Abstract
Thymic dendritic cells (DCs) form a discrete subset of bone marrow (BM)-derived cells, the function of which is to mediate negative selection of autoreactive thymocytes. The developmental origin of thymic DCs remains controversial. Although cell transfer studies support a model in which T cells and thymic DCs develop from the same intrathymic pluripotential precursor, it remains possible that these two types of cells develop from independent intrathymic precursors. Notch proteins are cell surface receptors involved in the regulation of cell fate specification. We have recently reported that T cell development in inducible Notch1-deficient mice is severely impaired at an early stage, before the expression of T cell lineage markers. To investigate whether development of thymic DCs also depends on Notch1, we have constructed mixed BM chimeric mice. We report here that thymic DC development from Notch1−/− BM precursors is absolutely normal (in terms of absolute number and phenotype) in this competitive situation, despite the absence of Notch1−/− T cells. Furthermore, we find that peripheral DCs and Langerhans cells are also not affected by Notch1 deficiency. Our results demonstrate that the development of DCs is totally independent of Notch1 function, and strongly suggest a dissociation between intrathymic T cell and DC precursors.
Keywords: Notch1, dendritic cell, cell fate, T cell, development
Introduction
Dendritic cells (DCs) play a key role in T cell immune responses by endocytosing, processing, and presenting foreign antigens to specific T cells 1 2. All DCs originate from bone marrow (BM) precursors, but it is not clear whether these precursors differentiate and mature to DCs in the BM itself, or whether they migrate to and differentiate within other organs.
Different lines of evidence point to the existence of two types of DCs in the mouse. Although all DCs express CD11c and MHC class II, the two subsets can be distinguished according to their differential expression of cell surface markers such as CD8α, Mac-1, and DEC-205. The CD8α+Mac-1−DEC-205+ subset of DCs is thought to be of lymphoid origin, whereas the CD8α−Mac-1+DEC-205− DCs are believed to be myeloid derived. In the spleen, where the function of DCs is to present foreign antigens to specific T cells, both subpopulations of DCs are present. However, thymic DCs whose function is to mediate negative selection of autoreactive thymocytes 3 constitute only the lymphoid-related subset 4 5 6.
The development of thymic DCs and T cells has been reported to be closely linked via an intrathymic common precursor 6 7. In support of this hypothesis, intrathymic transfer of either early T cell precursors (CD4loCD44+ CD25−CD117+) or progenitor T cells (CD4−CD44+ CD25+CD117+) into sublethally irradiated mice results in the production of both thymic DCs and mature T cells. However, the next downstream population (defined as CD4−, CD44−CD25+, CD117−) has lost DC potential and generates only T cells 8. These data are consistent with a lymphoid-related origin for thymic DCs, and support the existence of a common intrathymic T/DC precursor 7 8. Nevertheless, this hypothesis has not been proven at the clonal level, and it remains possible that the populations transferred contain distinct DC and T cell precursors that cannot be distinguished phenotypically.
Further evidence that DC and T cell development might be closely linked is derived from gene-targeted mice. Mice homozygous for an Ikaros dominant-negative mutation (deletion of the DNA-binding domain) lack all cells of lymphoid origin, such as T, B, and NK cells, as well as lymphoid- and myeloid-derived DCs 9. More interestingly, mice homozygous for an Ikaros null allele (deletion of the COOH terminus of Ikaros) lack B and NK cells, as well as lymph nodes, but maintain a certain T cell differentiation potential 10. Although no myeloid DCs were found in such mice, lymphoid-related DCs were present in the thymus, suggesting a correlation between T cell and thymic DC development 11.
Notch gene family members have been shown to play crucial roles in binary cell fate decisions in many developmental systems 12. Notch proteins are conserved transmembrane receptors containing EGF repeats in their ectodomain that are implicated in ligand binding. The cytoplasmic domain harbors six ankyrin repeats and is involved in intracellular signaling 13. To date, four mammalian Notch homologues (Notch1–4) that interact with transmembrane-bound ligands such as Jagged1, Jagged2, Delta1, and Delta-like3 have been identified 14 15 16 17 18 19 20 21 22 23 24.
Several reports suggest a role for Notch family members in T cell development. Notch1, 2, and 3 as well as the ligands Jagged1 and 2 have been shown to be expressed in thymocytes and thymic stromal epithelium 25 26 27. Expression of transdominant active forms of Notch genes in transgenic mice has implicated Notch signaling in T cell development at the level of both CD4 versus CD8 and α/β versus γ/δ cell fate decisions 28 29. Notch signaling may also regulate proliferation, survival, and/or apoptosis of developing T cells 30 31.
We recently reported the generation of mice in which the Notch1 gene was conditionally inactivated. Lethally irradiated recipients reconstituted with BM cells derived from induced Notch1−/− mice were devoid of T cells in both the thymus and periphery. Instead of T cells, immature B cells accumulated in the thymus of such BM chimeras, suggesting a possible role for Notch1 in regulating the T versus B lineage decision 32. In other studies, transfer of retrovirally infected BM-derived hematopoietic precursor cells expressing the cytoplasmic domain of Notch1 has recently been shown to impair early B cell development and promote ectopic T cell development in the BM 33. Taken together, these complementary reports support a role for Notch1 in regulating T versus B cell fate specification from a bipotent common lymphoid progenitor (CLP).
The putative ability of Notch1 to regulate the B/T lineage decision in a binary fashion is of considerable interest in relation to the developmental origin of thymic DCs. It is generally accepted that progenitor B cells differentiate from a CLP in the BM, whereas CLPs that reach the thymus can adopt a T cell fate under the influence of the thymic microenvironment 34. Since thymic DCs are believed to be derived from an intrathymic common T/DC precursor that is downstream of the T/B lineage choice 7 35, it is of obvious interest to investigate whether thymic DCs can arise from Notch1-deficient BM precursors.
Here, we show that although induced Notch1−/− hematopoietic precursors are unable to generate T cells, they retain their full potential to generate all other myeloid and lymphoid lineages, including thymic DCs.
Materials and Methods
Generation of Mice with a loxP-flanked Notch1 Allele, and Activation of the Cre Recombinase.
Notch1lox/lox and Notch1lox/lox MxCre mice were generated as described previously 32. These mice are positive for the common leukocyte antigen CD45.2. Activation of the Cre recombinase was performed as described previously 32. In brief, adult mice received five intraperitoneal injections of 250 μg polyI-polyC (Sigma Chemical Co.) at 2-d intervals. 2 d after the last injection, mice were killed, and the BM was prepared for BM transplantation by T cell depletion. Genomic DNA was prepared from a portion of the T cell–depleted BM cells for assessing Notch1 deletion efficiency by Southern blot, and was quantified using a PhosphorImager® (Molecular Dynamics).
Generation of Mixed BM Chimeras.
Mixed chimeras were generated using a 1:2 mixture (10 × 106 and 20 × 106) of T cell–depleted BM cells from polyI-polyC–treated CD45.1+ wild-type (wt) and CD45.2+ Notch1lox/lox or induced Notch1−/− mice. 10-wk-old hosts (C57BL/6, CD45.1+; purchased from The Jackson Laboratory) were given 1,000 rads γ-irradiation 24 h before receiving the BM transplant. Radiation chimeras were maintained on antibiotic water and analyzed after 3 mo.
Flow Cytometry and Antibodies.
Four-color FACS® staining was performed as described elsewhere 36 using the following mAbs: anti-CD45.1–FITC or –biotin, anti-CD45.2–FITC or –biotin, anti-CD11c–FITC or –PE, anti-CD3ε–FITC or –allophycocyanin (APC), anti-Gr1–APC, anti-CD11b–PE, anti-CD44–CyChrome, anti-CD8α–CyChrome or –APC, anti-CD4–CyChrome, anti-B220–CyChrome or –APC, and anti-NK1.1–PE (PharMingen). Anti-F4/80–biotin was purified and conjugated in this laboratory. Biotinylated antibodies were revealed with either Streptavidin–CyChrome (PharMingen) or Streptavidin–APC (Molecular Probes). All FACS® analysis was performed using four colors on a FACSCalibur™ flow cytometer (Becton Dickinson), and was analyzed using CELLQuest™ software (Becton Dickinson). Dead cells were excluded by live gating of forward scatter and side scatter, and 100,000–200,000 cells were analyzed in each file.
Isolation of DCs from the Thymus and Spleen.
DC-enriched cell suspensions were prepared as described previously 37. In brief, organs were cut into small pieces and digested with collagenase A (0.5 mg/ml Boehringer Mannheim) and DNase I (40 μg/ml; Boehringer Mannheim) in RPMI 1640 medium supplemented with 5% FCS for 10 min at 37°C with continuous agitation. Digested fragments were filtered through a sieve, and the cell suspension was washed twice in PBS supplemented with 5% FCS and 5 mmol/liter EDTA containing 5 μg/ml DNase I. The cells were then resuspended in cold isoosmotic Optiprep™ solution (Nyegaard Diagnostics), pH 7.2, d 1.061 g/cm3, containing 5 mmol/liter EDTA to dissociate DC–thymocyte complexes, and the low density fraction was obtained by centrifugation at 1,700 g for 10 min. This low density fraction was washed twice in PBS-EDTA-FCS. DCs were identified in the low density cell fraction as MHC class II+CD11c+B220−F4/80− cells.
Isolation of Langerhans Cells from the Skin.
Langerhans cell (LC) preparation was performed as described previously 37 38. In brief, ears were rinsed with 70% ethanol, split using forceps into dorsal and ventral halves, and incubated with 0.5% trypsin (Sigma Chemical Co.) in PBS containing 5% FCS for 30 min at 37°C to allow the separation of the epidermal sheets from dermis. Subsequently, epidermal sheets were cultured for 24 h in 24-well tissue culture plates in the presence of 100 ng/ml GM-CSF (provided by Immunex Corp., Seattle, WA). LCs, together with keratinocytes, were released into the culture medium. Epidermal cell suspensions were obtained by filtering the culture medium and epidermal sheets through a sieve, and were washed in PBS with 5% FCS. LCs cells were identified in the epidermal cell suspension as MHC class II+ and CD11c+ cells.
Analysis of DCs and Other Hematopoietic Lineages in the Mixed BM Chimeras.
Two groups of mixed BM chimeras were constructed, CD45.1+ wt/CD45.2+ Notch1lox/lox (n = 8) and CD45.1+ wt/CD45.2+ induced Notch1−/− (n = 15). Cells bearing DC markers were analyzed on DC-enriched preparations from pooled thymi and spleens derived from the BM chimeras. Analyses of other hematopoietic lineages such as granulocytes, macrophages, B cells, and NK cells were performed on BM and spleens from individual mice.
Results
Experimental Strategy.
As the block in thymocyte development after induced inactivation of Notch1 is at a very early stage 32, Notch1 may also be involved in thymic DC differentiation, affecting a putative intrathymic common T/DC precursor. Since the deletion efficiency of Notch1 (mediated by the IFN-α–inducible MxCre transgene) in the thymus is only ∼40%, but is close to 100% in the BM, we decided to investigate this question using BM chimeras. To this end, CD45.2+ Notch1lox/lox MxCre+/− (Notch1−/−), CD45.2+ Notch1lox/lox (control), and CD45.1+ wt mice were treated with the IFN-α inducer polyI-polyC five times at 2-d intervals to delete an essential portion of the Notch1 gene ( Fig. 1 A). 2 d after the last injection, BM was harvested and prepared for transfer. To assess the Notch1 deletion efficiency, genomic DNA was prepared from a sample of induced Notch1−/− or control BM and analyzed by Southern blot. The deletion efficiency in Notch1−/− BM was close to 100%, as expected ( Fig. 1 B). Lethally irradiated wt hosts (CD45.1+) were reconstituted with either CD45.2+ Notch1−/− BM or CD45.2+ control BM, each mixed in a 2:1 ratio with CD45.1+ wt BM (hereafter described as Notch1−/− or control chimeras, respectively), and analyzed 3 mo later for the different myeloid and lymphoid lineages derived from each donor population.
Figure 1.
Inducible targeting of the Notch1 gene. (A) Schematic representation of the murine Notch1 gene. The Notch1 protein contains a signal peptide, 36 EGF repeats followed by a cysteine-rich domain (LN) in the extracellular domain, a transmembrane domain (TM), cytoplasmic ankyrin repeats (Cdc10), and a PEST sequence. The genomic organization of the Notch1 locus derived from control mice is partially shown 1; the exon coding for the leader peptide (filled square) is flanked by two loxP sequences (gray triangles), followed by the exon coding for the first EGF repeat (gray box). After induction of the Cre recombinase, the genomic portion harboring the exon coding for the leader peptide is deleted 2. Arrows indicate EcoRI fragments that differ in size between the genomic locus derived from control mice and the locus after deleting the loxP-flanked gene segment (induced Notch1−/−). (B) Southern blot analysis of EcoRI-digested genomic DNA derived from the BM used for setting up the mixed chimeric mice. The probe indicated in A reveals a 5.8-kb fragment for the allele derived from control animals 1, whereas deletion of the flanked loxP segment gives rise to a 2.3-kb fragment 2. Deletion efficiency was calculated after PhosphorImager® analysis to be >98%.
Notch1−/− BM Precursors Fail to Generate Thymic or Peripheral T Cells.
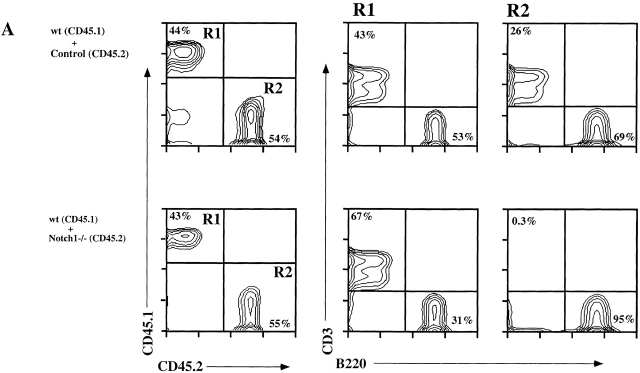
The distribution of the two allelic markers CD45.1 and CD45.2, which denote the donor origin of the cells, showed an equivalent contribution of leukocytes from each donor population in the blood of both control and Notch1-deleted chimeras ( Fig. 2 A). In the control chimeras, B220+ (B) and CD3+ (T) cells derived from both CD45.1+ and CD45.2+ BM were present, as expected. However, in the Notch1-deleted chimeras, although B220+ B cells were derived from both donor populations, >99% of CD3+ T cells were derived from the CD45.1+ wt BM, and <0.3% from the CD45.2+ Notch1-deficient BM ( Fig. 2 A).
Figure 2.
Induced Notch1−/− BM is unable to generate T cells in mixed BM chimeras. Two groups of mixed chimeric mice (wt control and wt induced Notch1−/−) were analyzed 3 mo after reconstitution with a 1:2 mixture of wt (CD45.1) and control (Notch1lox/lox, CD45.2) or induced Notch1−/− (CD45.2) BM-derived cells. Representative data are shown from each group. (A) FACS® profiles of PBLs stained with anti-CD45.1, anti-CD45.2, anti-CD3, and anti-B220 antibodies. On the left is shown the relative contribution of the competing CD45.1 (R1) versus CD45.2 (R2) BM-derived populations. In the middle and on the right are shown the distribution of CD3+ and B220+ cells within the gated CD45.1+ and CD45.2+ cell populations. (B) FACS® profiles of thymocytes stained with anti-CD45.1, anti-CD45.2, anti-CD44, and anti-B220 antibodies. On the left is shown the relative contribution of the competing CD45.1 versus CD45.2 BM-derived populations. Absolute cell numbers (×106) for thymocyte subsets gated by CD4 and CD8 expression into CD4 single positive (SP; CD4+CD8−), CD8 single positive (CD4−CD8+), double positive (DP; CD4+CD8+), and double negative (DN; CD4− CD8−) are shown in the bar diagram. The bars represent average values of pooled thymi derived from eight BM chimeras from each group. In the bar diagram, gray bars correspond to wt (CD45.1), and stippled bars correspond to control (Notch1lox/lox, CD45.2) or induced Notch1−/− (CD45.2) derived cell populations. On the right is shown the distribution of CD44+ and B220+ in the double-negative (CD4−CD8−) compartment of mixed BM chimeras constructed with wt (CD45.1) and induced Notch1−/− (CD45.2) BM cells.
In the thymus of control chimeras, reconstitution was similar to that observed in blood and other peripheral organs, with an equal contribution from both CD45.1+ and CD45.2+ BM donor populations. In contrast, in the Notch1-deleted chimeric thymus, 98% of cells were derived from CD45.1+ wt BM ( Fig. 2 B), in spite of the fact that absolute thymocyte numbers were similar in both control and Notch1-deleted chimeras (∼90 × 106). Furthermore, most of the residual CD45.2+ cells of Notch1−/− donor origin were CD4−CD8−B220+ B cells ( Fig. 2 B), confirming that Notch1-deficient precursors are unable to generate T cells in a competitive situation 32.
Notch1−/− BM Precursors Have the Full Potential for Generating Thymic DCs.
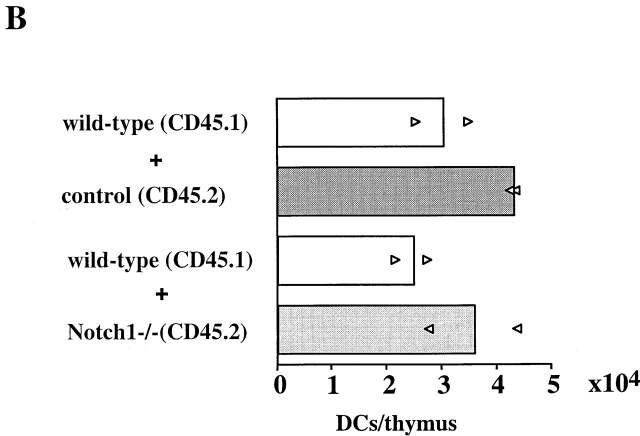
To determine whether induced Notch1−/− BM precursors are able to generate thymic DCs, the low density thymocyte fraction was prepared from control and Notch1−/− chimeras, and was stained with a combination of cell surface markers specific for DCs. As expected, a characteristic thymic DC population with the phenotype MHC class II+CD11c+CD8α+ derived from both CD45.1+ and CD45.2+ BM origin was detected in control chimeras ( Fig. 3 A). Surprisingly, in the competitive situation where T cells could not be generated, a phenotypically identical population of CD45.2+ DCs of Notch1−/− donor origin was detected in addition to the wt CD45.1+ DCs ( Fig. 3 A). Even the absolute numbers of thymic DCs derived from CD45.2+ control or CD45.2+ Notch1−/− BM were similar compared with DCs derived from wt BM ( Fig. 3 B). This result clearly demonstrates that thymic DC development is Notch1 independent, and furthermore, strongly suggests a developmental dissociation between thymic DC and T cell precursors.
Figure 3.
Induced Notch1−/− BM generates thymic DCs in mixed BM chimeras. (A) Mixed chimeric mice were analyzed 3 mo after reconstitution with a 1:2 mixture of wt (CD45.1) and control (Notch1lox/lox, CD45.2) or induced Notch1−/− (CD45.2) BM-derived populations. Thymi of these chimeric mice were pooled, and the DC fraction was enriched as described in Materials and Methods. The DC-enriched fraction was stained using anti-F4/80, anti-CD11c, anti–MHC class II, and anti-CD8α antibodies. The graph shows a FACS® analysis of CD11c versus MHC class II gated on F4/80− cells, as well as histograms for the expression of CD8α gated on CD11c+MHC class II+ cells. (B) Absolute cell numbers of thymic DCs derived from control and Notch1−/− chimeric mice. The bars represent average values per thymus of two independent experiments, and the triangles represent the average number of DCs per thymus from pooled thymi. n = 4 for wt (CD45.1) plus control (CD45.2), and n = 8 and 7 for wt (CD45.1) plus Notch1−/− (CD45.2) chimeras.
Notch1−/− BM Precursors Are Capable of Generating Peripheral DCs and LCs.
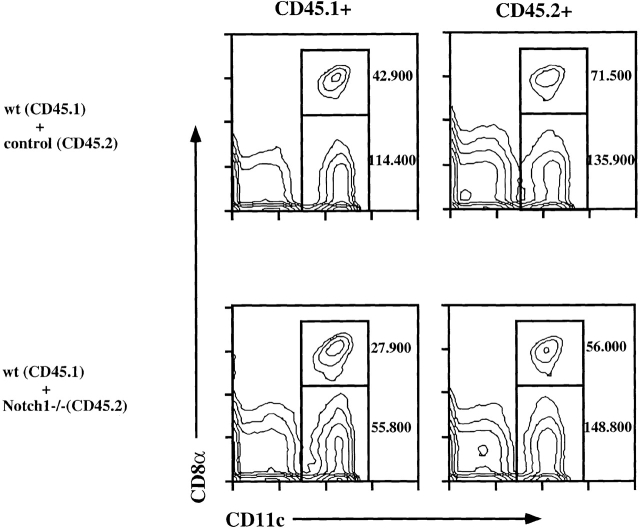
To investigate whether Notch1 deficiency could also affect the generation of myeloid-derived DCs, we analyzed splenic DCs in the same chimeric mice. As shown in Fig. 4, both lymphoid-related DCs (characterized by the expression of CD8α) and myeloid-related DCs (which are CD8α−) of both CD45.1+ and CD45.2+ origin were present in similar numbers in the spleens of control and Notch1−/− BM chimeras. This result indicates that Notch1 deficiency does not affect the generation of either myeloid- or lymphoid-related splenic DCs.
Figure 4.
Induced Notch1−/− BM generates splenic DCs in mixed BM chimeras. The BM chimeric mice described in Fig. 3 were also analyzed for splenic DC development. Spleens of chimeric mice were pooled, and the DC fraction was enriched as described in Materials and Methods. The DC-enriched fraction was stained using either anti-CD45.1 or anti-CD45.2, together with anti-F4/80, anti-CD11c, and anti-CD8α antibodies. The graph shows a FACS® analysis of CD11c versus CD8α gated on F4/80− cells for the two different groups of chimeric mice. Absolute numbers of the myeloid-related (CD11c+CD8α−) and lymphoid-related (CD11c+CD8α+) DCs per spleen are indicated next to the quadrants.
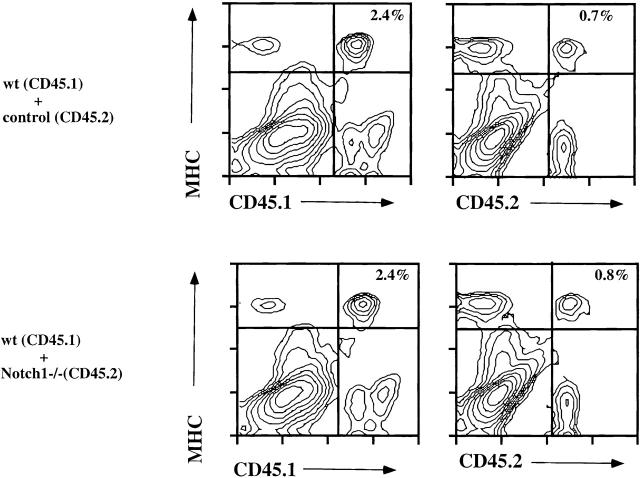
Skin DCs, known as LCs, are immature DCs that differentiate into mature DCs after antigenic stimulation and migrate to the T cell areas of the draining lymph nodes 1. To investigate if the generation of LCs was affected by the loss of Notch1, LCs were isolated from the epidermis of mouse ears from both control and Notch1−/− chimeric mice. As shown in Fig. 5, MHC class II–expressing LCs derived from Notch1−/− BM were present to a similar extent as LCs derived from control BM. Therefore, development of this form of immature DC likewise appears to be unaffected by the absence of Notch1.
Figure 5.
The generation of LCs is Notch1 independent. The same chimeric mice as described in the legend to Fig. 4 were analyzed for the presence of LCs, which were prepared from epidermal sheets of mouse ears as described in Materials and Methods. The cell populations were stained for the expression of CD45.1, CD45.2, and MHC class II. Percentages of LCs are indicated in the upper right quadrant.
Notch1 Deficiency Does Not Influence Granulocyte, Macrophage, NK Cell, or B Cell Development.
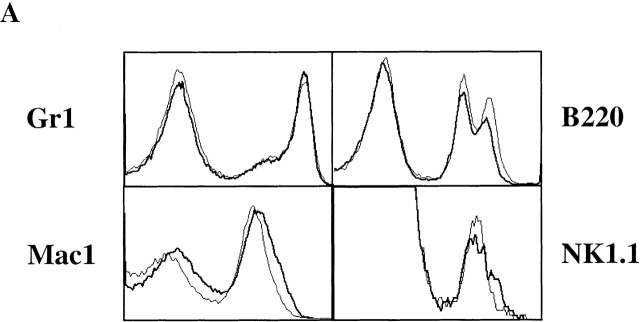
To investigate whether the absence of Notch1 affects other myeloid or lymphoid lineages, control or Notch1-deficient chimeric BM was analyzed for the presence of granulocytes, macrophages, and immature B cells by staining for the expression of Gr1, Mac-1, or B220, together with the corresponding allelic markers CD45.1 and CD45.2. As shown in Fig. 6 A, the staining patterns for Gr1, Mac-1, and B220 were indistinguishable between CD45.2+ BM cells of control or Notch1−/− donor origin. Similar results were obtained when the spleens of the chimeras were analyzed for the presence of NK cells using the marker NK1.1 ( Fig. 6 A). Importantly, calculation of absolute cell numbers in individual chimeric mice did not reveal any significant differences ( Fig. 6 B), indicating that Notch1 is dispensable for the development of granulocytes, macrophages, B cells, and NK cells.
Figure 6.
The development of granulocytes, macrophages, B cells, and NK cells is Notch1 independent. (A) Chimeric BM was analyzed for the presence of granulocytes, macrophages, and B cells by staining with mAbs specific for Gr1, Mac-1, and B220. Chimeric spleens were analyzed for the presence of NK cells by staining NK1.1 gated on CD3− cells. The histograms show the expression pattern of these markers in the CD45.2+ cells derived from either control (thin line) or induced Notch1−/− (bold line) BM cells. (B) Absolute cell numbers (×106) for Mac-1+, Gr1+, B220+, and NK1.1+(CD3−) cells in the CD45.2+ fraction derived from either control or induced Notch1−/− BM cells are calculated and shown as bar diagrams. The bars represent average values, and the triangles represent values from individual mice (n = 8 for both control and induced Notch1−/− chimeric mice).
Discussion
The data presented in this report demonstrate that Notch1 deficiency dissociates the development of T cells from all other known myeloid and lymphoid lineages. Thus, although T cell development from Notch1−/− precursors is arrested at a very early stage, the development of B cells, NK cells, macrophages, granulocytes, and DCs (including both thymic and splenic DCs, as well as LCs) is unaffected by Notch1 deficiency, even in a situation of competitive repopulation in mixed BM chimeras. Collectively, these results reveal an obligatory and selective role of Notch1 in T cell fate determination.
Implications for Myeloid Differentiation.
The finding that Notch1 is expressed on BM CD34+ progenitors 39, together with the observation that Notch ligands are expressed on BM stromal cells, initially led to the suggestion of a role for the Notch pathway in myeloid and/or erythroid differentiation 40 41. Further evidence for Notch function in myelopoiesis comes from studies using 32D cells, which are progenitor cells that can differentiate into granulocytes in the presence of a cytokine cocktail. Expression of a dominant active form of Notch or activation of full-length Notch1 by Jagged1 (a ligand for the Notch receptors) was found to inhibit differentiation of 32D cells in response to the cytokine cocktail 40 42. Our data indicating that Notch1 is dispensable for the development of DCs, granulocytes, and macrophages in vivo appear to conflict with previous results suggesting that constitutive Notch1 activity perturbs myeloid differentiation 40 43. This discrepancy may be due either to the experimental protocol used for studying myeloid differentiation, or simply to the fact that we cannot exclude the possibility that redundant signaling from other Notch gene family members may rescue the development of myeloid lineages. Nevertheless, our results are in agreement with a recent report in which retroviral infection of BM cells with a dominant active form of Notch1 did not affect myeloid differentiation, although it had profound effects on lymphoid differentiation 33.
Implications for B/T Cell Fate Determination.
Two independent lines of evidence implicate Notch1 in a binary lineage decision of a CLP to develop into a B cell or a T cell. First, as shown here and elsewhere 32, Notch1−/− BM precursors are unable to develop into T cells in the thymus of lethally irradiated wt recipients. However, instead of T cells, immature B cells of Notch1−/− origin (with a phenotype indistinguishable from those normally found in BM) accumulate in the irradiated wt thymus. In reciprocal experiments, Pui et al. 33 have shown that transferred BM cells overexpressing the constitutively active Notch1 intracellular domain do not develop into B cells in the BM of lethally irradiated recipients. Instead, immature T cells (mainly of the CD4+CD8+ phenotype) accumulate in the BM of these chimeric mice.
Taken together, these complementary “loss-of-function” and “gain-of-function” studies clearly indicate that varying levels of Notch1 expression not only control the development of T and B cells, but in addition are capable of promoting ectopic development of either lineage at the expense of the other. By analogy with the role of Notch genes in invertebrate systems 12, the simplest interpretation of these data would be that Notch1 provides a critical signal that determines a binary (T or B) cell fate decision by a CLP. Since the absence of Notch1 blocks T cell development and promotes ectopic B cell development, whereas Notch1 overexpression promotes ectopic T cell development at the expense of B cells, we would hypothesize that B cell development from the CLP represents the default pathway or primary cell fate that occurs in the absence of Notch1 signaling. According to this scenario, T cell development would be the secondary fate of a CLP, and would require Notch1 signaling.
Implications for the Existence of a Common Intrathymic T/DC Precursor.
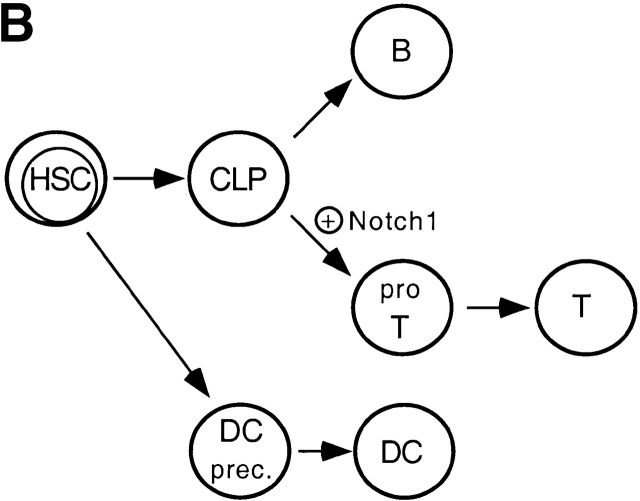
Two types of experiments have led to the widely held hypothesis ( Fig. 7 A) that thymic DCs are a lymphoid lineage derived from a common intrathymic precursor that also gives rise to T cells: (a) intrathymic transfer of sorted populations of CD44+CD4lo precursors or CD44+CD25+ progenitor T cells gives rise to both thymic DCs and T cells in sublethally irradiated recipients 7 8; and (b) the development of thymic DCs and T cells could not be clearly dissociated in gene-targeted mice deficient for either Ikaros or RelB transcription factors 10 11 44. However, both of these lines of evidence are ultimately based on correlations, and it remains possible that thymic DCs and T cells are derived from independent precursors that cannot be phenotypically distinguished.
Figure 7.
Two models for the development of thymic DCs from hematopoietic stem cells (HSC). In model A, a CLP first undergoes a B/T lineage choice. Committed T cell precursors (pro T) subsequently choose between a T and DC cell fate (for details, see references 6 and 35). In contrast, model B proposes that CLPs are exclusively directed towards a B or T cell fate depending on the absence or presence of Notch1 signaling, respectively. In this scenario, thymic DCs are derived from a precursor distinct from CLPs (see text for details).
The clear-cut dissociation of intrathymic T cell and DC development in chimeric mice reconstituted with Notch1−/− BM cells provides a serious challenge to the hypothesis that thymic DCs represent a lymphoid lineage derived from a common T/DC precursor. Indeed, as shown here and elsewhere 32, T cell development from Notch1−/− BM precursors is blocked at a very early stage (before expression of CD25), and preliminary results suggest that the few remaining CD44+ intrathymic cells of Notch1−/− origin do not express typical markers of CLPs such as Sca-1 and CD117 (data not shown). Nevertheless, thymic DC development is totally unaffected (in terms of both phenotype and absolute cell numbers) by Notch1 deficiency. Taken together with the evidence that Notch1 probably controls T/B fate specification in a binary fashion (described above), the simplest interpretation of our data would be to postulate that in the absence of Notch1 signaling, CLPs are deviated to the B cell lineage either before or shortly after their entry into the thymus. According to this scenario ( Fig. 7 B), Notch1–deficient thymic DCs could not be derived from a CLP, and hence would presumably originate from an independent thymic DC precursor population that has yet to be precisely identified.
Although we favor the hypothesis that T cells and thymic DCs are derived from distinct intrathymic precursors that differ in their developmental dependence on Notch1, other interpretations of our data cannot be excluded. For example, it is possible that thymic DCs arising from Notch1−/− precursors represent an aberrant developmental pathway in which CLPs are redirected to the DC fate in the absence of Notch1 signaling. According to this model, differentiation into thymic DCs could be considered as an alternative default pathway (or primary fate) of intrathymic CLPs. Nevertheless, the fact that thymic DC development from Notch1−/− precursors is not noticeably perturbed (with respect to either phenotype or absolute cell numbers) is difficult to reconcile with an altered T/DC cell fate specification, particularly when the effect of Notch1 deficiency on the T/B lineage decision is so dramatic.
Finally, the possibility cannot be formally excluded that other developmental pathways may lead to the production of thymic DCs in normal mice. For instance, thymic DCs may develop from a lymphoid precursor that is more closely related to B cells than to T cells. Such a putative B/DC precursor would presumably retain its bipotentiality in the absence of Notch1. Alternatively, thymic DC development may be a highly flexible process, with both lymphoid and nonlymphoid precursors having the potential to give rise to DCs depending on the availability of microenvironmental cues.
In conclusion, our data demonstrate that Notch1 is dispensable for the development of all myeloid and lymphoid lineages with the exception of T cells. Moreover, taken together with a recent report by Rodewald et al. 45, they seriously challenge the widely held hypothesis that thymic DCs and T cells share a common intrathymic precursor.
Acknowledgments
We thank Céline Marechal for technical help.
This work was supported in part by grants from the Swiss National Science Foundation (to M. Aguet) and the Human Frontier Science Program (to A. Wilson).
Footnotes
F. Radtke and I. Ferrero contributed equally to this work.
Abbreviations used in this paper: APC, allophycocyanin; BM, bone marrow; CLP, common lymphoid progenitor; DC, dendritic cell; LC, Langerhans cell; wt, wild-type.
References
- Cella M., Sallusto F., Lanzavecchia A. Origin, maturation and antigen presenting function of dendritic cells. Curr. Opin. Immunol. 1997;9:6–10 . doi: 10.1016/s0952-7915(97)80153-7. [DOI] [PubMed] [Google Scholar]
- Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252 . doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Brocker T., Riedinger M., Karjalainen K. Targeted expression of major histocompatibility complex (MHC) class II molecules demonstrates that dendritic cells can induce negative but not positive selection of thymocytes in vivo. J. Exp. Med. 1997;185:541–550 . doi: 10.1084/jem.185.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vremec D., Zorbas M., Scollay R., Saunders D.J., Ardavin C.F., Wu L., Shortman K. The surface phenotype of dendritic cells purified from mouse thymus and spleeninvestigation of the CD8 expression by a subpopulation of dendritic cells. J. Exp. Med. 1992;176:47–58 . doi: 10.1084/jem.176.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M., Inaba K., Hosono M., Kumamoto T., Ishida T., Muramatsu S., Masuda T., Ikehara S. Distinct mechanisms of neonatal tolerance induced by dendritic cells and thymic B cells. J. Exp. Med. 1991;173:549–559 . doi: 10.1084/jem.173.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardavin C. Thymic dendritic cells. Immunol. Today. 1997;18:350–361 . doi: 10.1016/s0167-5699(97)01090-6. [DOI] [PubMed] [Google Scholar]
- Ardavin C., Wu L., Li C.L., Shortman K. Thymic dendritic cells and T cells develop simultaneously in the thymus from a common precursor population. Nature. 1993;362:761–763 . doi: 10.1038/362761a0. [DOI] [PubMed] [Google Scholar]
- Wu L., Li C.L., Shortman K. Thymic dendritic cell precursorsrelationship to the T lymphocyte lineage and phenotype of the dendritic cell progeny. J. Exp. Med. 1996;184:903–911 . doi: 10.1084/jem.184.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos K., Bigby M., Wang J.H., Molnar A., Wu P., Winandy S., Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156 . doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- Wang J.H., Nichogiannopoulou A., Wu L., Sun L., Sharpe A.H., Bigby M., Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549 . doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- Wu L., Nichogiannopoulou A., Shortman K., Georgopoulos K. Cell-autonomous defects in dendritic cell populations of Ikaros mutant mice point to a developmental relationship with the lymphoid lineage. Immunity. 1997;7:483–492 . doi: 10.1016/s1074-7613(00)80370-2. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand M.D., Lake R.J. Notch signalingcell fate control and signal integration in development. Science. 1999;284:770–776 . doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Rebay I., Fleming R.J., Fehon R.G., Cherbas L., Cherbas P., Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrateimplications for Notch as a multifunctional receptor. Cell. 1991;67:687–699 . doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- Weinmaster G., Roberts V.J., Lemke G. A homolog of Drosophila Notch expressed during mammalian development. Development. 1991;113:199–205 . doi: 10.1242/dev.113.1.199. [DOI] [PubMed] [Google Scholar]
- Weinmaster G., Roberts V.J., Lemke G. Notch2a second mammalian Notch gene. Development. 1992;116:931–941 . doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- Reaume A.G., Conlon R.A., Zirngibl R., Yamaguchi T.P., Rossant J. Expression analysis of a Notch homologue in the mouse embryo. Dev. Biol. 1992;154:377–387 . doi: 10.1016/0012-1606(92)90076-s. [DOI] [PubMed] [Google Scholar]
- del Amo F.F., Gendron-Maguire M., Swiatek P.J., Jenkins N.A., Copeland N.G., Gridley T. Cloning, analysis, and chromosomal localization of Notch-1, a mouse homolog of Drosophila Notch. Genomics. 1993;15:259–264 . doi: 10.1006/geno.1993.1055. [DOI] [PubMed] [Google Scholar]
- Lardelli M., Lendahl U. Motch A and motch B—two mouse Notch homologues coexpressed in a wide variety of tissues. Exp. Cell Res. 1993;204:364–372 . doi: 10.1006/excr.1993.1044. [DOI] [PubMed] [Google Scholar]
- Lardelli M., Dahlstrand J., Lendahl U. The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech. Dev. 1994;46:123–136 . doi: 10.1016/0925-4773(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Uyttendaele H., Marazzi G., Wu G., Yan Q., Sassoon D., Kitajewski J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–2259 . doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- Lindsell C.E., Shawber C.J., Boulter J., Weinmaster G. Jaggeda mammalian ligand that activates Notch1. Cell. 1995;80:909–917 . doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- Shawber C., Boulter J., Lindsell C.E., Weinmaster G. Jagged2a serrate-like gene expressed during rat embryogenesis. Dev. Biol. 1996;180:370–376 . doi: 10.1006/dbio.1996.0310. [DOI] [PubMed] [Google Scholar]
- Bettenhausen B., Hrabe de Angelis M., Simon D., Guenet J.L., Gossler A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development. 1995;121:2407–2418 . doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- Dunwoodie S.L., Henrique D., Harrison S.M., Beddington R.S. Mouse Dll3a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development. 1997;124:3065–3076 . doi: 10.1242/dev.124.16.3065. [DOI] [PubMed] [Google Scholar]
- Hasserjian R.P., Aster J.C., Davi F., Weinberg D.S., Sklar J. Modulated expression of notch1 during thymocyte development. Blood. 1996;88:970–976 . [PubMed] [Google Scholar]
- Felli M.P., Maroder M., Mitsiadis T.A., Campese A.F., Bellavia D., Vacca A., Mann R.S., Frati L., Lendahl U., Gulino A., Screpanti I. Expression pattern of notch1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus componentsdistinct ligand-receptor interactions in intrathymic T cell development. Int. Immunol. 1999;11:1017–1025 . doi: 10.1093/intimm/11.7.1017. [DOI] [PubMed] [Google Scholar]
- Robey E. Regulation of T cell fate by Notch. Annu. Rev. Immunol. 1999;17:283–295 . doi: 10.1146/annurev.immunol.17.1.283. [DOI] [PubMed] [Google Scholar]
- Robey E., Chang D., Itano A., Cado D., Alexander H., Lans D., Weinmaster G., Salmon P. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 1996;87:483–492 . doi: 10.1016/s0092-8674(00)81368-9. [DOI] [PubMed] [Google Scholar]
- Washburn T., Schweighoffer E., Gridley T., Chang D., Fowlkes B.J., Cado D., Robey E. Notch activity influences the αβ versus γδ T cell lineage decision. Cell. 1997;88:833–843 . doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- Deftos M.L., He Y.W., Ojala E.W., Bevan M.J. Correlating Notch signaling with thymocyte maturation. Immunity. 1998;9:777–786 . doi: 10.1016/s1074-7613(00)80643-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehn B.M., Bielke W., Pear W.S., Osborne B.A. Protective effects of notch-1 on TCR-induced apoptosis. J. Immunol. 1999;162:635–638 . [PubMed] [Google Scholar]
- Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H.R., Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558 . doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- Pui J.C., Allman D., Xu L., DeRocco S., Karnell F.G., Bakkour S., Lee J.Y., Kadesch T., Hardy R.R., Aster J.C., Pear W.S. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308 . doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672 . doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Shortman K., Vremec D., Corcoran L.M., Georgopoulos K., Lucas K., Wu L. The linkage between T-cell and dendritic cell development in the mouse thymus. Immunol. Rev. 1998;165:39–46 . doi: 10.1111/j.1600-065x.1998.tb01228.x. [DOI] [PubMed] [Google Scholar]
- Wilson A., MacDonald H.R. A limited role for β-selection during γδ T cell development. J. Immunol. 1998;161:5851–5854 . [PubMed] [Google Scholar]
- Anjuere F., Martin P., Ferrero I., Fraga M.L., del Hoyo G.M., Wright N., Ardavin C. Definition of dendritic cell subpopulations present in the spleen, Peyer's patches, lymph nodes, and skin of the mouse. Blood. 1999;93:590–598 . [PubMed] [Google Scholar]
- Schuler G., Steinman R.M. Murine epidermal Langerhans cells mature into potent immunostimulatory dendritic cells in vitro. J. Exp. Med. 1985;161:526–546 . doi: 10.1084/jem.161.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner L.A., Kopan R., Martin D.I., Bernstein I.D. A human homologue of the Drosophila developmental gene, Notch, is expressed in CD34+ hematopoietic precursors. Blood. 1994;83:2057–2062 . [PubMed] [Google Scholar]
- Li L., Milner L.A., Deng Y., Iwata M., Banta A., Graf L., Marcovina S., Friedman C., Trask B.J., Hood L., Torok-Storb B. The human homolog of rat Jagged1 expressed by marrow stroma inhibits differentiation of 32D cells through interaction with Notch1. Immunity. 1998;8:43–55 . doi: 10.1016/s1074-7613(00)80457-4. [DOI] [PubMed] [Google Scholar]
- Milner L.A., Bigas A. Notch as a mediator of cell fate determination in hematopoiesisevidence and speculation. Blood. 1999;93:2431–2448 . [PubMed] [Google Scholar]
- Milner L.A., Bigas A., Kopan R., Brashem-Stein C., Bernstein I.D., Martin D.I. Inhibition of granulocytic differentiation by mNotch1. Proc. Natl. Acad. Sci. USA. 1996;93:13014–13019 . doi: 10.1073/pnas.93.23.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlesso N., Aster J.C., Sklar J., Scadden D.T. Notch1-induced delay of human hematopoietic progenitor cell differentiation is associated with altered cell cycle kinetics. Blood. 1999;93:838–848 . [PubMed] [Google Scholar]
- Wu L., D'Amico A., Winkel K.D., Suter M., Lo D., Shortman K. RelB is essential for the development of myeloid-related CD8α− dendritic cells but not of lymphoid-related CD8α+ dendritic cells. Immunity. 1998;9:839–847 . doi: 10.1016/s1074-7613(00)80649-4. [DOI] [PubMed] [Google Scholar]
- Rodewald H.R., Brocker T., Haller C. Developmental dissociation of thymic dendritic cell and thymocyte lineages revealed in growth factor receptor mutant mice. Proc. Natl. Acad. Sci. USA. 1999;96:15069–15073. doi: 10.1073/pnas.96.26.15068. [DOI] [PMC free article] [PubMed] [Google Scholar]