Abstract
The antigen receptor gene rearrangement at a given locus is tightly regulated with respect to cell lineage and developmental stage by an ill-defined mechanism. To study the possible role of precursor B cell antigen receptor (pre-BCR) signaling in the regulation of the ordered immunoglobulin (Ig) gene rearrangement during B cell differentiation, a newly developed system using μ heavy (H) chain membrane exon (μm)-deficient mice was employed. In this system, the antibody-mediated cross-linking of Igβ on developmentally arrested progenitor B (pro-B) cells mimicked pre-BCR signaling to induce early B cell differentiation in vivo. Analyses with ligation-mediated polymerase chain reaction revealed that the Igβ cross-linking induced the redirection of Ig gene rearrangements, namely, the suppression of ongoing rearrangements at the H chain locus and the activation of rearrangements at the light (L) chain locus. Upon the cross-linking, the κL chain germline transcription was found to be upregulated whereas the VH germline transcription was promptly downregulated. Notably, this alteration of the accessibility at the H and L chain loci was detected even before the induction of cellular differentiation became detectable by the change of surface phenotype. Thus, the pre-BCR signaling through Igβ appears to regulate the ordered Ig gene rearrangement by altering the Ig locus accessibility.
Keywords: pre–B cell receptor, surrogate light chain, allelic exclusion, B cell development, recombinase
Introduction
The diversity of antigen specificity among B cells is generated during their development through the rearrangement of gene segments encoding parts of the variable region of H and L chains 1 2. A common recombinase that recognizes a conserved recombination signal sequence (RSS) mediates all the rearrangement reactions 3. However, the rearrangement at a given locus is highly regulated during B cell development such that there is a clear-cut order of Ig gene rearrangements, namely, H chain gene rearrangements before L chain gene rearrangements 4 5. Once μH chain is expressed through a functional VHDHJH rearrangement in one allele of the H chain gene, the recombinase is directed towards the L chain locus, and the DHJH joint in the other allele of the H chain gene is not targeted for further VH to DHJH rearrangement. This regulation of recombinase targeting appears to be important for allelic exclusion, namely, the mechanism by which a single B cell has only one functionally rearranged H chain allele to ensure one immune specificity per cell 6 7. However, signals involved in this spatial and temporal regulation of the Ig gene rearrangements remained largely unknown. In this study, we examined the role of signaling through Igβ, a component of the pre-BCR, in this regulation by using a novel system to manipulate Ig gene rearrangements in vivo.
The pre-BCR is composed of the membrane-bound form of the μH chain, Vpre-B/λ5 surrogate L chain, and an Igα/Igβ heterodimer 8 9. Its expression is restricted to the early pre-B cell stage of B cell development in which the rearrangement of the H chain gene but not yet the L chain gene has been completed 10 11. The critical role of the pre-BCR in B cell generation was illuminated by creating pre-BCR–deficient mice in which B cell development was found to be severely impaired at the transition from the pro-B cell stage to the large pre-B cell stage 12 13. The importance of the pre-BCR was further confirmed in humans by finding agammaglobulinemia patients with mutations of either the μH chain gene or the λ5 gene 14 15. The pre-BCR has been suggested to regulate several hallmark events associated with progression from the pro-B to pre-B cell stage, including Ig gene rearrangements 16 17. Studies with transgenic mice indicated that allelic exclusion was mediated by the membrane-bound form of the μH chain 18 19 20. Indeed, allelic exclusion was found to be violated in heterozygous mice carrying a targeted disruption of the membrane exon of the μH chain in one allele and an unmutated gene in the other allele 21. Furthermore, it has been shown that μH chain genes were not allelically excluded at the pre-B cell stage in λ5-deficient mice 22. These results support the idea that the pre-BCR is involved in the process of allelic exclusion at the pre-B cell stage, even though an underlying mechanism including a signaling pathway remains to be elucidated. The membrane-bound form of the μH chain was also found to be functional in the induction of κL chain rearrangements when expressed in in vitro pro-B cell lines 23 24 25. On the other hand, studies with a series of knockout mice clearly demonstrated that neither μH chain nor surrogate L chain was a prerequisite for L chain gene rearrangement 13 21 26 27. Thus, the role of the pre-BCR in a L chain gene rearrangement remains an open question.
To assess the proposed roles of the pre-BCR in regulation of Ig gene rearrangements, it is essential to show whether signaling through the pre-BCR actually suppresses or induces rearrangements at the H and L chain loci in vivo. However, to our knowledge no study has directly addressed this issue so far. The extremely low level of pre-BCR surface expression has hampered biochemical studies of pre-BCR signaling 10 28, and a ligand triggering pre-BCR signaling remains unidentified. Moreover, transformed pre-B cell lines turned out not to be proper materials for analyzing the differentiation signals of the pre-BCR. To overcome these limitations, we have recently established a novel system that provides a superior way to analyze pre-BCR signaling by using bone marrow pro-B cells 29. The Igα/Igβ heterodimer was detected in association with calnexin on the surface of μH chain− pro-B cells, albeit at low levels, and was able to transduce signals across the cell membrane when cross-linked 29 30. In vivo treatment of recombination activating gene (RAG)-2–deficient mice with anti-Igβ mAb revealed that cross-linking Igβ on developmentally arrested pro-B cells induced early B cell differentiation from pro-B to small pre-B cells 29. Therefore, we concluded that the antibody-mediated cross-linking of Igβ on pro-B cells mimicked the signal that is normally transduced via the pre-BCR. As it is extremely difficult to analyze the differentiation signal through pre-BCR itself, our novel system should provide a powerful tool for analyzing how pre-BCR signaling regulates differentiation events such as Ig gene rearrangements. However, it is impossible to examine the effects of Igβ cross-linking on Ig gene rearrangements using RAG-2–deficient mice because the recombination machinery necessary for rearrangements does not work in these mice 31. Therefore, we used μm-deficient mice, in which B cell development was completely arrested at the pro-B cell stage, as in RAG-2–deficient mice, but the recombination machinery was kept intact 12. Our results indicate that the signaling through Igβ, a component of the pre-BCR, regulates locus accessibility for ordered Ig gene rearrangement.
Materials and Methods
Mice.
μm-deficient mice 12 and RAG-2–deficient mice 31 were provided by Dr. K. Rajewsky (University of Cologne, Germany) and Dr. F.W. Alt (Howard Hughes Medical Institute, Boston, MA, The Children's Hospital), respectively. They were bred and maintained under specific pathogen-free conditions in our animal facility and used for analysis at 8–12 wk of age. All of the experiments in this study were performed according to the Guidelines for Animal Use and Experimentation as set out by our Institutions.
Antibodies.
FITC-conjugated anti-CD45R/B220 (RA3-6B2), PE-conjugated anti-CD45R/B220 (RA3-6B2), and biotin-conjugated antibodies specific to CD25 (7D4), CD117 (3C1), BP-1 (6C3), CD2 (RM2-5), and κL chain (R5-240) were purchased from PharMingen. The mAb against λ5 (LM34 32) was purified and labeled with biotin by standard methods. PE-conjugated streptavidin was purchased from Southern Biotechnology Associates, and allophycocyanin (APC)-conjugated streptavidin was provided by PharMingen. The mAb against mouse Igβ (HM79) was described previously 30.
Cell Preparation and Flow Cytometry.
Bone marrow cells were isolated from femurs and tibias of mice and suspended in staining buffer (PBS containing 0.1% BSA and 0.1% NaN3). For the enrichment of B lineage cells, B220+ cells were purified from bone marrow by using anti-CD45R (B220)–conjugated microbeads according to the procedure of the manufacturer (Miltenyi Biotec). More than 90% of the cells in the enriched population were found to express B220 (data not shown). Staining of bone marrow cells was performed as described 10. Debris, erythrocytes, and dead cells were excluded from the analysis by forward and side scatter and propidium iodide gatings. For cytoplasmic staining, bone marrow cells were fixed and permeabilized with Cytofix/Cytoperm kit (PharMingen) according to the manufacturer's instructions. Stained cells were analyzed by FACSCalibur™ (Becton Dickinson). Cell sorting was performed with a FACS Vantage™ (Becton Dickinson).
PCR and Reverse Transcription PCR Analyses.
Genomic DNA was prepared from B220+ bone marrow cells and subjected to PCR to detect V-J joints of the κL chain gene. PCR conditions and primers were described previously 33. The PCR products were electrophoresed in 2% agarose gel, blotted onto nylon membranes, and then hybridized with 32P-labeled oligonucleotide probe Jκ2-P (5′-GTGCCTCCACCGAACGTCCA-3′). The radioactivity was quantified with a Bio-Image analyzer (model BAS 2500; Fuji Photo Film Co. Ltd.). For reverse transcription (RT)-PCR, total RNA was prepared from B220+ bone marrow cells using Isogen (Nippon gene) and treated with RNase-free DNase (GIBCO BRL) to remove contaminating genomic DNA. cDNA was prepared using Moloney murine leukemia virus (MMLV) Reverse transcriptase RNaseH− (ReverTra Ace; Toyobo Co., Ltd.) and oligo dT or VHJ558 antisense primer 34. RT was carried out at 50°C. For VHJ558 germline transcripts, samples were amplified as described previously 34. For other genes, PCR was carried out with 25 cycles of 20 s at 94°C, 15 s at 55°C, and 1 min at 72°C. Sequences of the primers used for μ0 germline transcripts, VHJ558 germline transcripts, κL chain germline transcripts, and hypoxanthine phosphoribosyl transferase (HPRT) have been described previously 34 35 36 37. Each PCR amplification was carried out within the logarithmic phase (data not shown). The PCR products were electrophoresed in 3% agarose gel, blotted onto nylon membranes, and hybridized with the following 32P-labeled probes: CH-P, 5′-GACAGCAACTGCGAGGTGGCT-3′; VHJ558-P, 5′-AAGGCCACACTGACTGCA-3′; Jκ1-P, 5′-GTGCCTCCACCGAACGTCCA-3′; and HPRT-P, 5′-GCTGGTGAAAAGGACCTCT-3′.
Ligation-mediated PCR Assays.
Ligation-mediated PCR (LMPCR) assays were carried out as described 38 39 with minor modification. In brief, genomic DNA from B220+ bone marrow cells was subjected to linker ligation at 16°C overnight in 10 μl ligation mixture (50 mM Tris [pH 7.4], 10 mM MgCl2, 20 mM dithiothreitol, 1 mM ATP, 5 μg/ml BSA, 10 pmol BW linker, and 2U T4 DNA ligase [Takara Shuzo]) and then heated to 95°C for 10 min to inactivate T4 ligase. Samples were subjected to nested PCR as described 38 39 after normalization for DNA content by PCR amplification of RAG-1 gene as described previously 35. The PCR products were electrophoresed in 3% agarose gel, blotted onto nylon membranes and hybridized with the following 32P-labeled oligonucleotide probes: 5′DFL16.1-P, 5′-GCCCTGGCCAGTATGGTCTCTGGT-3′; 5′Jκ1-P, 5′-CAGTGAGGAGGGTTTTTGTACAGCCAGACAG-3′; or RAG-1–P, 5′-AGGTAGCTTAGCCAACATGGCACG-3′.
Results
B Cell Differentiation Is Induced in μm-deficient Mice When Treated with Anti-Igβ mAb.
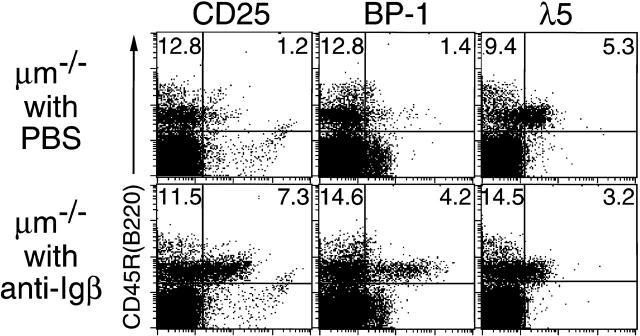
μm-deficient mice were treated by a single injection of 1 mg of anti-Igβ mAb HM79. Flow cytometric analysis of their bone marrow cells isolated on day 6 after injection revealed that the expression of CD25 (Tac), BP-1, and CD2 on CD45R (B220)+ B lineage cells was upregulated, whereas that of λ5 and CD117 (c-kit) was downregulated in the treated mice compared with control mice (Fig. 1, not all data shown). The average size of the CD45R (B220)+ cells was smaller than that of corresponding cells in untreated mice (data not shown). These phenotypic changes were very similar to those observed at the pro-B to small pre-B transition in normal B cell development. Thus, B cell differentiation could be induced by the cross-linking of Igβ on pro-B cells in μm-deficient mice, as previously observed in RAG-2–deficient mice 29, as long as judged by the phenotypic changes.
Figure 1.
The in vivo treatment with anti-Igβ mAb induced phenotypic changes of bone marrow pro-B cells in μm-deficient mice. μm-deficient (μm−/−) mice (10-wk-old) were injected intraperitoneally with PBS or 1 mg of anti-Igβ mAb HM79. Bone marrow cells were isolated on day 6 after the injection and stained with FITC–anti-CD45R (B220) and biotin-conjugated antibodies specific to either CD25 (Tac), BP-1, or λ5 in conjunction with PE-streptavidin. Data shown are representative of six repeated analyses of two-color flow cytometry. The percentage of cells for a given phenotype is shown. Treatment with control antibody (normal hamster IgG) did not cause any phenotypic change of pro-B cells (data not shown).
Igβ Cross-Linking on Pro-B Cells Induces the Activation of Rearrangements at the L Chain Locus.
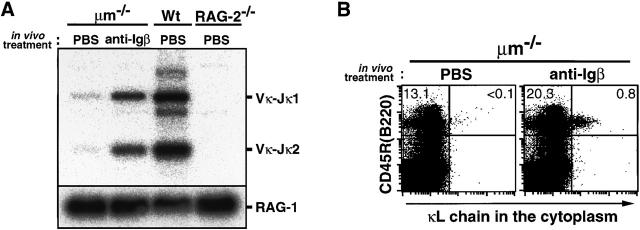
We next addressed the question whether the signaling through Igβ could also have any effect on Ig gene rearrangements. First, to examine the configuration of κL chain genes, CD45R (B220)+ bone marrow cells were isolated from μm-deficient mice 6 d after the treatment with anti-Igβ mAb or PBS, and subjected to PCR analysis with a pair of primers specific for Vκ and Jκ2. Though low levels of PCR products corresponding to Vκ-Jκ1 and Vκ-Jκ2 joints could be detected even in untreated or PBS-treated mice, as described previously 21 26, their levels were found 20 and 40 times higher, respectively, in the anti-Igβ–treated mice (Fig. 2 A). This indicates that the cross-linking of Igβ on pro-B cells promotes the production of Vκ-Jκ joints. To confirm the effect of Igβ cross-linking on L chain gene rearrangement, the expression of κL chain protein was analyzed by flow cytometry. In accord with the PCR analysis, κL chain was detectable in the cytoplasm of ∼4% of CD45R (B220)+ bone marrow cells 6 d after the anti-Igβ treatment while hardly seen in those from control mice treated with PBS (Fig. 2 B). Thus, the cross-linking of Igβ on pro-B cells appeared to promote κL chain gene rearrangements.
Figure 2.
The cross-linking of Igβ on pro-B cells in μm-deficient mice promoted the production of Vκ-Jκ joints and κL chain proteins. μm-deficient mice (10-wk-old) were injected intraperitoneally with either 1 mg of anti-Igβ mAb, normal hamster IgG, or PBS. On day 6 after injection, bone marrow cells were isolated from the mice. (A) B220+ cells were isolated from their bone marrow cells by magnetic cell sorting using B220-specific beads. Equivalent amounts of DNA extracted from B220+ cells were amplified by PCR with a combination of Vκ degenerate primers and a primer downstream of Jκ2. Southern blot of the PCR products was hybridized with oligonucleotide probe specific to the Jκ2 gene. As positive and negative controls, samples prepared from normal (Wt) and RAG-2–deficient (RAG-2−/−) mice in the same way were analyzed in parallel. Positions of the amplified fragments corresponding to Vκ-Jκ1 and Vκ-Jκ2 joints are shown on the right side. Treatment with normal hamster IgG gave the same result as treatment with PBS (data not shown). RAG-1 gene was amplified in parallel to confirm that each sample contained approximately the same amount of DNA. (B) Bone marrow cells were stained with PE–anti-B220 on the surface, followed by cytoplasmic staining with biotin–anti-κL chain (revealed by APC-streptavidin). Two-color analyses of cells present in the lymphocyte gate are shown. Percentage of κL chain–positive and –negative B220+ cells is shown.
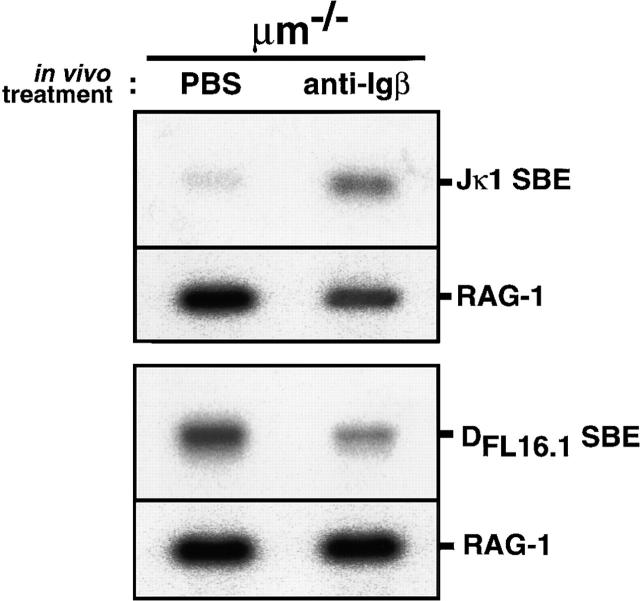
However, the observed increase in Vκ-Jκ joints and κL chain proteins among the B lineage population in the treated mice might not be due to the activation of rearrangements. It could be the consequence of preferential expansion of a minor population of cells that had already completed κL chain gene rearrangements before the treatment. Therefore, we used the LMPCR assay, which reflects more accurately the amount of active V(D)J recombination at a particular locus because it detects intermediates in the recombination reaction rather than its end-products 38. DNA was prepared from CD45R (B220)+ bone marrow cells 6 d after the treatment with anti-Igβ mAb or PBS, and subjected to ligation with double strand linkers followed by nested PCR using a linker-specific primer and a pair of primers specific for the broken-ended RSS upstream of Jκ1 38 39. As shown in Fig. 3 (top), the frequency of intermediates of Vκ-Jκ rearrangement carrying broken-ended RSS upstream of Jκ1 greatly increased in the anti-Igβ–treated μm-deficient mice compared with the PBS-treated mice. This clearly indicated that the antibody-mediated cross-linking of Igβ on pro-B cells indeed activated the recombination at the κL chain loci.
Figure 3.
The cross-linking of Igβ on pro-B cells in μm-deficient mice activated rearrangements at the L chain locus but suppressed those at the H locus. DNA extracted from B220+ bone marrow cells as in the legend to Fig. 2 A was subjected to LMPCR to detect signal broken ends upstream of the Jκ1 segment (Jκ1 SBE, top) and those upstream of the DFL16.1 (DFL16.1 SBE, bottom). RAG-1 gene was amplified in parallel to confirm that each sample contained approximately the same amount of DNA.
Igβ Cross-Linking on Pro-B Cells Induces the Suppression of Rearrangements at the H Chain Locus.
The LMPCR assay was further applied to examine recombination activity at the H chain locus by using primers specific for the broken-ended RSS upstream of DFL16.1 38 39. The frequency of intermediates of VH to DHJH rearrangements carrying the broken-ended RSS upstream of DFL16.1 was found to be much lower in CD45R (B220)+ bone marrow cells from the anti-Igβ–treated μm-deficient mice than in those from PBS-treated control mice (Fig. 3, bottom). This clearly indicated that the antibody-mediated cross-linking of Igβ on pro-B cells suppressed the VH to DHJH recombination at the H chain locus.
Igβ Cross-Linking on Pro-B Cells Induces the Alteration of Accessibility at the H and L Chain Loci before the Induction of Surface Phenotypic Changes.
It has been shown that small pre-B cells in bone marrow of normal mice have a high activity of recombination at the L chain locus, but no activity at the H chain locus 39. Therefore, the alteration of recombination activity at the H and L chain loci observed in the CD45R (B220)+ bone marrow cells from μm-deficient mice might not be a primary effect of the Igβ cross-linking. It could be simply the consequence of the differentiation from pro-B cells to small pre-B cells induced by the anti-Igβ treatment. To address this issue, we analyzed the kinetics of the surface phenotypic changes of CD45R (B220)+ bone marrow cells after the treatment with anti-Igβ mAb, compared with the kinetics of the germline transcription at the H and L chain loci, which has been demonstrated to correlate strongly with rearrangement at those loci 40 41.
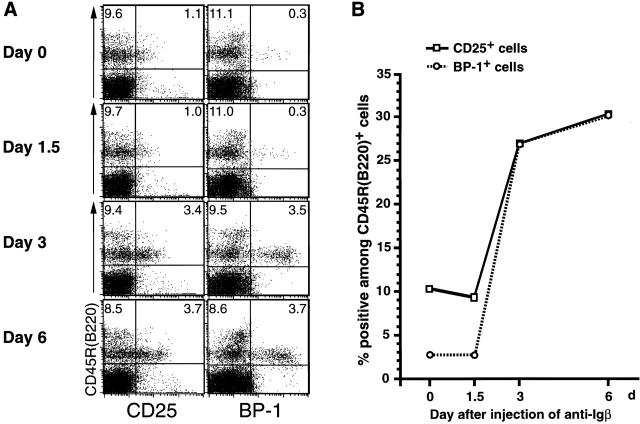
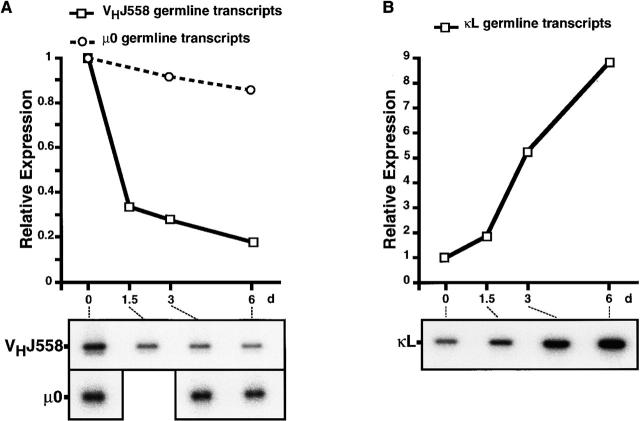
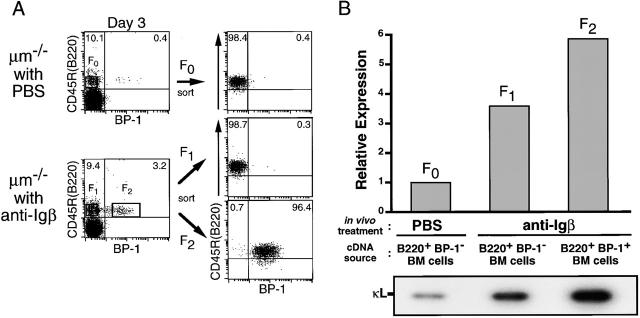
As shown in Fig. 4 A, no apparent change was detected on day 1.5 after injection in the expression of CD25 and BP-1 on CD45R (B220)+ bone marrow cells, whereas their expression was upregulated on days 3 and 6. The kinetics of the proportion of CD25+ or BP-1+ cells among CD45R (B220)+ B lineage cells is summarized in Fig. 4 B. In parallel with the analysis of these surface markers, total RNA was prepared from CD45R (B220)+ bone marrow cells on days 0, 1.5, 3, and 6 after injection. Semiquantitative RT-PCR with specific primers revealed that the cross-linking of Igβ on pro-B cells induced a drastic reduction of VHJ558 germline transcripts derived from unrearranged VH segments, although μ0 germline transcripts stayed at almost the same levels (Fig. 5 A). On day 6, the amount of VHJ558 germline transcripts was diminished to approximately one fifth of that on day 0. Notably, threefold reduction of VHJ558 germline transcripts was detected as early as on day 1.5 after injection, even though no apparent change in the surface phenotype of pro-B cells was observed at that time (compare Fig. 4 B with Fig. 5 A). In contrast, the level of κL chain germline transcripts derived from an unrearranged κL chain gene was greatly upregulated as much as ninefold by day 6 after injection (Fig. 5 B). An approximately twofold increase of κL chain germline transcripts was detected as early as day 1.5 after injection. To clarify the significance of this increase in the κL chain germline transcripts before the induction of cellular differentiation, the level of κL chain germline transcripts was further examined in BP-1− versus BP-1+ fraction sorted from CD45R (B220)+ bone marrow cells of μm-deficient mice 3 d after the treatment with anti-Igβ mAb or PBS (Fig. 6). As expected, BP-1+ differentiated cells from the anti-Igβ–treated mice expressed 5.9 times higher levels of κL chain germline transcripts compared with BP-1− cells from the PBS-treated control mice (F2 versus F0 in Fig. 6 B). Intriguingly, a 3.6-fold increase of κL chain germline transcripts was detected even in BP-1− undifferentiated cells from the anti-Igβ–treated mice compared with the corresponding cells from the control mice (F1 versus F0 in Fig. 6 B). Therefore, the upregulation of κL chain germline transcripts appeared to start before the differentiation from BP-1− cells to BP-1+ cells. These results indicate that upon the anti-Igβ treatment, the κL chain locus in pro-B cells became open and more accessible for recombinases, whereas the H chain locus became less accessible, even before the sign of cellular differentiation became detectable on the cell surface.
Figure 4.
Kinetics of the surface phenotypic changes of CD45R+ bone marrow cells in μm-deficient mice treated with anti-Igβ mAb. (A) μm-deficient mice (10-wk-old) were injected intraperitoneally with 1 mg of anti-Igβ mAb. Bone marrow cells were isolated at the indicated time points after the injection and stained with FITC–anti-CD45R (B220) and biotin-conjugated antibodies specific to either CD25 (Tac) or BP-1 in conjunction with PE-streptavidin. Data shown are representative of three repeated analyses of two-color flow cytometry. The percentage of cells for a given phenotype is shown. (B) Kinetics of the proportion of CD25+ or BP-1+ cells among CD45R (B220)+ B lineage cells in bone marrow is shown.
Figure 5.
The effect of the Igβ cross-linking on germline transcription from Ig genes. μm-deficient mice (10-wk-old) were injected intraperitoneally with 1 mg of anti-Igβ mAb. On days 0, 1.5, 3, and 6 after injection, B220+ cells were enriched from their bone marrow as in the legend to Fig. 2 A. DNase-treated RNA purified from the B220+ cells was reverse transcribed and amplified by PCR with primers specific to VHJ558 or μ0 germline transcripts (A) and specific to κL chain germline transcript (B). Southern blot of PCR products was hybridized with oligonucleotide probes specific to each gene. Radioactivity was quantified with a Bio-Image analyzer and normalized based on the expression of the HRPT gene. The kinetics of expression levels of each transcript is shown. Relative radioactivity is plotted on the y-axis (top). The signal in the B220+ cells on day 0 is set to 1.0 in each figure. Treatment with normal hamster IgG as control did not elicit any change in the gene expression (data not shown).
Figure 6.
The cross-linking of Igβ on pro-B cells upregulated κL chain germline transcripts even before the induction of surface phenotypic changes. (A) μm-deficient mice (10-wk-old) were injected intraperitoneally with PBS or 1 mg of anti–Igβ mAb. On day 3 after injection, bone marrow cells were isolated and stained with FITC–anti-CD45R (B220) and biotin-conjugated anti–BP-1 in conjunction with APC-streptavidin. BP-1− B220+ cells (F0 and F1 fractions, left) and BP-1+ B220+ cells (F2 fraction) were sorted separately by a cell sorter and reanalyzed for the expression of BP-1 and B220 (right). (B) RNA extracted from cells in F0, F1, or F2 fraction was subjected to RT-PCR and Southern blot analyses to detect κL chain germline transcripts (bottom) as in the legend to Fig. 5 B. Relative expression of the transcripts in each fraction was calculated from radioactivity and is shown in the top. The signal in the BP-1− B220+ cells from the PBS-treated μm-deficient mice (F0) is set to 1.0.
Discussion
This study demonstrated that the antibody-mediated Igβ cross-linking on pro-B cells induced their differentiation to the small pre-B cell stage in μm-deficient mice as if it mimicked pre-BCR signaling. This has allowed us to directly examine the possible effects of signaling through Igβ, a signal-transducing component of the pre-BCR, on the Ig gene rearrangements in early B cell precursors. We found that the Igβ signaling induced a drastic change in the targeting of V(D)J recombinase activity, from being predominantly active at the H chain locus to being restricted to the L chain locus. Importantly, we demonstrated that the change of locus accessibility indicated by the upregulation of κL chain germline transcripts and the downregulation of VH germline transcripts was detectable even before the cellular differentiation indicated by the change of surface phenotype. Therefore, the alteration of locus accessibility induced by the Igβ signaling is not simply a consequence of the differentiation of pro-B cells to small pre-B cells, where decreased accessibility at the H chain locus and increased accessibility at the L chain locus have been reported 38 39 42. Our results strongly suggest that the signaling through the Igα/Igβ heterodimer of the pre-BCR is involved in the regulation of ordered Ig gene rearrangement by altering the accessibility of the H and L chain loci.
Previous studies with a series of knockout mice demonstrated that neither μH chain nor surrogate L chain was required for L chain gene rearrangement 13 21 26 27. In accord with this, low levels of κL chain germline transcripts and some κL chain gene rearrangements could be detected in bone marrow pro-B cells from μm-deficient mice. However, interestingly we found that the Igβ cross-linking on pro-B cells drastically increased κL chain germline transcripts as well as the occurrence of κL chain gene rearrangements. The induction of such κL chain germline transcription was also observed in pro-B cells of RAG-deficient mice when the μH chain transgene was expressed 43 44. Therefore, pre-BCR signaling via Igβ appears to facilitate opening the L chain loci for efficient access of recombinase even though it is not necessarily a prerequisite for the induction of L chain gene rearrangements.
The importance of Igβ in the allelic exclusion was previously demonstrated by creating mice carrying the transgene encoding the chimeric molecule between the μH chain and the Igβ cytoplasmic tail 45. Consistent with this, in our system used in this study, the in vivo cross-linking of Igβ on pro-B cells resulted in the suppression of rearrangements at the H chain locus as detected by LMPCR. We further demonstrated that VHJ588 germline transcripts in pro-B cells were downregulated to one third as early as day 1.5 after the anti-Igβ treatment, namely before cellular differentiation. This prompt closing of the H chain locus would be a mechanism of allelic exclusion at the H chain locus observed in normal B cell development, in that as soon as μH chain is produced through a functional VHDHJH rearrangement in one allele of the H chain gene, the rearrangement at the other allele is supposed to be prevented. Thus, the signaling through Igβ appears to be involved in allelic exclusion through altering the accessibility at the H chain locus. Since VH but not μ0 germline transcription was suppressed by Igβ cross-linking, the inaccessibility appears to be restricted to VH segments at the H chain locus. This is consistent with previous observations in μH chain transgenic mice 38 42.
Another level of regulation in allelic exclusion has been proposed. It has been shown that RAG-2 proteins are quickly degraded during the S, G2, and M phases of the cell cycle 46 47. Since the pre-BCR signal drives the cell cycle for several rounds of division 10, RAG-2 proteins produced during the pro-B cell stage for H gene rearrangement would be degraded rapidly in pre-BCR–expressing large pre-B cells. It seems reasonable that the pre-BCR signal leads to closing of the H chain locus and opening of the L chain locus while it downregulates the RAG activity. When the expression of surrogate L chain is shut off, most likely by pre-BCR signaling itself, and the RAG activity increases again 35, V(D)J recombinase targets the L chain locus but no longer the H chain locus. Thus, the pre-BCR appears to guarantee the allelic exclusion doubly by regulating both the recombinase and its target loci.
There has been much debate about what triggers pre-BCR signaling in normal B cell development, whether binding of a putative ligand or self-aggregation of the pre-BCR 17. Whether the surface expression of the pre-BCR is essential for its function is another important question 48 49, since the expression level of the pre-BCR on the surface of pre-B cells is extremely low compared with that of the BCR, and the vast majority of the pre-BCR was detected intracellularly 10 28. This study did not address those issues. Nevertheless, it clearly demonstrated that the Igβ complex on pro-B cells is fully competent to transduce signals across the cell membrane for cell differentiation and Ig gene rearrangements, though its expression level on the cell surface is extremely low and almost undetectable even by flow cytometry, as in the case of the pre-BCR 29. This favors the idea that the pre-BCR exerts its function on the cell surface in spite of its paucity.
The system described in this study is unique in that the ordered program of Ig gene rearrangements can be triggered in vivo by one shot of the antibody injection. Abelson murine leukemia virus (A-MuLV)–transformed pro-B and pre-B cell lines as well as Ig transgenic mice have been widely used to study the mechanism of Ig gene rearrangements 4 18 19 20 36 38 42 43 44 45. Our system has an advantage over those studies in that the antibody-mediated elicitation of transient and synchronous responses of pro-B cells allows us to examine the kinetics of each process in the rearrangement program, such as the alteration of chromatin structure and transcription in a particular locus. This approach should facilitate the elucidation of the mechanism by which the ordered Ig gene rearrangement is regulated during B cell development.
Acknowledgments
We thank Drs. K. Rajewsky and F.W. Alt for providing μm-deficient and RAG-2–deficient mice, respectively.
This work was supported in part by grants from the Ministry of Education, Science, Sports and Culture (Japan).
Footnotes
Abbreviations used in this paper: APC, allophycocyanin; BCR, B cell antigen receptor; HPRT, hypoxanthine phosphoribosyl transferase; LMPCR, ligation-mediated PCR; pre-B, precursor B; pro-B, progenitor B; RAG, recombination activating gene; RSS, recombination signal sequence; RT, reverse transcription.
References
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Alt F.W., Blackwell T.K., Yancopoulos G.D. Development of the primary antibody repertoire. Science. 1987;238:1079–1087. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- Schatz D.G., Oettinger M.A., Schlissel M.S. V(D)J recombinationmolecular biology and regulation. Annu. Rev. Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- Alt F., Rosenberg N., Lewis S., Thomas E., Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cellsrearrangement of heavy but not light chain genes. Cell. 1981;27:381–390. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- Reth M.G., Ammirati P., Jackson S., Alt F.W. Regulated progression of a cultured pre-B-cell line to the B-cell stage. Nature. 1985;317:353–355. doi: 10.1038/317353a0. [DOI] [PubMed] [Google Scholar]
- Rolink A., Melchers F. Molecular and cellular origins of B lymphocyte diversity. Cell. 1991;66:1081–1094. doi: 10.1016/0092-8674(91)90032-t. [DOI] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Karasuyama H., Kudo A., Melchers F. The proteins encoded by the VpreB and λ5 pre-B cell–specific genes can associate with each other and with μ heavy chain. J. Exp. Med. 1990;172:969–972. doi: 10.1084/jem.172.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubata T., Reth M. The products of pre-B cell–specific genes (λ5 and VpreB) and the immunoglobulin μ chain form a complex that is transported onto the cell surface. J. Exp. Med. 1990;172:973–976. doi: 10.1084/jem.172.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasuyama H., Rolink A., Shinkai Y., Young F., Alt F.W., Melchers F. The expression of Vpre-B/λ5 surrogate light chain in early bone marrow precursor B cells of normal and B cell-deficient mutant mice. Cell. 1994;77:133–143. doi: 10.1016/0092-8674(94)90241-0. [DOI] [PubMed] [Google Scholar]
- Lassoued K., Nunez C.A., Billips L., Kubagawa H., Monteiro R.C., LeBlen T.W., Cooper M.D. Expression of surrogate light chain receptors is restricted to a late stage in pre-B cell differentiation. Cell. 1993;73:73–86. doi: 10.1016/0092-8674(93)90161-i. [DOI] [PubMed] [Google Scholar]
- Kitamura D., Roes J., Kuhn R., Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin μ chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- Kitamura D., Kudo A., Schaal S., Muller W., Melchers F., Rajewsky K. A critical role of λ5 protein in B cell development. Cell. 1992;69:823–831. doi: 10.1016/0092-8674(92)90293-l. [DOI] [PubMed] [Google Scholar]
- Yel L., Minegishi Y., Coustan-Smith E., Buckley R.H., Trubel H., Pachman L.M., Kitchingman G.R., Campana D., Rohrer J., Conley M.E. Mutations in the mu heavy-chain gene in patients with agammaglobulinemia. N. Engl. J. Med. 1996;335:1486–1493. doi: 10.1056/NEJM199611143352003. [DOI] [PubMed] [Google Scholar]
- Minegishi Y., Coustan-Smith E., Wang Y.H., Cooper M.D., Campana D., Conley M.E. Mutations in the human λ5/14.1 gene result in B cell deficiency and agammaglobulinemia. J. Exp. Med. 1998;187:71–77. doi: 10.1084/jem.187.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers F., Karasuyama H., Haasner D., Bauer S., Kudo A., Sakaguchi N., Jameson B., Rolink A. The surrogate light chain in B-cell development. Immunol. Today. 1993;14:60–68. doi: 10.1016/0167-5699(93)90060-X. [DOI] [PubMed] [Google Scholar]
- Karasuyama H., Rolink A., Melchers F. Surrogate light chain in B cell development. Adv. Immunol. 1996;63:1–41. doi: 10.1016/s0065-2776(08)60853-6. [DOI] [PubMed] [Google Scholar]
- Weaver D., Costantini F., Imanishi-Kari T., Baltimore D. A transgenic immunoglobulin mu gene prevents rearrangement of endogenous genes. Cell. 1985;42:117–127. doi: 10.1016/s0092-8674(85)80107-0. [DOI] [PubMed] [Google Scholar]
- Nussenzweig M.C., Shaw A.C., Sinn E., Danner D.B., Holmes K.L., Morse H.C., III, Leder P. Allelic exclusion in transgenic mice that express the membrane form of immunoglobulin μ. Science. 1987;236:816–819. doi: 10.1126/science.3107126. [DOI] [PubMed] [Google Scholar]
- Manz J., Denis K., Witte O., Brinster R., Storb U. Feedback inhibition of immunoglobulin gene rearrangement by membrane μ, but not by secreted μ heavy chains. J. Exp. Med. 1988;168:1363–1381. doi: 10.1084/jem.168.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura D., Rajewsky K. Targeted disruption of μ chain membrane exon causes loss of heavy-chain allelic exclusion. Nature. 1992;356:154–156. doi: 10.1038/356154a0. [DOI] [PubMed] [Google Scholar]
- Loffert D., Ehlich A., Muller W., Rajewsky K. Surrogate light chain expression is required to establish immunoglobulin heavy chain allelic exclusion during early B cell development. Immunity. 1996;4:133–144. doi: 10.1016/s1074-7613(00)80678-0. [DOI] [PubMed] [Google Scholar]
- Reth M., Petrac E., Wiese P., Lobel L., Alt F.W. Activation of Vκ gene rearrangement in pre-B cells follows the expression of membrane-bound immunoglobulin heavy chains. EMBO (Eur. Mol. Biol. Organ.) J. 1987;6:3299–3305. doi: 10.1002/j.1460-2075.1987.tb02649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias A., Kopf M., Williams G.S., Buhler B., Kohler G. Molecular requirements for the μ-induced light chain gene rearrangement in pre-B cells. EMBO (Eur. Mol. Biol. Organ.) J. 1991;10:2147–2155. doi: 10.1002/j.1460-2075.1991.tb07749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubata T., Tsubata R., Reth M. Crosslinking of the cell surface immunoglobulin (μ-surrogate light chain complex) on pre-B cells induces activation of V gene rearrangements at the immunoglobulin κ locus. Int. Immunol. 1992;4:637–641. doi: 10.1093/intimm/4.6.637. [DOI] [PubMed] [Google Scholar]
- Ehlich A., Schaal S., Gu H., Kitamura D., Muller W., Rajewsky K. Immunoglobulin heavy and light chain genes rearrange independently at early stages of B cell development. Cell. 1993;72:695–704. doi: 10.1016/0092-8674(93)90398-a. [DOI] [PubMed] [Google Scholar]
- Chen J., Trounstine M., Alt F.W., Young F., Kurahara C., Loring J.F., Huszar D. Immunoglobulin gene rearrangement in B cell deficient mice generated by targeted deletion of the JH locus. Int. Immunol. 1993;5:647–656. doi: 10.1093/intimm/5.6.647. [DOI] [PubMed] [Google Scholar]
- Winkler T.H., Rolink A., Melchers F., Karasuyama H. Precursor B cells of mouse bone marrow express two different complexes with the surrogate light chain on the surface. Eur. J. Immunol. 1995;25:446–450. doi: 10.1002/eji.1830250221. [DOI] [PubMed] [Google Scholar]
- Nagata K., Nakamura T., Kitamura F., Kuramochi S., Taki S., Campbell K.S., Karasuyama H. The Igα/Igβ heterodimer on μ-negative proB cells is competent for transducing signals to induce early B cell differentiation. Immunity. 1997;7:559–570. doi: 10.1016/s1074-7613(00)80377-5. [DOI] [PubMed] [Google Scholar]
- Koyama M., Ishihara K., Karasuyama H., Cordell J.L., Iwamoto A., Nakamura T. CD79α/CD79β heterodimers are expressed on pro-B cell surfaces without associated μ heavy chain. Int. Immunol. 1997;9:1767–1772. doi: 10.1093/intimm/9.11.1767. [DOI] [PubMed] [Google Scholar]
- Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M., Alt F.W. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- Karasuyama H., Rolink A., Melchers F. A complex of glycoproteins is associated with VpreB/λ5 surrogate light chain on the surface of μ heavy chain–negative early precursor B cell lines. J. Exp. Med. 1993;178:469–478. doi: 10.1084/jem.178.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell R.G., Wilson J.B., Anderson S.J., Longnecker R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998;9:405–411. doi: 10.1016/s1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- Corcoran A.E., Riddell A., Krooshoop D., Venkitaraman A.R. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391:904–907. doi: 10.1038/36122. [DOI] [PubMed] [Google Scholar]
- Grawunder U., Leu T.M., Schatz D.G., Werner A., Rolink A.G., Melchers F., Winkler T.H. Down-regulation of RAG1 and RAG2 gene expression in preB cells after functional immunoglobulin heavy chain rearrangement. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- Schlissel M.S., Corcoran L.M., Baltimore D. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med. 1991;173:711–720. doi: 10.1084/jem.173.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki K., Sunaga S., Ikuta K. The V-J recombination of T cell receptor-γ genes is blocked in interleukin-7 receptor–deficient mice. J. Exp. Med. 1996;184:2423–2427. doi: 10.1084/jem.184.6.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M., Constantinescu A., Morrow T., Baxter M., Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- Constantinescu A., Schlissel M.S. Changes in locus-specific V(D)J recombinase activity induced by immunoglobulin gene products during B cell development. J. Exp. Med. 1997;185:609–620. doi: 10.1084/jem.185.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos G.D., Alt F.W. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- Schlissel M.S., Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 1989;58:1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- Stanhope-Baker P., Hudson K.M., Shaffer A.L., Constantinescu A., Schlissel M.S. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85:887–897. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- Spanopoulou E., Roman C.A., Corcoran L.M., Schlissel M.S., Silver D.P., Nemazee D., Nussenzweig M.C., Shinton S.A., Hardy R.R., Baltimore D. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 1994;8:1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- Young F., Ardman B., Shinkai Y., Lansford R., Blackwell T.K., Mendelsohn M., Rolink A., Melchers F., Alt F.W. Influence of immunoglobulin heavy- and light-chain expression on B-cell differentiation. Genes Dev. 1994;8:1043–1057. doi: 10.1101/gad.8.9.1043. [DOI] [PubMed] [Google Scholar]
- Papavasiliou F., Misulovin Z., Suh H., Nussenzweig M.C. The role of Igβ in precursor B cell transition and allelic exclusion. Science. 1995;268:408–411. doi: 10.1126/science.7716544. [DOI] [PubMed] [Google Scholar]
- Lin W.C., Desiderio S. Regulation of V(D)J recombination activator protein RAG-2 by phosphorylation. Science. 1993;260:953–959. doi: 10.1126/science.8493533. [DOI] [PubMed] [Google Scholar]
- Lin W.C., Desiderio S. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc. Natl. Acad. Sci. USA. 1994;91:2733–2937. doi: 10.1073/pnas.91.7.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y., Isselbacher K.J., Cherayil B.J., Pillai S. Tyrosine phosphorylation of Blk and Fyn Src homology 2 domain-binding proteins occurs in response to antigen-receptor ligation in B cells and constitutively in pre-B cells. Proc. Natl. Acad. Sci. USA. 1994;91:4204–4208. doi: 10.1073/pnas.91.10.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y., Isselbacher K.J., Pillai S. Bruton tyrosine kinase is tyrosine phosphorylated and activated in pre-B lymphocytes and receptor-ligated B cells. Proc. Natl. Acad. Sci. USA. 1994;91:10606–10609. doi: 10.1073/pnas.91.22.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]