Abstract
MHC class II molecules and invariant chain assemble at a neutral pH in the endoplasmic reticulum and are transported to a low pH compartment where the invariant chain is trimmed to the class II–associated invariant chain peptide (CLIP). For many major histocompatibility complex class II molecules, DM is required for rapid removal of CLIP, which allows binding of antigenic peptides. Since I-Ag7 confers susceptibility to type I diabetes in NOD mice, the biochemical requirements for peptide loading were examined using soluble I-Ag7 expressed in insect cells. I-Ag7 formed long-lived complexes with naturally processed peptides from transferrin and albumin, whereas several peptides that represent T cell epitopes of islet autoantigens were poor binders. I-Ag7–peptide complexes were not sodium dodecyl sulfate (SDS) resistant, indicating that SDS sensitivity may be an intrinsic property of I-Ag7. Complexes of I-Ag7 and CLIP formed at a neutral pH, but rapidly dissociated at pH 5. This rapid dissociation was due to a poor fit of M98 of CLIP in the P9 pocket of I-Ag7, since substitution of M98 by a negatively charged residue greatly enhanced the stability of the complex. These biochemical properties of I-Ag7 result in the rapid generation of empty molecules at an endosomal pH and have a global effect on peptide binding by I-Ag7.
Keywords: major histocompatibility complex, autoimmunity, antigen presentation, I-Ag7, type I diabetes
Genome-wide analyses in families with type I diabetes and in nonobese diabetic (NOD)1 mice demonstrated that multiple genes contribute to susceptibility and that the MHC is an important susceptibility locus (1–5). In humans, susceptibility is associated with the MHC class II region, in particular with the DR4/DQ8 and DR3/DQ2 haplotypes. The risk for the development of type I diabetes is particularly high in subjects who carry both the DR4/DQ8 and DR3/DQ2 haplotype or who are homozygous for either haplotype (1, 6). It is generally thought that the DQ molecules (DQ8 and DQ2) are involved in the disease process, but particular DR4 alleles also contribute to susceptibility (for review see reference 7). Both DQ8 and DQ2 are characterized by the absence of aspartic acid at β57 (alanine in both alleles). The DR2/DQ6 haplotype (DQB1*0602) confers dominant protection from the disease.
Two features characterize the MHC class II region of NOD mice. NOD mice express only I-Ag7 but not I-E molecules, due to a deletion in the Eα promoter. This lack of I-E expression is important since transgenic expression of I-E or other I-A molecules protects from spontaneous disease (5, 8–11). The I-A molecule of NOD mice, I-Ag7, has the same α chain as I-Ad but a unique β chain sequence. The most striking characteristic of the I-Ag7 β chain is the presence of histidine and serine at β56 and β57, instead of proline and aspartic acid as in other murine I-A alleles (12). Crystallographic studies of human and murine MHC class II molecules demonstrated that β57 is located in the P9 pocket of the peptide binding site. In most MHC class II molecules, β57 forms a salt bridge with α76, which is missing in I-Ag7 (13–16).
It has recently been proposed that I-Ag7 molecules are poor peptide binders. This hypothesis is based on the observation that only a small fraction of immunoprecipitated I-Ag7 molecules are SDS resistant. Also, binding experiments with affinity-purified, detergent-solubilized I-Ag7 molecules demonstrated little or no binding of peptides in that study (17). To examine the biochemical properties of I-Ag7 in the absence of detergent, we expressed I-Ag7 as a soluble protein in Drosophila Schneider cells. I-Ag7 formed long-lived complexes with naturally processed peptides from transferrin and serum albumin, while several peptides that represent T cell epitopes of islet antigens were poor binders. I-Ag7 formed a complex with the class II–associated invariant chain peptide (CLIP) at a neutral pH, but these complexes rapidly dissociated at pH 5. These biochemical properties of I-Ag7 result in the rapid generation of empty molecules at the pH of the peptide loading compartment and have a global effect on peptide binding by this MHC class II molecule.
Materials and Methods
Synthetic Peptides.
Peptides were synthesized by Quality Controlled Biochemicals, Inc. and subjected to quality control by reverse-phase HPLC and mass spectrometry.
cDNA Constructs.
cDNA constructs were made by reverse transcriptase PCR using RNA from NOD spleens. Fos and Jun dimerization domains were attached to the 3′ end of the I-Ag7 α and β extracellular domains, respectively, through a seven-amino acid linker [Val-Asp-(Gly)5] that contained a SalI restriction site. The cDNAs encoding the signal peptides and extracellular domains of I-Ag7 α and β chains were amplified first and cloned separately into the EcoRI/SalI site of the pRmHa-3 vector under the control of the inducible metallothionein promoter (18). The Fos and Jun segments were excised with SalI from similar constructs made for HLA-DR2 (19) and cloned into the SalI site of these vectors (I-Aα-Fos, I-Aβ-Jun). The orientation of the Fos and Jun segments and the sequence of the constructs were confirmed by dideoxy-sequencing. The following oligonucleotides were used for the amplification of I-Aα and I-Aβ segments. Forward primer for I-Aα, 5′ AAA AAA gAA TTC ATg CCg TgC AgC AgA gCT CTg 3′; reverse primer for I-Aα, 5′ AAA AAA gTC gAC TTC TgT CAg CTC TgA CAT gg 3′; forward primer for I-Aβ, 5′ AAA AAA gAA TTC ATg gCT CTg CAg ATC CCC AgC 3′; reverse primer for I-Aβ, 5′ AAA AAA gTC gAC CTT gCT CCg ggC AgA CTC ggA 3′. The EcoRI and SalI sites in the forward and reverse primers, respectively, are underlined.
Transfection of Drosophila Schneider Cells.
Drosophila melanogaster S2 cells (4 × 106) were transfected with 20 μg of each plasmid, 0.5 μg of hygromycin selection vector pUC-hygMT, and 20 μl of liposomes (Invitrogen) in 1 ml medium without FCS for 4 h. After transfection, cells were grown in Schneider medium (Sigma Chemical Co.) supplemented with 10% of FCS. Selection was performed by addition of hygromycin to a final concentration of 0.1 mg/ml. Approximately 2 wk after transfection, cells were cloned by limiting dilution in 96-well plates. Clones were expanded and tested for secretion of I-Ag7 by Western blot analysis after induction of expression with copper sulfate (1 mM). The clone with the highest expression level was scaled up in roller bottles using Schneider medium supplemented with 5% FCS and 0.1 mg/ml of hygromycin. The supernatant was harvested 4 d after induction of protein expression with copper sulfate at 1 mM (Sigma Chemical Co.).
Purification of I-Ag7.
Supernatants were concentrated by ultrafiltration using a YM30 spiral membrane cartridge (Amicon). The I-A–specific 10-2.16 mAb (20; hybridoma from the American Type Culture Collection) was coupled to cyanogen bromide–activated Sepharose 4B beads (Pharmacia Biotech). The concentrated supernatant was passed through a Sepharose 4B precolumn and the 10-2.16 column at a flow rate of 9 ml/h. The column was then washed with 100 mM phosphate buffer, pH 8.0, and the protein was eluted with 2 bed volumes of 50 mM glycine, pH 11.5. Fractions were immediately neutralized by addition of 2 M Tris, pH 8.8, and dialyzed against PBS. Protein concentration was determined with the Coomassie Plus protein assay (Pierce).
Western Blot Analysis and HPLC Gel Filtration.
Antisera specific for the Fos and Jun dimerization domains were generated by immunizing Lewis rats with synthetic peptides (provided by Dr. P. Kim, Whitehead Institute, Cambridge, MA). Fos peptide sequence, Ac-CGGTDTLQAETDQLEDEKYALQTEIANLLKEKEKL-CONH2; Jun peptide sequence, Ac-CGGIARLEEKVKTLKAQNYELASTANMLREQVAQL-CONH2.
For Western blot analysis, proteins were separated under reducing conditions by SDS-PAGE (12%) and transferred to a polyvinylidene difluoride membrane (Millipore Corp.). Membranes were blocked overnight in 3% non-fat dry milk in TBST-buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.2% Tween 20). Membranes were probed with Fos antiserum (1:1,000) and/or Jun antiserum (1:500). Bound Fos and Jun antibodies were detected by incubating the membranes with horseradish peroxidase–conjugated anti–rat IgG (1:3,000). The blots were developed by ECL (Amersham).
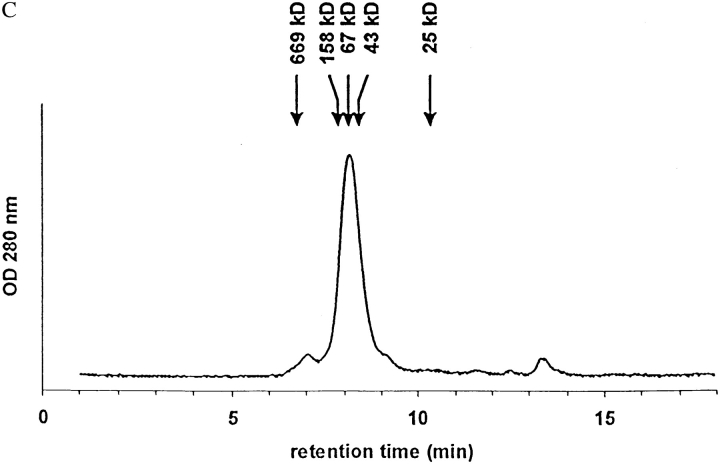
HPLC gel filtration analysis of I-Ag7 was performed using a Bio-Gel SEC 30-XL column (300 × 7.8 mm; Bio-Rad) with phosphate buffered saline, pH 7.8, at a flow rate of 0.8 ml/min. The following proteins were used as molecular mass standards: thyroglobulin (669 kD), aldolase (158 kD), BSA (67 kD), ovalbumin (43 kD), and chymotrypsinogen A (25 kD).
Peptide Binding Assays.
Purified I-Ag7 (0.6 μg) was incubated with biotinylated test peptide at 37°C for 18 h in a final volume of 50 μl. Buffers were either 20 mM sodium acetate/100 mM NaCl, pH 5.0, or PBS, pH 7.4, both containing 1 mM PMSF and 1 mM EDTA. In parallel, a 96-well plate (Fluoronunc; Nunc) was coated overnight at 4°C with 50 μl of 10 μg/ml of the I-A–specific antibody 10-2.16 in sodium bicarbonate buffer, pH 9.6. Nonspecific binding sites were blocked with 10% FCS in PBS/0.05% Tween 20 (PBS-Tween) for 2 h at room temperature. Wells were washed three times with PBS-Tween and I-Ag7–peptide samples were added to the wells. After a 1-h incubation, wells were washed four times with PBS-Tween and europium-labeled streptavidin (Wallac Oy, Finland) was added (1:2,000 in PBS/0.5% BSA, 100 μl/well) for detection of I-Ag7–bound biotinylated peptide. After a 1-h incubation at room temperature, wells were washed six times with PBS-Tween and Delfia enhancement solution (Wallac Oy) was added. Fluorescence was quantitated after 30 min in a fluorescence plate reader (Delfia 1234; Wallac Oy).
For competition assays, 0.6 μg of soluble I-Ag7 was incubated with biotinylated transferrin peptide (1 μM) at pH 5.0 in the presence of nonbiotinylated competitor peptide (8 nM–25 μM). I-Ag7–bound biotinylated transferrin peptide was quantitated as described above.
Peptide Dissociation Experiments.
For CLIP and CLIP 98D dissociation assays, soluble I-Ag7 was loaded with biotinylated CLIP or CLIP 98D (1 μM) at 37°C for 18 h at pH 7.4. An equal volume of 20 mM sodium acetate, pH 4.5, or PBS, pH 7.4, was added in the presence of nonlabeled mouse serum albumin peptide (100 μM). The final pH was either 5.0 or 7.4. At different time points (5–120 min), the reaction was stopped by freezing the samples at −20°C in the presence of BSA (200 μM). For quantification of bound biotinylated peptide, samples were mixed with an equal volume of 40 μM Tris, pH 7.4, and captured with 10-2.16 antibody–coated wells at 4°C for 1 h. All wells were developed by incubation for 1 h at 4°C with streptavidin-europium (dilution of 1:2,000). Fluorescence was quantitated as described above.
For other peptide dissociation assays, soluble I-Ag7 was incubated with biotinylated peptide (1 μM) at 37°C for 18 h at pH 5.0 in a final volume of 50 μl. Then the pH of the samples was either maintained at pH 5.0 or adjusted to pH 7.4 (by addition of Tris, pH 8.8). Nonlabeled mouse serum albumin peptide (100 μM) was added to prevent rebinding of biotinylated peptides. At different time points (5 min to 72 h), reactions were stopped by freezing the samples at −20°C in the presence of BSA (200 μM). Bound biotinylated peptides were then quantitated as described above.
Herpes Simplex Virus 2 VP16-specific T Cell Line and T Cell Proliferation Assay.
7-wk-old female NOD mice were immunized subcutaneously with 100 μg of Herpes simplex virus (HSV)-2 VP16 peptide in CFA. Draining lymph nodes were removed 10 d after immunization and lymph node cells were seeded at 5 × 105 per well in 96-well U-bottomed plates in serum-free media (AIM-V; GIBCO BRL) containing 5.5 × 10−5 M of 2-ME. HSV-2 VP16 peptide was added to a final concentration of 5 μM. After 3 d of culture, the medium was replaced with RPMI supplemented with 10% FCS, 5 U/ml recombinant human IL-2 (Boehringer Mannheim), 2-ME, and nonessential amino acids (Sigma Chemical Co.). T cells were grown for 6–8 d in this media and cells were fed every third day. The T cell line was maintained in culture by repeated cycles of a 3-d restimulation with irradiated NOD spleen cells and 5 μM HSV-2 VP16 peptide in AIM-V, followed by a 7-d resting period in RPMI supplemented with 10% FCS and recombinant IL-2.
T cell proliferation assays were set up in triplicates in 96-well U-bottomed plates with 2.5 × 104 T cells and 2.5 × 105 irradiated NOD spleen cells per well in AIM-V containing 2-ME. For blocking experiments, antibodies were added to a final concentration of 10 μg/ml. After 72 h of culture, 1 μCi of [3H]thymidine (NEN) was added to each well and incorporated radioactivity was quantitated in a beta-scintillation counter after harvesting of cells onto glass fiber filters (Wallac Oy).
Results
Expression and Purification of Soluble I-Ag7.
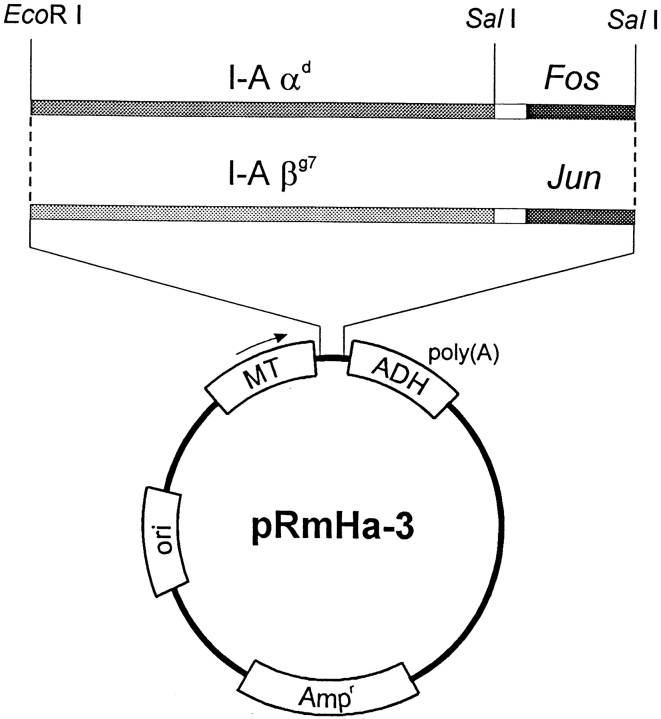
Soluble I-Ag7 was expressed in Drosophila Schneider cells by replacing the transmembrane and cytoplasmic segments of I-Aα and I-Aβ with leucine zipper dimerization domains from the transcription factors Fos and Jun, respectively. A short, flexible linker [Val-Asp-(Gly)5] that encoded a SalI restriction site was placed between the extracellular domains of I-Aα and I-Aβ and the Fos/Jun segments (Fig. 1). Leucine zipper dimerization domains were previously shown to promote the assembly of HLA-DR2 and I-Ad (19, 21).
Figure 1.
cDNA constructs for the expression of soluble I-Ag7. The signal peptides and extracellular domains of I-Ag7 α and β chains were fused in frame with the leucine zipper dimerization domains of the transcription factors Fos and Jun. A short flexible linker [Val-Asp-(Gly)5] was placed between the 3′ end of the I-A α/β extracellular domains and the Fos/Jun leucine zippers. The I-Aα–Fos and I-Aβ–Jun constructs were separately cloned into the pRmHa-3 vector under the control of the inducible metallothionein promoter. These constructs were transfected into Drosophila Schneider cells.
The I-Aα–Fos and I-Aβ–Jun constructs were separately cloned into the pRmHa-3 vector under the control of the copper-inducible metallothionein promoter (18). These constructs were cotransfected with a hygromycin selection plasmid into S2 cells and transfectants were cloned by limiting dilution after initial selection with hygromycin. Expression was induced by addition of copper sulfate to the growth medium and the protein was purified from concentrated supernatants by affinity chromatography using mAb 10-2.16. The yield was ∼0.7–1.6 mg/liter of culture.
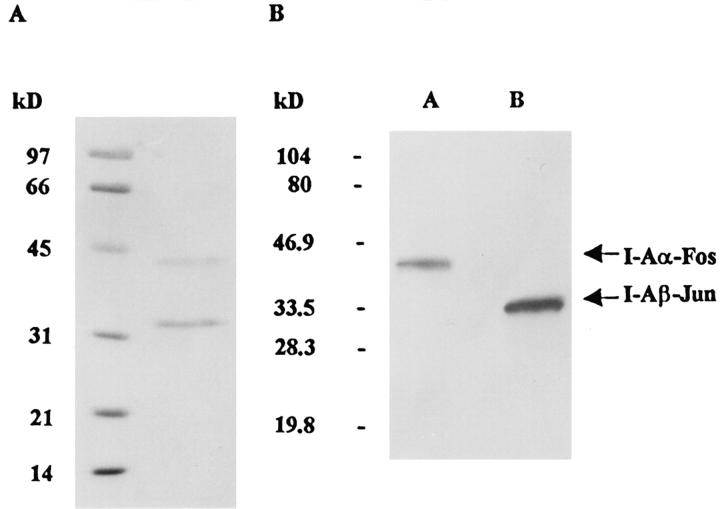
SDS-PAGE of the purified protein demonstrated two bands with an apparent molecular mass of 43.3 and 33 kD (Fig. 2 A). The identity of these bands as I-Aα–Fos and I-Aβ–Jun, respectively, was confirmed by Western blot analysis using antisera specific for the Fos and Jun leucine zippers (Fig. 2 B). The migration of the two chains was similar to that observed for α and β chains of HLA-DR2 expressed with Fos/Jun dimerization domains (19). Purified I-Ag7 eluted from a HPLC gel filtration column with a molecular mass appropriate for soluble I-Ag7 (Fig. 2 C). Importantly, the protein did not form high molecular mass aggregates, indicating that it was indeed soluble in the absence of detergent.
Figure 2.
Biochemical characterization of soluble I-Ag7. I-Ag7 was purified from concentrated supernatants by affinity chromatography using a mAb specific for the I-Aβ chain (10-2.16). (A) Purified I-Ag7 was analyzed by SDS-PAGE (12%) and stained with Coomassie Blue. (B) The identity of α and β chains was confirmed by Western blot analysis, using polyclonal antisera raised against the Fos and Jun leucine zipper segments. The migration of I-Aα–Fos and I-Aβ–Jun was similar to α and β chains of HLA-DR2 expressed with these leucine zipper dimerization domains (19). (C) HPLC gel filtration analysis of recombinant I-Ag7. 20 μg of I-Ag7 were separated on a Bio-Gel SEC 30-XL column (300 × 7.8 mm; Bio-Rad) with PBS at a flow rate of 0.8 ml/min and detection at 280 nm. The following proteins were used as molecular mass markers: thyroglobulin (669 kD), aldolase (158 kD), BSA (67 kD), ovalbumin (43 kD), and chymotrypsinogen A (25 kD).
Peptide Binding by Soluble I-Ag7.
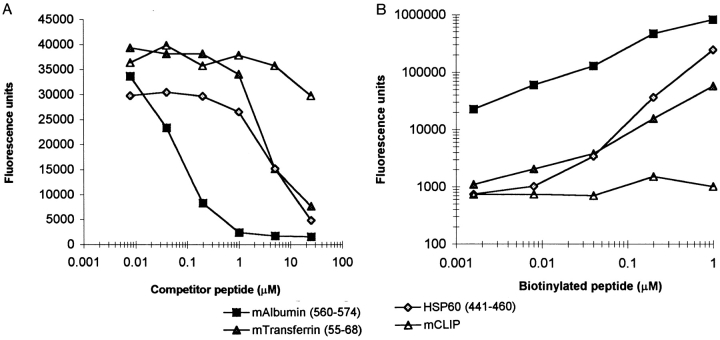
Soluble I-Ag7 was used to examine the binding of naturally processed I-Ag7 peptides as well as peptides from islet autoantigens under detergent-free conditions (Fig. 3, Table I). This is important because detergents can have a major effect on peptide binding by MHC class II molecules. For example, detergents with unbranched intermediate length hydrocarbon chains were found to promote dissociation of CLIP from HLA-DR (22). Two different binding assays were performed. In the direct binding assay, biotinylated peptides were incubated with soluble I-Ag7 and bound biotinylated peptides were quantitated with europium-labeled streptavidin after capture of complexes with an I-A–specific antibody. In the competition assay, unlabeled peptides were used at different concentrations to compete for binding of a biotinylated transferrin peptide to I-Ag7.
Figure 3.
Binding of naturally processed peptides and peptides from islet autoantigens to soluble I-Ag7. Binding of peptides to soluble I-Ag7 was examined both in a competition assay at pH 5 (A) and in a direct binding assay at pH 5 (B). (A) Unlabeled peptides were used to compete for the binding of biotinylated transferrin peptide (1 μM) to purified I-Ag7. (B) In the direct binding assay, biotinylated peptides (1.6 nM–1 μM) were incubated with soluble I-Ag7 (0.6 μg) at pH 5.0. I-Ag7–bound biotinylated peptides were quantitated with europium-labeled streptavidin after capture with mAb 10-2.16. Fluorescence was quantitated in a time-resolved fluorometer (Delfia; Wallac Oy).
Table I.
Peptides That Were Examined for Binding to Purified I-Ag7
| Peptides | Reference | Sequences | ||
|---|---|---|---|---|
| Murine serum albumin (560–574) | 23 | KPKATAEQLKTVMDD | ||
| Murine transferrin (55–68) | 23 | GHNYVTAIRNQQEG | ||
| Human HSP60 (441–460) | 41 | GCALLRCIPALDSLTPANED | ||
| Human HSP60 (169–180) | 42 | AQVATISANGDK | ||
| Murine insulin B (9–23) | 43 | SHLVEALYLVCGERG | ||
| Human GAD65 (509–528) | 44 | IPPSLRYLEDNEERMSRLSK | ||
| Human GAD65 (247–266) | 44 | NMYAMMIARFKMFPEVKEKG | ||
| Human GAD65 (524–543) | 44 | SRLSKVAPVIKARMMEYGTT | ||
| Human CPH (362–381) | 17 | KNSLINYLEQIHRGVKGFVR | ||
| Human CPH (440–464) | 17 | YFSPAVGVDFELESFSERKEEEKEEL | ||
| HSV-2 VP16 (430–444) | 34 | EEVDMTPADALDDFD | ||
| Murine CLIP (86–100) | PVSQMRMATPLLMRP |
A naturally processed peptide from mouse albumin (residues 560–574) was the best binder in both assays (reference 23, Fig. 3). This peptide competed for binding of the biotinylated transferrin peptide at very low peptide concentrations and also gave very high counts in the direct binding assay, even when the concentration of the biotinylated albumin peptide was <100 nM. Strong binding could also be demonstrated for another naturally processed peptide from transferrin (res. 55–68), as well as for a peptide from HSP60 (res. 441–460) (Fig. 3). The murine CLIP peptide showed little or no binding at this pH (pH 5). Several peptides that were identified as T cell epitopes of islet antigens competed for binding to I-Ag7 only at relatively high peptide concentrations, indicating that they were relatively poor binders. This included all three peptides from GAD 65 (res. 247– 266, 509–528, and 524–543) as well as HSP60 (169–180) and insulin (9–23). The HSP60 (169–180), insulin (9–23), and GAD65 (249–263) peptides were also synthesized with an NH2-terminal biotin group and tested in the direct binding assay. These peptides gave only low counts that were close to background (data not shown). It is important to note that the majority of the peptides that have been described as T cell epitopes actually represent the human rather than the murine sequences. Since there are sequence differences in the sequences of some of these peptides, it is possible that some of the murine peptides are stronger binders. The immunodominant myelin basic protein peptide (MBP, Ac1-11) that induces experimental autoimmune encephalomyelitis in mice with the H-2u haplotype is also a low affinity binder, whereas the immunodominant T cell epitope of MBP in mice with the H-2s haplotype (res. 87– 99) is a high affinity binder (24).
Rapid Dissociation of CLIP from I-Ag7 at an Endosomal pH.
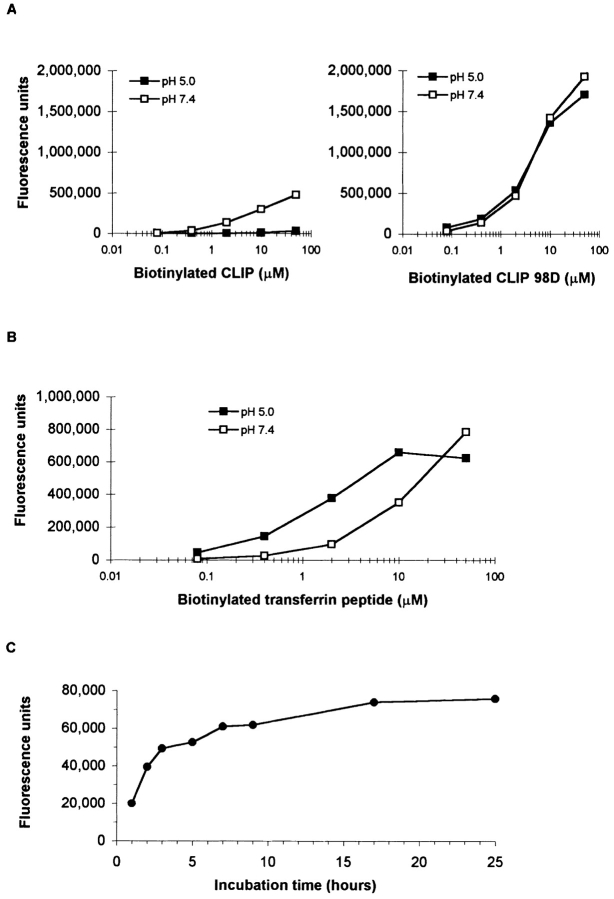
The transferrin peptide bound to I-Ag7 at both an acidic and a neutral pH (Fig. 4 B). The binding kinetics were characteristic for an “empty” MHC class II molecule and similar to those previously observed for soluble HLA-DR2 (Fig. 4 C; reference 19). In contrast, CLIP bound to I-Ag7 at a neutral pH; at an acidic pH little or no binding was detected (Fig. 4 A). Analysis of human and murine CLIP peptides has demonstrated a binding mode in which two methionine residues of CLIP occupy the P1 and P9 pockets of the MHC class II peptide binding site. These residues correspond to positions 90 and 98 of mouse as well as residues 91 and 99 of human invariant chain, respectively (25–27). Since the transferrin peptide carried a negative charge at the P9 position, the corresponding residue of CLIP (methionine 98) was substituted by aspartic acid (CLIP 98D). This single amino acid substitution resulted in strong binding both at a neutral and at an acidic pH (Figs. 4 and 5). These results demonstrated that the binding characteristics of CLIP to I-Ag7 were due to the interaction of methionine 98 with the P9 pocket of I-Ag7.
Figure 4.
Binding of CLIP and transferrin (55–68) to soluble I-Ag7. Peptide binding to I-Ag7 was examined using biotinylated peptides that were incubated with I-Ag7 at 37°C. I-A–peptide complexes were captured with immobilized 10-2.16 mAb and the amount of biotinylated peptide was quantitated with europium-labeled streptavidin. (A) Poor binding of CLIP to I-Ag7 at an endosomal pH. CLIP peptide bound to I-Ag7 at a neutral pH, but little or no binding was detected at pH 5.0. These binding properties of CLIP were due to a poor fit of methionine 98 in the P9 pocket of I-Ag7, as substitution of this residue by aspartic acid (CLIP 98D) greatly enhanced binding at both pH 5 and at pH 7.4. (B) Optimal binding of transferrin peptide to I-Ag7 at endosomal pH. The transferrin peptide bound to I-Ag7 both at pH 5.0 and at pH 7.4. Lower peptide concentrations were required for loading of I-Ag7 at the acidic pH. (C) Kinetics of transferrin peptide binding to I-Ag7. The transferrin peptide (1 μM) was incubated with I-Ag7 (0.2 μM) at 37°C for different periods of time. Half-maximal loading was observed after ∼2 h and a plateau was reached after ∼16 h. The kinetics of peptide binding were therefore similar to soluble HLA-DR2 (19).
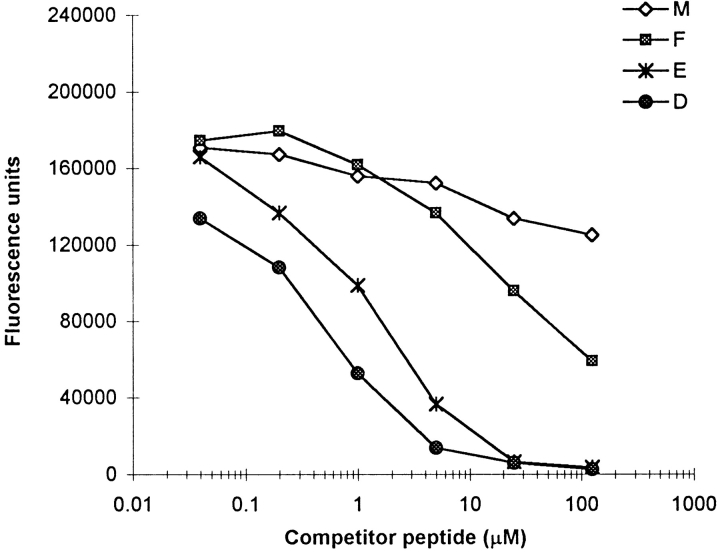
Figure 5.
Substitution of CLIP at the P9 position greatly enhances binding to I-Ag7. Single amino acid analogues of CLIP were tested for their ability to compete for binding of the biotinylated transferrin peptide at pH 5. Although the native CLIP peptide showed little or no competition, substitution of methionine at position 98 by aspartic acid or glutamic acid greatly increased the ability of the peptide to compete for binding.
MHC class II molecules assemble with invariant chain at a neutral pH in the endoplasmic reticulum and are then transported through the Golgi complex to an endosomal compartment that has a low pH. In this compartment, the invariant chain is proteolytically cleaved, leaving the CLIP peptide in the binding site. The removal of CLIP is catalyzed by H-2M, which allows binding of antigenic peptides (28–32). Therefore, it was important to examine the stability of complexes that were assembled at a neutral pH and then shifted to an acidic pH. For that purpose, I-Ag7 was incubated with biotinylated CLIP at a neutral pH at 37°C. An unlabeled competitor peptide was then added in order to prevent rebinding of biotinylated CLIP and the pH was changed for different periods of time (5–120 min) (Table II). After this incubation period, the sample was neutralized and the amount of bound biotinylated peptide was quantitated. This experimental design in which complexes were formed at a neutral pH and neutralized again after incubation at pH 5 for different periods of time allowed the kinetics of CLIP dissociation to be analyzed.
Table II.
Rapid Dissociation of CLIP from I-Ag7 at an Endosomal pH
| Conditions | CLIP | CLIP 98D | ||||||
|---|---|---|---|---|---|---|---|---|
| pH 7.4 | pH 5.0 | pH 7.4 | pH 5.0 | |||||
| Fluorescence U | ||||||||
| I-Ag7, no biotinylated peptide | 462 | 636 | ||||||
| Biotinylated CLIP or CLIP 98D* | 58762 | 670032 | ||||||
| Biotinylated CLIP or CLIP 98D plus competitor‡ | 12855 | 775282 | ||||||
| Additional incubation§ | ||||||||
| 5 min | 11190 | 712 | 750899 | 419044 | ||||
| 10 min | 10355 | 798 | 667831 | 425805 | ||||
| 20 min | 10244 | 813 | 739503 | 303485 | ||||
| 30 min | 8237 | 680 | 740163 | 342409 | ||||
| 60 min | 6783 | 587 | 788291 | 254762 | ||||
| 120 min | 3948 | 515 | 758073 | 206928 | ||||
Dissociation of CLIP was examined at pH 5.0 and pH 7.4. The biotinylated CLIP peptide was first bound to I-Ag7 at a neutral pH by incubation at 37°C. The complex was then shifted to pH 5.0 for 5–120 min at 37°C and an excess of unlabelled competitor peptide was added to prevent rebinding of biotinylated CLIP. The sample was then neutralized and the amount of bound biotinylated CLIP peptide was determined as described above. Even a brief incubation at pH 5.0 resulted in complete dissociation of CLIP from I-Ag7. At pH 7.4, dissociation was significantly slower and bound CLIP was still detectable after 2 h of incubation. Substitution of methionine 98 by aspartic acid (CLIP 98D) greatly enhanced binding, as well as the stability of the complex at a neutral or an acidic pH.
16 h incubation at 37°C.
16 h incubation at 37°C, followed by an addition of unlabeled competitor peptide.
Additional incubation at 37°C at the indicated pH in the presence of competitor peptide, followed by neutralization of the pH 5 samples. Length of incubation is given in min.
These experiments demonstrated that even a brief incubation (5 min) at pH 5.0 before neutralization resulted in complete dissociation of CLIP from I-Ag7 (Table II). Even with shorter incubation times at pH 5 (<2 min), no bound CLIP was detected (data not shown). When the complex was kept at pH 7.4, dissociation was considerably slower and bound CLIP was still detected after 120 min. The rapid, pH-dependent dissociation of CLIP was due to methionine 98 of CLIP, which occupies the P9 pocket of MHC class II molecules (25). Substitution by aspartic acid (CLIP 98D) greatly increased the stability of the complex (Table II). These results indicated that the stability of I-Ag7–CLIP complexes was pH dependent, and that the P9 pocket of I-Ag7 was critical in determining this property.
A possible explanation for the pH-dependent dissociation of CLIP was that the interaction of peptide side chains with the P9 pocket was pH dependent. Therefore, a panel of CLIP analogue peptides in which methionine 98 was substituted by all naturally occurring amino acids was examined in the competition assay (Table III). These experiments demonstrated three important properties of I-Ag7: (i) At both an acidic and a neutral pH, I-Ag7 has a strong preference for a negatively charged residue at P9. (ii) The interaction of I-Ag7 and CLIP is relatively weak at a neutral pH, and there is little or no interaction at an acidic pH. (iii) The interaction of some peptide side chains with the P9 pocket is pH dependent. Particularly striking is the observation that a CLIP analogue with histidine at P9 competes well at pH 7.4 (IC50 of 4 μM), but not at pH 5.0 (IC50 of >125 μM).
Table III.
pH-dependent Interaction of Different Peptide Side Chains with the P9 Pocket of I-Ag7
| Amino acid at position P9 of CLIP | IC50 | |||
|---|---|---|---|---|
| pH 5.0 | pH 7.4 | |||
| μM | ||||
| M | >125 | 50 | ||
| P | >125 | >125 | ||
| G | >125 | 10 | ||
| A | 15 | 2.0 | ||
| V | 17 | 50 | ||
| L | >125 | 40 | ||
| I | >125 | >125 | ||
| F | 50 | 80 | ||
| Y | 60 | >125 | ||
| W | >125 | 80 | ||
| H | >125 | 4.0 | ||
| K | >125 | >125 | ||
| R | >125 | >125 | ||
| D | 0.5 | 0.5 | ||
| E | 1.6 | 0.4 | ||
| Q | >125 | 25 | ||
| N | 125 | 80 | ||
| S | 20 | 40 | ||
| T | 40 | >125 | ||
| C | 20 | 40 | ||
Residue 98 of CLIP was substituted by all naturally occurring amino acids. These analogues were examined in the competition assay, at both an acidic and neutral pH. Peptides with a negative charge at P9 competed for binding both at pH 5.0 and pH 7.4 at low peptide concentrations. In contrast, binding of other analogues was strongly pH dependent. For example, the histidine analogue did not compete for binding at pH 5.0, but competed well at a neutral pH.
I-Ag7 Forms Long-lived Complexes with Naturally Processed Peptides, But Not SDS-resistant Dimers.
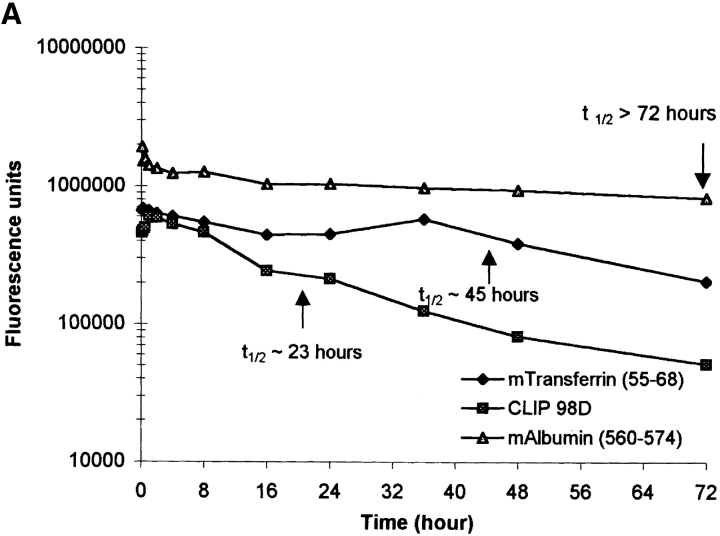
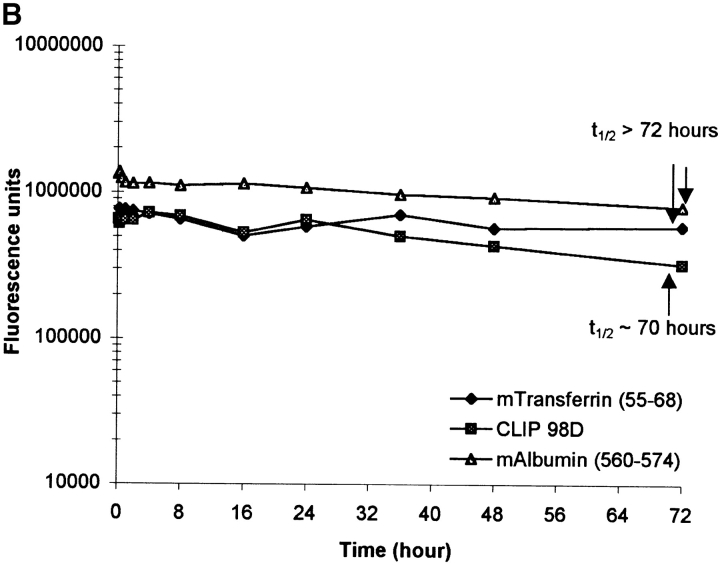
Since it has been hypothesized that the interaction of peptides with I-Ag7 is short lived (17), we determined the dissociation rates of several peptides, which included the naturally processed transferrin and serum albumin peptides, the CLIP 98D analogue, and peptides that have been identified as T cell epitopes in NOD mice (Fig. 6, Table IV). Purified I-Ag7 was first incubated with one of the biotinylated peptides (1 μM), then the pH was adjusted to pH 7.4 or maintained at pH 5.0 and mouse serum albumin peptide was added to a final concentration of 100 μM to prevent rebinding of dissociated biotinylated peptide. The GAD 65 peptide dissociated very rapidly and a t1/2 value could not be estimated. In contrast, complexes of I-Ag7 and several other peptides were long lived, particularly at a neutral pH. The estimated t1/2 value for the serum albumin peptide was >72 h and for the transferrin peptide ∼45 h at pH 5.0 and >72 h at a neutral pH. The dissociation of the CLIP 98D analogue was faster, with a t1/2 value of ∼23 h at pH 5.0 and ∼70 h at pH 7.4 (Fig. 6). These results demonstrated that I-Ag7 forms long-lived complexes with naturally processed peptides and that the dissociation rate of peptides was affected by pH.
Figure 6.
pH-dependent dissociation of peptides from I-Ag7. I-Ag7 (0.6 μg) was incubated with biotinylated peptides from mouse albumin and transferrin or the CLIP 98D analogue (all at 1 μM) at 37°C for 18 h at pH 5.0. The pH of the reaction was either kept at pH 5.0 (A) or adjusted to pH 7.4 (B). At this time, an excess of mouse serum albumin peptide (100 μM) was added as a competitor to prevent rebinding of biotinylated peptide. After incubation at the indicated pH for 5 min to 72 h, bound biotinylated peptide was quantitated as described in Materials and Methods. All three peptides dissociated slowly at a neutral pH, but dissociation of the transferrin and CLIP 98D analogue was more rapid at pH 5.0.
Table IV.
I-Ag7 Forms Long-lived Complexes with Certain Peptides
| Peptide | Dissociation time (t1/2) | |||
|---|---|---|---|---|
| pH 5.0 | pH 7.4 | |||
| Murine serum albumin (560–574) | >72 h | >72 h | ||
| Murine transferrin (55–68) | ∼45 h | >72 h | ||
| Human HSP60 (441–460) | ∼14 h | ∼25 h | ||
| Human GAD65 (249–263) | not detectable | not detectable | ||
| HSV-2 VP16 (430–444) | not detectable | not detectable | ||
| Murine CLIP | not detectable | ∼1 h | ||
| Murine CLIP 98D analogue | ∼23 h | ∼70 h | ||
The dissociation time (t1/2) of a panel of peptides was examined as described in Fig. 6. Several peptides formed long-lived complexes with I-Ag7, particularly at a neutral pH. The peptides included the naturally processed peptides from serum albumin and transferrin, as well as a peptide from human HSP60 (res. 441–460) that has been identified as a T cell epitope in NOD mice. The dissociation of the CLIP peptide at pH 5.0 was very rapid and a half-life could not be determined. These data demonstrate that I-Ag7–peptide complexes can be long lived, even though I-Ag7–peptide complexes are not SDS resistant.
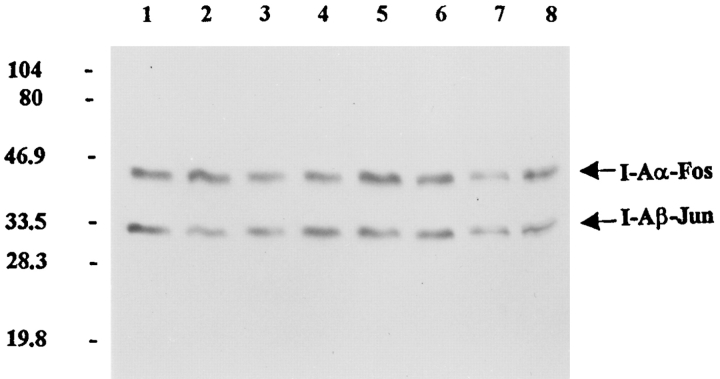
Nevertheless, I-Ag7–peptide complexes were not resistant to SDS (Fig. 7). I-Ag7 (0.6 μg) was loaded at pH 5.0 with four different peptides (transferrin, serum albumin, CLIP, or CLIP 98D analogue) by overnight incubation at 37°C. Fractions of I-Ag7–peptide complexes (0.1 μg) were examined for the formation of SDS-resistant dimers by Western blot analysis using polyclonal antisera specific for the Fos and Jun segments. An SDS-resistant dimer could not be detected with any of the peptides tested. Therefore, the SDS assay does not determine whether peptides were bound to I-Ag7 before addition of detergent. Soluble HLA-DR1 expressed in insect cells forms SDS-stable dimers with the influenza hemagglutinin (307–319) peptide (33). This indicates that SDS-sensitivity is not a general property of soluble MHC class II molecules expressed in insect cells.
Figure 7.
I-Ag7/peptide complexes are not SDS-resistant. I-Ag7 (0.6 μg) was loaded at pH 5.0 with four different peptides (transferrin, mouse serum albumin, CLIP, or CLIP 98D, each at 5 μM) by overnight incubation at 37°C. Fractions of I-Ag7–peptide complexes (0.1 μg) were examined for the formation of SDS-stable dimers by Western blot analysis using polyclonal antisera specific for the Fos and Jun segments. Samples were either not boiled (lanes 1–4) or boiled (lanes 5–8). Lanes 1 and 5, transferrin peptide; lanes 2 and 6, mouse serum albumin peptide; lanes 3 and 7, CLIP peptide; lanes 4 and 8, CLIP 98D analogue.
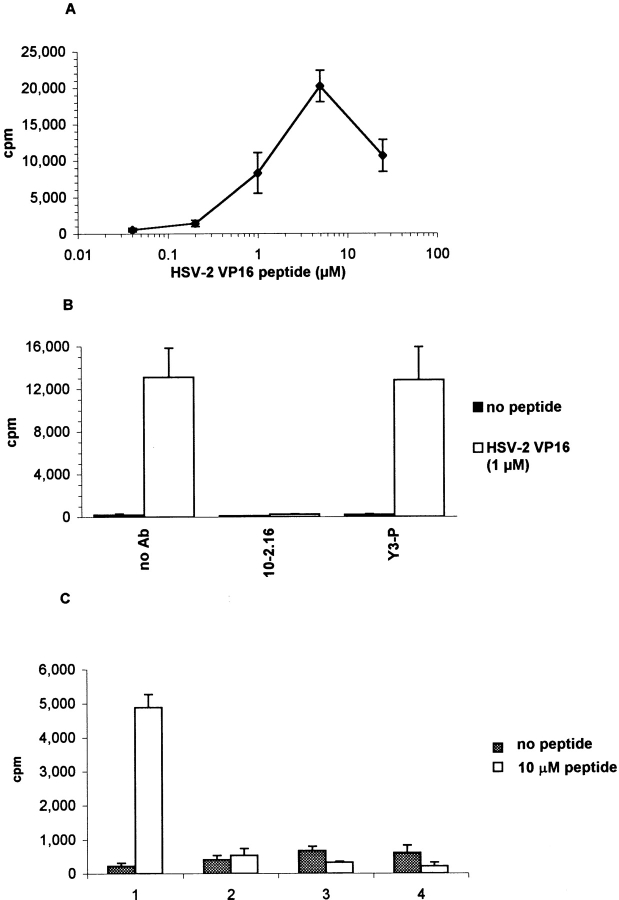
Since several of the peptides that were identified as T cell epitopes of islet autoantigens were poor binders, we examined if peptides with such binding properties could induce a T cell response. We chose the peptide from HSV-2 VP16, which has been reported to bind to HLA-DQ8 (34), since it represented a foreign antigen. This peptide showed little binding in the direct binding assay, but competed for binding of the biotinylated transferrin peptide at high peptide concentrations (data not shown). NOD mice were primed with this peptide in CFA and a T cell line specific for the HSV-2 peptide could be established from draining lymph nodes by restimulation with the HSV-2 peptide (Fig. 8 A). This T cell line was I-Ag7 restricted since the response could be completely blocked by mAb 10-2.16, but not by a control antibody (Y3-P) (Fig. 8 B). Stimulation of this T cell line required the continuous presence of the peptide in the culture. Even a single wash of the APCs after overnight incubation with the peptide completely eliminated the T cell response (Fig. 8 C, lane 2). These results demonstrate that T cells from NOD mice can be primed even by a peptide that is a poor binder for I-Ag7.
Figure 8.
NOD mice can be primed by a foreign peptide that is a weak binder to I-Ag7. A T cell line specific for HSV-2 VP16 (res. 430–444) was generated by immunizing NOD mice with this peptide in CFA. (A) Dose–response of the T cell line to the HSV-2 peptide using irradiated NOD spleen cells as APCs. T cell proliferation was determined by [3H]thymidine incorporation. (B) The HSV-2–specific T cell line was I-Ag7 restricted since proliferation was blocked by mAb 10-2.16 but not by a control antibody (Y3-P). (C) Stimulation of HSV-2–specific T cells required continuous presence of the peptide. Splenocytes from NOD mice were pulsed overnight ± 10 μM HSV-2 peptide in serum-free media (AIM-V) and then split into four aliquots: 1, No wash of APCs before coculture with T cells; 2, APCs washed once with 10 ml of RPMI; 3, APCs washed twice and incubated for 30 min between washes; and 4, APCs washed twice and incubated for 60 min between washes.
Discussion
Soluble I-Ag7 was used to define the biochemical requirements for peptide loading by this MHC class II molecule, which confers susceptibility to type I diabetes in NOD mice. Several peptides, including two naturally processed peptides from exogenous antigens, bound to I-Ag7. The interaction with the invariant chain–derived CLIP peptide was relatively weak and pH dependent. CLIP bound to I-Ag7 at a neutral pH, but rapidly dissociated at an endosomal pH. This property of CLIP was due to methionine 98, which occupies the P9 pocket of the MHC class II peptide binding site (25). Substitution of this residue of CLIP greatly increased the stability of the complex, both at an acidic and neutral pH. These characteristics of I-Ag7 result in the rapid generation of empty molecules at an endosomal pH.
Several mutant cell lines have been described that are defective in antigen presentation due to a lack of DM expression. In these cell lines, MHC class II–CLIP complexes accumulate because DM does not catalyze the removal of CLIP (32). For several human and murine MHC class II molecules, CLIP peptides are abundant among naturally processed peptides in normal cell lines, reflecting a relatively stable interaction (35, 36). For example, CLIP peptides have been eluted from purified I-Ab and I-Ad. CLIP was found to bind with a relatively high affinity to I-Ad (IC50 of 14 nM) and was one of the best binders identified for this class II molecule (36). These results are important, since I-Ag7 and I-Ad share the same I-A α chain (12). Thus, the binding properties of CLIP to I-Ag7 appear to be due to the unique I-Ag7 β chain.
The requirement for DM in the presentation of antigens by I-Ag7 has been analyzed using T cell hybridomas specific for chicken ovalbumin (37). For all T cell hybridomas, the antigen was presented by transfectants that expressed only I-Ag7 and invariant chain, but not H-2M, indicating that the murine DM homologue is not essential for antigen presentation by I-Ag7. For two out of four hybridomas with a distinct epitope specificity, presentation was enhanced by coexpression of H2-M. These results indicate that presentation of certain epitopes by I-Ag7 can be enhanced by H-2M. This may be due to “editing” of peptide content as well as stabilization of empty I-Ag7 molecules by H-2M, as described for human MHC class II molecules (30, 31, 38).
The recently solved crystal structure of I-Ad demonstrates several features of I-A–peptide interactions that are important in this context (16). Detailed analysis of the I-Ad peptide binding motif demonstrated that only two peptide residues in a 6-amino acid core were important for binding (39). These residues are located in the P4 and P9 pockets of the I-Ad crystal structure. I-Ad and I-Ag7 have the same α chain sequence, but differ at 17 positions of the β chain. An important difference is the presence of an interchain salt bridge between β57 Asp and α76 Arg in I-Ad, which is absent in I-Ag7 (β57 Ser). In I-Ag7–peptide complexes, a salt bridge may instead be formed by P9 Asp or P9 Glu of bound peptides and α76 Arg. This would explain the strong preference for negatively charged residues at the P9 position, which was first noted among naturally processed peptides eluted from I-Ag7 (23). In contrast, I-Ad has a preference for alanine or serine at the P9 position. An I-Ag7 peptide binding motif has been proposed, in which a preference for aliphatic residues was noted at P6 and for hydrophobic residues at P9 (40). Also, at P11 binding was enhanced by the presence of aspartic acid. It appears that these data are in general agreement with other data on I-Ag7– peptide interactions, provided that the relative positions are reassigned such that these data reflect the preferences of the P4, P7, and P9 pockets of I-Ag7.
Are I-Ag7 molecules poor peptide binders or are only some of the peptides that have been identified thus far poor ligands? This may be a separate issue from the lack of SDS resistance. The analysis of soluble I-Ag7 clearly demonstrates that this MHC class II molecule binds peptides. Two naturally processed peptides from extracellular antigens, mouse albumin and transferrin, are strong binders, indicating that I-Ag7 does not have a global defect in presentation of exogenous antigens. Peptide binding by purified I-Ag7 has only been reported in two studies that used detergent-solubilized molecules. In one of these studies, little or no specific peptide binding could be demonstrated (17). In the other study, high detergent concentrations (2% NP-40) were used in the peptide binding assay (40). Binding was demonstrated for a hen egg lysozyme peptide (res. 9–27), but not for the mouse albumin peptide that had been eluted from purified I-Ag7 and that represents the best binder for soluble I-Ag7 (23). It is possible that these difficulties in examining peptide binding by I-Ag7 are related to the isolation method, in particular to the use of certain detergents. The naturally processed transferrin and serum albumin peptides formed long-lived complexes with I-Ag7, in particular at a neutral pH. However, several peptides that have been identified as T cell epitopes of islet antigens were found to be poor binders. A similar observation has been made in the experimental autoimmune encephalomyelitis model, since the immunodominant MBP peptide (Ac1-11) in mice with the H-2u haplotype is a very weak binder (24). Taken together, our data indicate that some peptides form long-lived complexes with I-Ag7. However, it is possible that a smaller fraction of I-Ag7 molecules represent long-lived class II–peptide complexes and/or that the average affinity of I-Ag7 bound peptides is lower than that of peptides bound to other I-A molecules.
I-Ag7 immunoprecipitated from splenocytes of NOD mice does not form SDS-resistant dimers (17). This property was also observed for soluble I-Ag7, even when the molecules were loaded with peptides. Therefore, it appears that SDS-sensitivity of the dimer is an intrinsic property of I-Ag7 and does not directly reflect peptide content. SDS sensitivity of the I-Ag7 dimer could be due to polymorphic residues at the dimerization interface of I-Aα and I-Aβ. Mutational analysis of I-Ag7 indicated that β56 and β57 do not determine SDS-sensitivity (17). The absence of the salt bridge between β57 and α76 is therefore not responsible for the lack of SDS resistance. It is also important to keep in mind that instability of the dimer in SDS does not indicate that the dimer is also unstable under native conditions. In fact, analysis of I-Ag7 purified from B cells did not indicate that the dimer was unstable in the absence of SDS (17).
The pH-dependent interaction of I-Ag7 results in the rapid generation of empty molecules at an endosomal pH. These results demonstrate a novel mechanism by which the presentation of antigenic peptides can be affected by a disease-associated MHC class II molecule.
Acknowledgments
We wish to thank Dr. P. Kim for the generous gift of the Fos and Jun leucine zipper peptides and Dr. M. Lipes (Joslin Diabetes Center, Boston, MA) for generously providing RNA from NOD spleens.
This work was supported by a Career Development Award from the American Diabetes Association and by a grant from the National Institutes of Health (AI39619) to K.W. Wucherpfennig. D.H.F. Hausmann and S. Hausmann are recipients of postdoctoral fellowships from the Deutsche Forschungsgemeinschaft (DFG Ha2556/1-1) and the Deutscher Akademischer Austauschdienst, respectively.
Abbreviations used in this paper
- CLIP
class II–associated invariant chain peptide
- HSV
Herpes simplex virus
- MBP
myelin basic protein
- NOD
nonobese diabetic
Footnotes
D.H.F. Hausmann's present address is Universität Rostock, Klinik fur Innere Medizin, Abteilung f ür Gastroenterologie, Ernst-Heydemann Str. 6, 18055 Rostock, Germany, and S. Hausmann's present address is Universität Rostock, Institut für Medizinische Biochemie, Schillingallee 70, 18057 Rostock, Germany.
D.H.F. Hausmann and B. Yu contributed equally to this study.
References
- 1.Todd JA, Acha-Orbea H, Bell JI, Chao N, Fronek Z, Jacob CO, McDermott M, Sinha AA, Timmerman L, Steinman L, McDevitt HO. A molecular basis for MHC class II–associated autoimmunity. Science. 1988;240:1003–1009. doi: 10.1126/science.3368786. [DOI] [PubMed] [Google Scholar]
- 2.Todd JA, Aitman TJ, Cornall RJ, Ghosh S, Hall JRS, Hearne CM, Knight AM, Love JM, McAleer MA, Prins J-B, et al. Genetic analysis of autoimmune type 1 diabetes mellitus in mice. Nature. 1991;351:542–547. doi: 10.1038/351542a0. [DOI] [PubMed] [Google Scholar]
- 3.Nepom GT, Erlich H. MHC class-II molecules and autoimmunity. Annu Rev Immunol. 1991;9:493–525. doi: 10.1146/annurev.iy.09.040191.002425. [DOI] [PubMed] [Google Scholar]
- 4.Davies JL, Kawaguchi Y, Bennett ST, Copeman JB, Cordell HJ, Pritchard LE, Reed PW, Gough SCL, Jenkins SC, Palmer SM, et al. A genome-wide search for human type I diabetes susceptibility genes. Nature. 1994;371:130–136. doi: 10.1038/371130a0. [DOI] [PubMed] [Google Scholar]
- 5.Wicker LS, Todd JA, Peterson LB. Genetic control of autoimmune diabetes in the NOD mouse. Annu Rev Immunol. 1995;13:179–200. doi: 10.1146/annurev.iy.13.040195.001143. [DOI] [PubMed] [Google Scholar]
- 6.Nepom GT. Immunogenetics and IDDM. Diabetes Rev. 1993;1:93–103. [Google Scholar]
- 7.She J-X. Susceptibility to type I diabetes: HLA-DQ and DR revisited. Immunol Today. 1996;17:323–329. doi: 10.1016/0167-5699(96)10014-1. [DOI] [PubMed] [Google Scholar]
- 8.Lund T, O'Reilly L, Hutchings P, Kanagawa O, Simpson E, Gravely R, Chandler P, Dyson J, Picard JK, Edwards A, et al. Prevention of insulin-dependent diabetes mellitus in non-obese diabetic mice by transgenes encoding modified I-A β-chain or normal I-E α-chain. Nature. 1990;345:727–729. doi: 10.1038/345727a0. [DOI] [PubMed] [Google Scholar]
- 9.Quartey-Papafio R, Lund T, Chandler P, Picard J, Ozegbe P, Say S, Hutchings PR, O'Reilly L, Kioussis D, Simpson E, Cooke A. Aspartate at position 57 of nonobese diabetic I-Ag7β-chain diminishes the spontaneous incidence of insulin-dependent diabetes mellitus. J Immunol. 1995;154:5567–5575. [PubMed] [Google Scholar]
- 10.Wicker LS. Major histocompatibility complex–linked control of autoimmunity. J Exp Med. 1997;186:973–975. doi: 10.1084/jem.186.7.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serreze DV, Leiter EH. Genetic and pathogenic basis of autoimmune diabetes in NOD mice. Curr Opin Immunol. 1994;6:900–906. doi: 10.1016/0952-7915(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 12.Acha-Orbea H, McDevitt HO. The first external domain of the nonobese diabetic mouse class II I-Aβ chain is unique. Proc Natl Acad Sci USA. 1987;84:2433–2439. doi: 10.1073/pnas.84.8.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, Wiley DC. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 14.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban R, Strominger JL, Wiley DC. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 15.Fremont DH, Monnaie D, Nelson CA, Hendrickson WA, Unanue ER. Crystal structure of I-Akin complex with a dominant epitope of lysozyme. Immunity. 1998;8:305–317. doi: 10.1016/s1074-7613(00)80536-1. [DOI] [PubMed] [Google Scholar]
- 16.Scott CA, Peterson PA, Teyton L, Wilson IA. Crystal structures of two I-Ad-peptide complexes reveal that high affinity can be achieved without large anchor residues. Immunity. 1998;8:319–329. doi: 10.1016/s1074-7613(00)80537-3. [DOI] [PubMed] [Google Scholar]
- 17.Carrasco-Marin E, Shimizu J, Kanagawa O, Unanue ER. The class II MHC I-Ag7molecules from non-obese diabetic mice are poor peptide binders. J Immunol. 1996;156:450–458. [PubMed] [Google Scholar]
- 18.Bunch TA, Grinblat Y, Goldstein LSB. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogastercells. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalandadze A, Galleno M, Foncerrada L, Strominger JL, Wucherpfennig KW. Expression of recombinant HLA-DR2 molecules. Replacement of the hydrophobic transmembrane region by a leucine zipper dimerization motif allows the assembly and secretion of soluble DR αβ heterodimers. J Biol Chem. 1996;271:20156–20162. doi: 10.1074/jbc.271.33.20156. [DOI] [PubMed] [Google Scholar]
- 20.Oi VT, Jones PP, Goding JW, Herzenberg LA, Herzenberg LA. Properties of monoclonal antibodies to mouse Ig allotypes, H-2, and Ia antigens. Curr Top Microbiol Immunol. 1978;81:115–129. doi: 10.1007/978-3-642-67448-8_18. [DOI] [PubMed] [Google Scholar]
- 21.Scott CA, Garcia KC, Carbone FR, Wilson IA, Teyton L. Role of chain pairing for the production of functional soluble IA major histocompatibility complex class II molecules. J Exp Med. 1996;183:2087–2095. doi: 10.1084/jem.183.5.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avva RR, Cresswell P. In vivo and in vitro formation and dissociation of HLA-DR complexes with invariant chain-derived peptides. Immunity. 1994;1:763–774. doi: 10.1016/s1074-7613(94)80018-9. [DOI] [PubMed] [Google Scholar]
- 23.Reich E-P, von Grafenstein H, Barlow A, Swenson KE, Williams K, Janeway CA., Jr Self peptides isolated from MHC glycoproteins of non-obese diabetic mice. J Immunol. 1994;152:2279–2288. [PubMed] [Google Scholar]
- 24.Fairchild PJ, Wildgoose R, Atherton E, Webb S, Wraith DC. An autoantigenic T cell epitope forms unstable complexes with class II MHC: a novel route to escape from tolerance induction. Int Immunol. 1993;5:1151–1158. doi: 10.1093/intimm/5.9.1151. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh P, Amaya M, Mellins E, Wiley DC. The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3. Nature. 1995;378:457–462. doi: 10.1038/378457a0. [DOI] [PubMed] [Google Scholar]
- 26.Malcherek G, Gnau V, Jung G, Rammensee H-G, Melms A. Supermotifs enable natural invariant chain-derived peptides to interact with many major histocompatibility complex-class II molecules. J Exp Med. 1995;181:527–536. doi: 10.1084/jem.181.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weenink SM, Milburn PJ, Gautam AM. A continuous central motif of invariant chain peptides, CLIP, is essential for binding to various I-A MHC class II molecules. Int Immunol. 1997;9:317–325. doi: 10.1093/intimm/9.2.317. [DOI] [PubMed] [Google Scholar]
- 28.Busch R, Mellins ED. Developing and shedding inhibitions: how MHC class II molecules reach maturity. Curr Opin Immunol. 1996;8:51–58. doi: 10.1016/s0952-7915(96)80105-1. [DOI] [PubMed] [Google Scholar]
- 29.Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 30.Denzin LK, Hammond C, Cresswell P. HLA-DM interactions with intermediates in HLA-DR maturation and a role for HLA-DM in stabilizing empty HLA-DR molecules. J Exp Med. 1996;184:2153–2165. doi: 10.1084/jem.184.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber DA, Evavold BD, Jensen PE. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science. 1996;274:618–620. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]
- 32.Riberdy JM, Newcomb JR, Surman MJ, Barbosa JA, Cresswell P. HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature. 1992;360:474–477. doi: 10.1038/360474a0. [DOI] [PubMed] [Google Scholar]
- 33.Stern LJ, Wiley DC. The human class II MHC protein HLA-DR1 assembles as empty αβ heterodimers in the absence of antigenic peptide. Cell. 1992;68:465–477. doi: 10.1016/0092-8674(92)90184-e. [DOI] [PubMed] [Google Scholar]
- 34.Kwok WW, Domeier ME, Johnson ML, Nepom GT, Koelle DM. HLA-DQB1 codon 57 is critical for peptide binding and recognition. J Exp Med. 1996;183:1253–1258. doi: 10.1084/jem.183.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chicz RM, Urban RG, Gorga JC, Vignali DA, Lane WS, Strominger JL. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J Exp Med. 1993;178:27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunt DF, Michel H, Dickinson TA, Shabanowitz J, Cox AL, Sakaguchi K, Apella E, Grey HM, Sette A. Peptides presented to the immune system by the murine class II major histocompatibility complex molecule I-Ad . Science. 1992;256:1817–1820. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 37.Peterson M, Sant AJ. The inability of the nonobese diabetic class II molecule to form stable peptide complexes does not reflect a failure to interact productively with DM. J Immunol. 1998;161:2961–2967. [PubMed] [Google Scholar]
- 38.Kropshofer H, Arndt SO, Moldenhauer G, Hämmerling GJ, Vogt AB. HLA-DM acts as a molecular chaperone and rescues empty HLA-DR molecules at lysosomal pH. Immunity. 1997;6:293–302. doi: 10.1016/s1074-7613(00)80332-5. [DOI] [PubMed] [Google Scholar]
- 39.Sette A, Buus S, Colon S, Miles C, Grey HM. I-Ad-binding peptides derived from unrelated protein antigens share a common structural motif. J Immunol. 1988;141:45–48. [PubMed] [Google Scholar]
- 40.Harrison LC, Honeyman MC, Trembleau S, Gregori S, Gallazzi F, Augstein P, Brusic V, Hammer J, Adorini L. A peptide-binding motif for I-Ag7, the class II major histocompatibility complex (MHC) molecule of NOD and Biozzi AB/H mice. J Exp Med. 1997;185:1013–1021. doi: 10.1084/jem.185.6.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elias D, Marcus H, Reshef T, Ablamunits V, Cohen IR. Induction of diabetes in standard mice by immunization with the p277 peptide of a 60-kDa heat shock protein. Eur J Immunol. 1995;25:2851–2857. doi: 10.1002/eji.1830251021. [DOI] [PubMed] [Google Scholar]
- 42.Reizis B, Eisenstein M, Bockova J, Könen-Waisman S, Mor F, Elias D, Cohen IR. Molecular characterization of the diabetes-associated mouse MHC class II protein, I-Ag7 . Int Immunol. 1997;9:43–51. doi: 10.1093/intimm/9.1.43. [DOI] [PubMed] [Google Scholar]
- 43.Daniel D, Gill RG, Schloot N, Wegmann D. Epitope specificity, cytokine production profile and diabetogenic activity of insulin-specific T cell clones isolated from NOD mice. Eur J Immunol. 1995;25:1056–1062. doi: 10.1002/eji.1830250430. [DOI] [PubMed] [Google Scholar]
- 44.Kaufman DL, Clare-Salzler M, Tian J, Forsthuber T, Ting GSP, Robinson P, Atkinson MA, Sercarz EE, Tobin AJ, Lehmann PV. Spontaneous loss of T-cell tolerance to glutamic acid decarboxylase in murine insulin-dependent diabetes. Nature. 1993;366:69–72. doi: 10.1038/366069a0. [DOI] [PMC free article] [PubMed] [Google Scholar]