Abstract
The frequencies of the newly identified streptococcal superantigen genes smez, spe-g, and spe-h were determined in a panel of 103 clinical isolates collected between 1976 and 1998 at various locations throughout New Zealand. smez and spe-g were found in every group A Streptococcus (GAS) isolate, suggesting a chromosomal location. The spe-h gene was found in only 24% of the GAS isolates and is probably located on a mobile DNA element. The smez gene displays extensive allelic variation and appears to be in linkage equilibrium with the M/emm type. 22 novel smez alleles were identified from 21 different M/emm types in addition to the already reported alleles smez and smez-2 with sequence identities between 94.5 and 99.9%. Three alleles are nonfunctional due to a single base pair deletion. The remaining 21 alleles encode distinct SMEZ variants. The mosaic structure of the smez gene suggests that this polymorphism has arisen from homologous recombination events rather than random point mutation. The recently resolved SMEZ-2 crystal structure shows that the polymorphic residues are mainly surface exposed and scattered over the entire protein. The allelic variation did not affect either Vβ specificity or potency, but did result in significant antigenic differences. Neutralizing antibody responses of individual human sera against different SMEZ variants varied significantly. 98% of sera completely neutralized SMEZ-1, but only 85% neutralized SMEZ-2, a very potent variant that has not yet been found in any New Zealand isolate. SMEZ-specific Vβ8 activity was found in culture supernatants of 66% of the GAS isolates, indicating a potential base for the development of a SMEZ targeting vaccine.
Keywords: superantigen, streptococcal exotoxin, multiple allelic variation, gene mosaic, antigen variation
Introduction
Since the 1980s, there has been an alarming increase in group A Streptococcus (GAS)1-mediated severe invasive disease (SID) around the world 1 2 3 4 5. Streptococcal invasive disease may include soft tissue infections, such as cellulitis, and necrotizing fasciitis or deeper infections, such as septic scarlet fever (SF) and bacteremia 6. The most severe form of invasive infection results in clinical symptoms similar to those seen in patients with toxic shock syndrome (TSS) caused by Staphylococcus aureus and was therefore described as streptococcal toxic shock–like syndrome (STLS) 7 8. Characteristic symptoms of STLS include hypotension or shock, fever, rash, renal impairment, vomiting, and diarrhea 9. The multiorgan involvement in STLS suggests that a toxin produced by Streptococcus pyogenes might be involved in pathogenesis. Prime candidates are the streptococcal pyrogenic exotoxins (SPEs), a family of highly mitogenic proteins secreted individually or in certain combinations by many S. pyogenes strains. Together with the staphylococcal enterotoxins (SEs) and the toxic shock syndrome toxin (TSST), they belong to a large family of proteins, also known as bacterial superantigens (SAgs), with similar amino acid sequence, protein structure, and functional activities 10 11. SAgs simultaneously bind to MHC class II antigens and TCR molecules and thereby stimulate a large number of T cells in a TCR Vβ–specific mode. This event leads to the release of high systemic levels of cytokines, such as TNF-α and IL-1β, and of T cell mediators like IL-2 and IFN-γ 12 13 14.
It has also been suggested that bacterial SAgs are implicated in other diseases, such as acute rheumatic fever and Kawasaki disease (KD) 6 15 16. SPE-A has long been associated with the ability of GAS to cause SF and STLS, and is believed to be the primary cause of the skin rash in SF 8 17 18. Although it has been shown that SPEs can cause pathological conditions including fever and shock 19 and that severe streptococcal infections are associated with certain exotoxin-producing strains 17 20 21, a correlation between disease and a particular exotoxin and/or S. pyogenes strain remains elusive. Yu and Ferretti performed molecular epidemiologic analysis on 300 general S. pyogenes clinical isolates (from patients with a variety of diseases, except SF) and found the speA gene with a frequency of only 15% 21. Among 146 isolates from patients with SF, 45% were shown to contain speA. Hauser et al. analyzed 34 S. pyogenes isolates from patients with STLS and found the speA gene in 85%. The speC gene was found in only 21% of the isolates 20. Several investigators reported a correlation between STLS and serotypes M1, M3, T3, and M12 and that speA was mostly, but not exclusively, found in M1- and M3-type strains from patients with SF 17 20 21. On the other hand, a genetic and phenotypic diversity among S. pyogenes isolates from severe invasive infections and STLS was reported in the United States 22, and Forni et al. found an M-nontypable (MNT) strain to be common in STLS patients 9. Furthermore, Hsueh and co-workers found no association of severe invasive disease in Taiwan with any SPE 23.
Four allelic variants of the speA gene have been found in S. pyogenes so far 24. The speA1 allele occurs in many distinct clonal lines, but expression of SPE-A2 and SPE-A3, which are characterized by single amino acid substitutions, was shown in two single clones that have caused the majority of STLS episodes in the last decade 24 25. Musser et al. reported that all contemporary strains assigned to electrophoresis types (ETs) 1 and 2 (two highly pathogenic clones, which generally express serotype M1 and M3, respectively) have the speA2 and speA3 allelic variants, respectively 25. In contrast, ET1 and 2 isolates from disease episodes in the 1920s and 1930s contain the speA1 allele, suggesting temporal variation. Kline and Collins showed that SPE-A3 has a significantly increased mitogenic activity and MHC class II affinity compared with SPE-A1 and SPE-A2 26. These results suggest that SPE-A3 represents a more active form of SPE-A and might explain the fluctuation in severity and disease frequency of severe invasive infections.
The implication of other exotoxins, like SPE-C and SSA, in streptococcal infections is even less clear and is assumed on the basis that not all cultures from patients with STLS or other severe invasive diseases express SPE-A.
Since some cases of SID cannot be correlated to any of the known SPEs, there is a suggestion that one or more novel SPEs might play a role in the resurgence of SID in recent years. Recently, we reported the identification of the streptococcal exotoxins SMEZ-2, SPE-G, SPE-H, and SPE-J 27. SMEZ-2 is an allelic variant of the previously described SMEZ 28. Functional analysis using recombinant protein revealed that SPE-G and SPE-H target human T cells carrying the Vβ2 region, a specificity they share with SPE-C and TSST. SMEZ and SMEZ-2 target human Vβ4 and Vβ8 T cells, a unique specificity among streptococcal SAgs. A selective expansion of Vβ2 and Vβ8 T cells has been reported in patients with KD 15, but the potential role of SAgs in triggering this disease remains unclear.
In this report, we present evidence for wide genotypic variation in SMEZ among a panel of 103 clinical isolates selected to provide a spectrum of distinct M/emm types occurring in the community and to provide representation of subtypes within specific M types. Thus far, 24 smez alleles have been identified in 22 different M/emm types. Moreover, the frequency of smez in 95 analyzed GAS isolates approached 100%, making it the most common of all known sag genes, at least within New Zealand. Given the fact that SMEZ is also the most potent SAg produced by S. pyogenes, this has significant implications for its involvement in Streptococcus-mediated disease.
Materials and Methods
Bacterial Isolates.
The 103 streptococcal isolates were obtained from the Institute of Environmental Science and Research in Porirua, New Zealand. Group 1 consists of 52 isolates (including 5 β-hemolytic non-GAS strains) that were taken from various parts of New Zealand between 1976 and 1996. Group 2 contains 51 isolates (including 4 non-GAS) collected from South Auckland schools in 1998. The non-GAS were Streptococcus agalactiae (group B), Streptococcus equis (group C), and Streptococcus spp. (group G). Genotyping of all isolates was undertaken blinded to their strain definition. Two duplicates were included to ensure accuracy of the testing. The results of the duplicate isolates were not included in the overall statistics.
Genomic DNA Preparation.
All S. pyogenes isolates were grown overnight in 10 ml of Brain Heart Infusion medium (BHI; Difco) at 37°C in 15-ml Falcon tubes without agitation. The cells were pelleted and washed in 1 ml of lysis buffer (10 mM Tris, pH 8, 50 mM EDTA) in a 1.5-ml Eppendorf tube and finally resuspended in 400 μl lysis buffer. 4 U of mutanolysin (Sigma-Aldrich), 2 mg of lysozyme, and 40 μg of RNaseA (Sigma-Aldrich) were added. After incubating the samples for 1 h at 37°C with gentle agitation, 20 μg of proteinase K (Sigma-Aldrich) was added and incubated for an additional 30 min. Then, 40 μl of a 20% sarcosyl solution was added, and the DNA was extracted with phenol/chloroform and finally precipitated with ethanol. The DNA was resuspended in 50 μl of TE buffer.
Genotyping of S. pyogenes Isolates.
Purified genomic DNA from all S. pyogenes isolates was used for PCR with specific primers for the smez, spe-g, and spe-h genes as described previously 27. In addition, a primer pair specific to a DNA region encoding the 23S rRNA, oligo 23rRNA fw (GCTATTTCGGAGAGAACCAG) and oligo 23rRNA rev (CTGAAACATCTAAGTAGCTG), was designed and used for PCR as a positive control.
Southern Blot Analysis.
About 5 μg of genomic DNA was digested using restriction enzyme HindIII (GIBCO BRL) and loaded onto a 0.7% agarose gel. The DNA was transferred from the gel to a Hybond-N+ nylon membrane (Amersham Pharmacia Biotech) as described by Maniatis et al. 29. A 640-bp DNA fragment of the smez-2 gene was radiolabeled using the RadPrime Labeling System (GIBCO BRL) and [α-32P]dCTP (NEN Life Science Products). The nylon blots were hybridized with the radiolabeled probe in 2× SSC, 0.5% SDS, 5× Denhardt's overnight at 65°C. After washing twice in 0.2× SSC, 0.1% SDS at 65°C, the blots were analyzed on a Storm PhosphorImager® (Molecular Dynamics).
Cloning and Sequencing of smez Alleles.
All PCR products obtained from genotyping with different S. pyogenes isolates were enriched using the Wizard PCR DNA purification system (Promega) and cloned into T-tailed pBlueScript SKII vectors (Stratagene) followed by transformation into Escherichia coli XL1 blue.
The smez genes were analyzed by the dideoxy chain termination method using a Licor automated DNA sequencer (model 4200). Three subclones of every cloning experiment were analyzed to insure that no Taq polymerase–related mutations were introduced. The DNA sequences of the smez alleles have been annotated in EMBL/GenBank/DDBJ. The accession numbers are listed in Table .
Table 2.
List of All smez Variants Identified To Date
| Gene | Protein | pI | P50 | M/emm type | Accession no. |
|---|---|---|---|---|---|
| pg/ml | |||||
| smez-1 | SMEZ-1 | 5.4 | 0.1 | M1 | – |
| smez-2 | SMEZ-2 | 6.98 | 0.02 | n.d. | AF086626 |
| smez-3 | SMEZ-3 | 6.98 | 0.25 | MNT, emm12, M12 | AF143653 |
| smez-4 | SMEZ-4 | 5.73 | n.d. | PT180, M5 | AF143654 |
| smez-5 | SMEZ-5 | 5.17 | 0.7 | ST4547 | AF143655 |
| smez-6 | n.f. | – | – | MNT | AF143656 |
| smez-7 | SMEZ-7 | 6.14 | 0.3 | ST809 | AF143657 |
| smez-8 | SMEZ-8 | 5.17 | 0.25 | M4, M81 | AF143658 |
| smez-9 | SMEZ-9 | 8.86 | 0.2 | emm80, M80 | AF143659 |
| smez-10 | SMEZ-10 | 5.4 | n.d. | emm53, M53 | AF143660 |
| smez-11 | SMEZ-11 | 8.86 | n.d. | emm56 | AF143661 |
| smez-12 | SMEZ-12 | 5.17 | n.d. | emm59, M59 | AF143662 |
| smez-13 | SMEZ-13 | 6.14 | 0.2 | NZ1437 | AF143663 |
| smez-14 | SMEZ-14 | 6.14 | n.d. | NZ5118 | AF143664 |
| smez-15 | SMEZ-15 | 5.02 | n.d. | M41 | AF143665 |
| smez-16 | SMEZ-16 | 7.82 | n.d. | ST2267 | AF143666 |
| smez-17 | SMEZ-17 | 8.86 | n.d. | PT5757 | AF143667 |
| smez-18 | SMEZ-18 | 9.62 | n.d. | PT3875 | AF143668 |
| smez-19 | n.f. | – | – | emm44 | AF143669 |
| smez-20 | SMEZ-20 | 7.82 | 0.09 | PT2841 | AF143670 |
| smez-21 | SMEZ-21 | 6.14 | 0.3 | emm14 | AF143671 |
| smez-22 | SMEZ-22 | 6.98 | 0.3 | emm49 | AF143672 |
| smez-23 | SMEZ-23 | – | – | n.d. | AF143673 |
| smez-24 | SMEZ-24 | 8.85 | n.d. | ST3018 | AF143674 |
smez-6, smez-19, and smez-23 all have a single base pair deletion causing a frame shift. With the exception of smez-3, smez-4, and smez-8, all analyzed variants could be correlated to a single M/emm type. pI, calculated isoelectric point; P50, half-maximal mitogenic potency; n.d., not determined; n.f., nonfunctional (pseudogene).
Expression and Purification of Recombinant SMEZ Proteins.
Recombinant toxins were produced as described previously 27. In brief, individual smez genes were cloned from pBlueScript vector into the BamHI-EcoRI cloning site of pGEX-2T expression vector and transformed into E. coli DH5 cells. For expression of glutathione S-transferase (GST) fusion proteins, the DH5 cells were grown in tryptone phosphate medium at 30°C and induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at OD600 of 0.7–0.9. The GST fusion proteins were purified on glutathione (GSH) agarose. The mature proteins were then cleaved off from GST by trypsin digestion and further purified by two rounds of cation exchange chromatography. Due to their low isoelectric point, some SMEZ alleles could not be separated from GST by this method. In these cases, trypsin cleavage was performed on the GSH agarose and the mature toxins were eluted from the column.
Toxin Proliferation Assay.
Human PBLs were purified from blood of a healthy donor by Histopaque Ficoll (Sigma-Aldrich) fractionation. The PBLs were incubated in 96-well round-bottomed microtiter plates at 105 cells per well with RPMI-10 (RPMI with 10% FCS) containing varying dilutions of recombinant toxins. The dilution series was performed in 1:5 steps from a starting concentration of 10 ng/ml of toxin. After 3 d, 0.1 μCi [3H]thymidine was added to each well and cells were incubated for an additional 24 h. Cells were harvested and counted on a scintillation counter.
Jurkat Cell Assay.
Jurkat cells (a human T cell line) and LG-2 cells (a human B lymphoblastoid cell line, homozygous for HLA-DR1) were harvested in log phase and resuspended in RPMI-10. 100 μl of the cell suspension, containing 1 × 105 Jurkat cells and 2 × 104 LG-2 cells, was mixed with 100 μl of varying dilutions of recombinant toxins on 96-well plates. After incubating overnight at 37°C, 100 μl aliquots were transferred onto a fresh plate and 100 μl (1 × 104) of SeI cells (IL-2–dependent murine T cell line) per well was added. After incubating for 24 h, 0.1 μCi [3H]thymidine was added to each well and cells were incubated for an additional 24 h. Cells were harvested and counted on a scintillation counter. As a control, a dilution series of IL-2 was incubated with SeI cells.
For detection of native Vβ8-specific exotoxin expression in S. pyogenes, each isolate was grown in 10 ml of BHI medium overnight at 37°C. Cells were then removed by centrifugation and supernatants were diluted 1:10, 1:50, 1:250, and 1:1,000 in RPMI-10, and the assay was continued as described above.
Neutralization Assay.
PBLs were obtained and stimulated as described under Toxin Proliferation Assay above, with the exception that the 10% FCS was replaced by 10% human serum from various healthy donors. All stimulations were done using 40 pg/ml of recombinant toxin. PBLs from a single donor were used for all tests. The neutralizing response was determined by comparing the T cell proliferation with a control test using 10% FCS instead of 10% human serum. The mitogenic response obtained in the absence of human serum was set to 100%.
Results
Genotyping of S. pyogenes Isolates.
PCR-based genotyping was performed in order to determine the frequency of the recently identified sag genes smez, spe-g, and spe-h in streptococcal isolates (Table ). The PCR primers for smez were designed to anneal with both genes, smez and smez-2. The 103 isolates were collected in New Zealand between 1976 and 1998 from varying sites in patients with varying infections, although the majority were from sore throats. They comprised 94 GAS and 9 non-GAS, which were S. agalactiae (group B), S. equis (group C), and Streptococcus spp. (group G). There are 22 distinct M/emm types represented among the GAS isolates, 13 isolates are MNT, and in 2 cases the M type is unknown. The analysis was undertaken blinded to the details of each isolate, and two duplicate isolates were included (95/31 and 4202) to demonstrate the reproducibility of the testing procedure. The isolates are listed in two groups. Group 1 contained isolates collected within a large time frame (1976–1996) from all over New Zealand. Group 2 comprised isolates collected within a short time (1998) in a distinct area (South Auckland schools).
Table 1.
Genotyping of Streptococcal Isolates
| Group 1: Isolates collected from various parts of New Zealand between 1976 and 1996 | Group 2: Isolates collected from South Auckland schools in 1998 | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain no. | Group | M/ emm | Site | Disease | Rib. DNA | spe-g | spe-h | smez | Vβ8 | smez gene | Strain no. | Student ID | Group | M/ emm | Site | Disease | Rib. DNA | spe-g | spe-h | smez | Vβ8 | smez gene | |
| FP 1943 | A | M53 | ts | ST | + | + | − | + | − | n.d. | 4202 | 3310 | A | NZ5118 | ts | ST | + | + | − | + | + | smez-14 | |
| FP 2658 | A | M59 | ts | ST | + | + | − | + | − | smez-12 | 4202(2) | 3310 | A | NZ5118 | ts | ST | + | + | − | + | + | smez-14 | |
| FP 4223 | A | M80 | ts | ST | + | + | − | + | + | n.d. | 9606 | 2252 | A | MNT | ts | ST | + | + | − | + | − | smez-6 | |
| FP 5417 | A | M41 | ts | ST | + | + | − | + | + | smez-15 | 9639 | 2184 | A | MNT | ts | ST | + | + | + | + | + | smez-3 | |
| FP 5847 | A | M1 | ts | ST | + | + | − | + | + | n.d. | 9779 | 3230 | A | emm56 | ts | ST | + | + | − | + | + | smez-11 | |
| FP 5971 | A | M57 | ts | ST | + | + | + | + | − | n.d. | 9893 | 6144 | A | PT180 | ts | ST | + | + | + | + | + | smez-4 | |
| 1/5045 | A | M4 | ts | ST | + | + | − | + | + | smez-8 | 9894 | 6564 | A | emm59 | ts | ST | + | + | − | + | + | n.d. | |
| 79/1575 | A | M1 | ts | Tcarriage | + | + | + | + | + | smez-1 | 10019 | 6264 | A | emm44 | ts | ST | + | + | + | + | − | smez-19 | |
| 81/3033 | A | M12 | ts | ST | + | + | + | + | + | n.d. | 10028 | 9366 | A | emm41 | ts | ST | + | + | − | + | + | n.d. | |
| 82/20 | A | M4 | sk | Ulcer | + | + | − | + | + | n.d. | 10134 | 1880 | A | ST4547 | ts | ST | + | + | − | + | − | smez-5 | |
| 82/532 | A | M12 | ts | ST | + | + | + | + | + | n.d. | 10303 | 3564 | A | emm59 | ts | ST | + | + | − | + | − | smez-12 | |
| 82/675 | A | NZ1437 | ws | Wound | + | + | − | + | + | n.d. | 10307 | 4850 | A | NZ5118 | ts | ST | + | + | − | + | + | n.d. | |
| 84/141 | A | M12 | ts | ST | + | + | + | + | + | n.d. | 10438 | 4904 | A | ST3018 | ts | ST | + | + | − | + | + | smez-24 | |
| 84/1733 | A | M4 | ts | ST | + | + | − | + | + | n.d. | 10463 | TSP | A | emm49 | ts | ST | + | + | − | + | − | smez-22 | |
| 84/781 | A | NZ1437 | ts | ST | + | + | − | + | + | n.d. | 10649 | 11510 | A | ST2267 | ts | ST | + | + | − | + | + | smez-16 | |
| 85/1 | A | M12 | ts | ST | + | + | − | + | + | n.d. | 10730 | 11503 | A | MNT | ts | ST | + | + | − | + | − | n.d. | |
| 85/167 | A | M12 | ts | ST | + | + | + | + | + | n.d. | 10742 | 3374 | A | ST809 | ts | ST | + | + | − | + | + | n.d. | |
| 85/314 | A | NZ1437 | ws | Wound | + | + | − | + | + | n.d. | 10761 | 3254 | A | MNT | ts | ST | + | + | − | + | − | smez-6 | |
| 85/437 | A | M81 | ws | Inf. eczema | + | + | − | + | + | smez-8 | 10763 | 6614 | A | PT3875 | ts | ST | + | + | − | + | − | smez-18 | |
| 85/722 | A | n.d. | ? | ? | + | + | − | + | − | smez-23 | 10782 | 4850 | A | MNT | ts | ST | + | + | + | + | + | n.d. | |
| 86/435 | A | M4 | ts | RF | + | + | − | + | + | smez-8 | 10791 | 10290 | A | MNT | ts | ST | + | + | + | + | + | n.d. | |
| 87/169 | A | M12 | ts | ST | + | + | + | + | + | smez-3 | 10792 | 10308 | A | MNT | ts | ST | + | + | + | + | − | smez-6 | |
| 87/19 | A | M12 | ts | ST | + | + | + | + | + | smez-3 | 10846 | 8854 | A | NZ1437 | ts | ST | + | + | − | + | + | n.d. | |
| 87/781 | A | M12 | ts | ST | + | + | − | + | + | n.d. | 10902 | 6264 | A | NZ5118 | ts | ST | + | + | − | + | + | n.d. | |
| 88/627 | A | M12 | sk | Wound | + | + | − | + | − | n.d. | 10989 | 5194 | A | PT2841 | ts | ST | + | + | − | + | − | smez-20 | |
| 89/22 | A | M12 | ts | Fever | + | + | − | + | + | n.d. | 11070 | 1434 | A | emm65 | ts | ST | + | + | + | − | − | n.d. | |
| 89/25 | A | M12 | ur | Erysipelas | + | + | + | + | + | n.d. | 11072 | 1880 | A | ST4547 | ts | ST | + | + | − | + | − | smez-5 | |
| 89/26 | A | M1 | ts | AGN | + | + | − | + | + | n.d. | 11083 | 4358 | A | MNT | ts | ST | + | + | − | + | − | n.d. | |
| 89/54 | A | NZ1437 | ts | ST | + | + | − | + | + | n.d. | 11093 | 9791 | A | MNT | ts | ST | + | + | + | + | + | n.d. | |
| 90/306 | A | M5 | ear | Otorrhea | + | + | − | + | + | smez-4 | 11152 | 2030 | A | PT2612 | ts | ST | + | + | + | − | − | n.d. | |
| 90/424 | A | M4 | ts | ST | + | + | − | + | + | n.d. | 11222 | 4928 | A | NZ5118 | ts | ST | + | + | + | + | + | n.d. | |
| 91/542 | A | M12 | ts | ST | + | + | − | + | + | n.d. | 11227 | 8854 | A | emm14 | ts | ST | + | + | − | + | − | smez-21 | |
| 94/11 | A | NZ1437 | ps | Abscess | + | + | + | + | + | n.d. | 11244 | 2252 | A | ST4547 | ts | ST | + | + | − | + | − | smez-5 | |
| 94/229 | A | M49 | hvs | Endometr. | + | + | + | − | − | n.d. | 11276 | 4524 | A | MNT | ts | ST | + | + | − | − | − | n.d. | |
| 94/330 | A | M4 | ts | SF | + | + | − | + | + | n.d. | 11299 | 2950 | A | emm80 | ts | ST | + | + | − | + | + | smez-9 | |
| 94/354 | A | M12 | ts | ST | + | + | − | + | + | n.d. | 11574 | 3186 | A | ST809 | ts | ST | + | + | − | + | + | smez-7 | |
| 94/384 | A | M4 | sk | Wound | + | + | − | + | + | n.d. | 11580 | 3280 | A | emm53 | ts | ST | + | + | − | + | − | smez-10 | |
| 94/712 | A | NZ1437 | ws | Cellulitis | + | + | − | + | + | n.d. | 11610 | 2424 | A | emm57 | ts | ST | + | + | + | − | − | n.d. | |
| 95/127 | A | NZ1437 | bc | Cellulitis | + | + | − | + | + | n.d. | 11646 | 1880 | A | ST4547 | ts | ST | + | + | − | + | − | n.d. | |
| 95/31 | A | NZ1437 | ws | Abscess | + | + | − | + | + | smez-13 | 11681 | 3564 | A | emm12 | ts | ST | + | + | − | + | + | smez-3 | |
| 95/31(2) | A | NZ1437 | ws | Abscess | + | + | − | + | + | n.d. | 11686 | 5528 | A | PT5757 | ts | ST | + | + | − | + | + | smez-17 | |
| 95/361 | A | NZ1437 | ps | Abscess | + | + | − | + | + | n.d. | 11745 | 12397 | A | emm59 | ts | ST | + | + | − | + | − | n.d. | |
| 96/1 | A | n.d. | ? | ? | + | + | − | + | + | n.d. | 11789 | 1568 | A | MNT | ts | ST | + | + | − | + | − | smez-6 | |
| 96/364 | A | NZ1437 | bc | Burns | + | + | − | + | + | n.d. | 11802 | 3266 | A | MNT | ts | ST | + | + | − | + | − | n.d. | |
| 96/551 | A | M4 | eye | Eye infect. | + | + | − | + | + | n.d. | 11869 | 2950 | A | ST4547 | ts | ST | + | + | − | + | − | smez-5 | |
| 96/610 | A | M4 | ts | SF | + | + | − | + | + | n.d. | 11961 | 4916 | A | MNT | ts | ST | + | + | − | + | − | n.d. | |
| D21 | A | M1 | ts | Tcarriage | + | + | − | + | + | n.d. | 12015 | 12373 | A | emm59 | ts | ST | + | + | + | + | − | n.d. | |
| RC4063 | C | – | ts | ST | + | − | − | − | − | − | 7625 | 8215 | B | – | ts | ST | + | − | − | − | − | − | |
| SP9205 | C | – | ts | ST | + | − | − | − | − | − | 8011 | 3238 | B | – | ts | ST | + | − | − | − | − | − | |
| NI6174 | G | – | ts | ST | + | − | − | − | − | − | 10388 | 1653 | G | – | ts | ST | + | − | − | − | − | − | |
| NI6192 | B | – | ts | ST | + | − | − | − | − | − | OI2633 | 5395 | B | – | ts | ST | + | − | − | − | − | − | |
| VC4141 | G | – | ts | ST | + | − | − | − | − | − | |||||||||||||
The isolates were collected in New Zealand between 1976 and 1996 (group 1) and from South Auckland schools in 1998 (group 2) from patients with varying diseases. The results are based on PCR analysis using purified genomic DNA and specific primers for each of the sag genes. The non-GAS are: B, S. agalactiae; C, S. equis; G, Streptococcus spp. Site and disease abbreviations are as follows: ts, throat site; ws, wound site; sk, skin; ps, pus site; hvs, high vaginal site; bc, blood culture; ST, sore throat; RF, rheumatic fever; AGN, acute glomerulonephritis; T carriage, throat carriage. n.d., not determined.
All of the nine non-GAS isolates (belonging to groups B, C, and G) were negative for the tested sag genes. The frequencies for smez, spe-g, and spe-h within the GAS isolates were 95.6, 100, and 23.9%, respectively. A correlation between a certain M/emm type and the presence of the spe-h gene could not be established. The deficiency in this current set was that only five M/emm types were represented by more than one isolate. The most frequent serotype was M/emm12 with 13 isolates, from which 7 were positive and 6 were negative for spe-h, suggesting genetic diversity within the M/emm12 strain. In contrast, all 12 tested NZ1437/M89 isolates were negative for spe-h.
The high frequencies of smez and spe-g are of particular interest, as this has not been described for any other streptococcal sag gene thus far. Other spe genes, like speA, speC, and ssa, are found at much lower frequencies, and horizontal gene transfer might explain the varying frequencies of these genes in different strains. In contrast, both smez and spe-g were found in virtually all tested GAS isolates. Only four GAS isolates (11152, 11070, 94/229, and 11610) tested negative for smez. These were PT2612, emm65, M49, and emm57. Southern hybridization was performed to find out if the negative PCR results were due to lack of the smez gene or to lack/alteration of the primer binding site(s). HindIII-digested genomic DNA of selected streptococcal isolates was probed with a 640-bp radiolabeled smez-2 PCR fragment (Fig. 1). The smez gene is located on a 1953-bp HindIII DNA fragment in the M1 strain (700294; American Type Culture Collection) corresponding to fragment A in the Southern blot. In all four PCR-negative isolates, the smez probe hybridized to a HindIII fragment of ∼4 kb (fragment B), but not to the smez bearing fragment A (samples 17, 26, 57, and 67). In addition, the smez probe bound to a second DNA fragment of ∼4.2 kb (fragment C) in isolate 11152 (sample 17). In the M1 reference strain (lane 1) and in isolate 4202 (sample 50), the smez probe also bound to fragment B, in addition to fragment A. Fragment B in the M1 strain contains a 180-bp region that shares 97% sequence homology with the 3′ end of the smez gene. These results suggest that the four PCR-negative isolates possess a truncated smez gene or a smez-like sequence, but not a complete smez gene.
Figure 1.
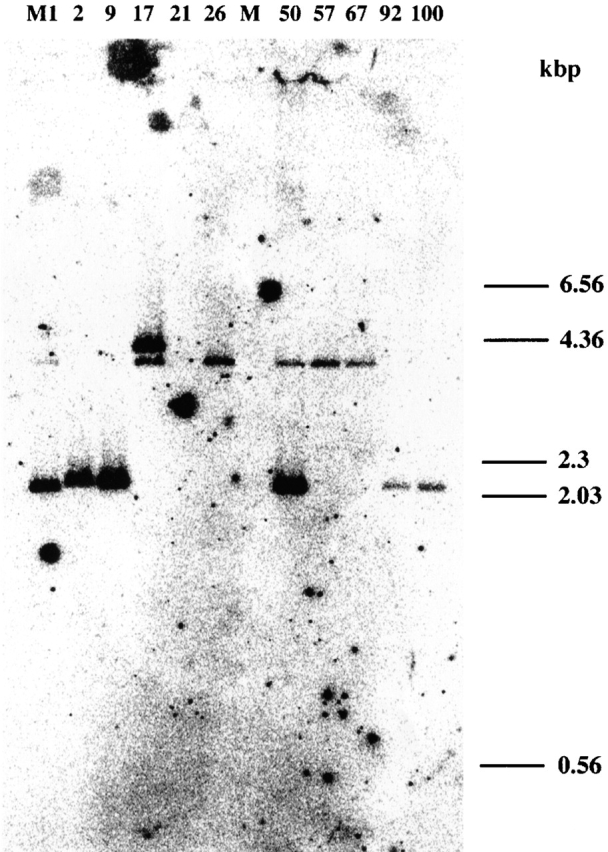
Southern blot analysis of genomic DNA with radiolabeled smez. HindIII-digested genomic DNA from various Streptococcus isolates was hybridized with a radiolabeled smez probe. Band A is a 1953-bp HindIII DNA fragment that carries the smez gene. Bands B and C are DNA fragments of ∼4 and 4.2 kbp, respectively, which both carry a smez-like region. M1, S. pyogenes reference strain (M1 type, 700294; American Type Culture Collection); 2, isolate 9639 (MNT); 9, isolate 11789 (MNT); 7, isolate 11152 (PT2612 type); 21, isolate RC4063 (group C Streptococcus); 26, isolate 11070 (emm65 type); M, DNA marker lane; 50, isolate 4202 (NZ5118/M92 type); 57, isolate 94/229 (M49 type); 67, isolate 11610 (emm57 type); 92, isolate 95/127 (NZ1437/M89 type); and 100, isolate 94/330 (M4 type).
Identification of Multiple smez Alleles.
DNA sequence analysis of the smez genes from different S. pyogenes isolates was performed. 37 isolates were analyzed, representing 22 M/emm types, 5 MNT isolates, and 2 isolates of unknown M type (Table ). 23 different alleles were identified, bringing the total number of smez alleles to 24 (smez is hereafter referred to as smez-1). The smez gene appears to be in linkage equilibrium with the M/emm type. Only three smez alleles (smez-3, smez-4, and smez-8) were found in more than one M/emm type. smez-3 was found in all three tested M/emm12 type isolates plus one MNT. smez-4 was found in one M5 and in one PT180. smez-8 was found in one M4 and in one M81 type strain. Four of the five MNT isolates carry smez-6 and one carries smez-3.
With the exception of smez-6, smez-19, and smez-23, which have individual single base pair deletions causing frame shifts, all smez alleles possess complete open reading frames. smez-2, which was isolated from a separate strain, was not found in any of the current panel of isolates. The 21 intact smez genes encode 21 different SMEZ variants.
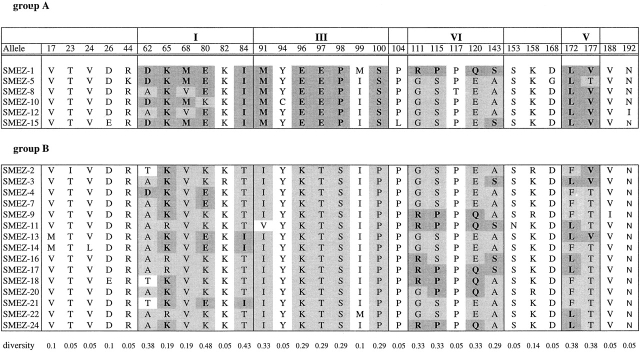
Sequence identities between smez alleles are between 94.5 (smez-1 and smez-2) and 99.9% (equivalent to one base pair difference), e.g., smez-4 and smez-7. A family tree of the 24 smez alleles was established based on DNA sequence homology (Fig. 2). The smez alleles were grouped into two clusters. Group A comprises smez-1, smez-5, smez-8, smez-10, smez-12, and smez-15. All other smez alleles are in group B. Sequence variation between all smez alleles is restricted to only 62 nucleotide positions, or 9.8% of the coding region for the mature protein (Table ). Most of the variations are G/A and T/C transitions, while transversions only occur at position 290 (A/C), 291 (A/T), and 358 (G/C). 31 of the variable positions (50%) result in amino acid changes; the other 50% are silent exchanges. The frequency of a nucleotide exchange at a particular position also varies significantly within the 24 smez alleles. 17 of the 62 sites are point mutations for single alleles, while the remaining 45 sites are point mutations common to 2 or more alleles. The most variable sites are positions 231, 234, and 238 and occur in 12 alleles (50%). Some of these highly variable nucleotides are linked together and build five distinct regions (Table ). One region (III) comprising 10 polymorphic sites between nucleotide 282 and 298 occurs in only 2 combinations within the 24 smez alleles. These combinations segregate the two main groups of the smez family tree. The other four blocks are less conserved, and sequence combinations between these blocks occur in several alleles, generating gene mosaics. Interestingly, the polymorphic sequence blocks are more frequent in the center of the gene, between nucleotide positions 231 and 427. Only one polymorphic block (V) is in the last third of the gene. No blocks occur in the first third. The sequence clusters and mosaic distribution are also reflected in the amino acid sequence (Table ). Interestingly, the five polymorphic amino acid positions within block III are all between amino acid positions 91 and 100 and are therefore clustered closer together than in the other blocks. The amino acid composition of this polymorphic block influences the isoelectric point of the SMEZ variants. All group A variants have a pI between 5.02 and 5.4, whereas the pIs of group B SMEZ are between 5.73 and 9.62 (Table ).
Figure 2.
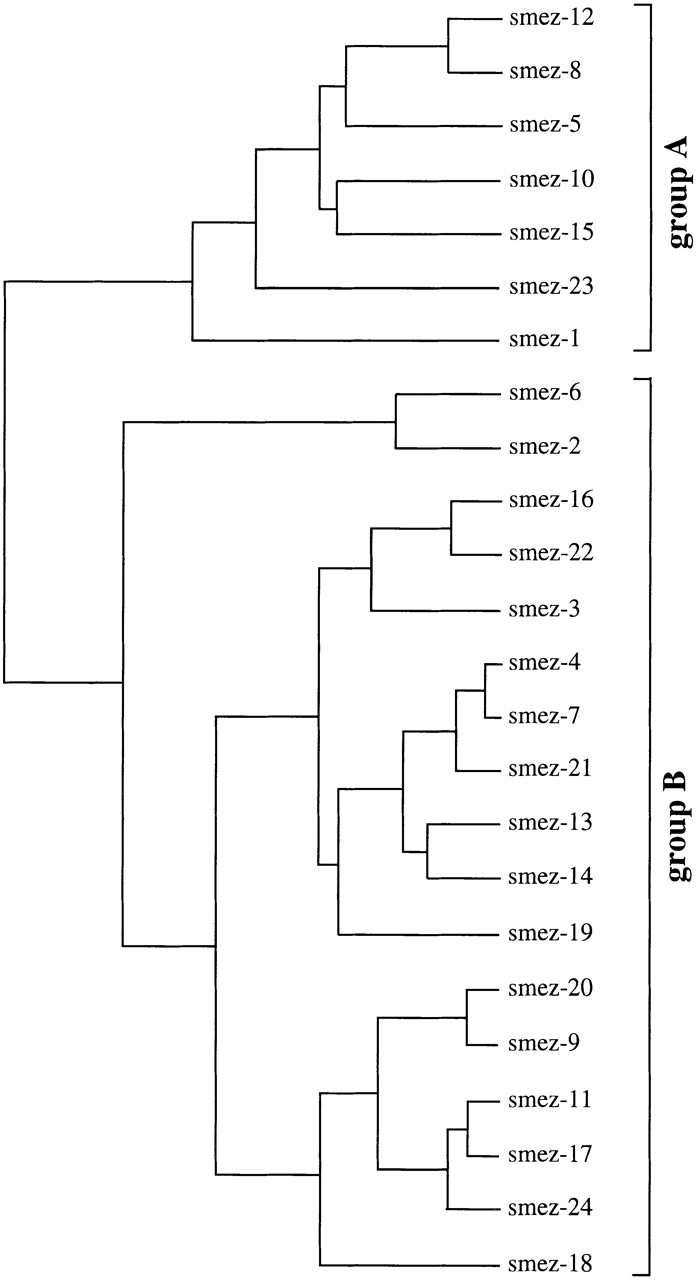
smez gene family tree. The family tree was created using the PileUp program v8 (Genetic Computer Group) and is based on nucleotide sequence homology. The smez family can be divided into two major groups based on a conserved DNA region (region III). smez-6, smez-19, and smez-23 are pseudogenes with a single base pair deletion causing a frame shift.
Table 3.
DNA Sequence Alignment of smez Alleles
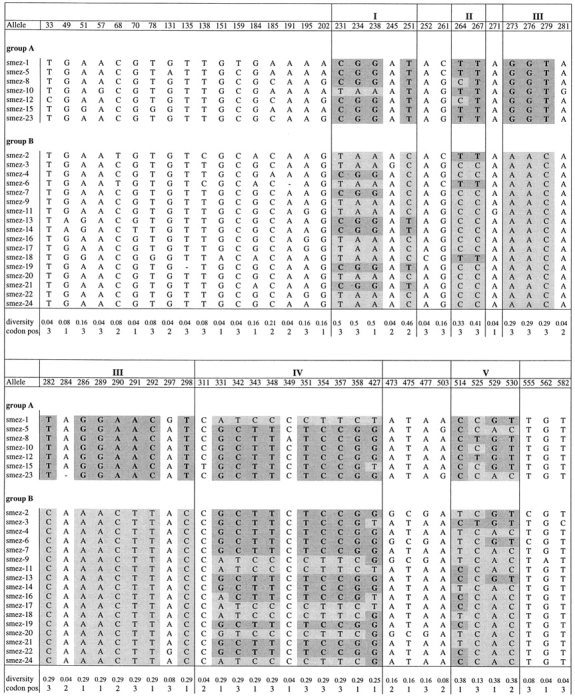
Only the 62 variable nucleotides (9.8% of the smez gene) are shown, of which 17 are point mutations for single alleles and 45 are common to 2 or more alleles. Some of the variable sites are linked together, building sequence blocks (shaded gray) within five distinct regions (I–IV). In region III, only two sequence combinations occur, correlating with the two major branches (groups A and B) of the smez family tree (see Fig. 2). Combinations between sequence blocks within the other four regions in different alleles generate gene mosaics.
Table 4.
Amino Acid Alignment of SMEZ Alleles
Only the 31 variable residues are shown (14.9% of the mature SMEZ protein). The sequence blocks and mosaic structure seen in the DNA sequence alignment (Table ) are also reflected in the amino acid sequence.
Expression of SMEZ Variants by S. pyogenes Isolates.
Both SMEZ-1 and SMEZ-2 specifically target human Vβ8-bearing T cells 27. Jurkat cells (a Vβ8 T cell line) were used to determine SMEZ expression from S. pyogenes culture supernatants from all 103 streptococcal isolates (see Materials and Methods). 62 isolates (66% of the GAS isolates) showed Vβ8-stimulating activity. No Vβ8 activity was detected from the nine non-GAS, the four smez PCR-negative GAS strains, and the isolates carrying the pseudogenes smez-6, smez-19, or smez-23. These results suggest that SMEZ is the only Vβ8-targeting SAg in S. pyogenes. Culture supernatant from 12 isolates with complete open reading frames for smez did not stimulate Jurkat cells. These isolates possess smez-5, smez-10, smez-12, smez-18, smez-20, smez-21, or smez-22. These results suggest the following possibilities: (a) those isolates do not express SMEZ under the selected growth conditions; (b) mutations outside the smez coding region repress gene expression; or (c) these isolates express SMEZ variants that have lost their specificity to target Vβ8 T cells. Recombinant SMEZ variants were generated in E. coli using the GST expression system.
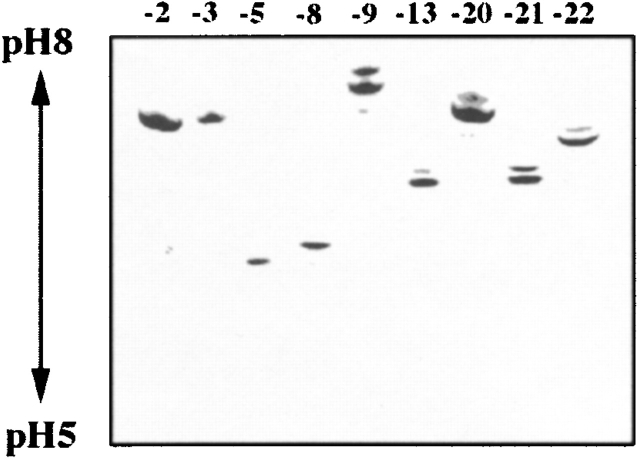
Successful recombinant expression was achieved for rSMEZ-3, rSMEZ-5, rSMEZ-7, rSMEZ-8, rSMEZ-9, rSMEZ-13, rSMEZ-20, rSMEZ-21, and rSMEZ-22. Fusion proteins were completely soluble and gave yields of ∼30 mg/liter. The purified recombinant toxins were applied to isoelectric focusing (Fig. 3). Isoelectric points between SMEZ variants were consistent with the calculated pIs (Table ).
Figure 3.
Isoelectric focusing gel. 2 μg of purified recombinant toxin was run on a 5.5% polyacrylamide isoelectric focusing gel (pH 5–8). The gel shows significant variation of the isoelectric point between SMEZ variants. Toxins with low isoelectric points, such as rSMEZ-5 and rSMEZ-8, belong to group A of the SMEZ family. Members of group B are characterized by a more basic isoelectric point.
Native conformation of the recombinant toxins was confirmed by human T cell proliferation assay. All purified SMEZ variants were active on human T cells with similar half-maximal responses (P50 values) between 0.09 and 0.3 pg/ml, except for rSMEZ-5 with a P50 of 0.7 pg/ml, and rSMEZ-10 and rSMEZ-12, which gave P50 values of 2 and 15 pg/ml, respectively (Table ).
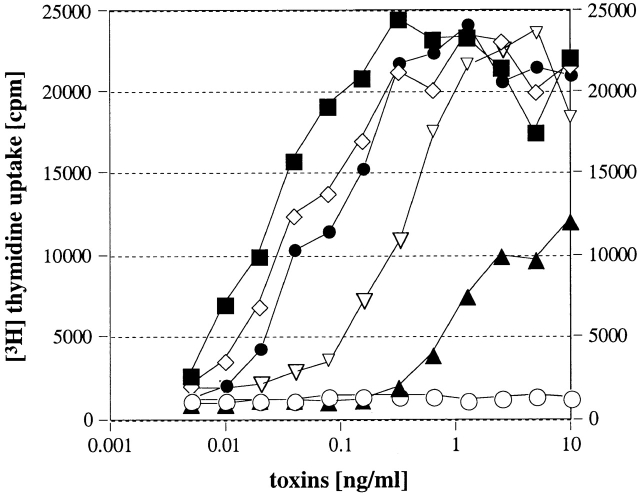
rSMEZ-5, rSMEZ-20, rSMEZ-21, and rSMEZ-22 all stimulated Jurkat cells (Fig. 4), thus confirming that the lack of Vβ8-stimulating activity of wild-type streptococcal culture supernatants is due to lack of expression rather than altered Vβ specificity of the SMEZ variants produced.
Figure 4.
Jurkat cell assay. Jurkat cells (bearing a Vβ8 TCR) and LG-2 cells were mixed with varying concentrations of recombinant toxin and incubated for 24 h, before SeI cells were added. After 1 d, 0.1 mCi [3H]thymidine was added and cells were counted after an additional 24 h. ○, unstimulated; ▿ , rSMEZ-1; ▴, rSMEZ-5; ▪, rSMEZ-20; ⋄, rSMEZ-21; and •, rSMEZ-22.
Antigenic Variation in SMEZ Variants.
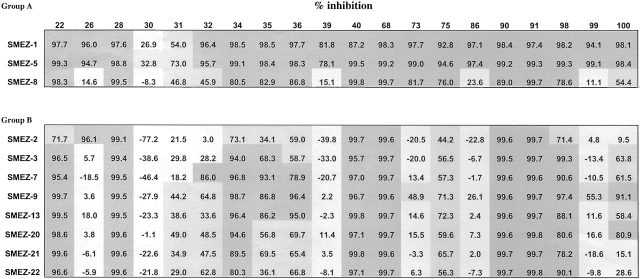
Human sera from 100 healthy donors were tested for the presence of naturally acquired neutralizing antibodies to SMEZ variants rSMEZ-1, rSMEZ-2, rSMEZ-3, rSMEZ-5, rSMEZ-7, rSMEZ-8, rSMEZ-9, rSMEZ-13, rSMEZ-20, rSMEZ-21, and rSMEZ-22. The neutralizing response was determined as the percentage of inhibition of T cell stimulation by SMEZ in the presence of 10% human serum versus 10% FCS. More than 75% inhibition was defined as full, 30–75% as moderate, and <30% as negative neutralization.
85 sera fully neutralized the activity of all 11variants tested. The remaining 15 sera displayed only partial neutralizing activity on a subset of the variants. 20 sera were selected for a repeat test, and the confirmed results are shown in Table . Seven sera (26, 32, 39, 73, 86, 99, and 100) displayed very different neutralizing response profiles, suggesting the existence of variant (epitope) specific antibodies. For example, serum 86 strongly neutralized rSMEZ-1 and rSMEZ-5, but none of the other variants. Interestingly, both SMEZ-1 and SMEZ-5 belong to group A and share a common amino acid stretch in region III (Table ). However, serum 86 did not effectively neutralize another group A variant, rSMEZ-8. The only amino acid differences between SMEZ-1, SMEZ-5, and SMEZ-8 that could explain this different neutralization are residues D62A, M68V (both in region I), and P117T (in region VI). Thus, for serum 86, the likely neutralizing epitopes must include one of these three residues.
Table 5.
Neutralization Assay
Human sera from 100 healthy New Zealand donors were used to study the presence of antibodies neutralizing the mitogenic activity of recombinant SMEZ variants. The figures given are the percentage of inhibition compared with a reference test in which human serum was replaced by FCS. The table shows the results obtained with 20 selected sera. The remaining 80 sera all showed high neutralizing responses for the whole panel of variants.
Serum 39 was similar to serum 86, and sera 73 and 75 also neutralized rSMEZ-8–stimulated T cells. SMEZ-1 and SMEZ-5 were commonly neutralized by most of the sera tested. Only two sera (30 and 31) displayed less than complete neutralization responses. In contrast, SMEZ-2 activity was not inhibited by eight sera, and another six sera showed only moderate inhibition. SMEZ-2 was therefore the least commonly neutralized variant.
Discussion
SMEZ is the most potent bacterial SAg so far discovered, with specificity towards Vβ4- and Vβ8-bearing human T cells. Unlike all other SAgs, SMEZ is highly polymorphic, having arisen from genetic mosaicism at a single genetic locus. 22 different smez alleles were found in 21 different M-type positive GAS isolates obtained over a 15-yr period throughout New Zealand, in addition to the 2 alleles, smez and smez-2, described previously 27 28. All of the 94 isolates tested positive for the smez gene by PCR or Southern analysis. Thus, smez is not only the most potent SAg, it is also the most prevalent. The polymorphism in the SMEZ protein results in antigenic variants that are not consistently neutralized by naturally occurring antibodies present in normal adult human sera, yet the human TCR β chain profile remains steadfastly Vβ4 and Vβ8.
Genetic Mosaicism.
The variation in smez has most likely arisen through gene mosaicism, and the strongest indicator for this is that 30 out of 31 polymorphic positions have only 2 possible amino acids. The mechanism for gene mosaics is homologous recombination between two copies of a gene, and Southern blot analysis shows that some isolates indeed carry a second smez gene or smez-like sequence. However, homologous recombination of smez-1 with the smez-like sequence on the M1 genome would result in another pseudogene due to a nonsense mutation within the smez-like sequence. Moreover, >2 homologous genes would be needed to generate the mosaic pattern seen in the 24 smez alleles. The different sequence segments appear to have originated from different strains, suggesting the possibility of intergenic recombination with a horizontally transferred homologous gene from another strain. Interstrain transfer followed by homologous recombination has been reported to play a key role in the evolution of emm-like genes 30, indicating that such a mechanism exists in S. pyogenes.
Sequence combination between different alleles is not the only mechanism to create variability in the smez gene. 11 of the 31 variable residues of the SMEZ variants correlate with individual point mutations for single alleles and must have occurred independently in the respective strains. This is also remarkable, given the fact that such a high mutation rate is not observed in any other sag genes, including spe-g, which was also found in all isolates tested. It is possible that point mutations in other sag genes are eliminated by selective pressure in order to maintain functionality of the toxin. However, this seems unlikely, since the variant residues in SMEZ do not contribute to the overall structure and are not conserved within the SAg family.
Allelic variation has been described for the staphylococcal enterotoxin C (SEC). SEC1 and SEC2 differ by only seven amino acid residues, but target a different set of TCR 31. Also, SEA and SEE, which still share 85% amino acid sequence identity, vary in their TCR specificity 32.
Protein expression of a Vβ8-stimulating toxin was observed in the culture supernatant of 66% of the GAS isolates. No Vβ8 activity was found in any of the isolates that carry a defective smez gene, suggesting that smez is the only Vβ8-specific toxin in S. pyogenes. Since the smez gene has been analyzed in only a fraction of the GAS panel, it is possible that more nonfunctional smez alleles occur in the natural S. pyogenes population. Of the identified complete smez genes, 7 out of 21 did not express the SMEZ protein, suggesting a tight control of gene transcription. However, it is important not to assume that expression under in vitro conditions always equates to expression in vivo. Interestingly, the SMEZ-producing strains include M1, M4, and M12 types. These are connected to STLS 33, whereas the SMEZ-13–producing NZ1437/M89 type strain is connected to rheumatic fever in New Zealand 34. A larger panel of GAS isolates, in particular those known to cause invasive diseases, must be analyzed for SMEZ expression to identify any connection between expression of a certain SMEZ variant and streptococcal diseases.
Antigenic Variation.
There are 31 variable amino acid positions in SMEZ. Analysis of a recent SMEZ-2 crystal structure shows that 26 of these residues are surface exposed and scattered across the entire protein. Only five of the variable positions have side chains that are buried, and all are conservative changes, indicating that there has been selective pressure to vary the surface of the molecule while preserving structural integrity of the molecule 34a. Widespread prevalence of SMEZ-producing strains of S. pyogenes in the normal adult human population was confirmed by seroconversion in 100/100 normal healthy young adults. However, 15% of the sera showed only partial neutralization profiles against a panel of recombinant SMEZ variants. SMEZ-1 was the most commonly neutralized, whereas SMEZ-2 was the least. SMEZ-2 is a rare variant that has not yet been found in any S. pyogenes isolate in New Zealand. SMEZ-2 was identified in a single isolate of unknown M/emm type from a patient with rheumatic fever in the United States, and is the most potent variant known to date 27. In contrast, SMEZ-1 was found associated with the M1 strain, a very common strain in New Zealand. These data suggest that certain SMEZ variants are better capable of generating a generic protective neutralizing response against all SMEZ variants than others, and that protective antibodies offer only partial protection against a subset of SMEZ variants. This raises an interesting question of whether individual SMEZ variants are linked to disease severity. Screening patients with severe streptococcal-mediated disease for lack of seroconversion against individual SMEZ variants might provide information in this regard.
The strong linkage between the smez gene and the emm gene, which codes for the major polymorphic surface antigen M protein, offers one explanation for the mechanisms of smez genetic variation. The smez gene is located 14 kb upstream from the Vir regulon. The M protein represents the major neutralizing surface antigen on Streptococci, and generation of diversity in the M protein antigenic determinants might also affect neighboring genes. Thus far, we have analyzed only 21 different M/emm types for the smez gene. It is likely that the actual number of smez alleles among the S. pyogenes population is much higher. This might be of particular interest, as the epidemiology of streptococcal disease appears to be connected to certain M types, and temporal shifts in prevalence of certain M types occur 35.
Role of SMEZ.
SAgs interfere with host adaptive immune responses, driving profound Th1-type responses in an antigen-nonspecific way characterized by high levels of INF-γ, IL-2, and nonspecific T cell proliferation. How this response improves survival of the bacteria has yet to be clearly established, but one might argue that the prime function is to prevent the production of high affinity cytotoxic antibodies by promoting a cellular, Th1-mediated response. The high degree of antigenic variation is good evidence that SMEZ provides a significant growth advantage for S. pyogenes and that the variation at this locus has been driven primarily by the need to escape antibody neutralization, rather than through a need to expand its T cell repertoire.
Retention of the TCR Specificity.
In contrast to the antigenic diversity, the TCR Vβ binding site of SMEZ appears to be unaffected by surrounding amino acid variation. One conclusion to draw from this is that Vβ4- and Vβ8-bearing T cell subsets are uniquely important in host defence against Streptococci. However, one could also argue that retention of this T cell specificity is due to the physical constraints of the SMEZ–TCR β chain interaction, so that other TCR β chains cannot be effectively accommodated without loss of SAg activity. However, the fact that SMEZ effectively targets murine TCR β chains 27 suggests that there are no special constraints on the SMEZ–TCR binding site. Why SMEZ should be privy to such extensive variation while all other SAgs are substantially invariant is unclear, but does point to an important role of this particular SAg in the survival of all group A Streptococci. There are examples of allelic variation in other streptococcal SAgs, such as SPEA 24, SPE-C 36, and SSA 37, but to a much lesser degree and without the mosaicism seen at the smez locus. Genes with mosaic structures are known in other pathogenic organisms such as streptokinase (skn 38), the emm-like genes 30, or the Streptococcus pneumoniae autolysin gene (lytA 39), and each codes for an important virulence factor.
The antigenic variation appears to have arisen through the selective advantage that SAg escape mutants provide to the species S. pyogenes. This is perhaps the best evidence yet that the SAgs confer an advantage on the strain that produces it. Moreover, the ubiquitous expression and genetic variation tend to suggest that smez, more than other SAgs, significantly enhances the survival of group A Streptococci in general and thus might be linked to increased virulence and streptococcal disease severity. The smez locus may provide an important genotyping locus to examine virulence and disease susceptibility, and may also offer a potential target for streptococcal vaccine development.
Acknowledgments
We wish to thank Dr. Lachy McLean for supplying us with human sera.
This study was supported by the Health Research Council of New Zealand.
Footnotes
Abbreviations used in this paper: ET, electrophoresis type; GAS, group A Streptococcus; GST, glutathione S-transferase; KD, Kawasaki disease; MNT, M-nontypable; SAg, superantigen; SE, staphylococcal enterotoxin; SF, scarlet fever; SID, severe invasive disease; SMEZ, streptococcal mitogenic exotoxin; SPE, streptococcal pyrogenic exotoxin; STLS, streptococcal toxic shock-like syndrome; TSS, toxic shock syndrome.
References
- Bronze M.S., Dale J.B. The reemergence of serious group A streptococcal infections and acute rheumatic fever. Am. J. Med. Sci. 1996;311:41–54. doi: 10.1097/00000441-199601000-00008. [DOI] [PubMed] [Google Scholar]
- Demers B., Simor A.E., Vellend H., Schlievert P.M., Byrne S., Jamieson F., Walmsley S., Low D.E. Severe invasive group A streptococcal infections in Ontario, Canada1987–1991. Clin. Infect. Dis. 1993;16:792–800. doi: 10.1093/clind/16.6.792. [DOI] [PubMed] [Google Scholar]
- Hoge C.W., Schwartz B., Talkington D.F., Breiman R.F., MacNeill E.M., Englender S.J. The changing epidemiology of invasive group A streptococcal infections and the emergence of streptococcal toxic shock-like syndrome. A retrospective population-based study. JAMA (J. Am. Med. Assoc.). 1993;269:384–389. [PubMed] [Google Scholar]
- Low D.E., Schwartz B., McGeer A. The reemergence of severe group A streptococcal diseasean evolutionary perspective. In: Scheld W.M., Armston D., Hughes J.M., editors. Emerging Infections. American Society for Microbiology; Washington, D.C.: 1997. pp. 93–123. [Google Scholar]
- Holm S.E., Norrby A., Bergholm A.M., Norgren M. Aspects of pathogenesis of serious group A streptococcal infections in Sweden, 1988–1989. J. Infect. Dis. 1992;166:31–37. doi: 10.1093/infdis/166.1.31. [DOI] [PubMed] [Google Scholar]
- Bisno A.L. Group A streptococcal infections and acute rheumatic fever. N. Engl. J. Med. 1991;325:783–793. doi: 10.1056/NEJM199109123251106. [DOI] [PubMed] [Google Scholar]
- Cone L.A., Woodard D.R., Schlievert P.M., Tomory G.S. Clinical and bacteriologic observations of a toxic shock-like syndrome due to Streptococcus pyogenes . N. Engl. J. Med. 1987;317:146–149. doi: 10.1056/NEJM198707163170305. [DOI] [PubMed] [Google Scholar]
- Stevens D.L., Tanner M.H., Winship J., Swarts R., Ries K.M., Schlievert P.M., Kaplan E. Severe group A streptococcal infections associated with a toxic shock-like syndrome and scarlet fever toxin A. N. Engl. J. Med. 1989;321:1–7. doi: 10.1056/NEJM198907063210101. [DOI] [PubMed] [Google Scholar]
- Forni A.L., Kaplan E.L., Schlievert P.M., Roberts R.B. Clinical and microbiological characteristics of severe group A streptococcus infections and streptococcal toxic shock syndrome. Clin. Infect. Dis. 1995;21:333–340. doi: 10.1093/clinids/21.2.333. [DOI] [PubMed] [Google Scholar]
- Alouf J.E., Knoell H., Koehler W. The family of mitogenic, shock-inducing and superantigenic toxins from staphylococci and streptococci. In: Alouf J.E., Freer J.H., editors. The Sourcebook of Bacterial Protein Toxins. Academic Press; San Diego: 1991. pp. 367–414. [Google Scholar]
- Marrack P., Kappler J. The staphylococcal enterotoxins and their relatives. Science. 1990;248:705–711. doi: 10.1126/science.2185544. [DOI] [PubMed] [Google Scholar]
- Fast D.J., Schlievert P.M., Nelson R.D. Toxic shock syndrome-associated staphylococcal and streptococcal pyrogenic toxins are potent inducers of tumor necrosis factor production. Infect. Immun. 1989;57:291–294. doi: 10.1128/iai.57.1.291-294.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman A., Kappler J.W., Marrack P., Pullen A.M. Superantigensmechanism of T-cell stimulation and role in immune responses. Annu. Rev. Immunol. 1991;9:745–772. doi: 10.1146/annurev.iy.09.040191.003525. [DOI] [PubMed] [Google Scholar]
- Kotzin B.L., Leung D.Y., Kappler J., Marrack P. Superantigens and their potential role in human disease. Adv. Immunol. 1993;54:99–166. doi: 10.1016/s0065-2776(08)60534-9. [DOI] [PubMed] [Google Scholar]
- Abe J., Kotzin B.L., Jujo K., Melish M.E., Glode M.P., Kohsaka T., Leung D.Y. Selective expansion of T cells expressing T-cell receptor variable regions Vβ2 and Vβ8 in Kawasaki disease. Proc. Natl. Acad. Sci. USA. 1992;89:4066–4070. doi: 10.1073/pnas.89.9.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.Y., Giorno R.C., Kazemi L.V., Flynn P.A., Busse J.B. Evidence for superantigen involvement in cardiovascular injury due to Kawasaki syndrome. J. Immunol. 1995;155:5018–5021. [PubMed] [Google Scholar]
- Talkington D.F., Schwartz B., Black C.M., Todd J.K., Elliott J., Breiman R.F., Facklam R.R. Association of phenotypic and genotypic characteristics of invasive Streptococcus pyogenes isolates with clinical components of streptococcal toxic shock syndrome. Infect. Immun. 1993;61:3369–3374. doi: 10.1128/iai.61.8.3369-3374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser J.M., Hauser A.R., Kim M.H., Schlievert P.M., Nelson K., Selander R.K. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseasesclonal diversity and pyrogenic exotoxin expression. Proc. Natl. Acad. Sci. USA. 1991;88:2668–2672. doi: 10.1073/pnas.88.7.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett S.P., Stevens D.L. Streptococcal toxic shock syndromesynthesis of tumor necrosis factor and interleukin-1 by monocytes stimulated with pyrogenic exotoxin A and streptolysin O. J. Infect. Dis. 1992;165:879–885. doi: 10.1093/infdis/165.5.879. [DOI] [PubMed] [Google Scholar]
- Hauser A.R., Stevens D.L., Kaplan E.L., Schlievert P.M. Molecular analysis of pyrogenic exotoxins from Streptococcus pyogenes isolates associated with toxic shock-like syndrome. J. Clin. Microbiol. 1991;29:1562–1567. doi: 10.1128/jcm.29.8.1562-1567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.E., Ferretti J.J. Molecular epidemiologic analysis of the type A streptococcal exotoxin (erythrogenic toxin) gene (speA) in clinical Streptococcus pyogenes strains. Infect. Immun. 1989;57:3715–3719. doi: 10.1128/iai.57.12.3715-3719.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussee M.S., Liu J., Stevens D.L., Ferretti J.J. Genetic and phenotypic diversity among isolates of Streptococcus pyogenes from invasive infections. J. Infect. Dis. 1996;173:901–908. doi: 10.1093/infdis/173.4.901. [DOI] [PubMed] [Google Scholar]
- Hsueh P.R., Wu J.J., Tsai P.J., Liu J.W., Chuang Y.C., Luh K.T. Invasive group A streptococcal disease in Taiwan is not associated with the presence of streptococcal pyrogenic exotoxin genes. Clin. Infect. Dis. 1998;26:584–589. doi: 10.1086/514567. [DOI] [PubMed] [Google Scholar]
- Nelson K., Schlievert P.M., Selander R.K., Musser J.M. Characterization and clonal distribution of four alleles of the speA gene encoding pyrogenic exotoxin A (scarlet fever toxin) in Streptococcus pyogenes . J. Exp. Med. 1991;174:1271–1274. doi: 10.1084/jem.174.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser J.M., Kapur V., Kanjilal S., Shah U., Musher D.M., Barg N.L., Johnston K.H., Schlievert P.M., Henrichsen J., Gerlach D. Geographic and temporal distribution and molecular characterization of two highly pathogenic clones of Streptococcus pyogenes expressing allelic variants of pyrogenic exotoxin A (Scarlet fever toxin) J. Infect. Dis. 1993;167:337–346. doi: 10.1093/infdis/167.2.337. [DOI] [PubMed] [Google Scholar]
- Kline J.B., Collins C.M. Analysis of the superantigenic activity of mutant and allelic forms of streptococcal pyrogenic exotoxin A. Infect. Immun. 1996;64:861–869. doi: 10.1128/iai.64.3.861-869.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft T., Moffatt S.L., Berkahn C.J., Fraser J.D. Identification and characterization of novel superantigens from Streptococcus pyogenes . J. Exp. Med. 1999;189:89–102. doi: 10.1084/jem.189.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamezawa Y., Nakahara T., Nakano S., Abe Y., Nozaki-Renard J., Isono T. Streptococcal mitogenic exotoxin Z, a novel acidic superantigenic toxin produced by a T1 strain of Streptococcus pyogenes . Infect. Immun. 1997;65:3828–3833. doi: 10.1128/iai.65.9.3828-3833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Fritsch E.F., Sambrook J. Molecular CloningA Laboratory Manual Cold Spring Harbor Laboratory ColdSpring Harbor, NY1989. [Google Scholar]
- Whatmore A.M., Kehoe M.A. Horizontal gene transfer in the evolution of group A streptococcal emm-like genesgene mosaics and variation in Vir regulons. Mol. Microbiol. 1994;11:363–374. doi: 10.1111/j.1365-2958.1994.tb00316.x. [DOI] [PubMed] [Google Scholar]
- Deringer J.R., Ely R.J., Stauffacher C.V., Bohach G.A. Subtype-specific interactions of type C staphylococcal enterotoxins with the T-cell receptor. Mol. Microbiol. 1996;22:523–534. doi: 10.1046/j.1365-2958.1996.1381506.x. [DOI] [PubMed] [Google Scholar]
- Hudson K.R., Robinson H., Fraser J.D. Two adjacent residues in staphylococcal enterotoxins A and E determine T cell receptor Vβ specificity. J. Exp. Med. 1993;177:175–184. doi: 10.1084/jem.177.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski C.A., Bardsley M., Beall B., Elliott J.A., Facklam R., Schwartz B., Farley M.M. Invasive group A streptococcal disease in metropolitan Atlantaa population-based assessment. Clin. Infect. Dis. 1998;27:150–157. doi: 10.1086/514632. [DOI] [PubMed] [Google Scholar]
- Martin D.R., Voss L.M., Walker S.J., Lennon D. Acute rheumatic fever in Auckland, New Zealandspectrum of associated group A streptococci different from expected. Ped. Infect. Dis. J. 1994;13:264–269. doi: 10.1097/00006454-199404000-00004. [DOI] [PubMed] [Google Scholar]
- Arcus V., Proft T., Sigrell J.A., Baker H.M., Fraser J.D., Baker E.N. Conservation and variation in superantigen structure and activity highlighted by 3-dimensional structures of two new superantigens from Streptococcus pyogenes . J. Mol. Biol. 2000;In press doi: 10.1006/jmbi.2000.3725. [DOI] [PubMed] [Google Scholar]
- Martin D., Kakani J., Szeto J. Clonal analysis of five M types causing most disease in New Zealand. Adv. Exp. Med. Biol. 1997;418:331–333. doi: 10.1007/978-1-4899-1825-3_79. [DOI] [PubMed] [Google Scholar]
- Kapur V., Nelson K., Schlievert P.M., Selander R.K., Musser J.M. Molecular population genetic evidence of horizontal spread of two alleles of the pyrogenic exotoxin C gene (speC) among pathogenic clones of Streptococcus pyogenes . Infect. Immun. 1992;60:3513–3517. doi: 10.1128/iai.60.9.3513-3517.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reda K.B., Kapur V., Goela D., Lamphear J.G., Musser J.M., Rich R.R. Phylogenetic distribution of streptococcal superantigen SSA allelic variants provides evidence for horizontal transfer of ssa within Streptococcus pyogenes . Infect. Immun. 1996;64:1161–1165. doi: 10.1128/iai.64.4.1161-1165.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur V., Kanjilal S., Hamrick M.R., Li L.L., Whittam T.S., Sawyer S.A., Musser J.M. Molecular population genetic analysis of the streptokinase gene of Streptococcus pyogenesmosaic alleles generated by recombination. Mol. Microbiol. 1995;16:509–519. doi: 10.1111/j.1365-2958.1995.tb02415.x. [DOI] [PubMed] [Google Scholar]
- Whatmore A.M., Dowson C.G. The autolysin-encoding gene (lytA) of Streptococcus pneumoniae displays restricted allelic variation despite localized recombination events with genes of pneumococcal bacteriophage encoding cell wall lytic enzymes. Infect. Immun. 1999;67:4551–4556. doi: 10.1128/iai.67.9.4551-4556.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]