Abstract
We describe the anatomical localization of three distinct dendritic cell (DC) subsets in the murine Peyer's patch (PP) and explore the role of chemokines in their recruitment. By two-color in situ immunofluorescence, CD11b+ myeloid DCs were determined to be present in the subepithelial dome (SED) region, whereas CD8α+ lymphoid DCs are present in the T cell–rich interfollicular region (IFR). DCs that lack expression of CD8α or CD11b (double negative) are present in both the SED and IFR. By in situ hybridization, macrophage inflammatory protein (MIP)-3α mRNA was dramatically expressed only by the follicle-associated epithelium overlying the SED, while its receptor, CCR6, was concentrated in the SED. In contrast, CCR7 was expressed predominantly in the IFR. Consistent with these findings, reverse transcriptase polymerase chain reaction analysis and in vitro chemotaxis assays using freshly isolated DCs revealed that CCR6 was functionally expressed only by DC subsets present in the SED, while all subsets expressed functional CCR7. Moreover, none of the splenic DC subsets migrated toward MIP-3α. These data support a distinct role for MIP-3α/CCR6 in recruitment of CD11b+ DCs toward the mucosal surfaces and for MIP-3β/CCR7 in attraction of CD8α+ DCs to the T cell regions. Finally, we demonstrated that all DC subsets expressed an immature phenotype when freshly isolated and maintained expression of subset markers upon maturation in vitro. In contrast, CCR7 expression by myeloid PP DCs was enhanced with maturation in vitro. In addition, this subset disappeared from the SED and appeared in the IFR after microbial stimulation in vivo, suggesting that immature myeloid SED DCs capture antigens and migrate to IFR to initiate T cell responses after mucosal microbial infections.
Keywords: chemokine, migration, T cell region, mucosal immunity, follicle-associated epithelium
Introduction
The precise role of dendritic cell (DC) populations in regulating the immune responses to orally administered antigens remains unclear. We have focused our studies on DCs in the Peyer's patch (PP), as this lymphoid tissue is the primary site for the induction of mucosal immune responses, such as the differentiation of B cells capable of producing secretory IgA. It is also likely the initial site for the generation of regulatory T cells producing IL-10 and TGF-β after low dose antigen feeding. In addition, prior studies suggested that PP DCs may be unique in their ability to prime T cells for providing help for IgA B cell differentiation 1.
In previous studies, we identified two distinct subsets of DCs in murine PP by immunohistochemical analysis 2. One population of DCs resides in the subepithelial dome (SED) region and is positioned to capture antigens transported by overlying M cells, whereas the other subset resides in the T cell–rich interfollicular region (IFR), where naive T cells are likely activated to become effector cells. Based on surface marker expression, we suggested that SED DCs may be less mature than IFR region DCs and speculated that antigens may be captured by cells in the SED and migrate to the IFR to prime T cells. More recently, we demonstrated that freshly isolated total PP DCs have distinct functions compared with splenic DCs with regard to their capacity to induce T helper cell differentiation in vitro 3. PP DCs were shown to prime naive CD4+ antigen-specific T cells to secrete IL-10, IL-4, and IFN-γ, whereas spleen DCs primed for secretion of predominantly IFN-γ. In addition, only PP DCs were shown to produce significant levels of IL-10 after stimulation in vitro with recombinant CD40 ligand. These data suggest that PP DCs have a particular capacity to induce the differentiation of T cells that produce IL-4 and IL-10 as well as TGF-β, cytokines that are important for IgA B cell differentiation and bystander suppression after oral antigen feeding.
It is now clear that there are at least two subclasses of DCs that are distinct in their phenotype and function and that may arise from distinct hematopoietic cell lineages. In the mouse, DCs derived from lymphoid precursors (lymphoid DCs) express the CD8αα homodimer, whereas those derived from myeloid precursors (myeloid DCs) express the CD11b molecule. Originally, lymphoid DCs were identified by Ardavin et al. as thymic DCs that share a close developmental pathway with T cell precursors 4. It has been argued that these DCs may be tolerogenic due to their limited capacity to induce T cell proliferation and their enhanced ability to induce Fas-mediated apoptosis of CD4+ T cells 5. In contrast, murine myeloid DCs induce vigorous proliferative responses of CD4+ T cells in vitro. Recently, two independent groups have reported that these two DC subsets may also have a differential capacity to induce T cell differentiation. Thus, it was shown that foot pad injection of splenic lymphoid (CD8α+) or myeloid (CD8α−) DCs, pulsed with antigen ex vivo, induced T cell responses skewed toward Th1 and Th2, respectively 6 7.
In prior studies, we characterized the expression of lineage-specific markers by DCs freshly isolated from the PP and spleen and found that myeloid (CD11b) and lymphoid (CD8α) markers were similarly expressed by DCs from the two lymphoid sites 3. We have now obtained data demonstrating that the particular ability of isolated total PP DC populations to produce IL-10 and skew T cell responses to Th2 differentiation, as discussed above, can be attributed to the CD11b+/CD8α− PP DC subset (Iwasaki, A., and B.L. Kelsall, manuscript in preparation). Thus, in addition to the differing capacities of lymphoid and myeloid DCs to induce distinct pathways of T cell differentiation, these data suggest that myeloid DCs from distinct tissues may also differ in this capacity.
We undertook the current study to gain a better understanding of the role of lymphoid and myeloid DCs in the complex environment of the PP. We initially determined the location of lymphoid and myeloid DC subsets in the PP by performing two-color immunofluorescence microscopy with antibodies against CD11c (which is expressed on both DC subsets) and lineage-related antigens CD8α, DEC-205, and CD11b. We found that myeloid DCs are located predominantly, if not exclusively, in the SED of the PP, whereas lymphoid DCs are located in the IFR. In addition, we identified a population of DCs at both sites that expresses neither CD11b nor CD8α.
We next explored the role of chemokines in the localization of these DC subsets to different regions of the PP. Chemokines are a superfamily of molecules originally described as proinflammatory chemotactic cytokines capable of inducing transendothelial migration of leukocytes into inflammatory sites. More recently, the functional scope of the chemokine family has been broadened by studies suggesting that chemokines also affect the normal trafficking of cells into distinct regions of uninflamed lymphoid organs 8. Little is known, however, about the role of specific chemokines in the migration and localization of DCs in vivo, primarily because most studies to date have only examined the responsiveness of in vitro–derived DCs to various chemokines. In the studies presented here, we focused on two chemokine/receptor pairs that have been suggested to play a role in DC trafficking to mucosal surfaces and lymphoid organs, namely, macrophage inflammatory protein (MIP)-3α/CCR6 and MIP-3β and secondary lymphoid organ chemokine (SLC)/CCR7, respectively 9.
We now present data demonstrating that mRNA for MIP-3α is dramatically expressed in follicle-associated epithelium (FAE). In addition, we show that myeloid SED DCs, but not lymphoid IFR DCs, express mRNA for CCR6 and migrate toward MIP-3α. In contrast, there is negligible expression of MIP-3α in the spleen, and unlike the PP myeloid DCs, splenic myeloid DCs do not migrate toward MIP-3α despite their expression of the receptor for MIP-3α, CCR6. Finally, we addressed the effect of maturation on DC subset stability and chemokine responsiveness. We determined that all DC subsets expressed a similarly immature phenotype when freshly isolated and maintained expression of subset markers upon maturation in vitro. In contrast, the expression of CCR7 was enhanced with maturation in vitro, and this correlated with the ability of PP myeloid DCs to migrate from the SED to the IFR after microbial stimulation in vivo. These are the first data to demonstrate the localization and chemotactic properties of DC subsets in the murine PP and to show that a physiologically relevant DC population is attracted to MIP-3α expressed by noninflamed mucosal tissue.
Materials and Methods
Animals.
6–8-wk-old female BALB/c mice were obtained from the National Cancer Institute (Frederick, MD).
Antibodies.
Identification of DC subsets was carried out using purified anti-CD11c (N418), anti-CD11b (M1/70), anti-CD8α (53-6.7), and anti-DEC-205 (NLDC-145). The above antibodies were purified from hybridoma supernatant grown in serum-free hybridoma media (Life Technologies) in the CELLMAX® artificial capillary system (Spectrum), except for 53-6.7, which was purchased from PharMingen. Flow cytometric sorting of DC subsets was carried out using either FITC- or PE-conjugated anti-CD11c (HL3), anti-CD11b (M1/70), anti-CD8α (53-6.7), and anti-CD45R (RA3-6B2). Before staining, Fc receptors (FcγRIII/II) were blocked using anti–mouse CD16/CD32 (2.4G2). The above antibodies were purchased from PharMingen. Isotype-matched controls used for staining DCs included rat IgG2a, κ (R35-95), rat IgG2b, κ (R35-38 or A95-1), and hamster IgG (G235-2356), which were all purchased from PharMingen.
Double Immunofluorescence Staining of PPs.
PPs were dissected from small intestines of either naive BALB/c mice or those injected intravenously with 25 μg of STAg (soluble Toxoplasma gondii tachyzoite antigen; provided by Dr. A. Sher, NIH, Bethesda, MD) 6 h earlier and were frozen in OCT medium (Sakura Finetek, U.S.A. Inc.). 8-μm sections were fixed in cold acetone and stained for DC markers using TSA®-Direct kit according to the manufacturer's instructions (NEN Life Science Products, Inc.). In brief, endogenous peroxidase activity was quenched with 3% H2O2 for 10 min. Sections were blocked with TNB buffer (NEN Life Science Products, Inc.), and 2 μg/ml (anti-CD11c) or 10 μg/ml (anti-CD11b, anti-CD8, and anti-DEC-205) of purified primary antibodies was applied for 1 h at room temperature. Slides were washed and incubated with horseradish peroxidase (HRP)-conjugated mouse F(ab′)2 anti–rat IgG (Jackson ImmunoResearch Labs., Inc.) for 30 min. DC lineage marker antibodies of the rat IgG isotype (CD11b, CD8α, and DEC-205) were detected with Cy3–Tyramide. Next, primary HRP was deactivated by treatment with 3% H2O2 for 10 min, and the sections were then incubated with biotinylated goat anti–hamster IgG (Vector Labs., Inc.). Slides were washed and incubated with streptavidin–HRP (NEN Life Science Products, Inc.). Staining by hamster anti-CD11c was visualized by amplification of the signal with FITC–Tyramide. Slides were mounted with Vectashield (Vector Labs., Inc.) and were analyzed by confocal microscopy with Zeiss Axioplan/BioRad MRC 1024 confocal laser microscope using a 40× objective with oil.
In Situ Hybridization.
In situ hybridization (ISH) was performed as previously described 10 by Molecular Histology, Inc. In brief, MIP-3α was amplified by PCR using forward (5′-CCGGAATTCTACATCAACTCCTGGAGCTG-3′) and reverse (5′-GCGGTGGCGGCCGCCTGTGTCCAATTCCATCCCA-3′) primers using Taq DNA polymerase (Takara). The PCR product containing EcoRI and NotI sites was inserted into pBluescript SKII (Stratagene, Inc.) at these sites. The CCR6 sequence was amplified from cDNA using the forward and reverse primers, 5′-GAATGAATTCCACAGAG-3′ and 5′-CAATGTTGCTTTGTGCTC-3′, respectively, and was inserted into PCR2.1-TOPO vector (Invitrogen Corp.). Both orientations were selected and linearized using HindIII for generation of sense and antisense probes. The CCR7 sequence was prepared by PCR amplification of cDNA from total mouse splenocytes using primer pairs (forward, 5′-CGCGCGGGATCCATGGACCAGGGGAAACCC-3′ and reverse, 5′-GCGCGCTCTAGACTACGGGGAGAAGGTTGT-3′) containing restriction enzyme sites BamHI and XbaI for inserting into the pGEM-11Zf(+) vector (Promega Corp.). The 35S-labeled sense and antisense riboprobes for MIP-3α, CCR6, and CCR7 were synthesized from these constructs containing full coding region sequences using T7, T3, or SP6 RNA polymerases. For ISH, paraffin-embedded sections of mouse spleen and PP were deparaffinized and pretreated with proteinase K at 37°C for 15 min. Nonspecific binding of probe was reduced by succinylation (1% succinic anhydride) and acetylation in 0.1 M triethanolamine. The slides were hybridized with a labeled probe at 1.6 × 105 cpm/ml incubated at 45°C overnight. The sections were washed and digested with RNase at 37°C for 40 min. Finally, slides were washed with 2× SCC at 60°C for 15 min and dehydrated with 0.3 M ammonium acetate in 70% ethanol for 5 min, followed by 0.3 M ammonium acetate in 95% ethanol for 5 min. The radioactive RNA probe bound to tissue section was detected by emulsion autoradiography using NTB-3 emulsion (Eastman Kodak Co.) and exposed at 4°C in the dark for 3–10 d.
Preparation of DCs.
DCs were prepared from PPs and spleens of naive 6–10-wk-old mice as previously described 3. In brief, PPs were treated with media containing dithiothreitol and EDTA for 90 min at 37°C to remove epithelial cells and were washed extensively with HBSS. PPs and spleens were digested with collagenase D and DNase and incubated in the presence of 5 mM EDTA at 37°C for 5 min. Single-cell suspension was prepared, and cells were incubated with anti–mouse CD11c-coated magnetic beads (Miltenyi Biotech) and selected on MACS™ separation columns. Cells selected on the basis of CD11c expression were then stained with PE-labeled DC lineage marker (CD11b or CD8α) antibody and FITC-conjugated anti-B220 antibody. Lineage marker–positive and –negative DCs (B220−) were isolated by flow cytometric sorting performed on a FACStar™ sorter (Becton Dickinson). Sorted DCs were routinely 98–100% positive for CD11c. The sorted populations were rigorously screened for contamination by B and T lymphocytes by performing reverse transcriptase (RT)-PCR on RNA derived from sorted DCs as described previously 3. None of the sorted DC populations contained macrophage contamination, as CD11c−/lo macrophages are excluded by sorting cells, which expressed only high levels of CD11c 11, and F4/80 staining was not observed on sorted DC populations. In some experiments, these purified DC subsets were stimulated overnight in vitro with CD40L trimer (10 μg/ml) provided by Immunex Corp.
Competitive RT-PCR for Chemokine Receptor Expression by DCs.
Total RNA was isolated from sorted DC populations by RNA isolation column (QIAGEN) and subsequently digested with RNase-free DNase (Life Technologies) to remove contaminating genomic DNA. Single-stranded cDNA was synthesized using SuperScript preamplification system (Life Technologies), and PCR was carried out for 35 cycles using primer pairs for CCR6 (forward, 5′-CCATGACTGACGTCTACCTGTTGAACA-3′; reverse, 5′-GAACAGCTCCAGTCCCATACCCAGCAG-3′), CCR7 (forward, 5′-GCTCAACCTGGCCGTGGCAGACATCC-3′; reverse, 5′-CCACTTGGATGGTGATCAAGGCCTCC-3′), and β2-microglobulin (β2m) (forward, 5′-TGACCGGCTTGTATGCTATC-3′; reverse, 5′-CAGTGTGAGCCAGGATATAG-3′). Oligonucleotides were synthesized by Operon Technologies. Competitive RT-PCR was carried out as previously described (reference 11a). In brief, unknown amounts of cDNA from sorted DCs were added to each PCR reaction tube. Serial dilutions (total of 10× four-fold dilutions) of known amounts of competitive plasmid pMCQ (provided by Dr. D. Shire, Sanofi Recherche, Montpellier, France) were added to the reaction tubes containing target cDNA. The competitive plasmid DNA contained the same primer templates for β2m as the target cDNA and served as internal standard. PCR was carried out for 35 cycles as described above, and the products were analyzed on 1% agarose gels. The amounts of cDNA that would generate equal band intensity to 1.5 pg of the competitive plasmid DNA were determined for each sorted DC population and were used for the amplification of chemokine receptor cDNA.
DC Chemotaxis Assay.
Chemotactic ability of FACS®-sorted DC subsets was analyzed using ChemoTx system according to the manufacturer's instructions (96-well ChemoTx chamber; Neuro Probe, Inc.). In brief, sorted DCs were resuspended in RPMI 1640 supplemented with 1% FCS and 25 mM Hepes. Chemokines at 1.2 μg/ml were placed in the lower chamber, and a filter with 5-μm pore size was placed on top. Aliquots of 20,000 cells/well were applied to the filter's top surface, and the plate was incubated at 37°C in 5% CO2 for 4 h. The cells migrating to the bottom chamber were counted with an inverted microscope in five or more nonoverlapping fields (magnification 40). Each assay was performed in duplicate, and the results were expressed as the mean ± SED of migrating cells per fields counted.
Statistical Analysis.
Normally distributed continuous variable comparisons were done using Student's t test.
Results
Localization of Myeloid and Lymphoid DCs within the Murine PP.
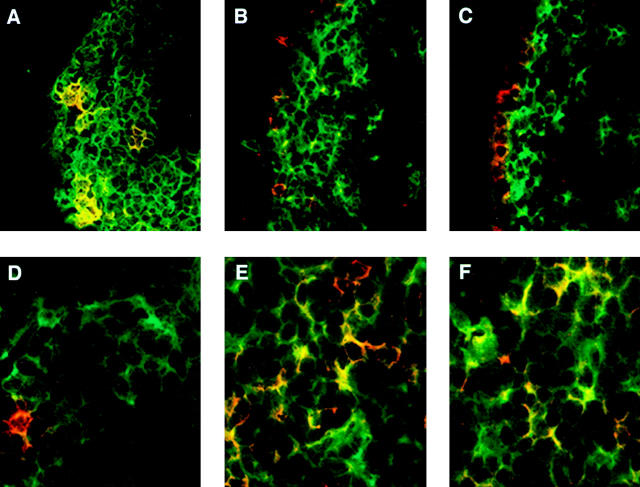
To localize lymphoid and myeloid DCs in the PP, frozen sections were stained with a pan-DC marker, anti-CD11c (green), in combination with lineage-specific markers anti-CD8α and anti-CD11b (red). As shown in Fig. 1 A, CD11b+/CD11c+ (yellow) myeloid DCs are clearly present within the SED region but are absent from the IFR. CD11b+/CD11c− (red) macrophages, however, are prominent in the lamina propria of the villi but are not found in the SED or the follicles and are found only occasionally in the T cell regions of PP. In contrast to myeloid DCs, the CD8α+ lymphoid DCs (Fig. 1 B, yellow cells) are detected exclusively in the IFR and not in the SED or follicle. We did find CD8α+ intraepithelial lymphocytes (red) within the FAE (Fig. 1 B). Moreover, we identified a novel subset of CD11c+ DCs that do not stain for either CD11b or CD8α (Fig. 1 and data not shown). These cells are located in both the SED and IFR (Fig. 1, green cells) and are also negative for the DEC-205 marker. To identify at a single-cell level the expression of lineage specific markers in situ, we carried out confocal analysis (Fig. 2). As noted above, CD11b+ DCs were found only in the SED region (Fig. 2 A) but not in the IFR (Fig. 2 D). In contrast, CD8α+ and DEC-205+ DCs were present in the IFR (Fig. 2E and Fig. F) but not in the SED region (Fig. 2B and Fig. C). As reported previously 12, DEC-205 was also expressed by the epithelial cells of the intestine (Fig. 2 C). Thus, myeloid and lymphoid DCs of the PPs localize in distinct regions, namely, the SED and IFR, respectively.
Figure 1.
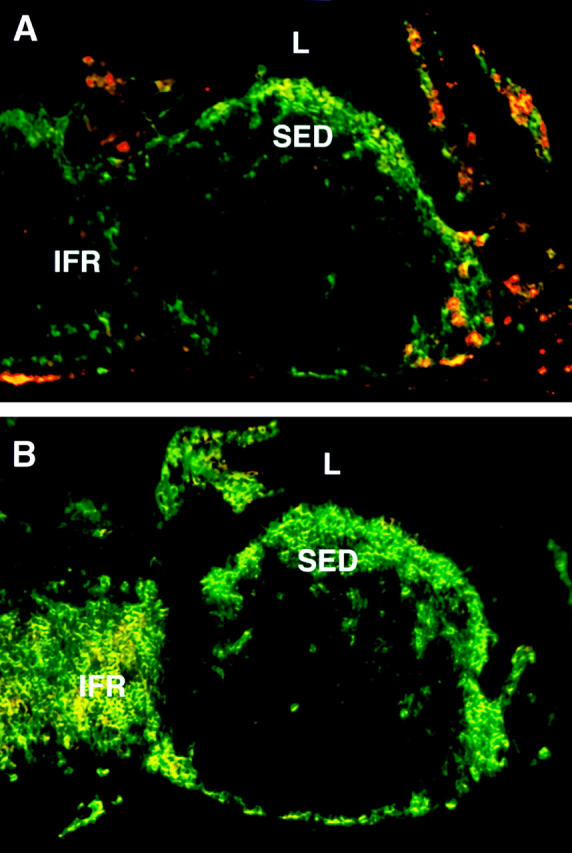
Localization of myeloid and lymphoid DCs in the PP. Frozen sections of PPs were doubly stained with antibodies against CD11c (green) in combination with (A) anti-CD11b (red) or (B) anti-CD8α (red). L, lumen.
Figure 2.
Confocal microscopic analysis of myeloid and lymphoid DCs in the PP. Cryostat sections of mouse PPs were doubly labeled with antibodies against CD11c (green) in combination with (A and D) anti-CD11b (red); (B and E) anti-CD8α (red); or (C and F) anti–DEC-205. SED regions (A–C) and IFRs (D–F) were analyzed by confocal microscopy.
Detection of MIP-3α, CCR6, and CCR7 mRNA Expression in the PP by ISH.
We next examined the role of chemokines in the recruitment of myeloid and lymphoid DCs to their respective locations within the PP in vivo. We analyzed by ISH the expression of mRNA for chemokine receptors CCR6 and CCR7, as well as for the CCR6 ligand MIP-3α. We focused on these chemokine/receptor pairs, as prior studies have suggested that these may be important for the localization of DCs within mucosal (CCR6/MIP-3α) and lymphoid (CCR7/MIP-3β) tissues 9. As shown in Fig. 3 C, MIP-3α mRNA was remarkably expressed in the FAE but not within any other regions of the PP or adjacent villi, including the villus-associated columnar epithelium. Correspondingly, mRNA for the MIP-3α receptor, CCR6, was strongly detected under the FAE within the SED and was detected at lower levels throughout the follicle (Fig. 3 A). CCR6 expression was not found in the T cell–rich IFR or the germinal center of the PP. In contrast, no appreciable level of expression of MIP-3α was detected in the spleen (Iwasaki, A., and B.L. Kelsall, unpublished observation).
Figure 3.
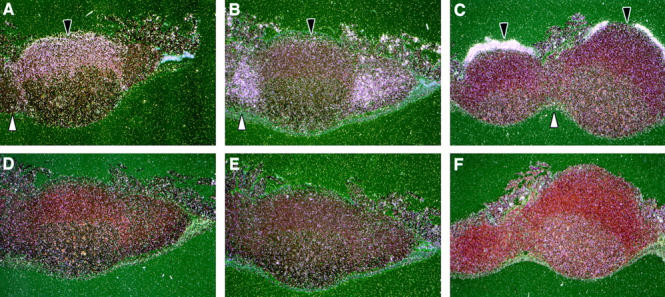
Expression of MIP-3α, CCR6, and CCR7 in the PP. Antisense and sense riboprobes were hybridized to paraffin-embedded sections of the PP. The radioactive RNA probe bound to tissue section was detected by emulsion autoradiography. Darkfield images are shown for antisense CCR6 (A), CCR7 (B), and MIP-3α (C) and sense CCR6 (D), CCR7 (E), and MIP-3α (F) in the PP. Black arrows indicate the SED, and white arrows point toward IFR.
In contrast to CCR6, CCR7 mRNA was found only within the IFR of the PP (Fig. 3 B). This localization of CCR7 mRNA overlaps with the previously reported expression pattern of its ligands MIP-3β and SLC 9 13 14 15 16. Moreover, we detected a sparse signal for the CCR7 within the SED region, which correlates with the distribution of myeloid DCs (Fig. 1 A). Thus, these results indicate that in the PP, cells present in the SED region express CCR6 and localize just under the FAE, where MIP-3α is dominantly expressed. In contrast, cells expressing high levels of CCR7 were found predominantly within the IFR of the PP, where its ligands MIP-3β and SLC are also found.
Expression of CCR6 and CCR7 by PP and Spleen DC Subsets.
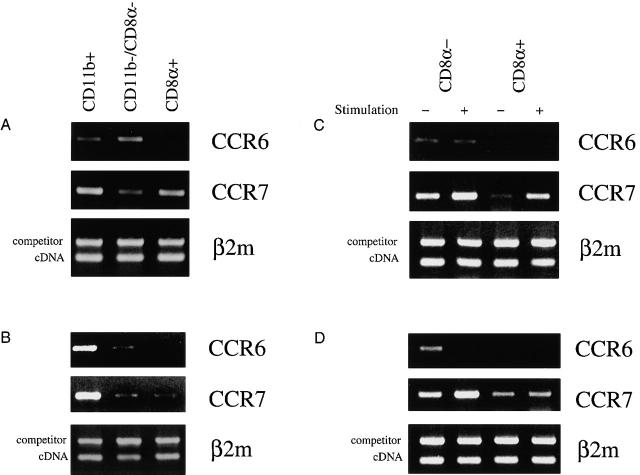
To assess whether the distinct distribution pattern of DC subsets in the PP may be attributed to differential chemokine receptor expression and recruitment, we first examined the expression of chemokine receptor mRNA by RT-PCR. Concurrently, we examined the expression of CCR6 and CCR7 by the splenic DC counterparts of the three distinct DC subsets found in the PP. Thus, CD11c+ DCs from PPs and spleens were freshly isolated by a previously published magnetic selection method 3. The DCs were further FACS® sorted to 98–100% purity into the three subsets described above, namely, CD11b+/CD8α−/CD11c+ (myeloid), CD11b−/CD8α+/CD11c+ (lymphoid), or double-negative (DN) CD11b−/CD8α−/CD11c+ DCs. Contamination by B cells, T cells, epithelial cells, and macrophages of purified DCs was excluded by rigorous screening with flow cytometry and RT-PCR (reference 3; data not shown). These purified DC subsets were assessed for the expression of mRNA for CCR6 and CCR7 by RT-PCR. The amounts of template cDNA used for each PCR were equalized by first performing competitive PCR using primers specific for β2m (Fig. 4). As depicted in Fig. 4 A, CD11b+ myeloid DCs from PP were found to express CCR6, whereas CD8α+ PP DCs did not. A moderate level of mRNA for CCR6 was also detected from DN (CD11b−/CD8α−) DCs. In contrast, CCR7 was expressed at similar levels by both myeloid and lymphoid DCs from the PP and to a lesser extent by DN DCs. In the spleen, mRNA for CCR6 was detected only in myeloid and to a lesser extent in DN DCs, but not in lymphoid DCs; CCR7 mRNA was detected in all subsets (Fig. 4 B). The level of mRNA for CCR7 was found to be consistently higher in the myeloid splenic DCs compared with the other subsets.
Figure 4.
Semiquantitative RT-PCR for CCR6 and CCR7 mRNA on FACS®-sorted fresh and stimulated DC subsets from the PP and spleen. Total RNA was isolated from FACS®-sorted CD11b+, CD8α+, or DN PP (A) or spleen (B) freshly isolated DCs. RNA was also isolated from either freshly isolated or in vitro–stimulated CD8α+ or CD8α− PP (C) or spleen (D) DCs. The cDNA was prepared, and the amount used for each PCR was equalized by competitive PCR using primers for the control gene (β2m) (bottom panels). The levels of CCR6 (top panels) and CCR7 (middle panels) mRNA expression were determined using specific primers as described in Materials and Methods. The amplification fragments were visualized on 1% agarose gel using ethidium bromide. Similar results were obtained from three independent experiments.
Thus, in the PP, myeloid DCs that were determined by immunofluorescence staining to be located in the SED express both CCR6 and CCR7, whereas lymphoid DCs found in the IFR express only CCR7. In the spleen, myeloid DCs, which localize in the marginal zone (reference 17 and data not shown), express both CCR6 and CCR7 mRNA, whereas lymphoid DCs that localize in the periarteriolar lymphoid sheath (reference 17 and data not shown) were found to express only CCR7. Taken together, the expression of chemokine receptor mRNA by the DC subsets as determined by RT-PCR and the localization of these cell subsets by immunofluorescence correlates with the chemokine receptor mRNA expression as determined in ISH.
Myeloid PP DCs Migrate toward MIP-3α.
In the next set of experiments, the migratory capacity of the three purified subsets of PP and splenic DCs toward MIP-3α, MIP-3β, and SLC was assessed by an in vitro chemotaxis assay. As shown in Fig. 5 A, the strongest migration toward MIP-3α was demonstrated by the myeloid CD11b+ PP DCs. In contrast, no statistically significant migration toward MIP-3α by the DN or lymphoid PP DCs was observed. Thus, the PP DC subset that migrated toward MIP-3α corresponded to that solely located just underneath the FAE, namely, the CD11b+ myeloid DCs. However, all PP DC populations migrated toward MIP-3β and SLC to a similar extent, consistent with the presence of CCR7 mRNA expressed by these subsets.
Figure 5.
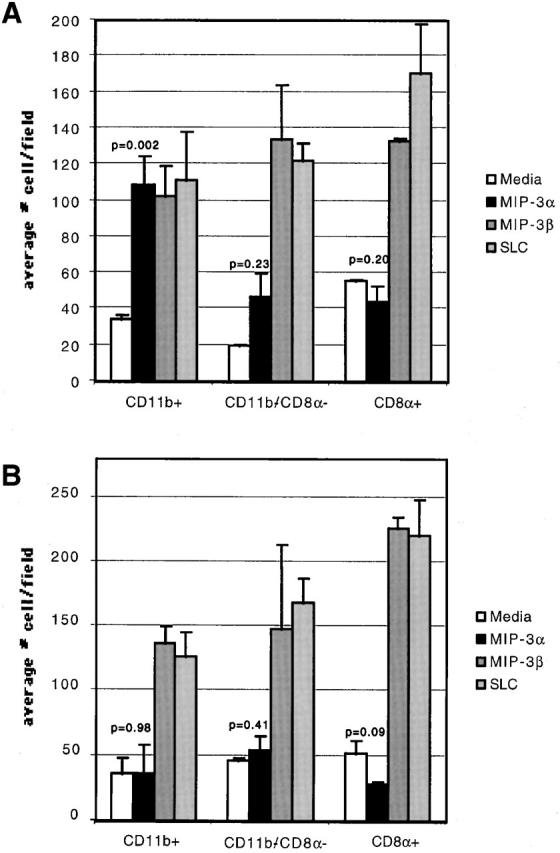
Chemotactic capacity of DC subsets toward MIP-3α and MIP-3β. FACS®-purified myeloid (CD11b+), DN (CD11b−/CD8α2), and lymphoid (CD8α1) DCs from PP (A) or spleen (B) were placed in the chemotaxis chamber in duplicates. Results are expressed as the average number of cells migrating to the bottom chamber per field. At least five independent fields were counted per bottom chamber. The P values for DC migration to MIP-3α compared with the media control are indicated. The data is representative of five independent experiments.
In the spleen, the myeloid DCs, although expressing high levels of CCR6 mRNA (Fig. 4 B), did not migrate toward MIP-3α. As in the PP, all subsets of splenic DCs migrated comparably to MIP-3β and SLC (Fig. 5 B). These data indicated that the myeloid DCs located in the PP SED are unique in their ability to migrate toward MIP-3α.
Thus, in the PP, the migratory capacity of myeloid and lymphoid DCs correlates with their selective chemokine receptor expression. Specifically, the CD11b+ PP DC subset expresses both CCR6 and CCR7 mRNA and migrates toward MIP-3α, MIP-3β, and SLC. In contrast, CD8α+ PP DCs express only CCR7 mRNA and migrate toward MIP-3β and SLC but not to MIP-3α. In the spleen, although the myeloid and DN DCs express CCR6 mRNA, all subsets of DCs migrated toward MIP-3β and SLC but not to MIP-3α.
PP Myeloid and Lymphoid DCs Maintain their Lineage Markers and Upregulate CCR7 Expression during In Vitro Maturation.
In a final series of studies, we addressed the effect of maturation status on DC subsets stability and chemokine responsiveness. In our previous study, we showed that overnight in vitro culture of total PP DCs, including those from the SED, induces differentiation into cells expressing the phenotype of IFR DCs, namely the expression of the DEC-205 molecule 2. This data implied that either SED DCs convert to IFR DC upon maturation or that DEC-205 expression is upregulated during maturation on both subsets of DCs. To distinguish between these possibilities, freshly isolated FACS®-purified CD8α− SED DCs and CD8α+ IFR DCs were stimulated overnight in vitro in the presence of recombinant murine CD40 ligand trimer and were assessed for their surface expression of lineage and activation markers. As depicted in Fig. 6, both CD8α+ and CD8α− DCs upregulated the expression of CD80, CD86, and MHC class II molecules to a similar degree. Moreover, although the expression of DEC-205 molecules on freshly isolated CD8α+ DCs was higher compared with CD8α− DCs, both of these DC subsets increased the expression of DEC-205 during the overnight culture. Thus, DEC-205 represents a maturation marker and is not restricted to the lymphoid lineage. Interestingly, the expression of CD8α and CD11b lineage markers by the two subsets was maintained upon maturation. Thus, the CD8α− DC subset did not downmodulate CD11b expression and become CD8α+ DCs, nor did CD8α+ DCs become CD11b+ during the maturation process. These data suggest that the SED myeloid DCs do not simply represent the immature stage of the IFR lymphoid DCs. Moreover, DN PP DCs were never found to express either CD8α or CD11b upon maturation in vitro (data not shown). Thus, the three subsets of DCs appear to belong to distinct classes that do not readily converge upon maturation.
Figure 6.
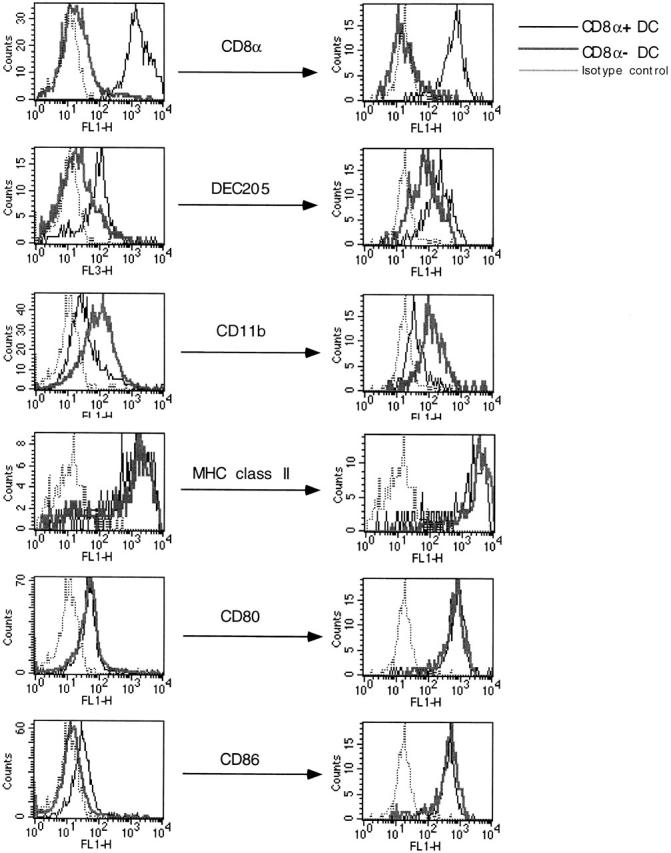
Flow cytometric analysis of CD8α+ and CD8α− PP DCs upon stimulation in vitro. Highly purified CD8α+ and CD8α− DCs from PP were isolated using flow cytometric sorting, and surface expression markers on freshly isolated (left) and in vitro–activated (right) DCs were determined by flow cytometry. The same concentrations of antibodies were used to stain the DCs at left and right. This figure is a representative of three similar experiments.
Next, we examined the regulation of chemokine receptor expression by myeloid and lymphoid DC subsets during overnight in vitro stimulation. The level of mRNA expression for CCR6 and CCR7 was determined by semiquantitative RT-PCR as described above from either freshly isolated or in vitro–stimulated FACS®-sorted DC populations. As shown in Fig. 4 D, CCR6 expression by the CD8α− DCs from spleen was diminished during the in vitro stimulation. The CCR6 mRNA level of PP CD8α− DCs was minimally altered upon maturation (Fig. 4 C). On the other hand, CCR7 expression was enhanced by both splenic and PP CD8α− DCs. The lymphoid CD8α+ DCs in both organs also enhanced the level of CCR7 upon maturation in vitro.
Myeloid PP DCs Migrate toward the T Cell Region upon Microbial Stimulation In Vivo.
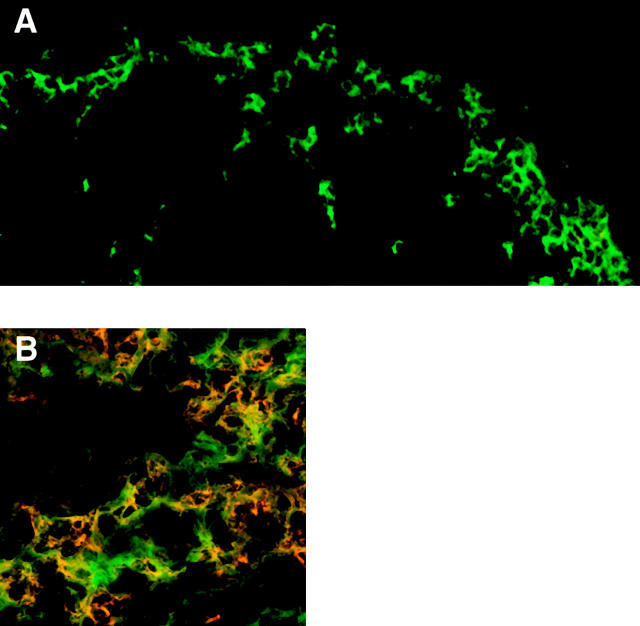
Finally, to examine the migration of DC subsets in the PP after microbial stimulation in vivo, BALB/c mice were given a systemic injection of STAg. In previous studies, STAg has been shown to induce rapid migration of splenic DCs to the inner T cell regions after systemic injection 18. When PPs from mice injected with STAg 6 h earlier were examined, there was a complete loss of CD11b+ DCs from the SED region (Fig. 7 A). This remarkable depletion of SED myeloid DCs was accompanied by the appearance of CD11b+ DCs in the IFR (Fig. 7 B). The CD8α+ DC population was expanded by the STAg injection but was still localized within the IFR (data not shown). Thus, in vivo, myeloid DCs appear to migrate from SED toward the T cell region of the PP upon microbial stimulation.
Figure 7.
In vivo migration of myeloid DCs from the SED to the IFR of the PP upon microbial stimulation. Cryostat sections of PPs were obtained from mice injected systemically with STAg 6 h earlier and were doubly labeled with antibodies against CD11c (green) in combination with anti-CD11b (red). The SED region (A) and IFR (B) were analyzed by confocal microscopy. The experiment was repeated twice with similar data.
Discussion
In this report, we describe the location of DC subsets in the murine PP. We show that in an unperturbed state, myeloid (CD11b+/CD8α−) and lymphoid (CD11b−/CD8α+) DCs are distributed to distinct regions, namely the SED and IFR, respectively. In addition, we demonstrate the presence of a significant novel population of CD11b−/CD8α− DCs at both sites. Next, we show that myeloid PP DCs express CCR6 and migrate toward MIP-3α, which is expressed predominantly by the FAE. This ability of myeloid DCs to migrate toward MIP-3α is unique to the PP, as the splenic myeloid DCs do not migrate toward this chemokine, although they express mRNA for CCR6. In contrast, lymphoid DCs from PP and spleen express only CCR7 and migrate toward MIP-3β and SLC but not to MIP-3α. Finally, in a last series of experiments, we addressed the effect of maturation status on DC subset stability and chemokine responsiveness. When freshly isolated, all DC subsets expressed a similarly immature phenotype. Upon differentiation in vitro, for all DC subsets, the levels of expression of CD8α and CD11b were unaltered, thus demonstrating the stability of these DC subsets. In contrast, expression of CCR7 was enhanced with maturation in vitro, and this correlated with the ability of PP myeloid DCs to migrate from the SED to the IFR after microbial stimulation in vivo.
The DC system is continuously being defined, and in both humans and mice, subsets of DCs with distinct phenotypes and functions have been described. However, as most studies to date have focused on DCs derived in vitro from cell precursors, the precise role of distinct DC subsets in vivo is not clear. In addition, there are significant differences between the functional phenotypes of DCs in mice and humans. In mice, two independent studies have demonstrated that subcutaneous injection of lymphoid DCs from the spleen pulsed with antigen induces Th1 responses, whereas splenic myeloid DCs induce Th2 responses in draining lymph nodes 6 7. In humans, however, the data concerning lymphoid and myeloid DC subsets is much less clear. Human plasmacytoid cells (lymphoid DCs) were originally shown to differentiate allogeneic T cells to secrete Th2 cytokines, whereas myeloid DCs were shown to induce Th1 cytokine secretion 19. In contrast, a recent report by Cella et al. showed that there is minimal difference in the capacity of myeloid versus lymphoid DCs to differentiate naive Th cells toward either Th1 or Th2 20. Clearly, more studies are needed to clarify the role of DC subsets in vivo.
The distinct localization of the myeloid and lymphoid DCs prompted us to investigate the role of chemokines in recruitment of these cells to their respective areas within the PP. Initially, we examined the expression pattern of the chemokine MIP-3α within the PP by ISH. A remarkable expression of MIP-3α was detected within the FAE, but not within the epithelium overlying the villi. These findings are somewhat inconsistent with a previous report describing the expression of MIP-3α mRNA in the non-FAE, as well as FAE of small intestine and colon in mice 21. This difference may be attributed to differences in the sensitivity of the probes employed for the ISH assays. In the spleen, no signal for MIP-3α mRNA was detected (Iwasaki, A., and B.L. Kelsall, unpublished observation), suggesting that MIP-3α may have a unique role in recruiting cells to mucosal surfaces. The mRNA of the receptor for MIP-3α, CCR6, was expressed in the SED as well as in the B cell follicle and mixed T cell and B cell region bordering the IFR and the follicles in the PP. CCR6 was not found to be highly expressed in the germinal center. In addition to its expression on CD11b+ and DN DCs, CCR6 may also be found on B cells and memory T lymphocytes in the PP, as the equivalent human cells have been shown to express CCR6 mRNA and protein 22. Thus, to address specifically the role of MIP-3α in DC recruitment in vivo, three distinct DC subsets were purified from the PP and spleen. Using semiquantitative RT-PCR analysis of mRNA from these purified DCs, we detected CCR6 mRNA from DCs residing in the SED (CD11b+ and CD11b−/CD8α−) but not from those present in the IFR (CD8α+). Moreover, correlating with the receptor mRNA expression pattern, the migratory capacity of DCs toward MIP-3α was restricted to the SED (myeloid) DCs and was not seen in the IFR (lymphoid) DCs. In the spleen, however, although a similar pattern of receptor mRNA expression was detected in the corresponding DC subsets, none of the subsets migrated toward MIP-3α. It is unclear why the splenic CD11b+ and the DN DCs of the spleen and PP, while expressing CCR6 mRNA, do not migrate toward MIP-3α. Whether posttranscriptional regulation of the CCR6 gene product or inhibitors of downstream signaling molecules are differentially regulated in the these DCs remains to be addressed. Taken together with the localization data, our data suggest that MIP-3α secreted by the FAE acts to recruit CD11b+ PP DCs to the SED region.
In contrast to CCR6 mRNA, the expression of CCR7 mRNA was confined mainly to the IFR of the PP (Fig. 3). As CCR7+ cells in the IFR may include CD4+ and CD8+ T cells as well as lymphoid DCs, we purified DC subsets to specifically analyze their migratory capacity. Interestingly, all DC subsets expressed CCR7 mRNA, as measured by RT-PCR, and migrated toward MIP-3β and SLC. It remains a mystery as to how CD11b+ DCs remain in the SED despite their ability to migrate toward SLC and MIP-3β, as assessed by our in vitro chemotaxis assay. One possibility is that in the PP, CD11b+ DCs are retained in the SED due to very high levels of MIP-3α secreted by the FAE compared with the levels of MIP-3β or SLC secreted by the cells in the IFR. Upon encounter with pathogens, however, these cells acquire selective ability to migrate toward the T cell region MIP-3β and SLC by upregulating the CCR7 molecule (Fig. 4 C). Indeed, we observed the migration of myeloid CD11b+ DCs from the SED to IFR after STAg stimulation in vivo (Fig. 7). This observation is supported by an earlier study of human CD34+ cell–derived DCs in which the immature DCs are shown to express CCR6 but not CCR7 mRNA and respond only to MIP-3α. However, upon stimulation with LPS or TNF-α, these cells now begin to respond to MIP-3β and SLC by de novo expression of CCR7 9.
Although a number of studies have investigated the expression pattern and migration toward MIP-3α by diverse cell subsets, the precise in vivo function of MIP-3α remains unclear. As discussed above, in human in vitro–cultured DCs, the expression of CCR6 mRNA and chemotaxis toward MIP-3α was found in immature CD34+ progenitor-derived or transiently adherent lung DCs but not in monocyte-derived DCs 9 22 23 24. In mice, Ogata et al. reported the lack of CCR6 mRNA in different in vitro–cultured, bone marrow–derived DC subsets tested 25. These data at first may seem to conflict with our result showing that myeloid DCs in the PP express CCR6 and migrate toward MIP-3α. However, a recent report by Yang et al. demonstrated that human monocyte-derived DCs can be made to express CCR6 mRNA and migrate toward MIP-3α by in vitro culture in the presence of TGF-β1 in addition to GM-CSF and IL-4 26. This implies that in an organ such as PP where high levels of TGF-β1, 2, and 3 are constitutively present (Iwasaki, A., S. Jakowlew, and B.L. Kelsall, manuscript in preparation), myeloid-derived DCs may be induced to express CCR6 and respond to MIP-3α.
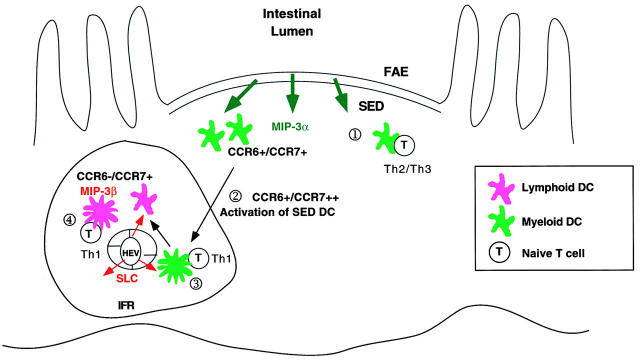
Taking into consideration the results described above, we propose the following mechanism for DC recruitment and migration within the PP and how distinct subsets of DCs may be involved in T cell differentiation (Fig. 8). First, both immature myeloid and lymphoid DCs are recruited to the PP by responding to MIP-3β and SLC secreted in the IFR. Although previous studies have suggested that CCR7 expression is only detected in “mature” DCs 9 27, it is still possible that “immature” DCs expressing moderate levels of CCR7 are recruited to the PP. Indeed, our data demonstrate that all freshly isolated PP and spleen DC populations express CCR7 and migrate toward SLC and MIP-3β, despite their lack of high levels of maturation markers such as CD80 and CD86 (Fig. 6). In addition, the level of CCR7 expression on freshly isolated DCs is enhanced upon maturation (Fig. 4). It is also possible, and perhaps more likely, that other undefined chemokine/receptor pairs play a role in immature DC recruitment to the PP. Once within the PP, the myeloid DCs acquire the ability to respond to MIP-3α due to the high level of TGF-β available in this microenvironment. They migrate to the SED region and remain there until encounter with antigens transported by M cells present within the FAE. At this time, the SED DCs may lose their ability to respond further to MIP-3α by surface receptor downregulation known as “desensitization” 28. If the antigen encountered is an innocuous protein antigen (food components), these DCs mediate the development of Th2/Th3 cells by secreting high levels of IL-10 and possibly TGF-β (Iwasaki, A., and B.L. Kelsall, manuscript in preparation). This process may occur in the SED (Fig. 8, ○1), where small numbers of CD4+ T cells present at this site are directly activated by myeloid DCs. This may be particularly relevant to low dose innocuous antigen feeding, where little antigen may reach the IFR, and there may be little activation of DCs for migration to the IFR. If the antigen is a microbial component, such as STAg, CD11b+ DCs are induced to undergo maturation. This process results in the enhanced CCR7 expression, which allows the DC to migrate toward the IFR, where high levels of MIP-3β and SLC are expressed (Fig. 8, ○2). En route, myeloid or DN DCs may differentiate into APCs capable of stimulating Th1 responses by secreting high levels of IL-12. Indeed, gastrointestinal pathogens known to induce strong Th1 responses at mucosal sites such as Salmonella typhimurium and Toxoplasma gondii are capable of stimulating the secretion of IL-12 from DCs 18 29. The CD11b+ DCs in the IFR stimulate T cells either directly (Fig. 8, ○3) or indirectly via transfer of antigen to CD8α+ DCs (Fig. 8, ○4), a process known as “cross-priming” 30 31 32. In either case, the result is the differentiation of T cells into Th1 cytokine-secreting phenotype. Studies to directly assess the phenotype and function of PP DCs with respect to both chemokine receptor expression and cytokine secretion after infection with mucosal pathogens will be important to test this model.
Figure 8.
Proposed mechanism of DC recruitment to the distinct regions of the PP. The CD11b+ DC (CCR6+/CCR7+) and CD8α+ DC (CCR6−/CCR7+) precursors from the blood enter the PP by responding to CCR7 ligands secreted by the IFR. The myeloid DCs acquire responsiveness to MIP-3α due to the high levels of TGF-β present in the PPs and are thus recruited toward the FAE secreting MIP-3α. On the other hand, lymphoid DCs remain in the IFR due to their expression of CCR7 but not CCR6. Upon encountering luminal antigen transported via M cells, SED DCs may undergo two distinct differentiation pathways. If the antigen encountered is an innocuous food protein, the default pathway for CD11b+ DCs is to generate Th2/Th3 responses through secretion of high levels of IL-10 and TGF-β and low levels of IL-12 in the SED ○1. However, upon encounter with microbial stimuli, such as double-stranded RNA or LPS (“danger signals”), conventional maturation of DCs is triggered ○2. This maturation of SED DCs leads to their migration toward the IFR by upregulating CCR7 expression. In the IFR, naive T cells are primed to secrete IFN-γ by antigen presented by the CD11b+ DCs directly ○3. Alternatively, antigens processed by CD11b+ DCs may be transferred to the Th1-inducing CD8α+ DCs for subsequent presentation to naive T cells by a process known as “cross-priming” ○4.
Collectively, the data presented here suggest a role for MIP-3α and MIP-3β and/or SLC in the recruitment of myeloid and lymphoid DCs to the SED and IFR of PP, respectively. As mRNA for MIP-3α was not detected in the spleen but selectively expressed by mucosal epithelium, this chemokine may be involved in the specific recruitment of DCs to mucosal surfaces. Deciphering the mechanism by which DCs traffic to and from mucosal lymphoid tissues will undoubtedly be important for understanding how immune responses are induced at these sites.
Acknowledgments
We thank Joshua Farber and Fang Liao for the riboprobes for CCR6 and MIP-3α and their helpful advice in reviewing the manuscript. We thank Jon Yewdell for performing confocal microscopy analysis of immunofluorescence sections, Ronald Germain and Jerome Delon for assistance with fluorescence microscopy, Marco Schito and Alan Sher for their helpful advice on the design and interpretation of the experiments involving STAg, Susan Barbieri for FACS® sorting, Marcia Taylor and Ronald Rabin for technical assistance with the chemotaxis assay, and Warren Strober and Stephen Straus for reviewing of the manuscript.
A. Iwasaki is the recipient of a Medical Research Council of Canada postdoctoral fellowship.
Footnotes
Abbreviations used in this paper: DC, dendritic cell; DN, double-negative; FAE, follicle-associated epithelium; HRP, horseradish peroxidase; IFR, interfollicular region; ISH, in situ hybridization; MIP, macrophage inflammatory protein; PP, Peyer's patch; SED, subepithelial dome; SLC, secondary lymphoid organ chemokine.
References
- Spalding D.M., Griffin J.A. Different pathways of differentiation of pre-B cell lines are induced by dendritic cells and T cells from different lymphoid tissues. Cell. 1986;44:507–515. doi: 10.1016/0092-8674(86)90472-1. [DOI] [PubMed] [Google Scholar]
- Kelsall B.L., Strober W. Distinct populations of dendritic cells are present in the subepithelial dome and T cell regions of the murine Peyer's patch. J. Exp. Med. 1996;183:237–247. doi: 10.1084/jem.183.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Kelsall B.L. Freshly isolated Peyer's patch, but not spleen, dendritic cells produce interleukin 10 and induce the differentiation of T helper type 2 cells. J. Exp. Med. 1999;190:229–240. doi: 10.1084/jem.190.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardavin C., Wu L., Li C.L., Shortman K. Thymic dendritic cells and T cells develop simultaneously in the thymus from a common precursor population. Nature. 1993;362:761–763. doi: 10.1038/362761a0. [DOI] [PubMed] [Google Scholar]
- Suss G., Shortman K. A subclass of dendritic cells kills CD4 T cells via Fas/Fas-ligand-induced apoptosis. J. Exp. Med. 1996;183:1789–1796. doi: 10.1084/jem.183.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Lopez R., De Smedt T., Michel P., Godfroid J., Pajak B., Heirman C., Thielemans K., Leo O., Urbain J., Moser M. CD8α1 and CD8α− subclasses of dendritic cells direct the development of distinct T helper cells in vivo. J. Exp. Med. 1999;189:587–592. doi: 10.1084/jem.189.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B., Smith J.L., Caspary G., Brasel K., Pettit D., Maraskovsky E., Maliszewski C.R. Distinct dendritic cell subsets differentially regulate the class of immune response in vivo. Proc. Natl. Acad. Sci. USA. 1999;96:1036–1041. doi: 10.1073/pnas.96.3.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Littman D.R. Chemokine receptors in lymphoid organ homeostasis. Curr. Opin. Immunol. 1999;11:319–325. doi: 10.1016/s0952-7915(99)80051-x. [DOI] [PubMed] [Google Scholar]
- Dieu M.C., Vanbervliet B., Vicari A., Bridon J.M., Oldham E., Ait-Yahia S., Briere F., Zlotnik A., Lebecque S., Caux C. Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J. Exp. Med. 1998;188:373–386. doi: 10.1084/jem.188.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C., Cottler-Fox M. In situ hybridization for the detection of HIV RNA in cells and tissues. In: Coligan J., Kruisbeck A., Margulies D., Shevach E., Strober W., editors. Current Protocols in Immunology. John Wiley & Sons Inc; New York: 1993. [Google Scholar]
- Metlay J.P., Witmer-Pack M.D., Agger R., Crowley M.T., Lawless D., Steinman R.M. The distinct leukocyte integrins of mouse spleen dendritic cells as identified with new hamster monoclonal antibodies. J. Exp. Med. 1990;171:1753–1771. doi: 10.1084/jem.171.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouaboula M., Legoux P., Pessegue B., Delpech B., Dumont X., Piechaczyk M., Casellas P., Shire D. Standardization of mRNA titration using a polymerase chain reaction method involving co-amplification with a multispecific internal control. J. Biol. Chem. 1992;267:21830–21838. [PubMed] [Google Scholar]
- Witmer-Pack M.D., Swiggard W.J., Mirza A., Inaba K., Steinman R.M. Tissue distribution of the DEC-205 protein that is detected by the monoclonal antibody NLDC-145. II. Expression in situ in lymphoid and nonlymphoid tissues. Cell. Immunol. 1995;163:157–162. doi: 10.1006/cimm.1995.1110. [DOI] [PubMed] [Google Scholar]
- Gunn M.D., Kyuwa S., Tam C., Kakiuchi T., Matsuzawa A., Williams L.T., Nakano H. Mice lacking expression of secondary lymphoid organ chemokine have defects in lymphocyte homing and dendritic cell localization. J. Exp. Med. 1999;189:451–460. doi: 10.1084/jem.189.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn M.D., Tangemann K., Tam C., Cyster J.G., Rosen S.D., Williams L.T. A chemokine expressed in lymphoid high endothelial venules promotes the adhesion and chemotaxis of naive T lymphocytes. Proc. Natl. Acad. Sci. USA. 1998;95:258–263. doi: 10.1073/pnas.95.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo V.N., Tang H.L., Cyster J.G. Epstein-Barr virus–induced molecule 1 ligand chemokine is expressed by dendritic cells in lymphoid tissues and strongly attracts naive T cells and activated B cells. J. Exp. Med. 1998;188:181–191. doi: 10.1084/jem.188.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willimann K., Legler D.F., Loetscher M., Roos R.S., Delgado M.B., Clark-Lewis I., Baggiolini M., Moser B. The chemokine SLC is expressed in T cell areas of lymph nodes and mucosal lymphoid tissues and attracts activated T cells via CCR7. Eur. J. Immunol. 1998;28:2025–2034. doi: 10.1002/(SICI)1521-4141(199806)28:06<2025::AID-IMMU2025>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Pulendran B., Lingappa J., Kennedy M.K., Smith J., Teepe M., Rudensky A., Maliszewski C.R., Maraskovsky E. Developmental pathways of dendritic cells in vivodistinct function, phenotype, and localization of dendritic cell subsets in FLT3 ligand-treated mice. J. Immunol. 1997;159:2222–2231. [PubMed] [Google Scholar]
- Sousa C.R., Hieny S., Scharton-Kersten T., Jankovic D., Charest H., Germain R.N., Sher A. In vivo microbial stimulation induces rapid CD40 ligand–independent production of interleukin 12 by dendritic cells and their redistribution to T cell areas. J. Exp. Med. 1997;186:1819–1829. doi: 10.1084/jem.186.11.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissoan M.C., Soumelis V., Kadowaki N., Grouard G., Briere F., de Waal Malefyt R., Liu Y.J. Reciprocal control of T helper cell and dendritic cell differentiation. Science. 1999;283:1183–1186. doi: 10.1126/science.283.5405.1183. [DOI] [PubMed] [Google Scholar]
- Cella M., Jarrossay D., Facchetti F., Alebardi O., Nakajima H., Lanzavecchia A., Colonna M. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat. Med. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Imai T., Baba M., Ishikawa I., Uehira M., Nomiyama H., Yoshie O. Selective expression of liver and activation-regulated chemokine (LARC) in intestinal epithelium in mice and humans. Eur. J. Immunol. 1999;29:633–642. doi: 10.1002/(SICI)1521-4141(199902)29:02<633::AID-IMMU633>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Liao F., Rabin R.L., Smith C.S., Sharma G., Nutman T.B., Farber J.M. CC-chemokine receptor 6 is expressed on diverse memory subsets of T cells and determines responsiveness to macrophage inflammatory protein 3 alpha. J. Immunol. 1999;162:186–194. [PubMed] [Google Scholar]
- Greaves D.R., Wang W., Dairaghi D.J., Dieu M.C., Saint-Vis B., Franz-Bacon K., Rossi D., Caux C., McClanahan T., Gordon S. CCR6, a CC chemokine receptor that interacts with macrophage inflammatory protein 3α and is highly expressed in human dendritic cells. J. Exp. Med. 1997;186:837–844. doi: 10.1084/jem.186.6.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power C.A., Church D.J., Meyer A., Alouani S., Proudfoot A.E., Clark-Lewis I., Sozzani S., Mantovani A., Wells T.N. Cloning and characterization of a specific receptor for the novel CC chemokine MIP-3α from lung dendritic cells. J. Exp. Med. 1997;186:825–835. doi: 10.1084/jem.186.6.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M., Zhang Y., Wang Y., Itakura M., Zhang Y.Y., Harada A., Hashimoto S., Matsushima K. Chemotactic response toward chemokines and its regulation by transforming growth factor-beta1 of murine bone marrow hematopoietic progenitor cell-derived different subset of dendritic cells. Blood. 1999;93:3225–3232. [PubMed] [Google Scholar]
- Yang D., Howard O.M., Chen Q., Oppenheim J.J. Cutting edgeimmature dendritic cells generated from monocytes in the presence of TGF-beta 1 express functional C-C chemokine receptor 6. J. Immunol. 1999;163:1737–1741. [PubMed] [Google Scholar]
- Saeki H., Moore A.M., Brown M.J., Hwang S.T. Cutting edgesecondary lymphoid-tissue chemokine (SLC) and CC chemokine receptor 7 (CCR7) participate in the emigration pathway of mature dendritic cells from the skin to regional lymph nodes. J. Immunol. 1999;162:2472–2475. [PubMed] [Google Scholar]
- Dieu-Nosjean M.C., Vicari A., Lebecque S., Caux C. Regulation of dendritic cell traffickinga process that involves the participation of selective chemokines. J. Leukoc. Biol. 1999;66:252–262. doi: 10.1002/jlb.66.2.252. [DOI] [PubMed] [Google Scholar]
- Marriott I., Hammond T.G., Thomas E.K., Bost K.L. Salmonella efficiently enter and survive within cultured CD11c+ dendritic cells initiating cytokine expression. Eur. J. Immunol. 1999;29:1107–1115. doi: 10.1002/(SICI)1521-4141(199904)29:04<1107::AID-IMMU1107>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Inaba K., Turley S., Yamaide F., Iyoda T., Mahnke K., Inaba M., Pack M., Subklewe M., Sauter B., Sheff D. Efficient presentation of phagocytosed cellular fragments on the major histocompatibility complex class II products of dendritic cells. J. Exp. Med. 1998;188:2163–2173. doi: 10.1084/jem.188.11.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K., Pack M., Inaba M., Sakuta H., Isdell F., Steinman R.M. High levels of a major histocompatibility complex II–self peptide complex on dendritic cells from the T cell areas of lymph nodes. J. Exp. Med. 1997;186:665–672. doi: 10.1084/jem.186.5.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman R.M., Inaba K. Myeloid dendritic cells. J. Leukoc. Biol. 1999;66:205–208. doi: 10.1002/jlb.66.2.205. [DOI] [PubMed] [Google Scholar]