Estimates put the origin of V(D)J recombination at ∼450 million years ago (for reviews, see references 1, 2). It has been speculated that it all started with a chance occurrence, the integration of a mobile element into a gene encoding an Ig domain 3. The acquisition of the V(D)J recombination system represented a major advance in the biological arms race between vertebrates and their pathogens, as the diversity created by the necessity of piecing antigen receptor genes back together again endowed the host with a new way to counter the onslaught of constantly mutating microbes. It appears that the V(D)J recombination system has, subsequent to its installation in the genome, played an integral and demonstrable role in reshaping Ig and TCR loci. Genetic repercussions in fact may continue even today.
As recently as only a few years ago, any real information bearing on the actual genesis of the V(D)J recombination system seemed to be irretrievably lost. However, as more was learned about the molecular genetic and biochemical properties of the V(D)J recombination proteins, termed recombination activating gene (RAG)-1 and RAG-2, tantalizing suggestions of a transposon origin began to emerge. These clues included the following: first, the fact that the genes encoding RAG-1 and RAG-2, which are unrelated in sequence, are tightly linked, and as such share this property with genes that are known to undergo horizontal transfer 4. Second, the chemical mechanism of the recombination reaction, where DNA strand breakage and rejoining is accomplished through one step transesterification reactions, was like that of several well-described mobile elements 5. Finally, a surprising finding further indicated that RAG-1 and RAG-2 might have once been part of a transposon when two groups independently demonstrated that purified RAG-1 and RAG-2 proteins have a latent ability to carry out the transposition of DNA 6 7.
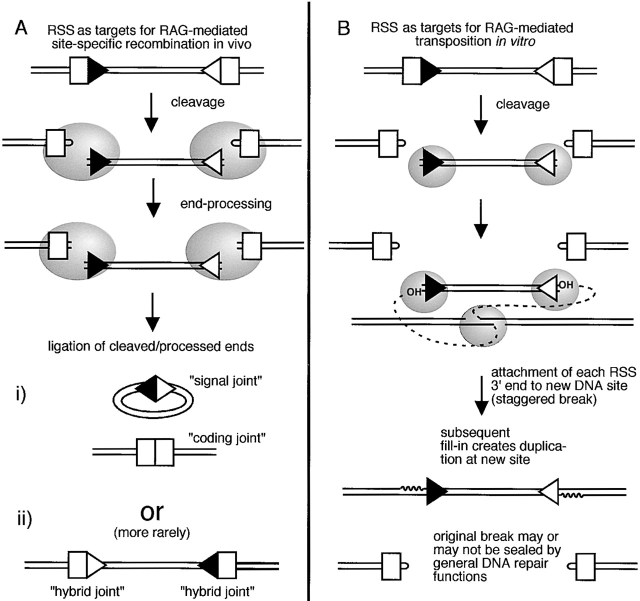
Why this latter observation was so important is that there was no a priori expectation that a protein that can perform V(D)J recombination through site-specific recognition of recombination signal sequences (RSS) should also be able to transpose RSS-terminated DNA fragments. A quick description of both types of rearrangement is needed to appreciate this point, and will also come to bear later in this commentary. As diagrammed in Fig. 1, the transpositional excision and reintegration of DNA (Fig. 1 B) is a fairly different transaction from the creation of site-specific connections in V(D)J recombination (Fig. 1 A). One difference is in the number and specificity of double strand DNA breaks; transposition not only entails the introduction of breaks at each of two RSS, as in V(D)J recombination, but also requires the generation of a third, non–sequence-specific cut at an undetermined integration site. The signature features of a transposition product versus those arising from site-specific V(D)J recombination are also quite distinct. After transposition, a transposed DNA fragment terminated by the RSS, is flanked by a five-nucleotide repeat created by a staggered cut at the target integration site (6 7; Fig. 1 B, wavy lines). In contrast, after V(D)J recombination, a signal joint and a coding joint are created. Signal joints are formed from exact RSS fusions, and coding joints from fusions of the associated coding segments. The latter characteristically contain small, irregular nucleotide deletions and insertions, reflecting various processing operations performed on the coding end intermediates as they undergo joining (Fig. 1 A). Thus, the unusual in vitro ability of RAG-1 and RAG-2 to do two quite different things, and in particular to transpose DNA, provided strong support for the original speculation that the V(D)J recombination system used to be a transposable element 3. The fact that the once portable genome now serves a different and highly utilitarian role in its new context suggests that the V(D)J recombination system stands as a prime example of the rehabilitation of “selfish DNA” (for a review, see reference 8).
Figure 1.
RAG-mediated rearrangement. (A) V(D)J recombination involves site-specific recognition and cleavage of a 12- and 23-spacer RSS (filled and open triangles). The steps that directly involve RAG-1 and RAG-2 are indicated by the gray ovals. These include strand cleavage and possibly some coding end processing. The gray ovals are not intended to specify actual protein position or size. The rejoining of the broken ends is achieved by other DNA repair functions. (i) The usual outcome is a signal joint (at most loci, this product is excised by recombination) and a coding joint. Mostly characteristic of the coding joint product, base loss and additions are also sometimes seen at signal joints. (ii) A less common outcome of V(D)J recombination is hybrid joint formation, where the RSS are exchanged between coding ends. This type of joint also exhibits base loss and addition. (B) RAG-mediated transposition involves site-specific recognition and cleavage of a 12- and 23-spacer RSS, followed by attachment of the RSS 3′ ends to a staggered break introduced at a nonspecific target site. Fill-in synthesis of the staggered break completes the formation of a 5-bp target site duplication, and this, along with the resealing of DNA ends, remaining at the exit site, presumably could be carried out by non-RAG functions. RAG-mediated transposition has not yet been observed in vivo.
Ig and TCR loci come in an extraordinary variety of shapes and sizes. The many variations boil down to differences in the arrangement and multiplicity of RSS-containing units (for reviews, see 1, 2). Because movement of RSS-containing units is emblematic of RAG-mediated events, germline RAG activation is suspected of creating at least some of the differences in antigen receptor loci.
Ultimately, each Ig or TCR locus is designed to conform to the same canon; namely, coding segments become joined by site-specific recombination of their attached RSS. The RSS are simple sequence motifs comprised of CACAGTG, a 12- or 23-bp unspecified spacer, and ACAAAAACC. Coding segments with 12-spacer RSS are joined to those with 23-spacer RSS. Beyond these common characteristics, almost anything goes, and different features include (a) the multiplicity of gene segments (one to several hundred); (b) the orientation of the gene segments (so that though joining is usually deletional, at some loci it occurs by inversion); (c) the presence or absence of D segments; (d) the type of gene segment (V, D, and J) associated with each type of RSS (12- or 23-spacer); (e) the overall arrangement of the locus (whether gene segments occur in an extended array with multiple Vs followed by Ds and/or Js followed by a single-copy C region, or as repeated units of a functional cluster comprised of a V, zero to two Ds, a J, and a C); and (f) the presence of prejoined germline genes 1 2.
The first hard evidence that germline V(D)J recombination was probably responsible for some of these differences surfaced with analyses of cartilaginous fish. In the horned shark and little skate, IgH genes occur as grouped (V-D1-D2-J and C) clusters (Fig. 2B iii), and it was observed that a significant fraction of these miniloci were partially or fully assembled (as V DD-J//C, VDJ//C, or V D1-D2J//C; 2). Unjoined (V-D-D-J//C) clusters were also present in the germline. These and other observations indicated that an RSS-directed rearrangement had altered the Ig loci in Chondrichthyes, and when first described in 1988, the most parsimonious evolutionary scheme appeared to be one in which joined clusters were derived from unjoined clusters rather than vice versa (for a review, see reference 2). At the time, there was no evidence that a joined VDJ exon might be split by integration of an RSS-terminated DNA segment; the supposition that a joined sequence might be altered through RSS integration seemed both ad hoc and unprecedented.
Figure 2.
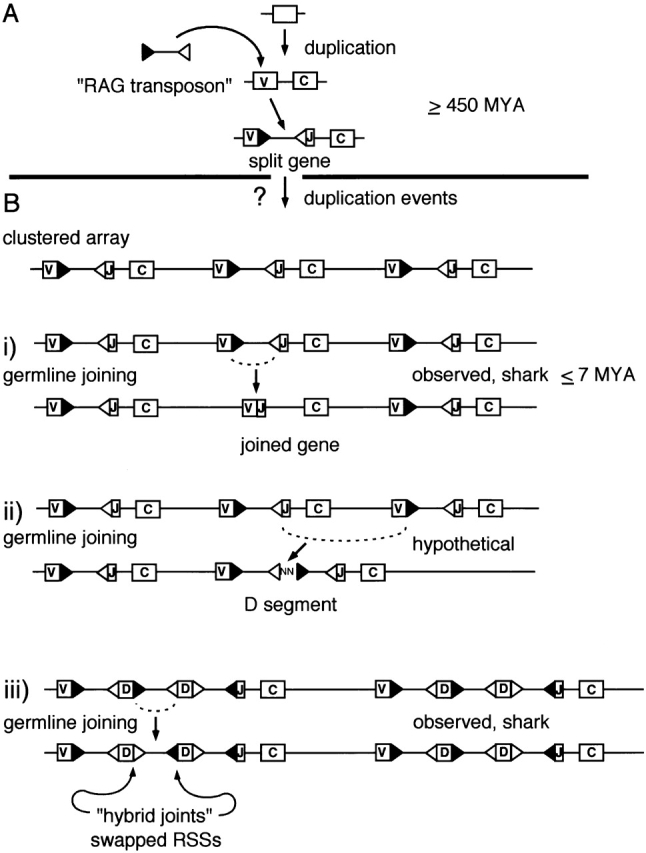
Postulated intermediates in the molecular evolution of the Ig and TCR loci. (A) One theory is that the event responsible for the existence of the modular recombination units characterizing today's Ig and TCR loci was a singularity that took place early in vertebrate evolution. With the integration of a mobile element, an ancient gene encoding an Ig superfamily domain was split into two parts. The mobile element imported RAG recombinase, its RSS, and possibly other element-related features, into the vertebrate genome. Reassembly of a functional exon required site-specific excision of the introduced mobile element, through a RAG-mediated recombination event targeting the RSS motifs. (B) After the RAG transposon integrated, the interrupted gene was duplicated. It has been suggested by Litman et al. (reference 2) that the presence of clustered arrays of duplicated genes, as found today in sharks, resembles an early locus configuration. On this view, V(D)J recombination activity in germline tissues could have led to the following derived features: (i) joined copies, as seen in cartilaginous fish, arose through a “standard” V(D)J joining event resulting in coding joint formation; (ii) the de novo creation of D segments could have arisen from intercluster recombinations resulting in the formation of signal joints with junctional insertions; (iii) the substitution of a 12-spacer RSS for a 23-spacer RSS or vice versa may have resulted from the “hybrid joint” outcome of germline joining. All of the postulated manipulations are RAG mediated and site specific. Events shown in i and iii are supported by evidence from sharks. MYA, million years ago.
One aspect of an early comparative study added further to the puzzle. As sketched in Fig. 2 B iii, two V-D-D-J-C clusters were seen to be related to one another by inversion of the RSS -terminated intervening sequence between the D1 and D2 gene segments 9. The outcome of the putative germline recombination event was not a typical coding joint (or signal joint) product. However, it is now well established that the V(D)J recombination system will in fact create a variant type of “hybrid joint” in somatic cells that exactly predicts the observed inversion (Fig. 1 A ii and Fig. 2 B iii; for a review, see reference 10). Therefore, in this one case it is almost certain that RAG-1 and RAG-2 acted as a site-specific recombinase, and not a transposase in the inversion of the interstitial region between D segments of the variant cluster. The significance of this, as has been discussed previously 11, is that it provided one simple mechanism whereby the “swapping” of 12- and 23-spacer RSS could have taken place during evolution, thus accounting for one type of organizational variation (see the list above) observed in Ig and TCR loci today.
In this issue, a study by Lee et al. 12 further investigates whether joined genes—later interrupted by a transposon insertion—or unjoined genes—subsequently connected by site-specific recombination—came first in the evolution of the Ig light chain locus in the nurse shark. There is increased interest in this issue, not only because it reveals the ways in which RAG-mediated events have reorganized Ig and TCR loci, but also because RAG-mediated transposition, though demonstrated in a purified in vitro system, has not yet been observed in any in vivo setting.
Lee et al. took advantage of the fact that certain Ig light chain genes in two shark species were apparent orthologs. Whereas all of the type III L chain genes in the horned shark were unjoined, the corresponding NS4 genes in the nurse shark occurred in both joined and unjoined form. A key feature of the analysis was that the NS4 genes in the nurse shark were highly homologous to one another, allowing evolutionary relationships to be established with some confidence. In combination, these circumstances enabled the authors to do two things: construct a phylogenetic tree of the NS4 family sequences, and provide through DNA sequence analysis a means to distinguish between the transposon integration and site-specific recombination scenarios.
As mentioned above, a sequence that has been interrupted by RAG-mediated transposition is expected to exhibit (relative to the uninterrupted form) a 5-bp target site duplication (or more rarely a 4- or 3-bp duplication; Fig. 1 B, wavy lines) on either side of an RSS-bordered insertion 6 7. A gene that has instead been joined through site-specific recombination is expected to exhibit (relative to the unjoined form) a loss of a small, unfixed number of bp from the ends of the joined coding sequences, along with the acquisition of junctional insertions (of two classes: one random in sequence, and termed an N insertion, another occurring only at ends that escaped trimming, and bearing a palindromic relationship to the cut end, termed a P insert; for a review, see reference 10). The two approaches taken by Lee et al. 12 returned the same answer: the joined NS4 genes arose through site-specific V(D)J recombination and not through germline RAG-mediated transposition. Their phylogenetic analyses indicated that germline joining occurred more than once, and in every case the unjoined form lacked any evidence of the DNA sequence duplications predicted for transposition. Instead joined genes exhibited junctions that appeared to reflect processing accompanying V(D)J recombination: trimming and P nucleotide addition.
A very intriguing aspect of the report of Lee et al. 12 is that it raises the possibility that germline RAG recombination is ongoing. The most recent of V(D)J recombination events to have taken place in the NS4 loci happened perhaps no more than 7 million years ago. The fact that no polymorphisms in terms of joined and unjoined light chain loci were discovered among the sampled nurse sharks (which were not close relatives judging from their MHC haplotypes) allowed the authors to surmise that such germline recombination events are probably infrequent. Nevertheless, site-specific V(D)J recombination did alter the nurse shark genome, and the variant, recombinant clusters have apparently become widespread in the shark population.
The possibility that germline V(D)J recombination has contributed in a positive way to the evolution of the Ig and TCR loci in many species seems quite likely. The mechanics of how RAG-mediated site-specific rearrangement might have created some of the quirks in Ig and TCR locus organization are shown in Fig. 2. In addition to the generation of joined gene clusters (Fig. 2B i), germline V(D)J recombination activity might more generally account for the generation of D segments. There is evidence that D segments are a derived feature and have arisen multiple times in evolution 2. Although one possibility is that for each instance, D segments were created by a pair of closely juxtaposed transposon insertions 6, it seems at least as likely that intercluster recombination could have generated D segments as shown in Fig. 2B ii. As mentioned above, another significant difference between loci is the variant disposition of 12- and 23-spacer RSS (Fig. 2B iii): there is a documented case of germline hybrid joint formation where V(D)J recombination apparently exchanged one type of RSS for another 9.
Obviously, if RAG-generated germline modifications are of evolutionary significance, they must be imagined to sometimes confer an advantage to the organism. Some such benefits are fairly easy to envision. The “invention” of D segments may have primarily been selected as an advantageous way to increase junctional diversity 2. RSS swapping could have aided in the evolutionary diversification of different loci 9. However, the advantages of possessing preassembled germline genes, which effectively limits diversity, is perhaps less immediately evident.
One possibility is that the germline-joined genes in various cartilaginous fish provide one way to ensure the production of antigen receptors with predetermined specificities in early fetal and/or neonatal life. In every type of animal for which the early and adult repertoires have been compared, the early repertoire is less complex. Xenopus is a classic example, where the tadpole repertoire contains only a subset of the antibodies found in the mature adult repertoire. The strict link between repertoire diversity and development was established by preventing metamorphosis. The antibody repertoire in these treated tadpoles retained tadpole characteristics in spite of their increase in size (to that of an adult frog) and their age when compared with their metamorphosed siblings 13. A second example is the mouse, where the molecular mechanism underlying the restriction is somewhat better understood. Here, the lack of terminal deoxynucleotidyl transferase activity in the fetus limits the insertion of N regions in H chain genes, with the result that a simpler set of CDR3 regions is created. Additional predetermination of the mouse fetal repertoire is achieved through preferential usage of particular VH gene segments early in mouse development.
The reason for a restricted early repertoire is thought to be to help protect against pathogens in the neonatal period. In the mouse, this possibility was tested experimentally by causing terminal deoxynucleotidyl transferase activity to be abnormally expressed during fetal development. The affected mice failed to generate certain Ig H and L chain genes and were highly susceptible to Streptococcus pneumoniae 14. In an analogous way, the presence of fused genes may represent a shark-specific solution to the same problem. As Lee et al. report, the prejoined Igλ genes are expressed in pups, but not in adult sharks 12.
A final, more overarching benefit to germline V(D)J recombination is that of evolutionary experimentation. As suggested by Lee et al., perhaps this activity has allowed for the generation of Ig-like genes encoding proteins with novel functions 12. One specific possibility is that VpreB, which is linked to the λ locus in mice 15, might represent one such example. The interesting possibility of the relationship between VpreB and split Ig genes has also been presented in reverse, that VpreB is an uninterrupted descendent from the original Ig domain 2. Whichever is correct (and, optimistically, we may one day be able to distinguish between these possibilities), the fact remains that RAG-mediated germline remodeling has clearly been an innovative evolutionary force.
It remains to be pointed out that some of the most extensive changes that have occurred during the evolution of Ig and TCR loci cannot be explained by some combination of the operations shown in Fig. 2. The conversion of “cluster” type loci to the “extended” form is not easily understood, nor is the huge variation in multiplicity of gene segments at different loci, or the manner in which, for some loci, gene segments were first flipped into an opposite transcriptional orientation. While some or all of these modifications may be due to more conventional types of rearrangement, and need not be at all related to any RAG-mediated event, there is still the possibility that they are manifestations of a transpositional type of RAG function. To date, the relationship between V(D)J recombination and a mobile element has been discussed largely in terms of familiar retroviruses and of transposons such as Mu and Tn10 5 6 7 16, but another chapter to the story will unfold when/if we can discover more about the exact sort of mobile element the “RAG transposon” actually was. Perhaps it was designed to mobilize or rapidly generate flocks of genes rather than singular units, or perhaps at some point in its evolution, the RAG transposon was a conglomerate generated in a pile-up of more than one type of mobile element (e.g., reference 17). Meanwhile, sharks, as animals in which such RAG-mediated experimentation is still apparently ongoing, are likely to prove a particularly fruitful system in which to further explore such questions.
References
- Marchalonis J.J., Schluter S.F., Bernstein R.M., Shen S., Edmundson A.B. Phylogenetic emergence and molecular evolution of the immunoglobulin family. Adv. Immunol. 1998;70:417–506. doi: 10.1016/s0065-2776(08)60392-2. [DOI] [PubMed] [Google Scholar]
- Litman G.W., Anderson M.K., Rast J.P. Evolution of antigen binding receptors. Annu. Rev. Immunol. 1999;17:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- Sakano H., Huppi K., Heinrich G., Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- Oettinger M.A., Schatz D.G., Gorka C., Baltimore D. Rag-1 and Rag-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., Mizuuchi D., Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- Agrawal A., Eastman Q.M., Schatz D.G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- Hiom K., Melek M., Gellert M. DNA transposition by the RAG1 and RAG2 proteinsa possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- Britten R.J. Mobile elements inserted in the distant past have taken on important functions. Gene. 1997;205:177–182. doi: 10.1016/s0378-1119(97)00399-5. [DOI] [PubMed] [Google Scholar]
- Kokubu F., Litman R., Shamblott M.J., Hinds K., Litman G.W. Diverse organization of immunoglobulin VH gene loci in a primitive vertebrate. EMBO (Eur. Mol. Biol. Organ.) J. 1988;11:3413–3422. doi: 10.1002/j.1460-2075.1988.tb03215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.M. The mechanism of V(D)J joininglessons from molecular, immunological, and comparative analyses. Adv. Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- Lewis S., Hesse J.E., Mizuuchi K., Gellert M. Novel strand exchanges in V(D)J recombination. Cell. 1988;55:1099–1107. doi: 10.1016/0092-8674(88)90254-1. [DOI] [PubMed] [Google Scholar]
- Lee S.S., Fitch D., Flajnik M.F., Hsu E. Rearrangement of immunoglobulin genes in shark germ cells. J. Exp. Med. 2000;191:1637–1647. doi: 10.1084/jem.191.10.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu E., Du Pasquier L. Changes in the amphibian antibody repertoire are correlated with metamorphosis and not with age or size. Dev. Immunol. 1992;2:1–6. doi: 10.1155/1992/28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict C.L., Kearney J.F. Increased junctional diversity in fetal B cells results in a loss of protective anti-phosphorylcholine antibodies in adult mice. Immunity. 1999;10:607–617. doi: 10.1016/s1074-7613(00)80060-6. [DOI] [PubMed] [Google Scholar]
- Carson S., Wu G.E. A linkage map of the mouse immunoglobulin lambda light chain locus. Immunogenetics. 1989;29:173–179. doi: 10.1007/BF00373642. [DOI] [PubMed] [Google Scholar]
- Santagata S., Besmer E., Villa A., Bozzi F., Allingham J., Sobacchi C., Haniford D.B., Vezzoni P., Nussenzweig M.C., Pan Z.-Q. The RAG1/RAG2 complex constitutes a 3′ flap endonucleaseimplications for junctional diversity in V(D)J and transpositional recombination. Mol. Cell. 1999;4:935–947. doi: 10.1016/s1097-2765(00)80223-3. [DOI] [PubMed] [Google Scholar]
- Nakajima Y., Hashido K., Tsuchida K., Takada N., Shiino T., Maekawa H. A novel tripartite structure comprising a mariner-like element and two additional retrotransposons found in the Bombyx mori genome. J. Mol. Evol. 1999;48:577–585. doi: 10.1007/pl00006500. [DOI] [PubMed] [Google Scholar]