Abstract
Here we show a correlation between telomere length and organismal sensitivity to ionizing radiation (IR) in mammals. In particular, fifth generation (G5) mouse telomerase RNA (mTR)−/− mice, with telomeres 40% shorter than in wild-type mice, are hypersensitive to cumulative doses of gamma rays. 60% of the irradiated G5 mTR−/− mice die of acute radiation toxicity in the gastrointestinal tract, lymphoid organs, and kidney. The affected G5 mTR−/− mice show higher chromosomal damage and greater apoptosis than similarly irradiated wild-type controls. Furthermore, we show that G5 mTR−/− mice show normal frequencies of sister chromatid exchange and normal V(D)J recombination, suggesting that short telomeres do not significantly affect the efficiency of DNA double strand break repair in mammals. The IR-sensitive phenotype of G5 mTR−/− mice suggests that telomere function is one of the determinants of radiation sensitivity of whole animals.
Keywords: telomerase, radiosensitivity, DNA DSB repair, cancer, radiotherapy
Introduction
Ionizing radiation (IR) is a potent inducer of DNA double strand breaks (DSBs), which, if unrepaired, may either lead to cell death or trigger neoplastic transformation 1. Studies in mammals and yeast have shown that mutations in genes responsible for DNA DSB repair result in increased IR sensitivity 2 3. Two major mechanisms are involved in repairing DSB in mammalian cells: nonhomologous end joining (NHEJ) and homologous recombination (HR) 4 5.
Telomeres are specialized protective structures at the ends of chromosomes 6. There is increasing evidence that proteins involved in DSB DNA repair may have a role at the telomeres. Yeast telomeres serve as storage sites for DNA repair proteins such as yKu80 and Sir3p, which, upon induction of DSB, are mobilized from the telomeres to the sites of damage 7 8. In mammals, Ku proteins bind to telomeric sequences 9 10 and protect telomeres from end–end fusions 11, and the DNA repair complex RAD50/MRE11/NBS1 associates with telomeric protein TRF2 12. Telomere length can also influence IR sensitivity, in particular, mutations in DNA repair proteins yKu80, yKu70, Mre11, Rad50, Xrs2, Sir2, and Sir3 result in yeast strains with altered telomere length and altered IR sensitivities 13. In Caenorhabditis elegans, DNA repair mtr-2 mutant shows telomere shortening and IR hypersensitivity 14. An association between telomere length and IR has not been established yet in mammals. Furthermore, it is not clear how short telomeres influence IR sensitivity in mammals. Here, we study the radiation sensitivity of mice lacking the RNA component of telomerase (mouse telomerase RNA [mTR]−/−) 15. These mice lack telomerase activity and show telomere shortening at a rate of 4–5 kb per mouse generation 15. Telomere shortening in these mice is accompanied by an increase in the number of chromosome ends without detectable telomeres and in the frequency of chromosome fusions 15 16. Our results indicate that short telomeres result in organismal hypersensitivity to IR and in an increased DNA damage after irradiation. Furthermore, we show that this increased radiation-induced DNA damage in late generation mTR−/− mice does not seem to be the direct consequence of a defect in a known DNA DSB repair pathway.
Materials and Methods
Mice.
Wild-type (wt) and different generations of mTR−/− mice were generated as described 15. The genetic background was 60% C57BL6, 37.5% 129Sv, and 2.5% SJL 15 16 17.
Statistical Analysis.
Sample means, standard deviations, chi-squares, and Student's t tests were calculated using Microsoft Excel.
Gamma Irradiation.
Single doses of 1.75 Gy were administered to age-matched 5-mo-old wt and mTR−/− mice weekly using a 137Cs source (MARK 1-30 Irradiator; Shepherd & Associates) at a rate of 1.14 Gy/min.
Histopathological Analysis.
Sections from different organs from nonirradiated and irradiated wt and mTR−/− mice were fixed in 10% buffered formalin and stained in parallel with hematoxylin–eosin.
Telomere Length.
To measure telomere length, FLOW-FISH (fluorescence in situ hybridization) was carried out on fresh bone marrow (BM) cells (a total of 1,500–4,000 nuclei for each sample were analyzed) as described 17. For FLOW-FISH on blood cells, blood (100–200 μl) was obtained before and after irradiation from the retroorbital plexus. Red blood cells were lysed by osmotic shock. Lymphocytes were resuspended in FCS, and an equal volume of FCS plus 20% DMSO was added dropwise. The cells were frozen gradually at −80°C and kept in liquid nitrogen. After collection of all blood samples, cells were thawed at 37°C, and an equal volume of 8% dextran (10% Rheomacrodex; Pharmacia Antibioticos Farma, S.A.) and 5% human albumin was added. Cells were centrifuged at 900 g for 20 min at 4°C and were resuspended in RPMI 1640 containing 10% FBS and 0.55 mM β-mercaptoethanol. Approximately 0.5 × 106 cells were used per sample, and FLOW-FISH was carried out as described 17.
Cytogenetic Determination of Radiation-induced Chromosomal Damage in wt and mTR−/− Mice.
Direct BM chromosome preparations were made from unirradiated control and gamma-irradiated mice as described 18 and stained with concentrated paint probes for chromosomes 1, 2, and 3 (Cambio) simultaneously. Slides were pretreated, denatured 19, and hybridized with a total of 10 μl of hybridization mix containing 2 μl of biotin-labeled chromosome 3 probe, 2 μl of FITC-labeled chromosome 2 probe, 1 μl each of biotin- and FITC-labeled chromosome 1 probe, 0.5 μg of mouse CoT 1 DNA (Life Technologies), and 10 μg of human total genomic DNA. After 48-h hybridization at 42°C, slides were washed in 2× SSC, followed by two 15-min washes in 0.5× SSC/50% formamide at 42°C and a final wash in 2× SSC. Biotin-labeled probes were detected with alternating layers of avidin–Texas Red and biotinylated goat anti–avidin D antibody (Vector Laboratories), and FITC-labeled probes were detected with alternating layers of rabbit anti-FITC–IgG (Dako) and FITC-labeled goat anti–rabbit IgG (Sigma-Aldrich) 20. Three layers of each fluorescent detecting reagent were applied. Slides were mounted in Vectashield (Vector Laboratories) containing 400 ng/ml DAPI (4,6-diamino-2-phenylindole) counterstain and scored using a Nikon epifluorescence microscope equipped with appropriate filter sets. Aberrations involving painted chromosomes were scored. Events that were clearly clonal in origin were counted only once in the determination of aberration frequencies. Quantitative comparison of all chromosome-type aberrations taken together (chromosome-type aberration score) employed a modified version 19 of the PAINT classification system 21 in which the number of color junctions arising from translocations, dicentrics, etc. and the number of deletions are summed and expressed per 100 cells.
Annexin V Labeling of Fresh Splenocytes to Determine In Vivo Apoptosis upon Irradiation in wt and G2 and G5 mTR−/− Mice.
The following reagents were used: FITC-conjugated annexin V (Immunotech), biotinylated rat anti–mouse B220 (Southern Biotechnology Associates), PE-conjugated hamster anti–mouse CD3 (Southern Biotechnology Associates), and spectral red label (SPRD)-conjugated streptavidin (Southern Biotechnology Associates). 24 h after irradiation, mouse splenocytes were incubated with biotinylated anti-B220 and SPRD-conjugated streptavidin, followed by incubation with FITC-conjugated annexin V and TRI-COLOR flow cytometry analysis.
Apoptosis and Cell Cycle Profile in Proliferating Splenocytes.
24 h after irradiation at the indicated gamma ray dose, splenocytes were isolated, and a total of 0.8 × 106 splenocytes were incubated in RPMI 1640 containing 10% FCS, 0.55 mM β-mercaptoethanol, and 10 mg/ml LPS (Sigma-Aldrich). After 48 h of culture, 0.250 × 106 cells were incubated with rat FITC-conjugated anti–mouse CD45R/B220 (BD PharMingen) and 20 mg/ml propidium iodide and analyzed by flow cytometry.
Sister Chromatid Exchange Determination.
Sister chromatid exchange (SCE) frequencies were determined in splenocytes. Spleens were removed, disaggregated in RPMI 1640/20% FCS, and stored on ice for 24 h. 5-ml cultures at 2 × 106 cells/ml in RPMI 1640/20% FBS were stimulated with 2 μg/ml Con A and 25 μg/ml LPS and grown in the presence of 10 μM bromodeoxyuridine for 48 h at 37°C. Cultures were irradiated with a 60Co gamma source (0.75 Gy/min) after 24 h of culture. Cells were harvested by hypotonic KCl/methanol–acetic acid fixation after colcemid treatment (0.004 μg/ml) for the final 2 h of culture. Slides were prepared and stained 22, and 25 metaphases were scored per treatment.
V(D)J Recombination Experiments.
PCR primers used were DHall: 5′-GGGA/CTTTTTGTC/GAAGGG/TATCTACTACTGTG-3′; JH4: 5′-AAAGACCTGCAGAGGCCATTCTTACC-3′; VH7183: 5′-CGCGAAGCTTCGTGGAGTCTGGGGGA-3′; DR218: 5′- ACCCCAGTAGTCCATAGCATAGTAAT-3′; Vβ8.1, 2, 3: 5′-GAGGAAAGGTGACATTGAGC-3′; Jβ2.6: 5′-GCCTGGTGCCGGGACCGAAGTA-3′; Vβ8 probe: 5′-GGGCTGAGGCTGATCCATTA-3′; 5′-intervening sequence (IVS): 5′-gtaagaatggcctctccaggt-3′; and 3′-IVS: 5′-GACTCAATCACTAAGACAGCT-3′.
Single-cell suspensions from lymphoid organs and BM from 6-wk-old mice were obtained. DNA was prepared for PCR as described 23. Semiquantitative PCR was done as described 23. PCR reactions (20 μl) contained 2–8 μl of template (∼10,000–40,000 genomes, which correspond to 50–200 ng of DNA), 160 ng of each primer, 1 U of Taq polymerase (PerkinElmer), and the reaction buffer provided by the manufacturer containing 1.5 mM MgCl2. Amplification (30 cycles) was performed at an annealing temperature of 65°C. The reaction was analyzed in 2.5% agarose gel electrophoresis and was transferred onto a nylon membrane (Hybond-N+). Oligo probes were labeled with T4 polynucleotide kinase and γ-[32P] (Amersham Pharmacia Biotech), and the probe was separated from unincorporated γ-[32P]ATP in a NAP™ 5 column (Amersham Pharmacia Biotech). For analysis of Ig coding joints, two sets of primers were used. To evaluate the D-JH rearrangement, the pair included the degenerate primer DHall and JH4 23; for V-DJH rearrangement, the JH4 primer was combined with VH7183 24. A JH4-specific probe (DR218) was used for the detection of Ig coding joint PCR products as described elsewhere 24. The TCR coding joints Vβ8-DJβ2.6 were detected using the primers Vβ8.1, 2, 3 and Jβ2.6 25 and probed with the Vβ8 probe 25. A sequence from the heavy chain locus that is not affected by Ig rearrangement was amplified as a control (see IVS in Fig. 6 A) using 5′- and 3′-IVS primers 26.
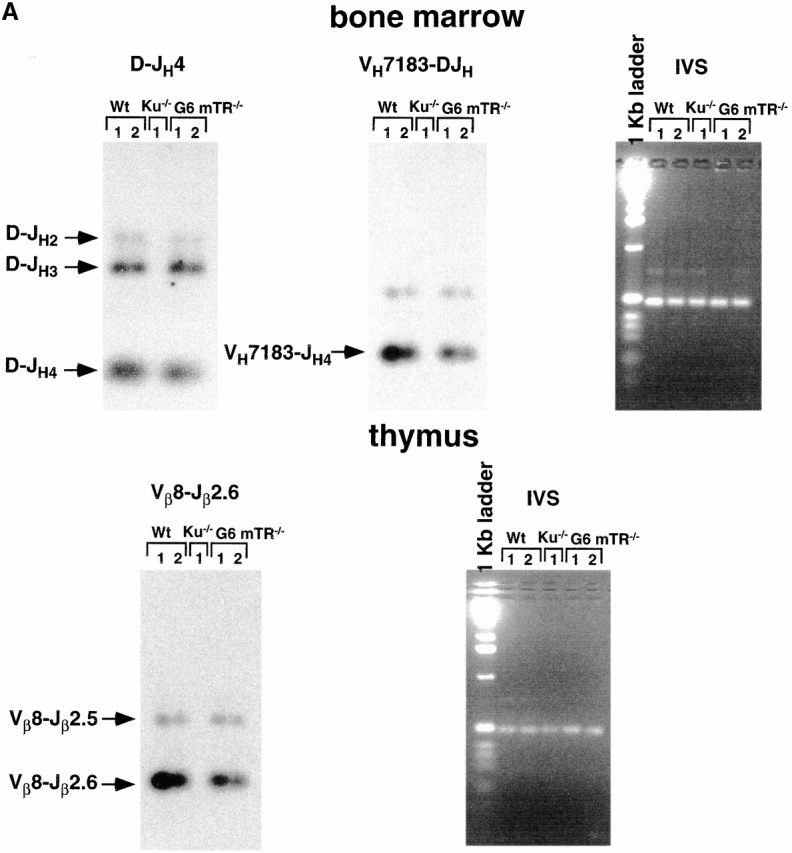
Figure 6.
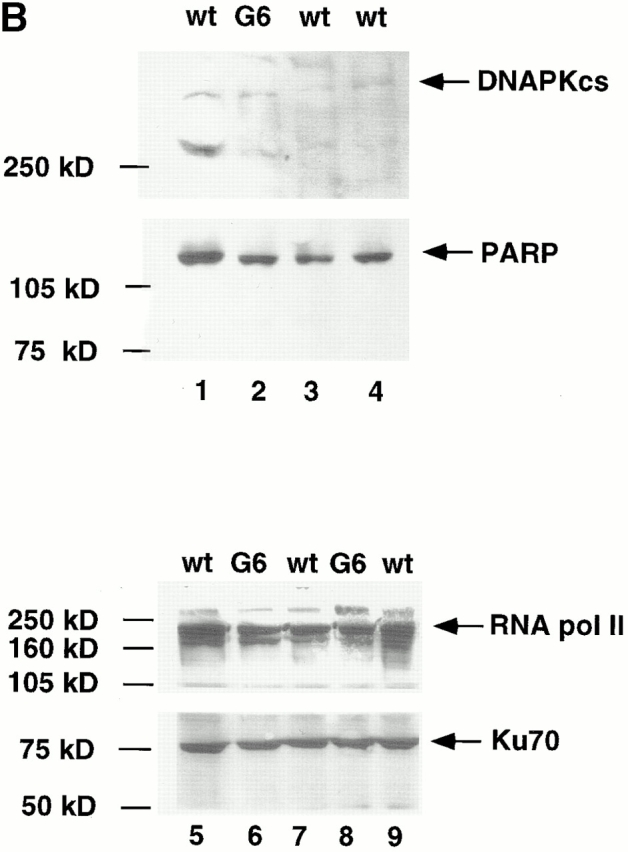
(A) Joint formation in two wt (1 and 2), one Ku86−/− 1, and two G6 mTR−/− mice (1 and 2). For analysis of Ig coding joins, two sets of primers were used: D-JH rearrangement, primers DHall and JH4; V-DJH rearrangement, primers JH4 and VH7183. The JH4-specific probe (DR218) was used for the detection of Ig coding joints. The TCR coding joins Vβ8-DJβ2.6 were detected using the primers Vβ8.1, 2, 3 and Jβ2.6 for PCR and the Vβ8 probe for detection. An IVS from the heavy chain locus that is not affected by Ig rearrangement was amplified as a control for PCR reaction (IVS) using 5′-IVS and 3′-IVS primers. (B) Western blots to detect endogenous levels of DNAPKcs (∼470 kD), Ku70 (∼70 kD), and PARP (∼110 kD). Western blots were carried out as described in Materials and Methods. The positions of the different specific bands are indicated by arrows. As loading control, RNA pol II levels were detected in the different extracts.
Western Blots.
Nuclear extracts were prepared from passage two wt, Ku86−/−, or different generation mTR−/− mouse embryonic fibroblasts as described 27. Nuclear extracts were separated on SDS/7.5% polyacrylamide gels for detection of Ku70 and RNA pol II and on SDS/5% polyacrylamide gels for detection of poly(ADP-ribose)polymerase (PARP) and catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs). PARP: mAb from Labgen; DNA-PKcs: rabbit anti–human DNA-PKcs polyclonal antibody from Oncogene Research Products; Ku70 mAb from Kamiya Biomedical Company; RNA polymerase II: rabbit anti–mouse polyclonal antibody from Santa Cruz Biotechnology, Inc.
Results and Discussion
mTR−/− Mice with Short Telomeres Are Hypersensitive to IR.
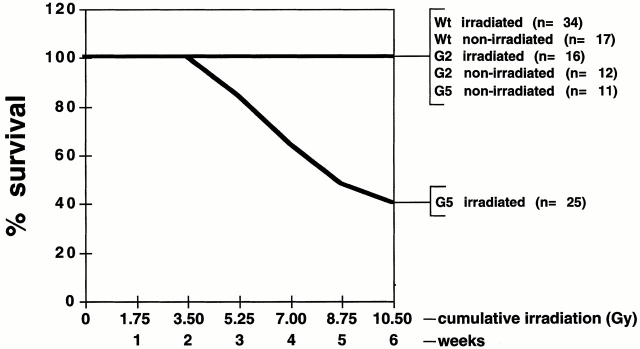
To assess the role of telomere length in organismal sensitivity to IR, wt, second generation (G2), and fifth generation (G5) mTR−/− mice were irradiated with cumulative weekly doses of gamma rays (1.75 Gy each dose). We purposely used G5 rather than G6 mTR−/− mice because G5 mTR−/− mice have 40% shorter telomeres than wt mice but do not show proliferative defects or pathologies, except for increased infertility 15 16 28. Thus, pathologies appearing in irradiated G5 mTR−/− mice will be a direct consequence of radiation. wt and G2 mTR−/− mice did not show radiation-induced loss of viability after six doses of 1.75 Gy gamma rays (Fig. 1), and these mice survived up to 2 mo after the sixth radiation dose, showing no signs of disease. In contrast, irradiated G5 mTR−/− mice showed mortality after the third dose of radiation (Fig. 1), with 60% of the irradiated G5 mTR−/− mice dead after the sixth irradiation. None of the control nonirradiated age-matched wt and G5 mTR−/− mice showed mortality during the course of the irradiation experiment or in the following 2 mo (Fig. 1). Similarly, none of the irradiated G5 mTR−/− that survived the six radiation doses showed signs of disease during the subsequent 2 mo (not shown). A Student's t test value of P = 0.0487 indicates that the mortality curve of the irradiated G5 mTR−/− mice is significantly different from that of wt irradiated, wt nonirradiated, G2 mTR−/− irradiated, G2 mTR−/− nonirradiated, and G5 mTR−/− nonirradiated mice. A recent study using G6 mTR−/− instead of G5 mTR−/− mice also described IR hypersensitivity of these mice as compared with wt 29.
Figure 1.
Viability of wt (17 nonirradiated and 34 irradiated) and G2 (12 nonirradiated and 16 irradiated) and G5 mTR−/− (11 nonirradiated and 25 irradiated) mice after administration of cumulative doses of gamma rays (1.75, 3.50, 5.25, 7.00, 8.75, and 10.50 Gy). The graph shows survival until 1 wk after the administration of the sixth dose. The wt and G2 and G5 mTR−/− survivors survived at least up to 2 mo after the sixth irradiation dose, at which point the mice were killed.
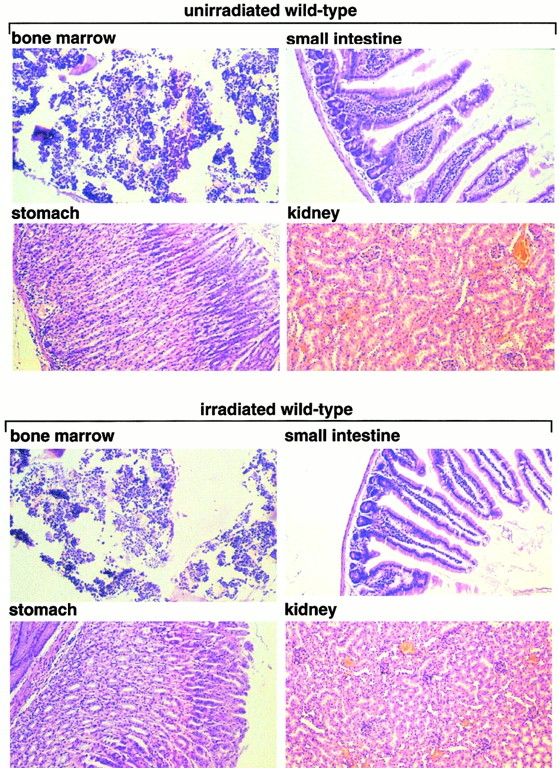
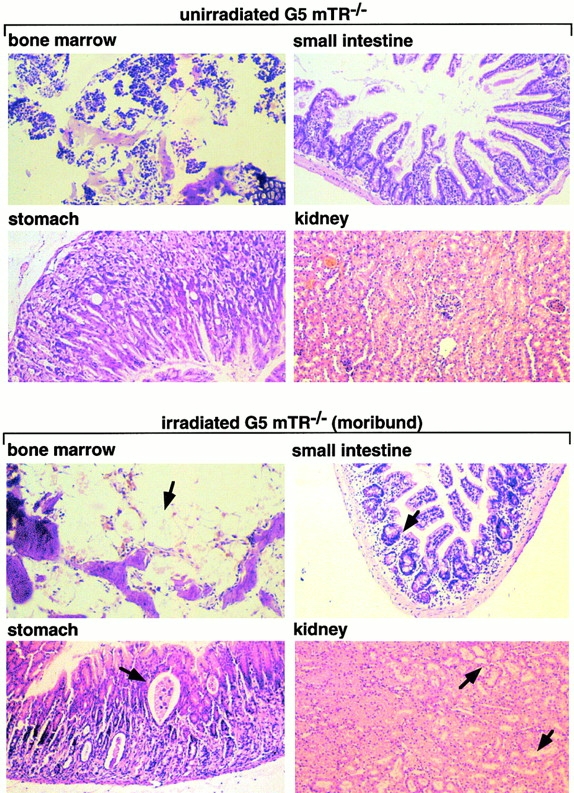
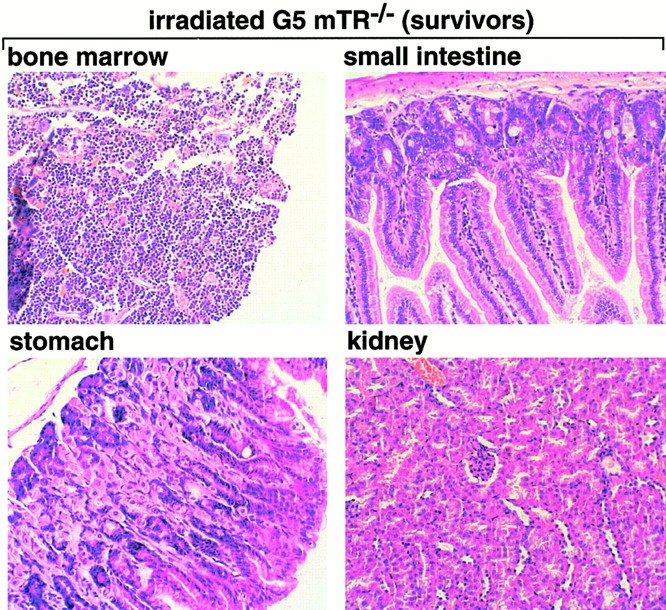
To determine the cause of death, irradiated G5 mTR−/− mice showing signs of poor health (moribund) were killed and examined in parallel with age-matched nonirradiated G5 mTR−/− mice and with similarly irradiated wt and G5 mTR−/− mice showing no signs of disease (survivors). Heart, liver, bladder, salivary glands, and lungs were histologically normal (Table ). Spleen and BM were depleted of cells in moribund irradiated G5 mTR−/− mice but not in similarly irradiated wt and G5 mTR−/− survivors or in nonirradiated G5 mTR−/− mice (Table ). Fig. 2 shows normal histology of BM, stomach, small intestine, and kidney from irradiated wt and G5 mTR−/− survivors as well as in the nonirradiated controls. In contrast, these organs showed severe defects in the irradiated G5 mTR−/− moribunds. In particular, irradiated G5 mTR−/− moribunds showed severe BM aplasia (Fig. 2, arrows), parietal cell degeneration, and cystic formation in the stomach mucosa (Fig. 2, arrows), degeneration of villi and crypts in the small intestine (Fig. 2, arrows), and acute atrophy of kidney tubules (Fig. 2, arrows), a hallmark of radiation nephropathy 30. Similar radiation toxicity has been described mice deficient in ATM (ataxia telangiectasia mutated) and PARP, both proteins that are important in the DNA damage response 31 32.
Table 1.
Summary of Pathologies
| Tissue | wt (N) | wt (I) | G5 mTR−/− (N) | G5 mTR−/− (I) (moribund) |
|---|---|---|---|---|
| BM | Normal | Normal | Slightly decreased cellularity | Severe aplasia |
| Spleen | Normal size (65–154 × 106 cells) | Normal size (3.5–35 × 106 cells) | Normal size (3.5–20 × 106 cells) | Small size (<0.25 × 106 cells) |
| Lymph nodes | Normal | Normal | Normal | No lymphoid follicles, plasmocytosis, histiocytosis |
| Salivary glands | Normal | Normal | Normal | Normal |
| Testis | Normal | Normal | Atrophy | Atrophy |
| Small intestine | Normal | Normal | Slight villi atrophy | Severe villi atrophy |
| Stomach | Normal | Normal | Normal | Parietal cell degeneration and cystic formation in the stomach mucosa |
| Kidney | Normal | Normal | Normal | Severe necrosis of tubules |
| Bladder | Normal | Normal | Normal | Normal |
| Lung | Normal | Normal | Normal | Normal |
| Liver | Normal | Normal | Normal | Normal |
I, irradiated; N, nonirradiated.
Figure 2.
Histopathological analysis of nonirradiated and irradiated wt and G5 mTR−/− mice. Bone, stomach, small intestine, and kidney histological sections corresponding to nonirradiated and irradiated wt and G5-mTR−/− mice are shown. Only the irradiated moribund G5 mTR−/− mice showed radiation-induced pathologies such as dramatic BM aplasia, degeneration of parietal cells, and cystic formation in the stomach, acute atrophy of the villi with crypt degeneration in the small intestine, and acute degeneration of kidney tubules (see arrows indicating abnormalities). Differences in staining are due to slight differences in the thicknesses of the sections. All images were taken at the same magnification (99×) except BM (150×). Sections were fixed in 10% buffered formalin and stained with hematoxylin–eosin.
Increased Apoptosis and Normal Cell Cycle Profile in Irradiated G5 mTR−/− Mice.
Annexin V labels cells undergoing apoptosis in vivo. We measured the percentage of annexin V–labeled spleen B cells (B220+ cells) isolated from nonirradiated and irradiated wt and G2 and G5 mTR−/− mice. In the absence of irradiation, there were no differences in annexin V staining between genotypes: 18.4, 12.7, and 12.9 arbitrary fluorescence units for wt, G2, and G5 mTR−/− mice, respectively (Table ). After irradiation at different doses, G5 mTR−/− spleens contained more annexin V–positive B cells than similarly irradiated wt mice, 28.8 and 137.9 arbitrary units for 1.75 and 4 Gy, respectively, in the case of the G5 mTR−/− mice as compared with 12.5 and 16.8 arbitrary units in the case of wt (Table ). To determine if IR hypersensitivity of G5 mTR−/− mice could also be the consequence of a cell cycle defect associated with short telomeres, we analyzed by flow cytometry the cell cycle profile of spleen B cells (B220+/CR45R+) that were induced to enter the cell cycle upon stimulation with LPS (a B cell–specific mitogen). In the absence of irradiation, there were no statistically significant differences in cell cycle profile between genotypes (Table ). Upon irradiation with a single gamma dose of 4 Gy, G5 and G6 mTR−/− splenocytes showed higher apoptosis than those from similarly irradiated wt mice; the percentages of apoptotic cells were 63.9 ± 8.9 and 63.8 ± 2.1 for G5 and G6 mTR−/− splenocytes, respectively, as compared with 36.0 ± 6.2 for similarly irradiated wt. All together, these results indicate increased IR-induced apoptosis in the G5 and G6 mTR−/− mice (Table ). This increase in apoptosis in G5 and G6 mTR−/− mice was statistically significant, as indicated by Student's t test values of P = 0.0007 and 0.0003, respectively. Importantly, G0-G1 and S-G2 values were similar in all genotypes after irradiation (Table ), indicating no defects in proliferation or cell cycle checkpoints in late generation mTR−/− cells.
Table 2.
Increased Cell Death in Irradiated G5 mTR−/− Splenocytes (B Cells)
| Genotype | No irradiation | 1.75 Gy | 4 Gy |
|---|---|---|---|
| wt | 18.4 | 12.5 | 16.8 |
| G2 mTR−/− | 12.7 | 19.6 | 19.5 |
| G5 mTR−/− | 12.9 | 28.8 | 137.9 |
Mouse splenocytes were incubated with biotinylated anti-B220 (B cell marker) and SPRD-conjugated streptavidin, which was followed by incubation with FITC-conjugated annexin V and flow cytometric analysis. The intensity of annexin V staining in the B220+ cells was determined and expressed in arbitrary units.
Table 3.
Apoptosis and Cell Cycle Profile in LPS-stimulated Unirradiated and Irradiated wt and G2, G5, and G6 mTR−/− B Splenocytes
| 48 h after stimulation | ||||
|---|---|---|---|---|
| Genotype (dose) | n | Apoptosis | G1/G0 | S/G2 |
| % | % | % | ||
| wt (0 Gy) | 5 | 9.6 ± 2.0 | 56.9 ± 13.3 | 32.2 ± 13.4 |
| wt (1.75 Gy) | 5 | 26.4 ± 8.3 | 46.8 ± 10.9 | 26.7 ± 12.2 |
| wt (4 Gy) | 5 | 36.0 ± 6.2 | 43.8 ± 6.2 | 19.9 ± 9.7 |
| G2 mTR−/− (0 Gy) | 3 | 5.1 ± 3.7 | 58.0 ± 19.2 | 36.8 ± 16.6 |
| G2 mTR−/− (1.75 Gy) | 3 | 26.3 ± 14.9 | 43.3 ± 7.1 | 29.9 ± 21.9 |
| G2 mTR−/− (4 Gy) | 3 | 49.2 ± 15.9 | 32.9 ± 8.9 | 17.5 ± 12.3 |
| G5 mTR−/− (0 Gy) | 5 | 10.1 ± 4.6 | 55.8 ± 11.6 | 34.0 ± 8.2 |
| G5 mTR−/− (1.75 Gy) | 6 | 39.6 ± 17.4 | 36.0 ± 11.0 | 24.3 ± 12.5 |
| G5 mTR−/− (4 Gy) | 5 | 63.9 ± 8.9 | 25.9 ± 7.7 | 10.1 ± 5.4 |
| G6 mTR−/− (0 Gy) | 2 | 9.7 ± 8.8 | 55.7 ± 9.5 | 34.7 ± 2.5 |
| G6 mTR−/− (1.75 Gy) | 2 | 40.0 ± 25.7 | 39.9 ± 15.1 | 19.6 ± 8.3 |
| G6 mTR−/− (4 Gy) | 2 | 63.8 ± 2.1 | 26.0 ± 2.8 | 10.3 ± 2.1 |
24 h after mice were irradiated at the indicated gamma ray dose, splenocytes were isolated and a total of 0.8 × 106 splenocytes were incubated in RPMI 1640 containing 10% FBS, 0.55 μM β-mercaptoethanol, and 10 μg/ml LPS. After 48 h of culture, 0.250 × 106 cells were incubated with rat FITC-conjugated anti–mouse CD45R/B220 and 20 μg/ml PI and analyzed by flow cytometry.
Inverse Correlation between Telomere Length and Organismal IR Sensitivity.
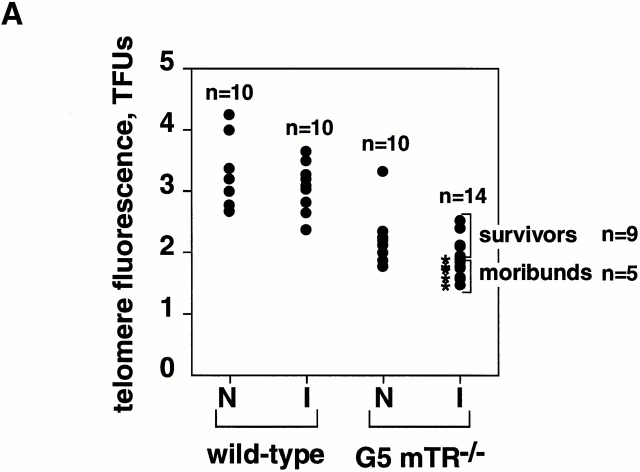
The fact that G5 but not G2 mTR−/− mice showed decreased viability and increased apoptosis after gamma irradiation suggests an inverse correlation between telomere length and organismal sensitivity to IR in mammals. We measured telomere length in fresh BM cells in groups of nonirradiated and irradiated wt and G5 mTR−/− cohorts using FLOW-FISH (reference 7; Fig. 3 A). Telomere fluorescence was significantly higher in BM cells from unirradiated wt mice than in those from unirradiated G5 mTR−/− cohorts (3.351 ± 0.548 and 2.236 ± 0.420 arbitrary fluorescence units, respectively; Fig. 3 A; Student's t test, P = 9.07659 × 10−5), in agreement with telomere loss occurring with increasing mTR−/− generations, as previously described 15 16 28. Importantly, telomere fluorescence was lower in the group of irradiated G5 mTR−/− mice that showed signs of radiosensitivity (Fig. 3 A, asterisk, moribunds) than in the irradiated surviving G5 mTR−/− group (survivors; 1.636 ± 0.119 and 2.088 ± 0.233 arbitrary fluorescence units, respectively; Fig. 3 A). A Student's t test value of P = 0.00173146 indicates that the difference in telomere length between both groups of mice is statistically significant. The telomere fluorescence intensity was also lower in the irradiated moribund G5 mTR−/− group than in the group of nonirradiated G5 mTR−/− mice (1.636 ± 0.119 and 2.236 ± 0.420 arbitrary fluorescence units, respectively; Fig. 3 A). A Student's t test value of P = 0.00136244 indicates that it is a statistically significant difference. It is important to point out that there is no statistically significant difference in telomere length between nonirradiated G5 mTR−/− mice and the irradiated G5 mTR−/− survivors (Student's t test, P = 0.34908841), indicating that the apparent elongation of telomeres in the surviving irradiated G5 mTR−/− mice (Fig. 3 A) is not statistically significant.
Figure 3.
(A) Telomere length analysis using FLOW-FISH in fresh BM cells from nonirradiated (N) and irradiated (I) wt (n = 10 of each) and G5 mTR−/− (N, n = 10; and I, n = 14) cohorts. FLOW-FISH was carried out as described 17. Telomere length is represented as arbitrary units of fluorescence. Average telomere fluorescence values and their corresponding SDs are indicated in the text. Mice that died upon irradiation are indicated with an asterisk (moribund). (B) Telomere length analysis using FLOW-FISH in blood cells from the same mouse before (B) or after (A) irradiation. wt (n = 6) and G2 (n = 6) and G5 mTR−/− (n = 9) cohorts were used. Each nonirradiated or irradiated mouse is indicated by the same symbol before (B) and after (A) the completion of the irradiation protocol. TFU, arbitrary telomere fluorescence units.
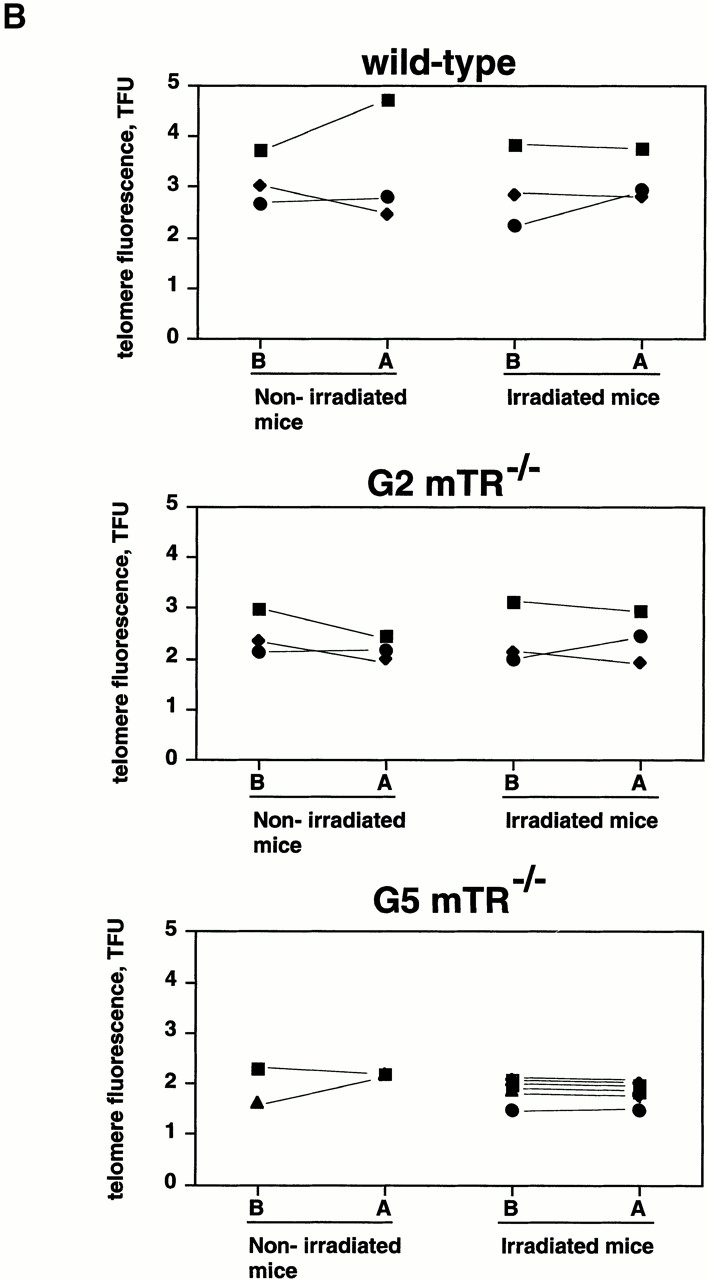
To address whether loss of viability in irradiated G5 mTR−/− mice could be due to an additional radiation-induced telomere loss, we used FLOW-FISH to measure telomere length in blood cells taken from the same animal before and after whole body irradiation. Fig. 3 B shows that in individual wt and G2 and G5 mTR−/− mice, telomeres did not significantly change as a consequence of irradiation. Student's t test analysis showed that telomeres were not significantly different in blood cells extracted from the same animal before or after it was irradiated. Thus, irradiation does not trigger significant changes in telomere length: wt mice before versus after irradiation, P = 0.49800000 (no statistically significant difference in telomere length); G2 mTR−/− mice before versus after irradiation, P = 0.95686187 (no statistically significant difference in telomere length); and G5 mTR−/− mice before versus after treatment, P = 0.06622803 (no statistically significant difference in telomere length).
Increased Radiation-induced Chromosomal Damage in IR-sensitive G5 mTR−/− Mice.
A reduction in, or lack of, recovery after split/fractionated doses of radiation, as observed here in 60% of the G5 mTR−/− mice, may be indicative of a higher radiation-induced DNA damage. To address this indication, we performed a cytogenetic analysis of unirradiated and gamma irradiated wt and G2 and G5 mTR−/− mice. For this, fresh BM chromosome preparations were made and stained with concentrated paint probes for chromosomes 1, 2, and 3 simultaneously (Fig. 4). The frequency of aberrations involving chromosomes 1, 2, and 3 was determined by chromosome painting. As previously described, we detected an spontaneous frequency of end–end fusions in unirradiated G5 mTR−/− animals (Table ) 15 16. Gamma radiation exposure elevated the frequency of aberrations in all genotypes (Table ). Importantly, whereas wt and G2 mTR−/− mice did not show any end–end fusions or deletions upon irradiation (zero Robertsonian fusions, dicentrics, and deletions per 100 metaphases; Table ), the frequency of these chromosomal aberrations was significantly increased in the group of irradiated G5 mTR−/− mice as compared with nonirradiated controls. For statistical analysis, a chi-squared test was performed: dicentrics, none induced in wt or G2 mTR−/− mice; G5 mTR−/− (unirradiated, 1/347; irradiated, 9/259), chi-squared = 9.11, 1 degree of freedom (df), P < 0.01, statistically significant; deletions, significant only in G5 mTR−/− mice (unirradiated, 0/347; irradiated, 6/259), chi-squared = 8.06, 1 df, P < 0.01, statistically significant; Robertsonian-like fusions (p-arm events), none scored in wt or G2 mTR−/− mice; G5 mTR−/− (unirradiated, 2/347; irradiated, 28/259), chi-squared = 31.39, 1 df, P < 0.001, statistically significant; telomeric associations + Robertsonian-like fusions (p-arm events), wt and G2 mTR−/− none scored, and G5 mTR−/− (unirradiated, 27/347; irradiated, 49/259), chi-squared = 14.67, 1 df, P < 0.001, statistically significant. In contrast, the frequency of chromatid-type aberrations such as chromatid breaks, isochromatid breaks, and chromatid exchanges was not significantly elevated in irradiated G5 mTR−/− mice (2.4 per 100 metaphases) as compared with similarly irradiated wt (0.5 per 100 metaphases) or G2 mTR−/− mice (2.2 per 100 metaphases) (Table ).
Figure 4.
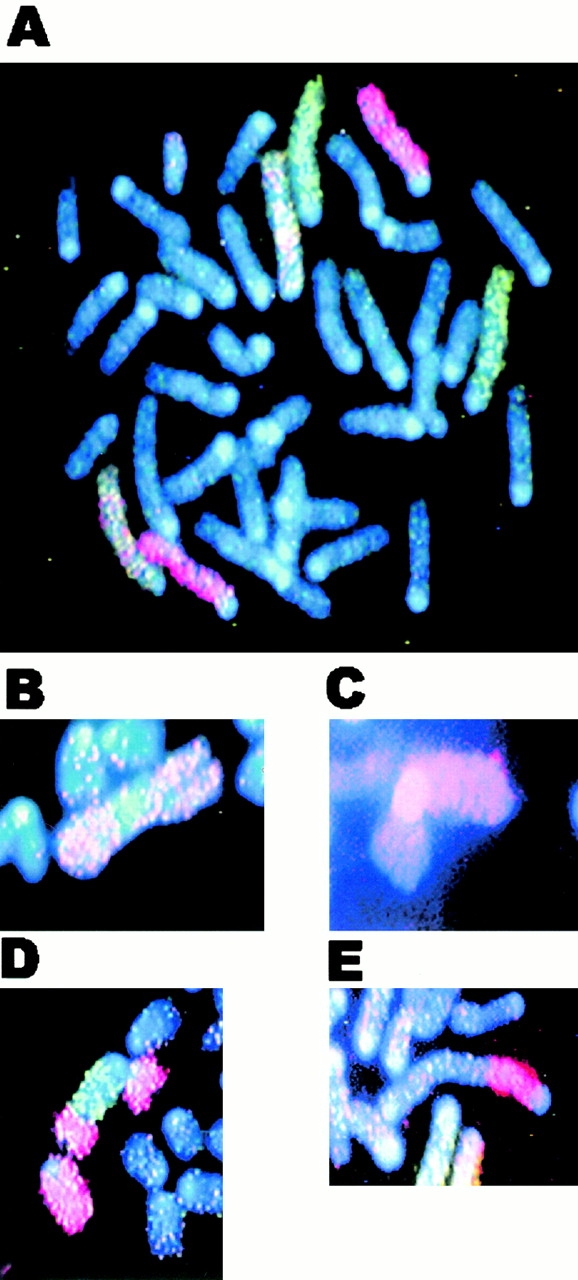
Representative chromosomal aberrations in irradiated G5 mTR−/− BM cells. (A) Control, unirradiated wt cell with no aberrations in chromosomes 3 (red), 2 (green), or 1 (red/green). (B–E) Aberrations observed in irradiated G5 mTR−/− cells: interhomologue Robertsonian fusion of chromosome 1 (B), interchromosome Robertsonian fusion involving chromosome 3 and an unidentified chromosome (C), translocation between chromosome 2 (green) and chromosome 3 (red) (D), and q-q arm dicentric between chromosome 3 and an unidentified chromosome (E). See Table for complete cytogenetic analysis of irradiated control and mTR−/− mice.
Table 4.
Normalized Frequencies of Chromosomal Aberrations in BM Cells of wt and G2 and G5 mTR−/− Mice
| Genotype: | wt | G2 mTR−/− | G5 mTR−/− | |||
|---|---|---|---|---|---|---|
| Treatment | Control | Irradiated | Control | Irradiated | Control | Irradiated |
| Telomeric associations | 0 | 0 | 0 | 0 | 7.2 | 8.1 |
| End–end fusions | ||||||
| Robertsonian fusions | ||||||
| Interhomolgue | 0 | 0 | 0 | 0 | 0 | 7 |
| Interchromosome | 0 | 0 | 0 | 0 | 0.6 | 3.9 |
| Dicentrics (q-q arm) | 0 | 0 | 0 | 0 | 0.3 | 3.5 |
| Total | 0 | 0 | 0 | 0 | 0.9 | 14.4 |
| Other chromosome-type aberrations | ||||||
| Reciprocal translocations | 0 | 2 | 0 | 2.2 | 0 | 2.3 |
| Terminal translocations | 0 | 0.5 | 0 | 2.2 | 0.9 | 1.2 |
| Complex translocations | 0 | 0 | 0 | 0 | 0.3 | 0 |
| Deletions | 0.7 | 1 | 0 | 0 | 0 | 2.3 |
| Bicolored fragments | 0 | 0 | 0 | 0 | 0 | 0.4 |
| Total | 0.7 | 3.5 | 0 | 4.4 | 1.2 | 4.2 |
| Chromatid-type aberrations | ||||||
| Chromatid breaks | 0.7 | 0.5 | 0 | 0 | 0.3 | 1.2 |
| Isochromatid breaks | 0 | 0 | 0 | 2.2 | 0 | 0.8 |
| Chromatid exchanges | 0 | 0 | 0 | 0 | 0 | 0.4 |
| Intra-arm intrachanges | 0 | 0 | 0 | 0 | 0.6 | 0 |
| Total | 0.7 | 0.5 | 0 | 2.2 | 0.9 | 2.4 |
| Whole chromosome losses/gains | ||||||
| −1 | 4.3 | 8 | 0 | 8.7 | 6.6 | 12.4 |
| +1 | 0.7 | 0.5 | 0 | 0 | 0 | 3.5 |
| +2 | 0 | 0 | 0 | 0 | 0 | 1.2 |
| Total | 5.0 | 8.5 | 0 | 8.7 | 6.6 | 17.1 |
| No. cells scored | 146 | 200 | 25 | 46 | 347 | 259 |
| No. animals | 2 | 2 | 1 | 1 | 4 | 4 |
DNA DSB Repair in wt and Different Generation mTR−/− Mice.
IR is a potent inducer of DSB in the DNA, and many radiosensitive phenotypes are associated with defects in DSB repair. There are two major mechanisms responsible for DSB repair in mammalian cells, NHEJ and HR. To investigate if any of these pathways is affected in late generation mTR−/− mice, we determined: (a) SCE frequencies in wt and G6 mTR−/− primary splenocytes as an indication of HR frequencies 33 and (b) V(D)J recombination in thymus and BM from wt and G6 mTR−/− mice 34 as an indication of end-joining activities.
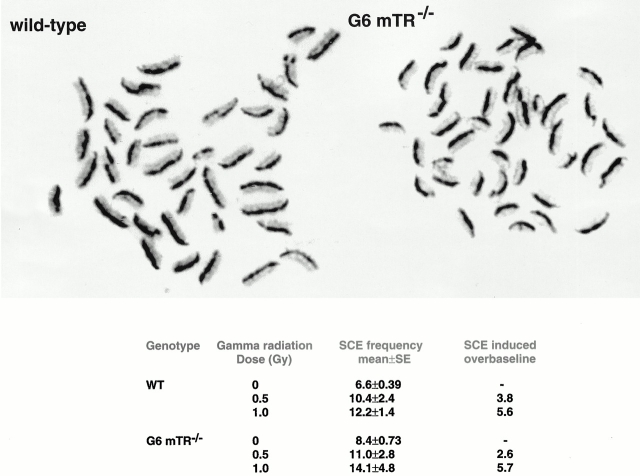
SCE Frequencies.
Baseline SCE frequency in unirradiated G6 mTR−/− splenocytes was slightly elevated compared with wt (8.4 ± 0.73 and 6.6 ± 0.39, respectively). Student's t test P < 0.05 indicates a statistically significant difference; Fig. 5 shows representative examples and quantification of SCE frequencies at different doses. Radiation exposure elevated SCE frequencies in both genotypes in a dose-dependent fashion (10.4 ± 2.4 and 12.2 ± 1.4 for 0.5 and 1.0 Gys, respectively, in the case of wt cells and 11.0 ± 2.8 and 14.1 ± 4.8 for 0.5 and 1.0 Gys, respectively, in the case of G6 mTR−/− cells; Fig. 5). The absolute frequencies of SCE attained in G6 mTR−/− cells were higher than in wt, but the differences were not statistically significant (Student's t test, P > 0.05). The numbers of SCEs induced over baseline were comparable (Fig. 5). As SCEs arise largely as a consequence of HR 33, these data indicate that HR is active in G6 mTR−/− cells. The slight elevation of baseline SCE may indicate an elevated HR activity in G6 mTR−/− cells; similar elevations have been noted in cells deficient in components of the NHEJ pathway, such as in Ku86−/− cells 34.
Figure 5.
Examples of spleen cell metaphases stained to visualize sister chromatid exchanges. wt metaphase, unirradiated. G6 mTR−/− metaphase, unirradiated. SCEs can be seen at points at which dark and light staining exchanges between chromatids. Frequencies of baseline and gamma radiation–induced SCE in wt and G6 mTR−/− splenocytes are also shown. 25 metaphases scored per treatment.
V(D)J Recombination.
The variable region genes of Igs and TCR molecules are assembled from germline segments during development of B and T lymphocytes by a DNA rearrangement process known as V(D)J recombination 35. The V(D)J recombination process involves a step of cleavage or introduction of DSB at specific sequences and a joining step. Several mutations that affect V(D)J recombination also affect NHEJ DSB repair, such as Ku70−/−, Ku86−/−, and DNA-PKcs 35, indicating that a defective V(D)J recombination could be indicative of a DSB NHEJ repair defect. To determine whether G6 mTR−/− mice showed a defective V(D)J recombination, we first studied the formation of Ig heavy chain rearrangements in the B cell lineage using a semiquantitative PCR assay on genomic DNA from BM cells. Two different wt and two G6 mTR−/− mice showed similar formation of DH-JH4 and VH7183-DJH coding joints (Fig. 6 A). As expected, Ku86−/− mice did not show any of the two rearrangements (Fig. 6 A). A positive control for input DNA is also shown (Fig. 6 A). To detect coding joints formed by TCR rearrangement, semiquantitative PCR analysis of thymocyte DNA preparations was performed to detect Vβ8-DJβ2.6 joints. Similar amounts of PCR products of the appropriate sizes were detected in two independent wt and G6 mTR−/− mice but not in the Ku86−/− mice, as expected (Fig. 6 A). These results clearly show that late generation mTR−/− mice do not have a defect in V(D)J recombination, indicating that the DSB DNA repair activities involved in this process are fully functional in the late generation mTR−/− mice (Fig. 6 A). To rule out that a defect in NHEJ-mediated DSB repair in late generation mTR−/− mice could be due to degradation of some of the major DNA repair proteins, we studied DNA-PKcs, Ku70, and PARP status in wt and G6 mTR−/− nuclear extracts. As shown in Fig. 6 B, all of these DNA repair proteins were present in both wt and G6 mTR−/− extracts to similar levels (Fig. 6 B).
Conclusions.
The study of telomerase-deficient mice with different telomere lengths has provided strong evidence that telomere length and hypersensitivity to IR are connected in mammalian cells. The absence of telomerase activity per se does not affect radiation sensitivity when telomeres are sufficiently long (G2 mTR−/− mice). Importantly, gamma IR per se does not result in significant telomere shortening in telomerase-deficient mTR−/− mice, suggesting that exhaustion of proliferative potential of cells is not likely to be the cause of the phenotype. Indeed, we show that fractionated gamma radiation induces a high level of chromosomal damage in the G5 mTR−/− mice as compared with wt or G2 mTR−/− mice. Importantly, similar SCE frequencies and V(D)J recombination efficiency of wt and late generation mTR−/− mice suggest that decreased HR or NHEJ efficiency is not the direct cause of higher DNA damage present in irradiated G5 mTR−/− mice. Thus, short telomeres do not seem to be affecting the efficiency of the DSB repair machinery as we would have predicted from studies in yeast 7 8. Indeed, the radiation-sensitive phenotype of G5 mTR−/− mice is consistent with a direct role of telomeres in the DNA damage response. It is likely that the DNA repair machinery regards short telomeres as DNA breaks, triggering end–end fusions such as the Robersonian fusions and dicentrics found in irradiated G5 mTR−/− mice. We believe that the results presented here could have important implications for both radiotherapy and radiation protection. Telomerase inhibition in tumors (i.e., chemotherapy with telomerase inhibitors) could result in telomere loss, which in turn is likely to render the tumor more sensitive to radiotherapy. Similarly, telomere length may be one of the factors contributing to interindividual variation in radiation sensitivity.
Acknowledgments
We are indebted to G. Taccioli for V(D)J primers, P. Bonner and R. Hidalgo for gamma irradiations, E. Santos for mice genotyping, and M. Serrano for discussions.
F.A. Goytisolo and E. Samper have fellowships from the regional government of Madrid (Comunidad Autónoma de Madrid). Research at the laboratory of M.A. Blasco is funded by The Swiss Bridge Foundation and by grant PM97-0133 from the Ministry of Science and Technology, Spain, grant 08.1/0030/98 from CAM, grants EURATOM/991/0201 and FIGH-CT-1999-00002 from the European Union, and by the Department of Immunology and Oncology, Centro Nacional de Biotecnología-CSIC. This work was supported by EU Concerted Action F14PCT90066.
Footnotes
Abbreviations used in this paper: BM, bone marrow; DSBs, double strand breaks; FISH, fluorescence in situ hybridization; HR, homologous recombination; IR, ionizing radiation; IVS, intervening sequence; mTR, mouse telomerase RNA; NHEJ, nonhomologous end joining; PARP, poly(ADP-ribose)polymerase; SCE, sister chromatid exchange; wt, wild-type.
References
- Vamvakas S., Vock E.H., Lutz V.K. On the role of double-strand breaks in toxicity and carcinogenesis. Crit. Rev. Toxicol. 1997;27:155–174. doi: 10.3109/10408449709021617. [DOI] [PubMed] [Google Scholar]
- Kanaar R., Hoeijmakers J.H. Recombination and joiningdifferent means to the same ends. Genes Funct. 1997;1:165–174. doi: 10.1046/j.1365-4624.1997.00016.x. [DOI] [PubMed] [Google Scholar]
- Jeggo P.A. DNA-PKat the cross-roads of biochemistry and genetics. Mutat. Res. 1997;384:1–14. doi: 10.1016/s0921-8777(97)00009-8. [DOI] [PubMed] [Google Scholar]
- Jeggo P.A. DNA breakage and repair. Adv. Genet. 1998;38:185–218. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- Johnson R.D., Liu N., Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- Martin S.G., Laroche T., Suka N., Grunstein M., Gasser S.M. Relocalization of telomeric Ku and SIR proteins in response to DNA double-strand breaks. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- Mills K.D., Sinclair D.A., Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- Bianchi A., de Lange T. Ku binds telomeric DNA in vitro. J. Biol. Chem. 1999;274:21223–21227. doi: 10.1074/jbc.274.30.21223. [DOI] [PubMed] [Google Scholar]
- Hsu H.-L., Gilley D., Backburn E., Chen D.J. Ku is associated with the telomere in mammals. Proc. Natl. Acad. Sci. USA. 1999;96:12454–12458. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper E., Goytisolo F., Slijepcevic P., van Buul P., Blasco M.A. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-hang overhang. EMBO Reports. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.-D., Küster B., Mann M., Petrini J.H.J., de Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 at human telomeres. Nat. Genet. 2000;25:347–351. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- Haber J.E. The many interphases of Mre11. Cell. 1998;97:829–832. [Google Scholar]
- Ahmed S., Hodgkin J. MTR-2 check point protein is required from germline immortality and telomere replication in C. elegans . Nature. 2000;403:159–164. doi: 10.1038/35003120. [DOI] [PubMed] [Google Scholar]
- Blasco M.A., Lee H.-W., Hande P., Samper E., Lansdorp P., DePinho R., Greider C.W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Lee H.-W., Blasco M.A., Gottlieb G.J., Horner J.W., Greider C.W., DePinho R.A. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- Rufer N., Dragowska W., Thornbury G., Roosnek E., Lansdorp P.M. Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nat. Biotechnol. 1998;16:743–747. doi: 10.1038/nbt0898-743. [DOI] [PubMed] [Google Scholar]
- Lee J.J., Warburton D., Robertson E.J. Cytogenetic methods for the mousepreparation of chromosomes, karyotyping, and in situ hybridization. Anal. Biochem. 1990;189:1–17. doi: 10.1016/0003-2697(90)90036-9. [DOI] [PubMed] [Google Scholar]
- Bouffler S.D., Meijne E.I.M., Morris D.J., Papworth D. Chromosome 2 hypersensitivity and clonal development in murine radiation acute myeloid leukaemia. Int. J. Radiat. Biol. 1997;72:181–189. doi: 10.1080/095530097143400. [DOI] [PubMed] [Google Scholar]
- Finnon P., Lloyd D.C., Edwards A.A. Fluorescence in situ hybridization detection of chromosomal aberrations in human lymphocytesapplicability to biological dosimetry. Int. J. Radiat. Biol. 1995;68:429–435. doi: 10.1080/09553009514551391. [DOI] [PubMed] [Google Scholar]
- Tucker J.D., Morgan W.F., Awa A.A., Bauchinger M., Blakey D., Cornforth M.N., Littlefield L.G., Natarajan A.T., Shasserre C. A proposed system for scoring structural aberrations detected by chromosome painting. Cytogenet. Cell Genet. 1995;6:211–221. doi: 10.1159/000133916. [DOI] [PubMed] [Google Scholar]
- Alves P., Jonasson J. New staining method for the detection of sister-chromatid exchanges in BrdU-labelled chromosomes. J. Cell Sci. 1978;32:185–195. doi: 10.1242/jcs.32.1.185. [DOI] [PubMed] [Google Scholar]
- Taccioli G.E., Amatucci A.G., Beamish H.J., Gell D., Xiang X.H., Torres Arzayus M.I., Priestley A., Jackson S.P., Rothstein A.M., Jeggo P.A. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 1998;9:355–366. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- Zhu C., Bogue M.A., Lim D.S., Hasty P., Roth D.B. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- Bogue M.A., Zhu C., Aguilar-Cordova E., Donehower L.A., Roth D.B. p53 is required for both radiation-induced differentiation and rescue of V(D)J rearrangement in scid mouse thymocytes. Genes Dev. 1996;10:553–565. doi: 10.1101/gad.10.5.553. [DOI] [PubMed] [Google Scholar]
- Ouyang H., Nussenzweig A., Kurimasa A., da Costa Soares V., Li X., Cordón Cardó C., Li W., Cheong N., Nussenzweig M., Iliakis G. Ku70 is required for DNA repair but not for T cell antigen receptor gene recombination in vivo. J. Exp. Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A., Young C.S.H. A modified single-strand annealing model best explains the joining of DNA double strand breaks in mammalian cells and cell extracts. Mol. Cell Biol. 1994;14:170–180. doi: 10.1093/nar/23.6.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph K.L., Chang S., Lee H.-W, Blasco M., Gottlieb G., Greider W.C., DePinho R.A. Longevity, stress response, and cancer in aging telomerase deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- Wong K.K., Chang S., Weiler S.R., Ganesan S., Chaudhuri J., Zhu C., Artandi S.E., Rudolph K.L., Gottlieb G.J., Chin L. Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation. Nat. Genet. 2000;1:85–88. doi: 10.1038/79232. [DOI] [PubMed] [Google Scholar]
- Robbins M.E., Bonsib S.M. Radiation nephropathya review. Scanning Microsc. 1995;9:535–560. [PubMed] [Google Scholar]
- Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D. Atm-deficient micea paradigm of Ataxia telangiectasia . Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Menissier de Murcia J., Niedergang C., Trucco C., Ricoul M., Dutrillaux B., Mark M., Oliver F.J., Masson M., Dierich A., LeMeur M. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Sasaki M.S., Morrison C., Yamaguchi-Iwai Y., Takata M., Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.C., Ouyang H., Li X., Nagasawa H., Little J.B., Cheu D.J., Ling C.C., Fuks Z., Cordon-Cardo C. Ku80a candidate tumor suppressor gene for murine T cell lymphoma. Mol. Cell. 1998;2:1–8. doi: 10.1016/s1097-2765(00)80108-2. [DOI] [PubMed] [Google Scholar]
- Lewis S.M. The mechanism of V(D)J joininglessons from molecular, immunological, and comparative analyses. Adv. Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]