepared by harvesting supernatant 48 h after T cell addition. Conditioned medium was then used to support a second round of T cell activation. Mitogen and other additives were prepared in tryptophan-free buffers.
A chemically defined, serum-free medium (19) selectively deficient in tryptophan was prepared using tryptophan-free RPMI 1640 (Select-amine kit; GIBCO BRL) supplemented with insulin (10 μg/ml), iron-saturated transferrin (5 μg/ml), and BSA (1 mg/ml ultra-pure grade; measured concentration of free tryptophan <5 nM). Preliminary validation experiments confirmed that T cell proliferation in this medium was undetectable but was comparable to serum-based medium when tryptophan was added. To study T cells in the absence of Møs, T cells were activated using anti-CD3 mAb adsorbed onto plastic tissue culture wells (0.5 μg/cm2 in bicarbonate buffer, pH 9) plus soluble anti-CD28 mAb (1 μg/ml; PharMingen).
Tryptophan and IDO Assays.
The tryptophan-degrading activity of Møs reflects a multifactored combination of IDO expression, tryptophan transport into the cells, and intracellular conditions that posttranslationally affect enzyme activity (20). Therefore, when tryptophan depletion was the outcome of interest, we measured the rate of disappearance of tryptophan from culture supernatants over time. Tryptophan was assayed using the method of Bloxam and Warren (21). Proteins were precipitated with 10% TCA and free tryptophan assayed after conversion to norharman using formaldehyde and FeCl3. The reaction product was measured spectrofluorometrically (excitation 360 nm, emission 460 nm) and compared against a standard preparation of tryptophan. Validation studies showed this assay to be linear in the range of 0.1–100 μM, with an estimated threshold sensitivity of 0.05 μM.
Where it was desirable to show that tryptophan depletion in cultures was due to IDO activity, culture supernatants were assayed by HPLC for the presence of kynurenine. IDO catalyzes the oxidation of tryptophan to N-formylkynurenine, which in Møs is rapidly converted into kynurenine (22) and then to other downstream metabolites (7). With the exception of tryptophan oxygenase, which is found only in hepatocytes, IDO is the only enzyme capable of degrading tryptophan along the kynurenine pathway (8). Thus, the appearance of kynurenine in cultures was unambiguous evidence of functional IDO activity. However, because kynurenine can be converted into other downstream metabolites, this assay was not quantitative. Where quantitative data were required, the tryptophan depletion assay described above was used.
HPLC assays were performed by the Medical College of Georgia Molecular Biology Core Facility. Samples were prepared by extracting 150 μl culture supernatant with 1 ml methanol. Precipitated proteins were removed by centrifugation and the supernatant dried under vacuum. Samples were resuspended in 100 μl initial mobile phase (deionized water) and an aliquot injected onto a C-18 column (Phenomenex Luna C-18; 250 × 4.6 mm; 5 μm). Samples were eluted with a linear gradient of acetonitrile in water (0–80% over 20 min), and absorbance was measured at 254 nm. Standards for tryptophan, kynurenine, and 1-methyl-tryptophan were run with each assay to establish retention times. In preliminary validation studies, the identity and purity of each peak was confirmed by mass spectroscopy.
Protein Synthesis and Amino Acid Analysis.
Total protein synthesis was measured as incorporation of tritiated leucine (4 μCi/ml) over 24 h. TCA-insoluble proteins were precipitated and washed three times in 5% TCA, and the precipitate was analyzed by liquid scintillation counting. Amino acid concentrations in culture supernatants were measured by HPLC in our clinical Neonatal Nutrition Laboratory.
RT-PCR.
Møs were harvested with EDTA and total RNA prepared. Sample RNA (1 μg) was reverse transcribed with avian myeloblastosis virus (AMV)-RT, and a 182-bp fragment amplified with the following primers: forward, bp 237–254 of the published sequence (23); reverse, bp 402–418, spanning exons 3–4. Product formation was assessed by agarose gel electrophoresis and ethidium bromide staining. PCR product was isolated from the gel and reamplified with internal primers to confirm specificity.
Flow Cytometry.
Two-color FACS® analysis was performed using directly conjugated mAbs as previously described (24). T lymphocytes were identified by gating on CD3-positive cells, and expression of CD69, CD25, and CD71 was measured in the second color.
Statistics.
Experiments for all figures were replicated at least three times, and representative data are shown. Data points were measured in triplicate and the mean reported. Error bars show standard deviation. Where SD was <10%, error bars have been omitted for clarity. Comparisons of multiple groups within a single experiment were by ANOVA.
Results
Cocultures of Møs and T Cells Are Selectively Depleted of Tryptophan.
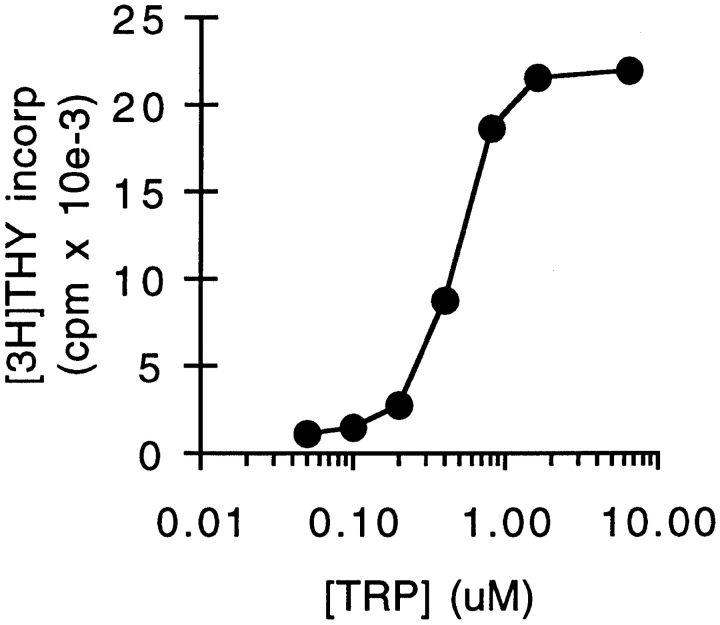
Supernatants were harvested from cocultures of Møs and mitogen-activated T cells after 48 h. Fresh lymphocytes were suspended in conditioned medium and activated with additional mitogen. Fig. 1 shows that conditioned medium completely failed to support T cell proliferation (<1% of the proliferation in fresh medium). However, the addition of tryptophan to conditioned medium fully restored its ability to support T cell proliferation, indicating that tryptophan was the only component that had been depleted. Consistent with this finding, amino acid analysis of conditioned media showed that all other essential amino acids were present, and only tryptophan was undetectable (data not shown). Titration of reagent tryptophan into conditioned medium gave a half-maximal concentration for T cell proliferation of 0.5–1 μM (Fig. 2), compared with a measured concentration of tryptophan in coculture-conditioned medium of <50 nM (the detection limit of our assay). Control-conditioned media from Møs alone, from cocultures of Møs + T cells without mitogen, or from T cells activated with fresh monocytes instead of Møs all supported T cell proliferation comparably to fresh medium (90–140% of control; n = 3–4/group).
Figure 1.

Coculture-conditioned medium is selectively depleted of tryptophan. Human monocytes were allowed to differentiate for 5 d in MCSF. Then, T cells were added and activated with anti-CD3 mAb. Conditioned medium was harvested from cocultures after 48 h and then used to support a second round of activation with fresh T cells. Replicate cultures were supplemented with individual amino acids to the concentrations normally found in RPMI 1640. Control cultures received either fresh medium (CTL) or no supplement (PBS). Proliferation was measured by thymidine incorporation after 72 h.
Figure 2.
Dose–response relationship to tryptophan for T cell proliferation. Tryptophan was titrated in coculture-conditioned medium (prepared as described in Fig. 1) and proliferation of T cells measured after 72 h.
Expression of IDO by MCSF-derived Møs.
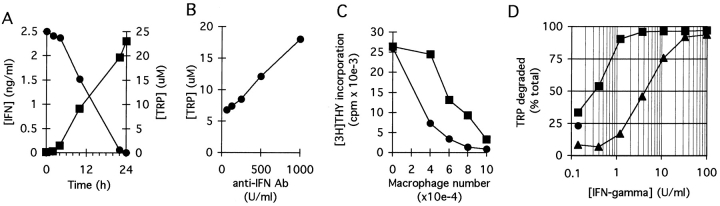
The kinetics of tryptophan elimination were measured by coincubating Møs and T cells with mitogen for 24 h to allow upregulation of the tryptophan depletion pathway and then adding fresh tryptophan and following its disappearance. As shown in Fig. 3 A, tryptophan was eliminated by first-order kinetics with a half-life of 2–3 h. The initial rate of elimination when tryptophan was not limiting was up to 20,000 pmol/ 106 cells/h. This far exceeded the consumption attributable to cellular metabolism (see control, Fig. 3 A), as Møs without activated T cells depleted tryptophan at a rate of 300 ± 130 pmol/106 cells/h (cumulative measurement obtained over 7 d; data not shown). This implied that the majority of tryptophan depletion by activated Møs was due to an inducible system, which we suspected was IDO.
Figure 3.

Elimination kinetics of tryptophan in cocultures and expression of IDO by MCSF-derived Møs. (A) MCSF-derived Møs were cultured for 24 h with autologous T cells, either with (•) or without (▪) anti-CD3 mAb. The medium was then replaced with fresh medium and supernatant from replicate cultures harvested at the times shown. Tryptophan concentration was assayed spectrofluorometrically as described in Materials and Methods. (B) IFN-γ–inducible IDO mRNA in MCSF-derived Møs. RT-PCR showing IDO expression in MCSF-derived Møs before (lane 4) and after (lanes 1–3) activation for 24 h with recombinant IFN-γ. Starting RNA for the reverse transcriptase reaction in lanes 1–3 was from 20,000, 2000, and 200 activated Møs, respectively, and from 20,000 unactivated Møs in lane 4. Lane 5 shows amplification of human IDO plasmid template giving the expected 182-bp product. (C) HPLC analysis of Mø culture supernatants showing degradation of tryptophan and production of kynurenine. MCSF-derived Møs were preactivated for 24 h with IFN-γ to induce IDO expression, and then the spent medium was replaced 90:10 with fresh medium. Trace 1 shows the analysis of supernatant immediately after adding fresh medium (time 0); trace 2 shows the conditioned medium 24 h later. The number of Møs in these experiments was kept low so that some tryptophan would be detectable at the end of the assay. The traces shown represent the portion of the elution gradient between 28 and 42% acetonitrile (minutes 7.00–10.50), during which kynurenine (K) and tryptophan (T) appeared. The peak labels are positioned at the points at which the purified standards eluted, which were within ±3 s of the corresponding sample peak. Compounds present in culture medium that also absorbed at OD254 (unlabeled peaks) were readily resolved from tryptophan and kynurenine, and the T and K peaks were confirmed by mass spectroscopy (see Materials and Methods). The experiment shown used purified Møs activated with recombinant ligands; identical results were obtained when Møs were activated in coculture with T cells plus mitogen. One of four experiments is shown.
Consistent with this finding, abundant IDO mRNA was detectable by RT-PCR in Møs after activation, whereas before activation, IDO message was undetectable (Fig. 3 B). To confirm the presence of IDO activity, culture supernatants were assayed for kynurenine. As shown in Fig. 3 C, depletion of tryptophan was accompanied by a corresponding increase in kynurenine production, confirming the presence of functional IDO activity.
Inhibition of IDO Prevents Mø-mediated Suppression of T Cells.
We next asked whether pharmacologic inhibition of IDO could prevent suppression of T cells in cocultures. The compound 1-methyl-tryptophan has been reported to be a potent competitive inhibitor of IDO activity when tested in vitro using purified enzyme (16, 17). To determine whether this agent could inhibit IDO activity in intact Møs, we added 1-methyl-tryptophan to activated Mø cultures. As shown in Fig. 4 A, the presence of 1-methyl-tryptophan markedly reduced the degradation of tryptophan by Møs, and this was accompanied by a corresponding inhibition of kynurenine production (e.g., compare the ratio of tryptophan to kynurenine after 24 h in Fig. 3 C), confirming that the target of the inhibitor was IDO.
Figure 4.
Inhibition of IDO activity prevents Mø-mediated suppression. (A) 1-methyl-tryptophan inhibits Mø IDO enzyme activity. MCSF-derived Møs were activated with IFN-γ for 24 h to induce IDO expression, and then fresh medium was added as described in Fig. 3, along with 1-methyl-tryptophan (1 mM). Supernatants were analyzed by HPLC for tryptophan (T) and kynurenine (K) immediately after the addition of fresh medium (trace 1) and 24 h later (trace 2). The 1-methyl-tryptophan peak (M) is off scale at the settings used. Control cultures for these experiments (Møs with IFN-γ but without 1-methyl-tryptophan) uniformly had >90% reduction in tryptophan at hour 24, with a corresponding increase in kynurenine, as shown in Fig. 3. The experiment shown used purified Møs activated with recombinant ligands; identical results were obtained when Møs were activated in coculture with T cells plus mitogen. (B) 1-methyl-tryptophan prevents T cell suppression in cocultures. T cells were added to MCSF-derived Møs and activated with anti-CD3 mAb. Replicate cultures were treated with varying concentrations of 1-methyl-tryptophan. Proliferation was measured after 72 h by thymidine incorporation. Controls (○) show proliferation by T cells without Møs at the highest concentration of inhibitor used (there was no effect of inhibitor on T cells alone throughout the range of concentrations shown). (C) A second inhibitor of IDO activity, 6-nitro-tryptophan, showed similar reversal of Mø-mediated inhibition of T cells. Experimental design as in B. (D) Supplementation with high concentrations of tryptophan prevents Mø-mediated suppression. Møs were seeded at low density (CC-lo; 5 × 104 cells/well) and high density (CC-hi; 2 × 105 cells/well) and the medium supplemented with 5× the normal tryptophan concentration. Proliferation was measured at hour 72. Controls show proliferation by Møs alone (M) and T cells alone (T).
Functionally, the addition of 1-methyl-tryptophan to cocultures abrogated the ability of Møs to suppress T cell proliferation in a dose-dependent manner (Fig. 4 B). Although this finding was consistent with the proposed role for IDO in Mø-mediated suppression, it might in theory indicate an unanticipated immunostimulatory role for 1-methyl-tryptophan itself. To exclude this possibility, we synthesized a second analogue of tryptophan, 6-nitro-tryptophan, which has also been reported to inhibit purified IDO enzyme in vitro (17). As shown in Fig. 4 C, 6-nitro-tryptophan also prevented Mø- mediated suppression in a dose-dependent fashion (Fig. 4 C). Finally, we tested the effects of supplemental tryptophan on suppression. As shown in Fig. 4 D, high levels of tryptophan did prevent suppression of T cells, provided that the number of Møs in cocultures was kept low. At our usual concentrations of Mø, it proved impossible to supplement with sufficient tryptophan to overcome its rapid degradation. Thus, by the use of two pharmacologic inhibitors of IDO and by tryptophan supplementation, the mechanism of T cell suppression in our system appeared to be depletion of tryptophan by IDO.
Tryptophan-degrading Activity Is Synergistically Induced by Early Signals of T Cell Activation.
Møs did not degrade tryptophan simply as a result of contact with T cells. Rather, there was an obligate requirement that the T cells attempt to activate (Fig. 3 A). In light of the existing studies implicating IFN-γ as an inducer of IDO (25–27), we suspected that IFN-γ from activating T cells might be the signal for IDO induction. Consistent with this idea, low but detectable levels of IFN-γ were present in cocultures within 4–6 h of T cell activation, coincident with the time that tryptophan degradation began (Fig. 5 A). Neutralizing antibodies against IFN-γ reduced the induction of tryptophan-degrading activity (Fig. 5 B) and reduced suppression of T cells by Møs (Fig. 5 C), supporting a role for IFN-γ in the signaling pathway. However, the dose–response relationship using recombinant IFN-γ revealed that relatively high concentrations of IFN-γ were required for full induction of tryptophan-degrading activity (Fig. 5 D). We therefore asked whether there was an additional signal that might act in concert with IFN-γ. CD40L is upregulated early in T cell activation and is known to act synergistically with IFN-γ to activate other Mø functions (28). Fig. 5 D shows that CD40L exerted marked synergy with IFN-γ, shifting the dose–response curve for IFN-γ one to two orders of magnitude so that significant tryptophan depletion began at IFN-γ concentrations of <1 U/ml.
Figure 5.
IFN-γ and CD40L act synergistically to induce IDO. (A) MCSF-derived Møs were cocultured with T cells and anti-CD3 mAb. Following lymphocyte addition, culture supernatants were harvested at the times shown and assayed for IFN-γ (▪, left axis) and tryptophan concentration (•, right axis). (B) Møs and T cells were cocultured with mitogen in the presence of various concentrations of neutralizing anti–IFN-γ antiserum. Tryptophan concentration in culture supernatants was determined after 18 h. A low density of Møs was used for these experiments so as not to obscure the effect of IFN-γ. (C) Møs were cultured at a range of seeding densities as shown, and then T cells and anti-CD3 mAb were added either with (▪) or without (•) neutralizing antibodies to IFN-γ (100 neutralizing U/ml). Antibodies to IFN-γ reduced the effectiveness of Møs in suppressing T cells, particularly when the number of Møs was limiting. (D) MCSF-derived Møs were cultured for 24 h with various concentrations of recombinant IFN-γ, either in the presence (▪) or absence (▴) of recombinant CD40L (500 ng/ml). At the end of the activation period, culture supernatants were assayed for the concentration of tryptophan remaining. The single round point shows tryptophan degradation in response to CD40L alone.
Effect of Tryptophan Deprivation on T Cell Protein and DNA Synthesis.
We have previously shown that T cells activated in coculture with MCSF-derived Møs initially enter the cell cycle but arrest before the first G1/S transition (4). We therefore asked whether a comparable phenomenon occurred when T cells were activated in the absence of tryptophan. Purified T cells (without monocytes or Møs) were cultured in tryptophan-free medium using immobilized anti-CD3 plus anti-CD28 mAb as activating stimuli. In this system, T cells stimulated in the presence of tryptophan activated normally, whereas T cells stimulated without tryptophan arrested before entry into the first S phase, as shown by the complete absence of DNA synthesis (Fig. 6). This arrest was not due to an absence of protein synthesis, as T cells without tryptophan successfully upregulated CD69, CD25 (high-affinity IL-2 receptor), and CD71 (transferrin receptor) (Fig. 7) and secreted IL-2 and IFN-γ (Fig. 8), all of which require new protein synthesis (29). Total protein synthesis, measured as incorporation of radiolabeled leucine during the first 24 h of activation, continued at a rate 40– 55% of controls (n = 3; see Materials and Methods), despite the absence of exogenous tryptophan. Nonetheless, no entry into S phase occurred. Thus, T cells activated in the absence of exogenous tryptophan arrested in a fashion similar to that which we had previously observed in coculture.
Figure 6.
T cells do not enter S phase in the absence of tryptophan. T cells were activated with immobilized anti-CD3 mAb plus anti-CD28, either in chemically defined tryptophan-free medium (▪) or in the same medium supplemented with 25 μM tryptophan (•). DNA synthesis was assayed by thymidine incorporation at the times shown.
Figure 7.
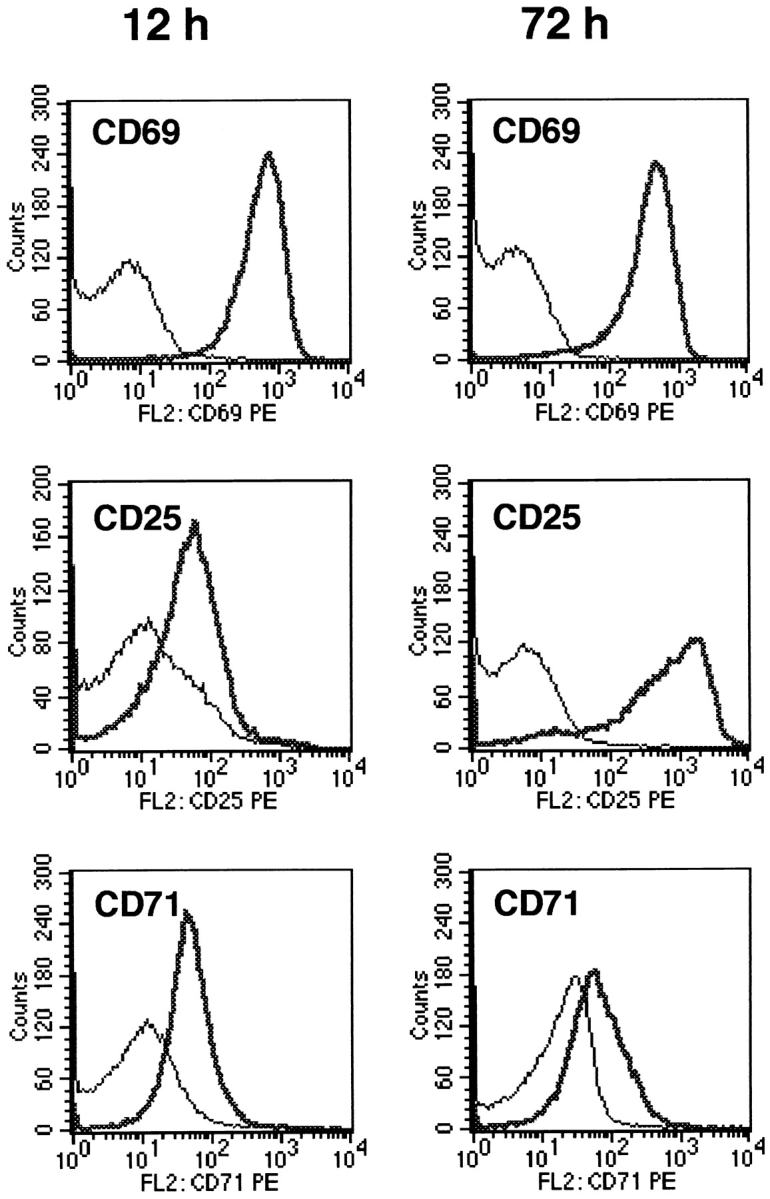
Expression of activation markers on T cells deprived of tryptophan. T cells were activated in tryptophan-free medium using immobilized anti-CD3/CD28 (heavy trace), or cultured under identical conditions but without anti-CD3/CD28 (light trace). At the times shown, both groups were harvested and stained for expression of activation markers as described in Materials and Methods.
Figure 8.
Production of IFN-γ and IL-2 by T cells deprived of tryptophan. T cells were activated with anti-CD3 mAb in tryptophan-free medium (▪) or in the same medium supplemented with 25 μM tryptophan (•) and the concentration of IFN-γ (A) and IL-2 (B) in culture supernatants determined at the times shown.
Identification of a Tryptophan-sensitive Arrest Point in Mid-G1.
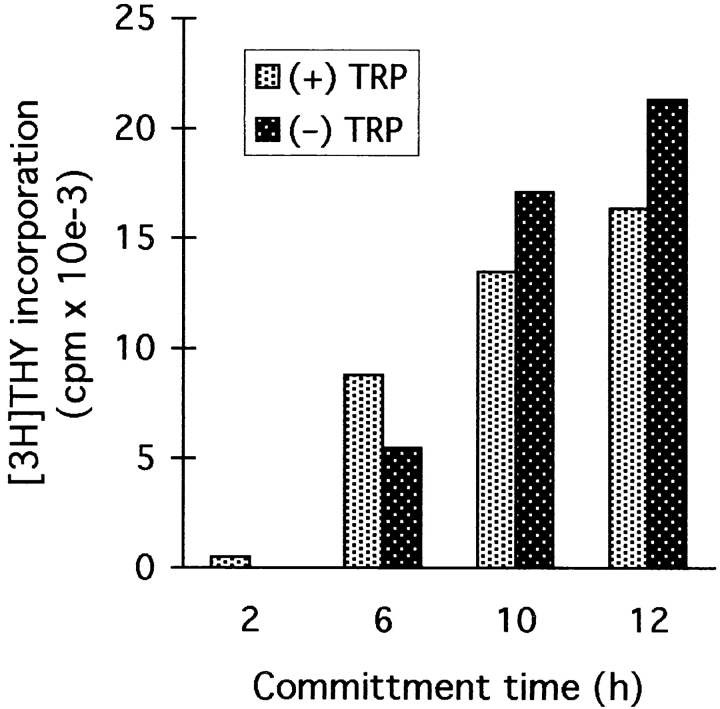
The upregulation of early G1 markers suggested that some portion of G1 was tryptophan independent. To test this hypothesis, T cells were activated for various times in the absence of tryptophan, and then tryptophan was added and the time to entry into S phase determined. Control cells, cultured with tryptophan throughout, reproducibly entered S phase 28–32 h after initial TCR engagement (times are reported as 4-h ranges to reflect the limit of precision of the assay). In contrast, T cells that had been preactivated under tryptophan-free conditions required only 12–16 h to enter S phase after tryptophan was added (Fig. 9 A), indicating that significant progression through G1 had occurred in the absence of tryptophan. The tryptophan-sensitive arrest point was stable, with T cells surviving >72 h in the absence of tryptophan with no loss of viability. When tryptophan was added to arrested cells, the time of entry into S phase was consistently 12–16 h, regardless of whether cells had been preactivated for 36, 48, or 72 h without tryptophan. This suggested that the arrest occurred at a specific point in G1 and that this position in the cell cycle was maintained until tryptophan was restored.
Figure 9.
T cells that have entered the tryptophan-sensitive arrested state retain their position in mid-G1. (A) T cells were activated in tryptophan-free medium using immobilized anti-CD3/CD28 (•). After a period of preactivation (24–72 h with similar results; 48 h in the experiments shown), tryptophan was added and the time to entry into S phase determined (defined as the initiation of thymidine incorporation). Replicate aliquots of cells were activated in tryptophan-containing medium without the 48-h preincubation period (▪). Lag time in each case was defined as the time to initiation of S phase from the point at which cells saw both tryptophan and anti-CD3. The arrow shows that the lag time to S phase was shortened by 12–16 h due to preactivation in the absence of tryptophan, suggesting that this portion of G1 had been accomplished before the point at which cells arrested. Representative of seven experiments at 36, 48, and 72 h, all showing the same lag time to S phase. (B) T cells were activated with anti-CD3/CD28 in the presence (▪) or absence (•) of tryptophan. After 14 h (the time of the putative arrest point estimated from A), tryptophan was added to the tryptophan-deficient cultures and entry into S phase determined. T cells rescued at hour 14 showed no delay compared with controls.
From the preceding experiments, we estimated that the tryptophan-independent portion of G1 was ∼14 h (calculated as the difference between the average time to S phase for resting T cells versus the time to S phase for preactivated cells). To test this estimate, we deprived T cells of tryptophan during the initial 14 h of activation, then added tryptophan just before the putative arrest point. As shown in Fig. 9 B, cultures deprived of tryptophan for the first 14 h entered S phase identically to T cells supplied with tryptophan throughout, supporting the hypothesis that the initial portion of G1 was independent of tryptophan. In additional experiments (not shown), delaying the addition of tryptophan beyond 14 h introduced a corresponding delay in entry into S phase, supporting the proposed localization of the arrest point close to hour 14.
T Cells Can Commit to Cell Division in the Absence of Tryptophan.
Resting (G0) T cells require TCR signaling in order to enter G1, but subsequent progression through the cell cycle rapidly becomes TCR independent (for a review see reference 29). In our system, commitment to TCR- independent cell division was first detectable ∼6 h after TCR engagement, and most cells were committed by hour 12. As shown in Fig. 10, this commitment occurred identically regardless of whether tryptophan was present or absent during the relevant time period. As long as the cells were not allowed to arrest (i.e., tryptophan was supplied before the tryptophan-sensitive checkpoint), commitment to cell division proceeded normally.
Figure 10.
T cells undergo normal commitment to TCR-independent activation in the absence of tryptophan. T cells were exposed to immobilized anti-CD3/CD28 for 2–12 h in the presence (light bars) or absence (dark bars) of tryptophan. At the times shown, cells were removed from contact with anti-CD3. After transfer, tryptophan was added to the tryptophan-deficient cultures, and all groups were continued out to hour 48. Cells were transferred in their own conditioned medium without washing and continued to receive anti-CD28 throughout. At hour 48, all groups were assayed for proliferation by thymidine incorporation. The 2-h time point (no proliferation after transfer) is included as a control to confirm that there was no carryover of anti-CD3 into the new cultures.
T Cells Reverse Their Commitment to Cell Cycle Progression upon Entering the Arrested State.
In contrast to the experiments shown in Fig. 10, however, once T cells entered the arrested state, simply restoring tryptophan was no longer sufficient to allow cell cycle progression. T cells were activated for 48 h in tryptophan-deficient medium using immobilized anti-CD3/CD28. The arrested cells were then removed from contact with anti-CD3, washed free of anti-CD28, and transferred to medium containing normal levels of tryptophan. As shown in Fig. 11, despite their previous 48-h exposure to anti-CD3, the arrested T cells still required additional TCR signaling plus the presence of tryptophan to exit the arrested state. Even costimulation via CD28 was not sufficient to promote cell cycle progression in the absence of TCR engagement.
Figure 11.
T cells require TCR signaling to exit the arrested state. T cells were activated for 48 h in the absence of tryptophan using immobilized anti-CD3/CD28. To simulate loss of contact with the APC, T cells were removed from the immobilized anti-CD3, washed, and returned to culture in medium containing 25 μm tryptophan. Upon replating, replicate cultures received immobilized anti-CD3, anti-CD28, or both.
Discussion
In this study, we show that tryptophan catabolism via IDO is the mechanism by which MCSF-derived Møs suppress T cell proliferation in vitro. We have recently tested this hypothesis of IDO-mediated T cell suppression in vivo using the model of allogeneic pregnancy. This model was chosen because it has long been recognized as paradoxical that the maternal immune system tolerates a genetically foreign fetus throughout gestation (30). IDO is known to be expressed in human placenta and has been reported to be localized to the zone of contact between fetal-derived tissues and the maternal immune system (31). Using 1-methyl-tryptophan (described in Fig. 4) as a pharmacologic inhibitor of IDO, we have demonstrated that IDO is a required component of the mechanism by which the allogeneic fetus protects itself from rejection by the maternal immune system and that inhibition of IDO breaks maternal tolerance to the allogeneic fetus (15). In the same report, we also showed that pharmacologic inhibition of IDO enhances the activation of autoreactive T cells. Thus, by two measures—breaking tolerance and enhancing autoreactivity— these data support a role for IDO in regulating T cell responses in vivo.
IDO has previously been viewed primarily as a host defense mechanism, inhibiting proliferation of intracellular pathogens (6, 9–13) or cancer cell lines (14) by depriving them of tryptophan (for a review see reference 8). In these settings, the proposed role of IDO has been to eliminate the cell's own stores of tryptophan. To our knowledge, no role for IDO in regulating the proliferation of adjacent cells has been suggested. However, both direct and indirect evidence indicates that IDO is widely expressed throughout the immune system (32, 33) and, specifically, that it is localized to a subset of cells with a Mø or dendritic cell morphology (33–35). These IDO-expressing cells are found at several putative sites of immune tolerance or privilege, including thymus, mucosa of the gut, epididymis, placenta, and the anterior chamber of the eye (32, 33, 36, 37). This pattern of widespread expression throughout the immune system is difficult to reconcile with a simple mechanism of host defense. We hypothesize that IDO expression by APCs functions to suppress undesirable T cell activation and thus helps maintain peripheral tolerance.
Two models might be proposed by which IDO could suppress T cells in vivo: it might catalyze the production of a suppressive metabolite of tryptophan, or it could deplete local tryptophan below some threshold level required for T cell activation. In repeated experiments, we have been unable to detect any evidence of an immunosuppressive metabolite in coculture supernatants (Figs. 1 and 2) (4). Furthermore, our experiments with isolated T cells imply a specific checkpoint in early T cell activation that is sensitive to low concentrations of tryptophan. For these reasons, we favor the tryptophan depletion hypothesis.
Implicit in this hypothesis is the assumption that cells expressing IDO in vivo could create a local microenvironment in which tryptophan is low, despite the availability of ample tryptophan elsewhere. In this regard, it is well established that delivery of a substrate into local microenvironments is sharply limited by the rate of diffusion (K d) through the interstitial space (38, 39). In the face of even normal metabolic demands, substrate concentrations rapidly fall to undetectable levels within a few cell diameters of the source of delivery (39). Because the rate of tryptophan consumption by IDO-expressing Møs is orders of magnitude greater than normal metabolic demands, it is plausible that such Møs could create local conditions of very low tryptophan concentrations. Although this hypothesis is now speculative with regard to tryptophan, the phenomenon is well documented with regard to, for example, the local hypoxic state created within muscle tissue during exercise.
Because tryptophan degradation by IDO is much greater than consumption by metabolic demands (Fig. 3), it is likely that IDO constitutes the major route of tryptophan depletion by activated Møs. However, IDO could act in combination with other pathways. Møs have a high rate of protein synthesis, and the incorporation of free tryptophan into proteins could contribute to local tryptophan depletion. Indeed, the tRNA synthetase for tryptophan (the WRS gene) is unique among tRNA synthetases in that it is massively induced in Mø lineage cell lines (but not lymphoid lines) by the same signals that induce IDO (40). It has been proposed that this induction allows Møs to compete preferentially for tryptophan when the concentration of substrate is low. Likewise, any pathway that transported tryptophan into Møs, whether for protein synthesis, degradation by IDO, or incorporation into other biosynthetic pathways, would also serve to deplete local tryptophan. Thus, IDO could act in concert with other catabolic pathways to render Møs an effective local “sink” for tryptophan.
The proposed tryptophan depletion model gains support from the apparent existence of a cell cycle arrest point sensitive to tryptophan concentration. Although the absence of any essential nutrient is, by definition, incompatible with long-term proliferation, the arrest point we describe appears more specific than simple protein starvation. First, although protein synthesis is reduced in the absence of exogenous tryptophan, it still occurs at a significant rate, presumably reflecting a combination of endogenous tryptophan stores and recycling of tryptophan from catabolism of endogenous and exogenous proteins (41). Yet despite ongoing protein synthesis, cell cycle progression is not simply delayed but rather is completely arrested. Second, the arrest induced by tryptophan deprivation occurs at a reproducible point in the cell cycle and remains stable once entered, suggesting a regulated process. Taken together, these attributes suggest a specific, tryptophan-sensitive cell cycle arrest point.
It has been noted by several groups that deprivation of certain amino acids—tryptophan in particular—exerts an inhibitory effect on cell cycle progression that cannot be explained by the effect on protein synthesis (42–45). For that reason, it has been suggested that levels of these amino acids may function as specific checkpoints regulating cell cycle progression. However, the biologic significance of such amino acid–specific checkpoints and the mechanism by which the levels of amino acids might be manipulated in order to regulate T cell activation has remained obscure. We now propose a system in which regulation of local tryptophan concentration functions as a means of communication between APCs and T cells, with APCs regulating the tryptophan level via IDO and T cells responding with either activation or arrest, depending on the level they detect.
As a strategy to inhibit T cell activation, arresting progression through the cell cycle is not unique to tryptophan metabolism. The immunosuppressive drugs mycophenolate, rapamycin, and leflunomide all induce a mid-G1 arrest in activating T cells, and this is believed to account in whole or part for their immunosuppressant action (46–48). Recent evidence suggests that T cells require one or more rounds of cell division to acquire a variety of effector functions (49–51), so inhibiting proliferation may also inhibit functional activity. In our system, it is currently unknown how T cells sense the level of tryptophan and trigger cell cycle arrest. Tryptophan-sensing systems in bacteria have been well described (52), but comparable systems in eukaryotes have not yet been identified. However, mammalian genes such as tryptophan oxygenase are known to be regulated by changes in tryptophan levels (53), so such sensing systems can be inferred to exist.
The requirement for a second signal from the TCR in order to exit the arrested state is an important finding in light of our proposed biologic model. Under this model, T cells that attempt to activate while in contact with an IDO-expressing APC are inhibited by the local absence of tryptophan. In theory, however, once such T cells were committed to cell division, they could migrate elsewhere and complete the activation process under tryptophan-sufficient conditions. The data presented in Fig. 11 show that once T cells have arrested, simply regaining tryptophan is no longer sufficient to allow continued activation. Despite the fact that T cells would normally have become independent of TCR signaling before the tryptophan-sensitive checkpoint (Fig. 10), once they enter the arrested state they apparently reverse this commitment and reimpose upon themselves a requirement for a second round of TCR signaling. From a biologic standpoint, this would mean that a T cell arrested by an IDO-expressing APC would be obliged to find a second, nonsuppressive APC presenting the same antigen in order to exit the arrested state.
What would be the fate of an arrested T cell if no such supportive APC could be found? In vitro, we find that arrested cells undergo progressive apoptosis after several days if not rescued by TCR engagement (4). Whether this means that they would likewise die in vivo, enter some form of anergy, or return to a resting state remains to be determined. However, the arrested state we describe differs from classical anergy (54) in several interesting respects. First, the cells retain their responsiveness to TCR engagement (Fig. 11). Second, costimulation via CD28 is not sufficient to rescue cells once they arrest. And third, arrested cells die if not rescued within a relatively brief window of time. Taken together, these attributes suggest that T cells arrested by tryptophan deprivation are not immediately deleted from the repertoire but that they must find a permissive APC and complete the activation process if they are to survive.
In conclusion, our hypothesis regarding the biologic role of IDO-expressing APCs is that they are involved in maintaining peripheral tolerance to self antigens. Our in vitro model has focused on MCSF-derived Møs as one example of immunosuppressive APCs, but dendritic cells or other APCs that possess inducible IDO could likewise be immunosuppressive. We speculate that tryptophan catabolism may constitute a previously unsuspected mechanism contributing to the regulation of peripheral T cell activation.
Acknowledgments
The authors thank J.-F. Tsai for expert technical assistance, T. Stoming for developing and performing the HPLC assays, and C. Rossignol and J. Bhatia for amino acid analysis.
This work was supported by the National Institutes of Health (grants K08 HL03395, R21 AI44759, and R01 HL60137 to D.H. Munn) and generous support from the Carlos and Marguerite Mason Trust.
Abbreviations used in this paper
- CD40L
CD40 ligand
- IDO
indoleamine 2,3-dioxygenase
- Mø
macrophage
- MCSF
macrophage colony-stimulating factor
References
- 1.Fazekas de St. Groth B. The evolution of self-tolerance: a new cell arises to meet the challenge of self-reactivity. Immunol Today. 1998;19:448–454. doi: 10.1016/s0167-5699(98)01328-0. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 3.Munn DH, Armstrong E. Cytokine regulation of human monocyte differentiation in vitro: the tumor-cytotoxic phenotype induced by macrophage colony-stimulating factor is developmentally regulated by interferon γ. Cancer Res. 1993;53:2603–2613. [PubMed] [Google Scholar]
- 4.Munn DH, Pressey J, Beall AC, Hudes R, Alderson MR. Selective activation-induced apoptosis of peripheral T cells imposed by macrophages: a potential mechanism of antigen-specific peripheral lymphocyte deletion. J Immunol. 1996;156:523–532. [PubMed] [Google Scholar]
- 5.Shimizu T, Nomiyama S, Hirata F, Hayaishi O. Indoleamine 2,3-dioxygenase: purification and some properties. J Biol Chem. 1978;253:4700–4706. [PubMed] [Google Scholar]
- 6.Carlin JM, Borden EC, Sondel PM, Byrne GI. Interferon-induced indoleamine 2,3-dioxygenase activity in human mononuclear phagocytes. J Leukoc Biol. 1989;45:29–34. doi: 10.1002/jlb.45.1.29. [DOI] [PubMed] [Google Scholar]
- 7.Werner ER, Bitterlich B, Fuchs D, Hausen A, Reibnegger G, Szabo G, Dierich MP, Wachter H. Human macrophages degrade tryptophan upon induction by interferon-γ. Life Sci. 1987;41:273–280. doi: 10.1016/0024-3205(87)90149-4. [DOI] [PubMed] [Google Scholar]
- 8.Taylor MW, Feng G. Relationship between interferon-γ, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 1991;5:2516–2522. [PubMed] [Google Scholar]
- 9.Pfefferkorn ER. Interferon γ blocks the growth of Toxoplasma gondiiin human fibroblasts by inducing the host cells to degrade tryptophan. Proc Natl Acad Sci USA. 1984;81:908–912. doi: 10.1073/pnas.81.3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta SL, Carlin JM, Pyati P, Dai W, Pfefferkorn ER, Murphy MJ. Antiparasitic and antiproliferative effects of indoleamine 2,3-dioxygenase enzyme expression in human fibroblasts. Infect Immun. 1994;62:2277–2284. doi: 10.1128/iai.62.6.2277-2284.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daubener W, Mackenzie C, Hadding U. Establishment of T-helper type 1- and T-helper type 2-like human Toxoplasmaantigen-specific T-cell clones. Immunol. 1995;86:79–84. [PMC free article] [PubMed] [Google Scholar]
- 12.Daubener W, Remscheid C, Nockemann S, Pilz K, Seghrouchni S, Mackenzie C, Hadding U. Anti-parasitic effector mechanisms in human brain tumor cells: role of interferon-γ and tumor necrosis factor-α. Eur J Immunol. 1996;26:487–492. doi: 10.1002/eji.1830260231. [DOI] [PubMed] [Google Scholar]
- 13.Nagineni CN, Pardhasaradhi K, Martins MC, Detrick B, Hooks JJ. Mechanisms of interferon-induced inhibition of Toxoplasma gondiireplication in human retinal pigment epithelial cells. Infect Immun. 1996;64:4188–4196. doi: 10.1128/iai.64.10.4188-4196.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aune TM, Pogue SL. Inhibition of tumor cell growth by interferon-γ is mediated by two distinct mechanisms dependent upon oxygen tension: induction of tryptophan degradation and depletion of intracellular nicotinamide adenine dinucleotide. J Clin Invest. 1989;84:863–875. doi: 10.1172/JCI114247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B, Brown C, Mellor AL. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191–1193. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- 16.Cady SG, Sono M. 1-methyl-d,l-tryptophan, β-(3-benzofuranyl)-d,l-alanine (the oxygen analog of tryptophan), and β-[3-benzo(b)thienyl]-d,l-alanine (the sulfur analog of tryptophan) are competitive inhibitors for indoleamine 2,3-dioxygenase. Arch Biochem Biophys. 1991;291:326–333. doi: 10.1016/0003-9861(91)90142-6. [DOI] [PubMed] [Google Scholar]
- 17.Southan MD, Truscott RJ, Jamie JF, Pelosi L, Walker MJ, Maeda H, Iwamoto Y, Tone S. Structural requirements of the competitive binding site of recombinant human indoleamine 2,3-dioxygenase. Med Chem Res. 1996;1996:343–352. [Google Scholar]
- 18.Moriya T, Hagio K, Yoneda N. A facile synthesis of 6-chloro-d-tryptophan. Bull Chem Soc Japan. 1975;48:2217–2218. [Google Scholar]
- 19.Munn DH, Cheung NK. Interleukin-2 enhancement of monoclonal antibody-mediated cellular cytotoxicity against human melanoma. Cancer Res. 1987;47:6600–6605. [PubMed] [Google Scholar]
- 20.Sono M, Taniguchi T, Watanabe Y, Hayaishi O. Indoleamine 2,3-dioxygenase: equilibrium studies of the tryptophan binding to the ferric, ferrous, and co-bound enzymes. J Biol Chem. 1980;255:1339–1345. [PubMed] [Google Scholar]
- 21.Bloxam DL, Warren WH. Error in the determination of tryptophan by method of Denckla and Dewey. A revised procedure. Anal Biochem. 1974;60:621–625. doi: 10.1016/0003-2697(74)90275-9. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto S, Hayaishi O. Tryptophan pyrrolase of rabbit intestine: d- and l-tryptophan-cleaving enzyme or enzymes. J Biol Chem. 1967;242:5260–5266. [PubMed] [Google Scholar]
- 23.Dai W, Gupta S. Molecular cloning, sequencing and expression of human interferon-γ-inducible indoleamine 2,3-dioxygenase cDNA. Biochem Biophys Res Commun. 1990;168:1–8. doi: 10.1016/0006-291x(90)91666-g. [DOI] [PubMed] [Google Scholar]
- 24.Munn DH, Bree AG, Beall AC, Kaviani MD, Sabio H, Schaub RG, Alpaugh RK, Weiner LM, Goldman SJ. Recombinant human macrophage colony-stimulating factor in nonhuman primates: Selective expansion of a CD16+monocyte subset with phenotypic similarity to primate natural killer cells. Blood. 1996;88:1215–1224. [PubMed] [Google Scholar]
- 25.Koide Y, Yoshida A. The signal transduction mechanism responsible for gamma interferon-induced indoleamine 2,3-dioxygenase gene expression. Infect Immun. 1994;62:948–955. doi: 10.1128/iai.62.3.948-955.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai W, Gupta SL. Regulation of indoleamine 2,3-dioxygenase gene expression in human fibroblasts by interferon-γ. J Biol Chem. 1990;265:19871–19877. [PubMed] [Google Scholar]
- 27.Chon SY, Hassanain HH, Gupta SL. Cooperative role of interferon regulatory factor 1 and p91 (STAT1) response elements in interferon-γ-inducible expression of human indoleamine 2,3-dioxygenase gene. J Biol Chem. 1996;271:17247–17252. doi: 10.1074/jbc.271.29.17247. [DOI] [PubMed] [Google Scholar]
- 28.Stout RD, Suttles J. The many roles of CD40 in cell-mediated inflammatory responses. Immunol Today. 1996;17:487–492. doi: 10.1016/0167-5699(96)10060-i. [DOI] [PubMed] [Google Scholar]
- 29.Crabtree GR. Contingent genetic regulatory events in T lymphocyte activation. Science. 1989;243:355–361. doi: 10.1126/science.2783497. [DOI] [PubMed] [Google Scholar]
- 30.Medawar PB. Some immunological and endocrinological problems raised by evolution of viviparity in vertebrates. Symp Soc Exp Biol. 1953;7:320–328. [Google Scholar]
- 31.Kamimura S, Eguchi K, Yonezawa M, Sekiba K. Localization and developmental change of indoleamine 2,3-dioxygenase activity in the human placenta. Acta Med Okayama. 1991;45:135–139. doi: 10.18926/AMO/32206. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida R, Nukiwa T, Watanabe Y, Fujiwara M, Hirata F, Hayaishi O. Regulation of indoleamine 2,3-dioxygenase activity in the small intestine and the epididymis of mice. Arch Biochem Biophys. 1980;203:343–351. doi: 10.1016/0003-9861(80)90185-x. [DOI] [PubMed] [Google Scholar]
- 33.Moffett J, Espey M, Namboodiri M. Antibodies to quinolinic acid and the determination of its cellular distribution within the rat immune system. Cell Tissue Res. 1994;278:461–469. doi: 10.1007/BF00331364. [DOI] [PubMed] [Google Scholar]
- 34.Espey M, Tang Y, Morse H, Moffett J, Namboodiri M. Localization of quinolinic acid in the murine AIDS model of retrovirus-induced immunodeficiency: implications for neurotoxicity and dendritic cell immunopathogenesis. AIDS. 1996;10:151–158. doi: 10.1097/00002030-199602000-00004. [DOI] [PubMed] [Google Scholar]
- 35.Espey M, Moffett J, Namboodiri M. Temporal and spatial changes of quinolinic acid immunoreactivity in the immune system of lipopolysaccharide-stimulated mice. J Leukoc Biol. 1995;57:199–206. doi: 10.1002/jlb.57.2.199. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida R, Urade Y, Nakata K, Watanabe Y, Hayashi O. Specific induction of indoleamine 2,3-dioxygenase by bacterial lipopolysaccharide in the mouse lung. Arch Biochem Biophys. 1981;212:629–637. doi: 10.1016/0003-9861(81)90406-9. [DOI] [PubMed] [Google Scholar]
- 37.Malina HZ, Martin XD. Indoleamine 2,3-dioxygenase: antioxidant enzyme in the human eye. Graefe's Arch Clin Exp Ophthalmol. 1996;234:457–462. doi: 10.1007/BF02539413. [DOI] [PubMed] [Google Scholar]
- 38.Casciari JJ, Sotirchos SV, Sutherland RM. Glucose diffusivity in multicellular tumor spheroids. Cancer Res. 1988;48:3905–3909. [PubMed] [Google Scholar]
- 39.Li CK. The glucose distribution in 9L rat brain multicell tumor spheroids and its effect on cell necrosis. Cancer. 1982;50:2066–2073. doi: 10.1002/1097-0142(19821115)50:10<2066::aid-cncr2820501017>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 40.Fleckner J, Martensen PM, Tolstrup AB, Kjeldgaard NO, Justesen J. Differential regulation of the human, interferon inducible tryptophanyl-tRNA synthetase by various cytokines in cell lines. Cytokine. 1995;7:70–77. doi: 10.1006/cyto.1995.1009. [DOI] [PubMed] [Google Scholar]
- 41.Smith CB, Deibler GE, Eng N, Schmidt K, Sokoloff L. Measurement of local cerebral protein synthesis in vivo: influence of recycling of amino acids derived from protein degradation. Proc Natl Acad Sci USA. 1988;85:9341–9345. doi: 10.1073/pnas.85.23.9341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dauphinais C, Waithe W. PHA stimulation of human lymphocytes during amino acid deprivation: protein, RNA, and DNA synthesis. J Cell Physiol. 1977;91:357–368. doi: 10.1002/jcp.1040910305. [DOI] [PubMed] [Google Scholar]
- 43.Woolley PV, Dion RL, Bono VH. Effects of tryptophan deprivation on L1210 cells in culture. Cancer Res. 1974;34:1010–1014. [PubMed] [Google Scholar]
- 44.Brunner M. Regulation of DNA synthesis by amino acid limitation. Cancer Res. 1973;33:29–32. [PubMed] [Google Scholar]
- 45.Tobey R, Ley K. Isoleucine-mediated regulation of genome replication in various mammalian cell lines. Cancer Res. 1971;31:46–51. [PubMed] [Google Scholar]
- 46.Cherwinski HM, Cohn RG, Cheung P, Webster DJ, Xu Y-Z, Caulfield JP, Young JM, Nakano G, Ransom JT. The immunosuppressant leflunomide inhibits lymphocyte proliferation by inhibiting pyrimidine biosynthesis. J Pharmacol Exp Ther. 1995;275:1043–1049. [PubMed] [Google Scholar]
- 47.Terada N, Takase K, Papst P, Nairn AC, Gelfand EW. Rapamycin inhibits ribosomal protein synthesis and induces G1 prolongation in mitogen-activated T lymphocytes. J Immunol. 1995;155:3418–3426. [PubMed] [Google Scholar]
- 48.Laliberte J, Yee A, Xiong Y, Mitchell BS. Effects of guanine nucleotide depletion on cell cycle progression in human T lymphocytes. Blood. 1998;91:2896–2904. [PubMed] [Google Scholar]
- 49.DeSilva DR, Urdahl KB, Jenkins MK. Clonal anergy is induced in vitro by T cell receptor occupancy in the absence of proliferation. J Immunol. 1991;147:3261–3267. [PubMed] [Google Scholar]
- 50.Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- 51.Oehen S, Brduscha-Riem K. Differentiation of naive CTL to effector and memory CTL: correlation of effector function with phenotype and cell division. J Immunol. 1998;161:5338–5346. [PubMed] [Google Scholar]
- 52.Babitzke P. Regulation of tryptophan biosynthesis: trp-ing the TRAP or how Bacillus subtilisreinvented the wheel. Molec Microbiol. 1997;26:1–9. doi: 10.1046/j.1365-2958.1997.5541915.x. [DOI] [PubMed] [Google Scholar]
- 53.Knox WE, Mehler AH. The adaptive increase of the tryptophan peroxidase-oxidase system of liver. Science. 1951;113:237–238. doi: 10.1126/science.113.2931.237. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz RH. Models of T cell anergy: is there a common molecular mechanism? . J Exp Med. 1996;184:1–8. doi: 10.1084/jem.184.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]