Abstract
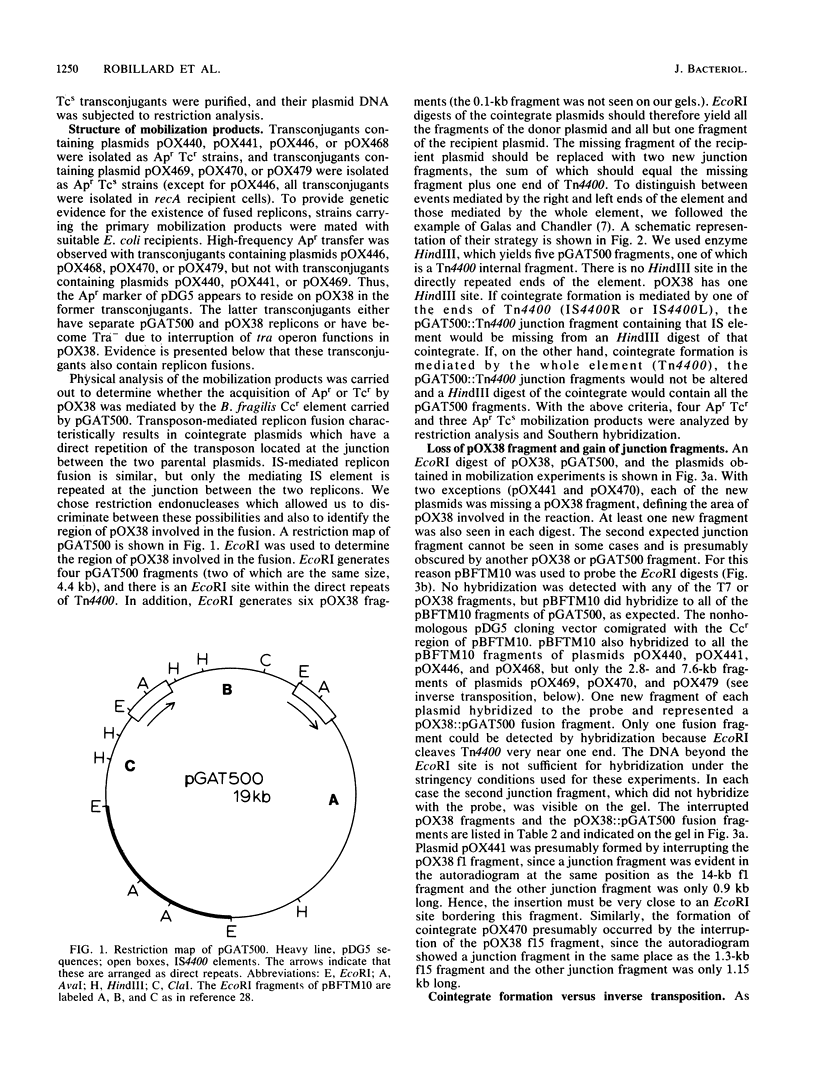
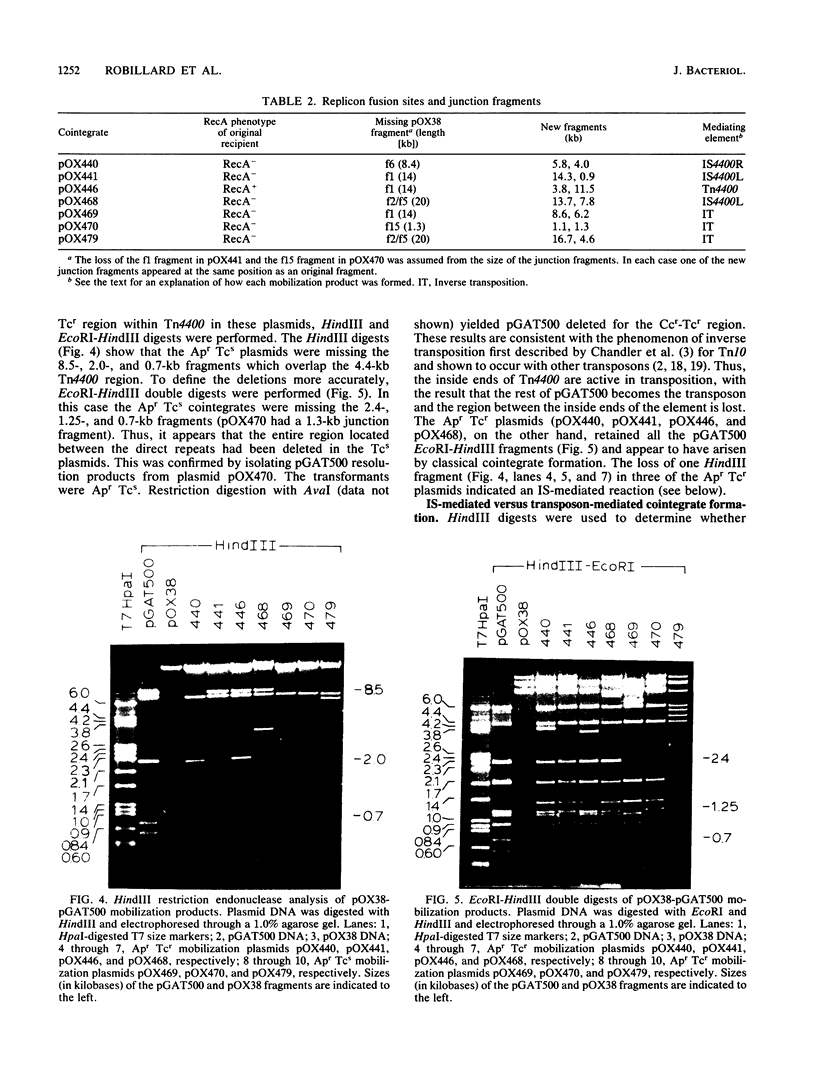
Transfer factor pBFTM10, isolated from the obligate anaerobic bacterium Bacteroides fragilis, carries a clindamycin resistance determinant which we have suggested is part of a transposable element. DNA homologous to this determinant is found in many Clnr Bacteroides isolates, either in the chromosome or on plasmids. We have now established that Ccr resides on a transposon, Tn4400. In addition to the Ccr determinant that functions under anaerobic conditions in B. fragilis, Tn4400 also carries a determinant for tetracycline resistance (Tcr) which only functions in Escherichia coli under aerobic conditions. The presence of Tn4400 on pBFTM10 does not confer tetracycline resistance on B. fragilis cells containing it. DNA from pBFTM10 was cloned in E. coli, with pDG5 as the cloning vector, to form pGAT500. Using a mobilization assay involving pGAT500 and an F factor derivative, pOX38, we determined that a 5.6-kilobase region of pBFTM10 DNA was capable of mediating replicon fusion and transposition. Most of the mobilization products resulted from inverse transposition reactions, while some were the result of true cointegrate formation. Analysis of the cointegrate molecules showed that three were formed by the action of one of the ends of Tn4400 (IS4400), and one was formed by the action of the whole element (Tn4400). The cointegrate molecule carrying intact copies of Tn4400 at the junction of the two plasmids could resolve to yield an unaltered donor plasmid (pGAT500) and a conjugal plasmid containing a copy of Tn4400 or a copy of one insertion sequence element (pOX38::Tn4400 or pOX38::IS4400). Thus, Tn4400 is a compound transposon containing active insertion sequence elements as directly repeated sequences at its ends.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M., Manning P. A., Kusecek B., Schwuchow S., Willetts N. A genetic analysis of F sex factor cistrons needed for surface exclusion in Escherichia coli. J Mol Biol. 1980 Apr 25;138(4):779–795. doi: 10.1016/0022-2836(80)90065-0. [DOI] [PubMed] [Google Scholar]
- Chandler M., Roulet E., Silver L., Boy de la Tour E., Caro L. Tn10 mediated integration of the plasmid R100.1 into the bacterial chromosome: inverse transposition. Mol Gen Genet. 1979 May 23;173(1):23–30. doi: 10.1007/BF00267687. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Deonier R. C., Mirels L. Excision of F plasmid sequences by recombination at directly repeated insertion sequence 2 elements: involvement of recA. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3965–3969. doi: 10.1073/pnas.74.9.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Chandler M. On the molecular mechanisms of transposition. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4858–4862. doi: 10.1073/pnas.78.8.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Chandler M. Structure and stability of Tn9-mediated cointegrates. Evidence for two pathways of transposition. J Mol Biol. 1982 Jan 15;154(2):245–272. doi: 10.1016/0022-2836(82)90063-8. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Hasegawa P., Davis C. E. Plasmid transfer from Escherichia coli to Bacteroides fragilis: differential expression of antibiotic resistance phenotypes. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7203–7206. doi: 10.1073/pnas.81.22.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Jr, Hasegawa P., Davis C. E. Expression in Escherichia coli of cryptic tetracycline resistance genes from bacteroides R plasmids. Plasmid. 1984 May;11(3):248–252. doi: 10.1016/0147-619x(84)90031-3. [DOI] [PubMed] [Google Scholar]
- Guiney D. G., Yakobson E. Location and nucleotide sequence of the transfer origin of the broad host range plasmid RK2. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3595–3598. doi: 10.1073/pnas.80.12.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S., Reed R. R., Steitz J. A., Low K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Johnson D. A., Willetts N. S. Construction and characterization of multicopy plasmids containing the entire F transfer region. Plasmid. 1980 Nov;4(3):292–304. doi: 10.1016/0147-619x(80)90068-2. [DOI] [PubMed] [Google Scholar]
- Kilbane J. J., Malamy M. H. F factor mobilization of non-conjugative chimeric plasmids in Escherichia coli: general mechanisms and a role for site-specific recA-independent recombination at orV1. J Mol Biol. 1980 Oct 15;143(1):73–93. doi: 10.1016/0022-2836(80)90125-4. [DOI] [PubMed] [Google Scholar]
- Malamy M. H., Tally F. P. Mechanisms of drug-resistance transfer in Bacteroides fragilis. J Antimicrob Chemother. 1981 Dec;8 (Suppl 500):59–75. doi: 10.1093/jac/8.suppl_d.59. [DOI] [PubMed] [Google Scholar]
- Marsh P. K., Malamy M. H., Shimell M. J., Tally F. P. Sequence homology of clindamycin resistance determinants in clinical isolates of Bacteroides spp. Antimicrob Agents Chemother. 1983 May;23(5):726–730. doi: 10.1128/aac.23.5.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison T. G., Malamy M. H. Comparisons of F factors and R factors: existence of independent regulation groups in F factors. J Bacteriol. 1970 Jul;103(1):81–88. doi: 10.1128/jb.103.1.81-88.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M. B., Malamy M. H. Role of the F factor oriV1 region in recA-independent illegitimate recombination. Stable replicon fusions of the F derivative pOX38 and pBR322-related plasmids. J Mol Biol. 1984 May 25;175(3):263–284. doi: 10.1016/0022-2836(84)90348-6. [DOI] [PubMed] [Google Scholar]
- Reif H. J. Genetic evidence for absence of transposition functions from the internal part of Tn981 a relative of Tn9. Mol Gen Genet. 1980;177(4):667–674. doi: 10.1007/BF00272678. [DOI] [PubMed] [Google Scholar]
- Rosner J. L., Guyer M. S. Transposition of IS1-lambdaBIO-IS1 from a bacteriophage lambda derivative carrying the IS1-cat-IS1 transposon (Tn9). Mol Gen Genet. 1980 Apr;178(1):111–120. doi: 10.1007/BF00267219. [DOI] [PubMed] [Google Scholar]
- Saedler H., Cornelis G., Cullum J., Schumacher B., Sommer H. IS1-mediated DNA rearrangements. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):93–98. doi: 10.1101/sqb.1981.045.01.017. [DOI] [PubMed] [Google Scholar]
- Shimell M. J., Smith C. J., Tally F. P., Macrina F. L., Malamy M. H. Hybridization studies reveal homologies between pBF4 and pBFTM10, Two clindamycin-erythromycin resistance transfer plasmids of Bacteroides fragilis. J Bacteriol. 1982 Nov;152(2):950–953. doi: 10.1128/jb.152.2.950-953.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker N. B., Guthrie E. P., Salyers A. A., Gardner J. F. Evidence that the clindamycin-erythromycin resistance gene of Bacteroides plasmid pBF4 is on a transposable element. J Bacteriol. 1985 May;162(2):626–632. doi: 10.1128/jb.162.2.626-632.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurray R. A., Nagaishi H., Clark A. J. Molecular cloning of DNA from F sex factor of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1976 Jan;73(1):64–68. doi: 10.1073/pnas.73.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J. Characterization of Bacteroides ovatus plasmid pBI136 and structure of its clindamycin resistance region. J Bacteriol. 1985 Mar;161(3):1069–1073. doi: 10.1128/jb.161.3.1069-1073.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. J., Macrina F. L. Large transmissible clindamycin resistance plasmid in Bacteroides ovatus. J Bacteriol. 1984 May;158(2):739–741. doi: 10.1128/jb.158.2.739-741.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tally F. P., Snydman D. R., Shimell M. J., Malamy M. H. Characterization of pBFTM10, a clindamycin-erythromycin resistance transfer factor from Bacteroides fragilis. J Bacteriol. 1982 Aug;151(2):686–691. doi: 10.1128/jb.151.2.686-691.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York M. K., Stodolsky M. Characterization of P1argF derivatives from Escherichia coli K12 transduction. I. IS1 elements flank the argF gene segment. Mol Gen Genet. 1981;181(2):230–240. doi: 10.1007/BF00268431. [DOI] [PubMed] [Google Scholar]