Abstract
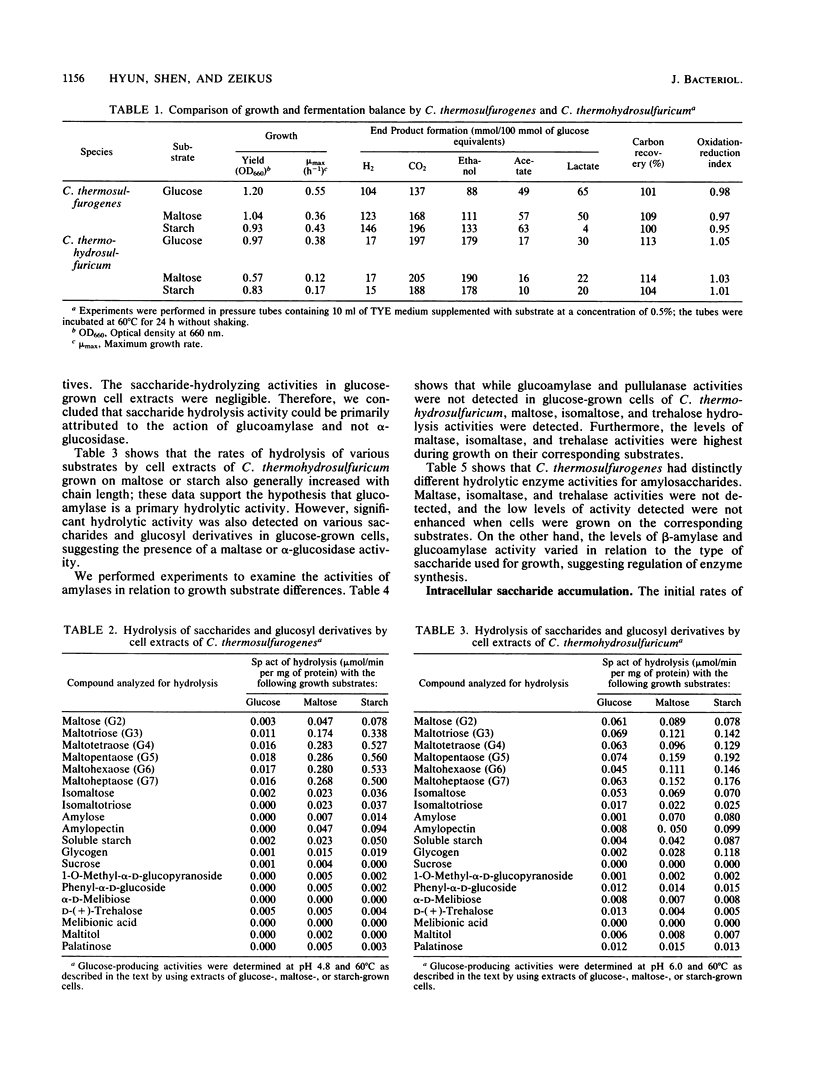
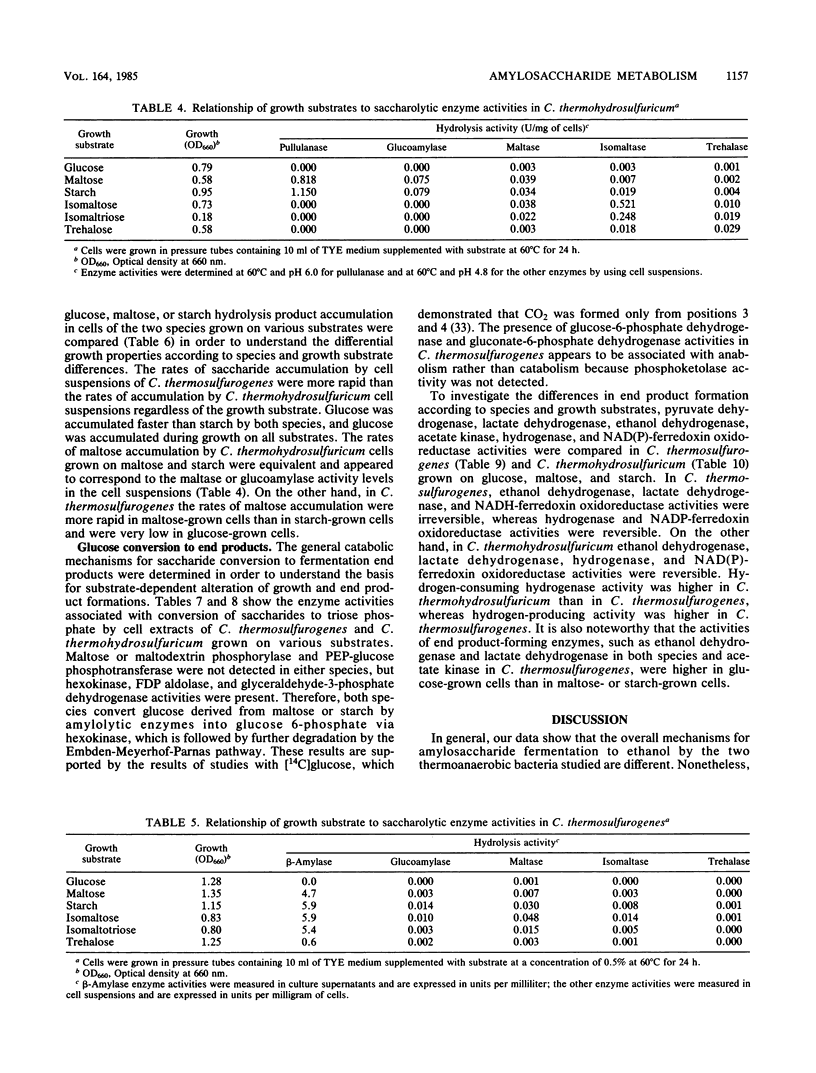
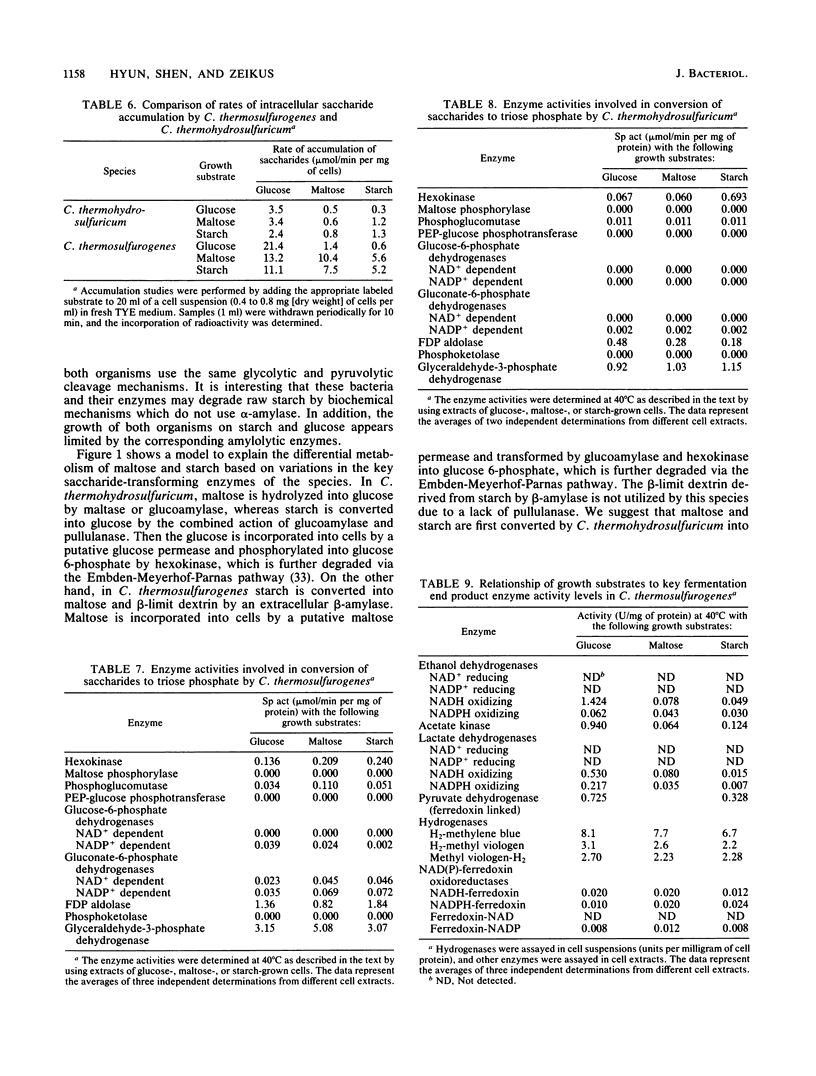
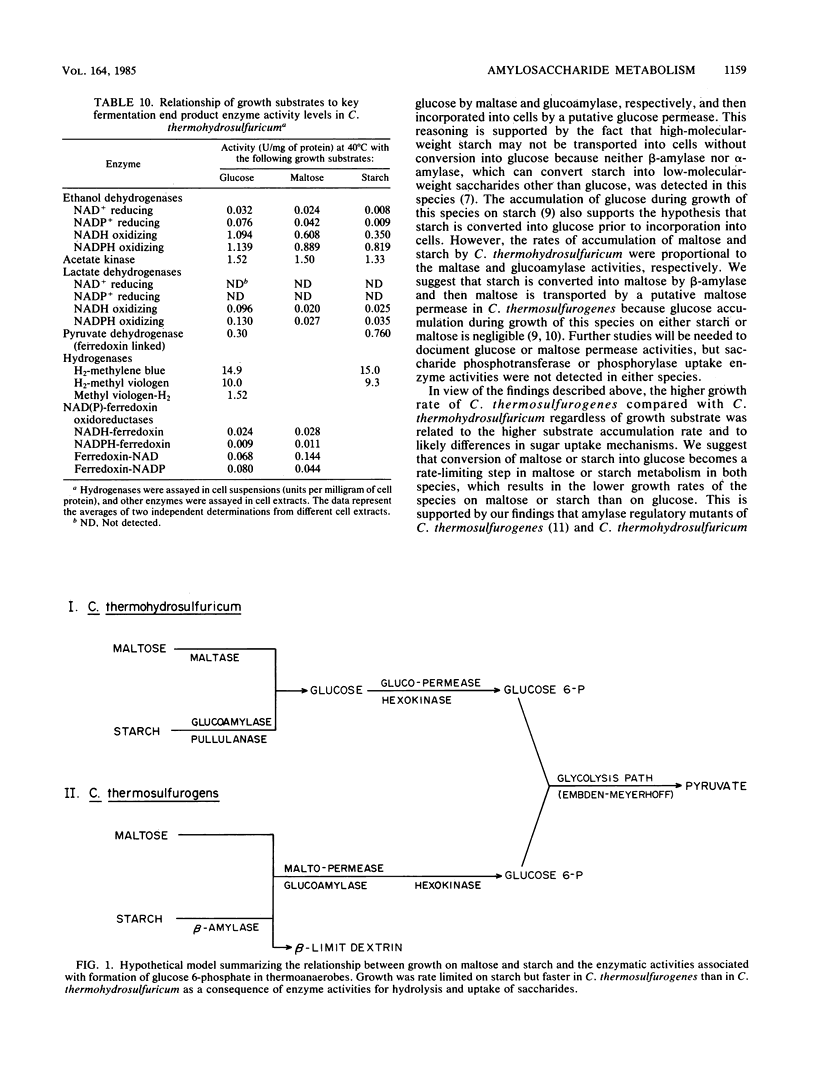
Clostridium thermosulfurogenes displayed faster growth on either glucose, maltose, or starch than Clostridium thermohydrosulfuricum. Both species grew faster on glucose than on starch or maltose. The fermentation end product ratios were altered based on higher ethanol and lactate yields on starch than on glucose. In C. thermohydrosulfuricum, glucoamylase, pullulanase, and maltase were mainly responsible for conversion of starch and maltose into glucose, which was accumulated by a putative glucose permease. In C. thermosulfurogenes, beta-amylase was primarily responsible for degradation of starch to maltose, which was accumulated by a putative maltose permease and then hydrolyzed by glucoamylase. Regardless of the growth substrate, the rates of glucose, maltose, and starch transformation were higher in C. thermosulfurogenes than in C. thermohydrosulfuricum. Both species had a functional Embden-Meyerhof glycolytic pathway and displayed the following catabolic activities: ferredoxin-linked pyruvate dehydrogenase, acetate kinase, NAD(P)-ethanol dehydrogenase, NAD(P)-ferredoxin oxidoreductase, hydrogenase, and fructose-1,6-diphosphate-activated lactate dehydrogenase. Ferredoxin-NAD reductase activity was higher in C. thermohydrosulfuricum than NADH-ferredoxin oxidase activity, but the former activity was not detectable in C. thermosulfurogenes. Both NAD- and NADP-linked ethanol dehydrogenases were unidirectional in C. thermosulfurogenes but reversible in C. thermohydrosulfuricum. The ratio of hydrogen-producing hydrogenase to hydrogen-consuming hydrogenase was higher in C. thermosulfurogenes. Two biochemical models are proposed to explain the differential saccharide metabolism on the basis of species enzyme differences in relation to specific growth substrates.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blusson H., Petitdemange H., Gay R. A new, fast, and sensitive assay for NADH--ferredoxin oxidoreductase detection in clostridia. Anal Biochem. 1981 Jan 1;110(1):176–181. doi: 10.1016/0003-2697(81)90132-9. [DOI] [PubMed] [Google Scholar]
- Hyun H. H., Zeikus J. G. General Biochemical Characterization of Thermostable Extracellular beta-Amylase from Clostridium thermosulfurogenes. Appl Environ Microbiol. 1985 May;49(5):1162–1167. doi: 10.1128/aem.49.5.1162-1167.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun H. H., Zeikus J. G. General Biochemical Characterization of Thermostable Pullulanase and Glucoamylase from Clostridium thermohydrosulfuricum. Appl Environ Microbiol. 1985 May;49(5):1168–1173. doi: 10.1128/aem.49.5.1168-1173.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun H. H., Zeikus J. G., Longin R., Millet J., Ryter A. Ultrastructure and extreme heat resistance of spores from thermophilic Clostridium species. J Bacteriol. 1983 Dec;156(3):1332–1337. doi: 10.1128/jb.156.3.1332-1337.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun H. H., Zeikus J. G. Regulation and genetic enhancement of beta-amylase production in Clostridium thermosulfurogenes. J Bacteriol. 1985 Dec;164(3):1162–1170. doi: 10.1128/jb.164.3.1162-1170.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun H. H., Zeikus J. G. Regulation and genetic enhancement of glucoamylase and pullulanase production in Clostridium thermohydrosulfuricum. J Bacteriol. 1985 Dec;164(3):1146–1152. doi: 10.1128/jb.164.3.1146-1152.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun H. H., Zeikus J. G. Simultaneous and Enhanced Production of Thermostable Amylases and Ethanol from Starch by Cocultures of Clostridium thermosulfurogenes and Clostridium thermohydrosulfuricum. Appl Environ Microbiol. 1985 May;49(5):1174–1181. doi: 10.1128/aem.49.5.1174-1181.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lamed R., Zeikus J. G. Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii. J Bacteriol. 1980 Nov;144(2):569–578. doi: 10.1128/jb.144.2.569-578.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamed R., Zeikus J. G. Glucose fermentation pathway of Thermoanaerobium brockii. J Bacteriol. 1980 Mar;141(3):1251–1257. doi: 10.1128/jb.141.3.1251-1257.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl L. G. Physiology of thermophilic bacteria. Adv Microb Physiol. 1979;19:149–243. doi: 10.1016/s0065-2911(08)60199-x. [DOI] [PubMed] [Google Scholar]
- Nelson D. R., Zeikus J. G. Rapid method for the radioisotopic analysis of gaseous end products of anaerobic metabolism. Appl Microbiol. 1974 Aug;28(2):258–261. doi: 10.1128/am.28.2.258-261.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. K., Ben-Bassat A., Zeikus J. G. Ethanol Production by Thermophilic Bacteria: Fermentation of Cellulosic Substrates by Cocultures of Clostridium thermocellum and Clostridium thermohydrosulfuricum. Appl Environ Microbiol. 1981 Jun;41(6):1337–1343. doi: 10.1128/aem.41.6.1337-1343.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. K., Zeikus J. G. Comparison of Extracellular Cellulase Activities of Clostridium thermocellum LQRI and Trichoderma reesei QM9414. Appl Environ Microbiol. 1981 Aug;42(2):231–240. doi: 10.1128/aem.42.2.231-240.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. K., Zeikus J. G. Differential metabolism of cellobiose and glucose by Clostridium thermocellum and Clostridium thermohydrosulfuricum. J Bacteriol. 1982 Jun;150(3):1391–1399. doi: 10.1128/jb.150.3.1391-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T. K., Zeikus J. G. Purification and characterization of an endoglucanase (1,4-beta-D-glucan glucanohydrolase) from Clostridium thermocellum. Biochem J. 1981 Nov 1;199(2):341–350. doi: 10.1042/bj1990341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAZUR J. H., KLEPPE K. The hydrolysis of alpha-D-glucosides by amyloglucosidase from Aspergillus niger. J Biol Chem. 1962 Apr;237:1002–1006. [PubMed] [Google Scholar]
- Reese E. T., Maguire A. H., Parrish F. W. Glucosidases and exo-glucanases. Can J Biochem. 1968 Jan;46(1):25–34. doi: 10.1139/o68-005. [DOI] [PubMed] [Google Scholar]
- Singleton R., Jr, Amelunxen R. E. Proteins from thermophilic microorganisms. Bacteriol Rev. 1973 Sep;37(3):320–342. doi: 10.1128/br.37.3.320-342.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOLIN M. J. FRUCTOSE-1,6-DIPHOSPHATE REQUIREMENT OF STREPTOCOCCAL LACTIC DEHYDROGENASES. Science. 1964 Nov 6;146(3645):775–777. doi: 10.1126/science.146.3645.775. [DOI] [PubMed] [Google Scholar]
- Wang L. H., Hartman P. A. Purification and some properties of an extracellular maltase from Bacillus subtilis. Appl Environ Microbiol. 1976 Jan;31(1):108–118. doi: 10.1128/aem.31.1.108-118.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Carlsson J. Regulation of lactate dehydrogenase and change of fermentation products in streptococci. J Bacteriol. 1975 Oct;124(1):55–61. doi: 10.1128/jb.124.1.55-61.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeikus J. G., Ben-Bassat A., Ng T. K., Lamed R. J. Thermophilic ethanol fermentations. Basic Life Sci. 1981;18:441–461. doi: 10.1007/978-1-4684-3980-9_26. [DOI] [PubMed] [Google Scholar]



