Abstract
Clonal composition and T cell receptor (TCR) repertoire of CD4+ and CD8+ T cells infiltrating actively demyelinating multiple sclerosis (MS) lesions were determined with unprecedented resolution at the level of single cells. Individual CD4+ or CD8+ T cells were isolated from frozen sections of lesional tissue by micromanipulation and subjected to single target amplification of TCR-β gene rearrangements. This strategy allows the assignment of a TCR variable region (V region) sequence to the particular T cell from which it was amplified. Sequence analysis revealed that in both cases investigated, the majority of CD8+ T cells belonged to few clones. One of these clones accounted for 35% of CD8+ T cells in case 1. V region sequence comparison revealed signs of selection for common peptide specificities for some of the CD8+ T cells in case 1. In both cases, the CD4+ T cell population was more heterogeneous. Most CD4+ and CD8+ clones were represented in perivascular infiltrates as well as among parenchymal T cells. In case 2, two of the CD8+ clones identified in brain tissue were also detected in peripheral blood. Investigation of the antigenic specificities of expanded clones may help to elucidate their functional properties.
Keywords: autoimmunity, demyelinating disease, T cell receptor β chain, gene rearrangement, peripheral blood
Introduction
Multiple sclerosis (MS) is an inflammatory demyelinating disease of the central nervous system that is generally believed to be of autoimmune origin. Destruction of myelin sheaths leads to the formation of demyelinated lesions in brain and spinal cord white matter (WM), which is associated with impaired nerve conduction and axonal loss resulting in neurological disability. In most cases, MS initially presents as a relapsing-remitting disease. However, with time, a majority of patients enter a secondary progressive course characterized by gradual clinical deterioration 1 2. While destruction of WM is ongoing, the lesions are histologically characterized by dense infiltrates of macrophages and T lymphocytes. These cells are found in dense aggregates around postcapillary venules, but are also scattered throughout the affected parenchyma 3 4. Even after myelin destruction has ceased, infiltrates may persist for months to years 3 5. At later stages, a glial scar may result with little or no signs of inflammation.
The etiology of MS remains enigmatic. Various clinical subtypes of the disease and distinct histopathological patterns have been described, indicating that MS may also be heterogeneous with respect to pathogenesis 3. Both environmental and genetic factors play a role 1 2 6. Using animal models of experimental autoimmune encephalomyelitis (EAE), it has been demonstrated that T cells, macrophages, and autoantibodies can contribute to immune-mediated myelin injury 4 7 8. However, disease-initiating roles of the various effector arms of the immune system remain to be proven in MS. Autoreactive T cells have been implicated in the pathogenesis, and several myelin but also nonmyelin antigens have been discussed as potential targets of an autoimmune attack 6 9. Oligoclonal expansions of CD4+ T cells were detected in cerebrospinal fluid (CSF [10, 11]), and an increased frequency of oligoclonal CD8+ T cells expressing particular TCR-Vβ gene segments were found in the blood of MS patients compared with normal controls 12.
Only limited information is available on the nature of T cells in MS lesions. The T cell infiltrates are mainly composed of TCR-α/β+CD4+ and TCR-α/β+CD8+ as well as TCR-γ/δ+ T cells in variable proportions 4 13. Several earlier studies have analyzed the TCR-α/β repertoire expressed in the lesions. By amplification of TCR gene rearrangements from reverse-transcribed mRNA extracted from whole tissue specimens, Wucherpfennig et al. 14 described polyclonal T cell populations contained in the lymphocytic infiltrate. Using similar approaches, other studies provided evidence for a restriction of the TCR repertoire expressed by infiltrating T cells 15 16 17. However, in these studies, sequence data could not be assigned to either CD4+ or CD8+ T cells. Therefore, TCR repertoire and clonal composition of the two subsets in MS lesions are still unknown. Another unresolved question is whether the T cells found within the destructed parenchyma differ in clonal composition and function from the large fraction of T cells aggregated around blood vessels, as suggested by results with murine EAE 18 19. Furthermore, little is known about potential differences in the composition of T cell populations in actively demyelinating versus inactive MS lesions.
This study addresses these questions by an analysis of clonal composition and TCR repertoire of the T cell infiltrate at the level of single cells. Individual CD4+ or CD8+ T cells located in perivascular infiltrates or in the parenchyma of actively demyelinating lesions were micromanipulated from tissue sections. Using a PCR-based technique described previously 20, TCR-β gene rearrangements were amplified from these single T cells and directly sequenced. V region sequence analysis revealed the presence of large clonal expansions primarily among CD8+ T cells but also in the CD4+ subset.
Materials and Methods
Patients and Tissue Samples.
Brain samples of case 1 were obtained from the Netherlands Brain Bank (Amsterdam, The Netherlands). The female patient died of cachexia at age 35, having suffered from relapsing-remitting and secondary-progressive MS for 10 yr. She received corticosteroid- and anti-TNF medication 3 and 2 yr before death, respectively, but received neither corticosteroids, anti-TNF, nor immunosuppressants during the last year of her life. Two blocks of periventricular WM frozen in liquid nitrogen upon autopsy were analyzed (see Fig. 1). HLA genotyping was performed in the group of Dr. Albert at the Laboratory for Immunogenetics (Ludwig-Maximilians-University, Munich, Germany) by sequencing (HLA-A 21), -B 22, and -C 23), PCR with sequence-specific primers (HLA-DRB 24, -DQB 25), or DNA typing with sequence-specific oligonucleotide probes (HLA-DQA 26). The patient was typed HLA-A*2402/*2601, HLA-B*1801/*39062, HLA-Cw*0701/*0702, HLA-DRB1*0101/*0801, HLA-DQA1*0101/*0401, HLA-DQB1* 0501/*0402.
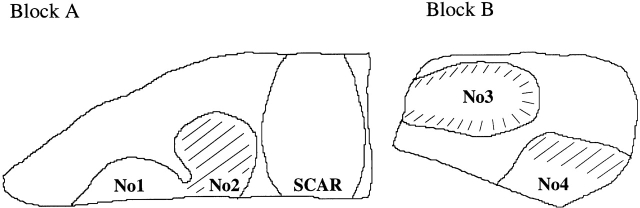
Figure 1.
Schematic overview of sections of the two blocks of brain tissue (A and B) analyzed for case 1. Nos. 1–4 designate MS lesions from which T cells were micromanipulated. Hatched lesional areas show signs of ongoing myelin destruction. Block A also contained an old lesion (SCAR) showing astroglial scarring, and only a little infiltration by T cells or other signs of inflammation.
Male patient 2 presented with recurrent episodes of left-sided hemianopsia at the age of 49. Cranial magnetic resonance imaging was indicative of a malignant glioma located in the right temporooccipital WM. Subsequently, the lesion was completely resected at the Department of Neurosurgery at the University of Bonn (Bonn, Germany) 2 wk after the first occurrence of symptoms. Histopathologically, a large area of inflammatory demyelination compatible with an MS lesion was found. Examination of CSF revealed the presence of oligoclonal immunoglobulins, further supporting the diagnosis of a first manifestation of MS. Within the following 3 yr (until submission of this manuscript), the patient suffered two relapses. Follow-up magnetic resonance imaging confirmed the occurrence of new demyelinated lesions. Brain tissue was frozen in liquid nitrogen immediately after resection, and blood was sampled 12 and 31 mo after surgery (6 and 17 mo after first and second relapse, respectively). The patient received IFN-β1a from the time of the second relapse until submission of this manuscript. He seems to be homozygous for the haplotype HLA-A*0101, HLA-B*0801, HLA-Cw*0701, HLA-DRB1*0301, HLA-DRB3*0101, HLA-DQA1*05, HLA-DQB1*0201, as only one allele was detected for each of the loci analyzed. As this haplotype is the most common in the Caucasian population, homozygosity is not uncommon. He gave informed consent to the use of brain material and blood for research purposes. In neither case was CSF available for analysis.
Immunohistochemistry, Micromanipulation, and Cell Counting.
Serial 10-μm–thick frozen sections were mounted onto glass slides. Every 15 sections, 3 sections were stained for luxol fast blue, Oil Red O (ORO), and hematoxylin and eosin according to standard procedures. Demyelinating activity of MS lesions was assessed by immunohistochemistry on acetone-fixed sections 27. Presence of myelin oligodendrocyte glycoprotein (MOG) and proteolipid protein (PLP) in macrophage endocytic vesicles and the acute stage inflammatory marker 27E10 on the macrophage surface indicates recent phagocytosis of myelin 3 27 28 29. ORO+ macrophages can be found in actively demyelinating lesions, but may still be detectable weeks to months after myelin destruction has ceased 3 5. Therefore, absence of ORO+ macrophages suggests that the lesion was inactive for at least some weeks before resection. The primary antibodies used were anti-MOG, anti-PLP (30; both provided by Dr. S. Piddlesden, University of Cardiff, Cardiff, UK), and anti-27E10 (purchased from BMA Biomedicals). Micromanipulation was performed on adjacent sections stained for CD8 (C8/144B; Dako), CD4 (MT310; Dako), CD3 (rabbit anti–human CD3; Dako), and human glial fibrillary acidic protein (GFAP, 6F2; Dako) as described 20 31. Staining for TCR-α/β+ and TCR-γ/δ+ T cells was done using antibodies βF1 (8A3; T Cell Diagnostics) and pan-γ/δ (Immu 510; Coulter-Immunotech Diagnostics), respectively. Biotinylated Fab fragments of a rabbit anti–mouse (E413; Dako) and a swine anti–rabbit (E431; Dako) antibody were used as secondary reagents.
Using hydraulic micromanipulators, cells were mobilized from surrounding tissue and transferred into PCR tubes as published 20 31.
For determination of CD8+/CD4+ ratios, 300–800 positive cells in the parenchymal and 100–400 positive cells in the perivascular location were counted per subset on serial sections. CD4+ cells displaying a macrophage morphology were excluded. In case 1, the CD8/CD4 ratio was determined in lesions 3 and 4 only, as hardly any CD4+ but numerous CD8+ T cells were detected in the parenchyma of lesions 1 and 2 (see Fig. 1 and legend to Table ). In both cases, counting of CD8+ and CD4+ T cells in perivascular infiltrates was hampered by the presence of CD4+ macrophages and high cellular density. Therefore, these values represent rough assessments.
Table 2.
Representation of CD4+ and CD8+ T Cell Clones in Perivascular and Parenchymal Locations of the Four Lesions Analyzed for Case 1
| Single parenchymal CD8+ T cells | Single parenchymal CD4+ T cells | Samples of perivascular cells | ||||
|---|---|---|---|---|---|---|
| Lesion | Clone no. | Frequency | Clone no. | Frequency | Clone no. | Frequency |
| No. 1 | 1 | 14/29 (48%) | 1 | 8/41 (20%) | ||
| 9 | 3/29 | 6 | 2/41 | |||
| 3, 4, 7, 15, 21 | 1/29 | 12 | 1/3 | 3, 4, 13,17 | 1/41 | |
| Sum: | 22/29 (76%) | Sum: | 14/41 (34%) | |||
| No. 2 | 1 | 6/28 (21%) | ||||
| 2 | 3/28 | 1, 8 | 3/25 | |||
| 9 | 2/28 | 2 | 2/25 | |||
| 3, 16, 18, 19 | 1/28 | 11 | 1/2 | 7, 10 16, 21 | 1/25 | |
| Sum: | 15/28 (54%) | Sum: | 12/25 (48%) | |||
| No. 3 | 1 | 8/24 (33%) | 1 | 4/14 | ||
| 16, 18 | 2/24 | 10, 11 | 2/19 | 4 | 2/14 | |
| 4, 14, 15, 17 | 1/24 | 12, 13 | 1/19 | 9, 14 | 1/14 | |
| Sum: | 16/24 (67%) | Sum: | 6/19 (32%) | Sum: | 8/14 | |
| No. 4 | 1 | 3/6 | ||||
| 10 | 1/13 | 9, 15 | 1/6 | |||
| Sum: | 5/6 | |||||
| Sum | 1 | 28/81 (35%) | ||||
| 9 | 5/81 | 1 | 18/86 (21%) | |||
| 2, 16, 18 | 3/81 | 10, 11 | 3/37 | 4, 8 | 3/86 | |
| 3, 4, 15 | 2/81 | 12 | 2/37 | 2, 6, 9 | 2/86 | |
| 7, 14, 17, 19, 21 | 1/81 | 13 | 1/37 | 3, 7, 10, 13, 14, 15, 16, 17, 21 | 1/86 | |
| Sum: | 53/81 (65%) | Sum: | 9/37 (24%) | Sum: | 39/86 (45%) | |
Clonal expansions were identified by amplification of an identical rearrangement from at least two different samples of cells micromanipulated from material of case 1, and were numbered arbitrarily. Except for clones 6 and 8, all clones could be assigned to the CD8+ or CD4+ subset, as the clonal V region sequence was obtained at least once from a single parenchymal CD8+ or CD4+ cell. Only potentially functional rearrangements are listed. Three clones defined by nonfunctional rearrangments accounted for 7 of the 24 nonfunctional rearrangments obtained in total. These clonal sequences could not be assigned to clones defined by potentially functional rearrangements. The population of parenchymal T cells was dominated by CD8+ T cells (CD8/CD4 ≈ 3, as determined in lesions 3 and 4). Likewise, perivascular CD8+ T cells outnumbered perivascular CD4+ T cells roughly by a factor of three.
Amplification of TCR-β Gene Rearrangements from Single Cells.
TCR-Vβ gene rearrangements were amplified by seminested PCR as described 20, with minor modifications specified below. In brief, a first round of amplification was performed using a mix of 24 Vβ gene family–specific and 8 Jβ-specific primers. In a second round of PCR, two to three of the same Vβ primers were used together with a mix of seven internal Jβ-specific primers in separate reactions. Compared with the primer mixes published previously 20, the Vβ25 primer was omitted, a new nested primer pair specific for Jβ2S3 (3′Jβ2S3: 5′-TCCCGGGGCGCCCCCTCCCCAGTT-3′ and 5′Jβ2S3: 5′-GAGCCCCCGCTTACCGAGCACTGTCA-3′) was employed, and the primers Vβ4, 3′Jβ2S7, and 5′Jβ2S7 were modified (Vβ4: 5′-TCCAGTGTCAAGTCGATAGCCAAGTC-3′; 3′Jβ2S7: 5′-TCCATCGTTCACCTTCTCTCTAAACA-3′; and 5′Jβ2S7: 5′-GCCCGAATCTCACCTGTGACCGTG-3′). Primers were purchased from Eurogentec (first round primers were “single cell quality”).
Sequence Analysis.
PCR products were gel purified from 2.5% agarose gels using the Qiaex® II gel extraction kit (QIAGEN) and directly sequenced using the ABI PRISM BigDye® Terminator Cycle Sequencing Ready Reaction Kit (PerkinElmer) and an automatic sequencer (ABI 377, PerkinElmer). V gene sequences were analyzed using DNASIS® software (Amersham Pharmacia Biotech). Sequence data is available from EMBL/GenBank/DDBJ under accession nos. A7405646–A7405872.
Sorting of Peripheral Blood T Cells.
For CDR3 spectratyping, PBMCs were purified from blood samples of case 2 obtained 12 and 31 mo after brain surgery by density gradient centrifugation using Ficoll-Paque (Amersham Pharmacia Biotech) and were stained with fluorescein-conjugated anti-CD3 (SK7; Becton Dickinson) and PE-conjugated anti-CD4 (MT310; Dako) or PE-conjugated anti-CD8 (DK25; Dako). CD3+CD4+ or CD3+CD8+ cells were sorted by FACS® using a FACS® 440 (Becton Dickinson) into Eppendorf tubes containing RPMI 1640 medium supplemented with 10% FCS. The tubes were cooled at 4°C during the sorting procedure. Dead cells were excluded from the analysis by staining with propidium iodide. PBMCs from blood sample “12 mo” had been cryopreserved before staining and sorting procedures. For clone-specific PCR, cryopreserved PBMCs obtained from blood drawn 12 and 31 mo after brain surgery were stained with PE-conjugated anti-CD8 and allophycocyanin-conjugated anti-CD45RO (UCHL-1; Becton Dickinson) antibodies, and 102 or 103 CD8+CD45RO+ cells were sorted into each of several PCR tubes. For single cell PCR controls, PBMCs were purified from blood of five healthy caucasian donors and stained with fluorescence-labeled antibodies. Single TCR-α/β+CD3+ cells were sorted into PCR tubes by FACS® as reported 20.
CDR3 Spectratyping of TCR-β Transcripts from Peripheral Blood T Cells.
Total RNA was prepared from 105 (sample “12 mo”), and 2 × 105 (sample “31 mo”) CD3+CD4+ and CD3+CD8+ cells, oligo-(dT)-primed cDNA was reverse transcribed, and CDR3 spectratype analysis was carried out as reported previously 32. In brief, cDNA was subjected to TCR-Vβ gene family–specific PCR in 26 separate reactions, each containing 1 of 26 Vβ family– and 1 Cβ-specific primer 12. Using 1 of 13 fluorescence-labeled Jβ-specific oligonucleotides 33, runoff products were generated from each Vβ-specific PCR product. Transcripts were resolved on a sequencing gel, and fluorescence intensities were measured with help of an automated DNA sequencer. Expanded candidate Vβ-Jβ subpopulations were subamplified from the initial Vβ-Cβ amplification product using Vβ- and Jβ-specific primers and were directly sequenced 32. A sequence readable in CDR3 results if the population of TCR-β mRNA molecules amplified by the Vβ-Jβ primer combination was dominated by a single clonal transcript.
Cloning of PCR Products.
The Vβ14-Jβ1S1 subpopulation was subamplified from cDNA derived from CD3+CD8+ cells with the respective Vβ-Jβ primer pair. 1 ng of the freshly amplified PCR product was cloned into pCR2.1 vector using the Original TA Cloning® Kit (Invitrogen). Colonies were randomly picked, and plasmid-DNA was purified using the QIAGEN Plasmid Mini Kit. Inserts were sequenced with the Vβ14-specific primer.
Generation of Jurkat Control Cells for Clone-specific PCR.
To establish and positively control clone-specific PCR (specific amplification of the V region sequences of clones 2, 3, and 8 identified in brain tissue of case 2), cells were generated that carried one copy of either TCR-β gene rearrangement as a stable genomic integration. Either rearrangement (PCR-product) was cloned into the pRc/CMV vector (Invitrogen). Integrity of primer binding sites (see below) was verified by sequencing and linearized vector (1 μg) was transfected into Jurkat cells (4 × 106; gift of Dr. M. Schreier, Novartis, Basel, Switzerland) using DMRIE-C reagent (Life Technologies) according to the manufacturer's instructions. Cells were seeded at 3 × 104 per well in 96-well plates and selected with 1 mg/ml G418 (Geneticin®; Life Technologies) for 5 wk to obtain stable integrants. Clones carrying a single copy of the transfected vector were identified by Southern blotting (data not shown).
Clone-specific PCR.
To determine efficiencies of clone-specific single target amplification (described above), even in a situation where only 1 of up to 103 cells carried the clonal target sequence, single Jurkat control cells were sorted into tubes containing 103 irrelevant TCR-α/β+CD3+ T cells from a healthy donor as described above. Upstream primers for V regions for first and second round amplification were Vβ4- (clone 2) and Vβ13-specific oligonucleotides (clones 3 and 8; described above). Nested primers (external primers for first round and internal primers for second round amplification) specific for clonal CDR3 regions were 5′-GTCAGGACGTTGGCCCCAGAAA-3′ (clone 2, external), 5′-AGGACGTTGGCCCCAGAAACCT-3′ (clone 2, internal), 5′-CCGAAGAACTGCTCATTGTAGGTG-3′ (clone 3, external), 5′-AGAACTGCTCATTGTAGGTGGCAA-3′ (clone 3, internal), 5′-CCCGAAGAACTGCTCATTGTATTG-3′ (clone 8, external), and 5′-GAAGAACTGCTCATTGTATTGCGC-3′ (clone 8, internal). After incubation with 0.5 mg/ml proteinase K (Life Technologies) for 2 h at 50°C followed by heat inactivation of the enzyme for 10 min at 95°C, samples were subjected to first-round amplification in the same reaction tube in 1× Thermophilic DNA Polymerase Buffer (Promega); 100 μM each of dATP, dGTP, dCTP, and dTTP; 42 nM of each primer; 1.5 mM MgCl2 in case of clone 2– and clone 3–specific PCR or 2 mM MgCl2 in case of clone 8–specific PCR; and 2.5 U Taq DNA Polymerase in Storage Buffer A (Promega). An initial cycle of 95°C for 2 min, a pause at 80°C during which Taq was added, 70°C for 30 s, and 72°C for 50 s was followed by 34 cycles of 95°C for 60 s, 70°C for 30 s, and 72°C for 50 s and a single 5-min incubation step at 72°C. Conditions for second-round amplification were as follows: 20 mM Tris-HCl, pH 8.4; 50 mM KCl; 100 μM each of dATP, dGTP, dCTP, and dTTP; 150 nM of each primer; 1.5 mM MgCl2 in case of clone 2– and clone 3–specific PCR or 4 mM MgCl2 in case of clone 8–specific PCR; 1 μl first-round reaction mixture; and 1.3 U Taq polymerase (Life Technologies) were subjected to 1 cycle of 95°C for 2 min, 70°C for 30 s, and 72°C for 90 s followed by 44 cycles of 95°C for 60 s, 70°C for 30 s, and 72°C for 60 s followed by 72°C for 5 min. When a single Jurkat control cell was analyzed together with 103 irrelevant T cells, 13 of 16, 13 of 16, and 13 of 25 such samples were positive in the specific amplification of V-regions of clones 2, 3, and 8, respectively. In no instance was a specific product obtained from samples of irrelevant T cells in the absence of a Jurkat control cell. For each of the three clone-specific amplifications, five arbitrarily chosen PCR products were sequenced; all represented the respective clonal sequence.
For analysis of blood samples of case 2, samples containing 102 or 103 CD8+CD45RO+ cells (described above) were analyzed by clone 2–, 3–, and 8–specific PCR in parallel with samples containing Jurkat cells carrying the respective rearrangement (two per tube) plus 102 or 103 CD8+CD45RO+ T cells from a healthy donor as positive controls. All PCR products obtained from test samples were sequenced.
Results
Selection of Brain Material and Design of the Single Cell PCR Analysis.
To identify actively demyelinating MS lesions in frozen sections of brain specimens, the molecular composition of myelin degradation products within macrophage vesicles 28 29 and the expression of 27E10, an inflammatory macrophage activation antigen, were determined by immunohistochemistry as described previously 3 27. Throughout this paper, lesions were considered “active” or “actively demyelinating” if signs of recent myelin phagocytosis and macrophage activation were detectable (described in Materials and Methods). All other lesions were considered “inactive,” irrespective of the presence of inflammatory infiltrates. In total, five lesions originating from two MS cases were selected for single cell PCR analysis. Four lesions were from autopsy material of case 1, including a lesion that was active over its entire area, an inactive lesion, and two lesions showing active as well as inactive areas (Fig. 1). A single lesion that showed signs of ongoing myelin destruction over its entire area was found in biopsy material of case 2 (not shown). It likely represented an early stage of lesion development, as the lesion was already resected 2 wk after clinical manifestation of the disease. All lesions contained large numbers of T cells located in perivascular aggregates (subsequently termed “perivascular cells”) or outside of these, scattered over the demyelinated parenchyma as solitary cells (“parenchymal cells”). In lesions of both cases, CD8+ T cells outnumbered CD4+ T cells by at least a factor of three (see legends to Table and Table ).
Table 3.
Representation of CD4+ and CD8+ T Cell Clones in Perivascular and Parenchymal Locations of the Single Lesion Analyzed for Case 2
| Single parenchymal CD8+ T cells | Single parenchymal CD4+ T cells | Samples of perivascular cells | |||
|---|---|---|---|---|---|
| Clone no. | Frequency | Clone no. | Frequency | Clone no. | Frequency |
| 2 | 4/24 (17%) | ||||
| 5 | 3/24 | 8 | 3/46 (7%) | ||
| 3, 6, 9 | 2/24 | 1, 2, 7 | 2/46 | ||
| 4, 8, 7, 10 | 1/24 | 13, 14, 15 | 2/52 | 3, 4, 10 | 1/46 |
| Sum: | 17/24 (71%) | Sum: | 6/52 (12%) | Sum: | 12/46 (26%) |
Clonal expansions were identified by amplification of an identical rearrangement from at least two different samples of cells micromanipulated from material of case 2, and were numbered arbitrarily. Except for clone 1, all clones could be assigned to the CD8+ or CD4+ subset, as the clonal V region sequence was obtained at least once from a single parenchymal CD8+ or CD4+ cell. Only potentially functional rearrangments are listed. One clone defined by a nonfunctional rearrangement accounted for 2 of the 17 nonfunctional rearrangements obtained in total. This clone could not be assigned to a clone defined by a potentially functional rearrangement. Within the parenchyma, CD8+ T cells outnumbered CD4+ T cells by a factor of five to six, and roughly by a factor of three in the perivascular location.
To analyze CD4+ and CD8+ T lymphocytes infiltrating active and inactive lesions for clonal composition and TCR repertoire, sections adjacent to those used for the mapping of lesional activity were stained for CD4, CD8, and CD3. Single parenchymal CD4+ or CD8+ and a small number of parenchymal CD3+ T cells were micromanipulated and transferred to PCR tubes. Within the perivascular infiltrates, reliable identification of individual CD4+ or CD8+ cells was difficult because of dense aggregation of the cells and leakage of the dye from positive cells into the surrounding tissue. Furthermore, micromanipulation of single cells from this location was hampered by the presence of connective tissue. Therefore, instead of single CD4+ and CD8+ cells, samples of 5–10 cells were micromanipulated from perivascular infiltrates and transferred to PCR tubes without paying attention to morphology or immunostaining of individual cells. These samples were mainly composed of T cells and macrophages.
TCR-β gene rearrangements were amplified from the genomic DNA of micromanipulated single parenchymal cells and samples of perivascular cells using a mixture of 25 Vβ family– and 8 Jβ-specific primers in the first of two rounds of amplification as described 20. PCR products were directly sequenced. Single astrocytes micromanipulated from adjacent sections (immunostained for the astrocyte marker GFAP), aliquots of the buffer covering the sections during the micromanipulation procedure (“buffer samples”), and water controls were analyzed in parallel as controls for cellular or PCR product contamination (Table ). Whereas, 229 of 798 single parenchymal cells (29%) and 86 of 140 samples of perivascular cells (61%) analyzed in total from both cases were positive for at least one specific PCR product, none of the negative control samples yielded a TCR gene rearrangement (Table ). The efficiency of single target amplification of TCR-β gene rearrangements was controlled by analysis of single TCR-α/β+CD3+ cells from blood of healthy donors sorted into PCR tubes by FACS®. Specific products could be amplified from 66 of 90 of such single cells (73%; Table ). These products were all unique and unrelated to those obtained from the micromanipulated cells. The results of the control amplifications are in accordance with our previous data, which suggest that the amplification of TCR-β gene rearrangements from single cells is reliable with respect to unbiased amplification of the majority of all possible rearrangements and the assignment of sequences to individual cells (20; data not shown). Additional evidence for the reliability of the combination of micromanipulation and single cell PCR was derived from the observation that rearrangements that had been obtained from micromanipulated CD8+ cells were never found in CD4+ cells or vice versa, although repeats were frequent within both the CD4+ and especially the CD8+ compartment.
Table 1.
Summary of Single Cell PCR Analysis of T Cells Micromanipulated from MS Lesions of Cases 1 and 2
| Samples analyzed | Positive for at least onespecific PCR product | Total no. of potentially functional rearrangements | |
|---|---|---|---|
| Case 1 | 547 single parenchymal T cells | 141/547 (26%) | 133 |
| 107 samples of perivascular cells | 60/107 (56%) | 86 | |
| Case 2 | 251 single parenchymal T cells | 88/251 (35%) | 76 |
| 33 samples of perivascular cells | 26/33 (79%) | 46 | |
| Controls | 90 micromanipulated GFAP+ astrocytes | 0/90 | — |
| 90 buffer samples | 0/90 | — | |
| 90 water controls | 0/90 | — | |
| 90 single FACS®-sorted T cells | 66/90 (73%) | 58 |
Frozen sections of MS lesions were used to micromanipulate samples of 5–10 cells from perivascular infiltrates and single parenchymal T cells located outside of perivascular infiltrates. TCR-β gene rearrangements were amplified from these samples. Single astrocytes micromanipulated from adjacent sections, aliquots of the buffer covering the sections during the micromanipulation procedure, and control tubes containing PCR buffer but no cells served as negative controls. Single T cells sorted from blood of healthy donors were used to control for the efficiency of single target amplification. Among the 448 rearrangements that were amplified in total from micromanipulated and control T cells and for which the reading frame could be unequivocally determined, 49 were nonfunctional (11%; i.e., either stop codon in CDR3, rearrangement of a pseudogene, or out-of-frame rearrangement). As this study focused on the detection of potentially functional rearrangements, additional bands amplified from single cells that may potentially represent nonfunctional rearrangements were not necessarily sequenced. Therefore, our results probably underestimate the prevalence of nonfunctional rearrangements in human T cells. To exclude the possibility that a substantial number of rearrangements was derived from γ/δ instead of α/β T cells (potentially functional TCR-β gene rearrangements were described in γ/δ T cells [reference 65]), adjacent tissue sections were stained for TCR-γ/δ or -α/β. In both cases, γ/δ T cells accounted for <3% of all T cells.
Oligoclonal Expansions Account for the Majority of CD8+ T Cells in Active and Inactive MS Lesions.
Table and Table assign potentially functional TCR-β gene rearrangements to the respective cell samples from which they had been amplified (parenchymal CD4+ and CD8+ or perivascular cells). Of the 81 single parenchymal CD8+ T cells from four different lesions of case 1 that yielded a potentially functional TCR-β gene rearrangement, 28 were unique; i.e., they were obtained from one particular cell sample only (Table ). However, 53 sequences (65%) were composed of 13 gene rearrangements, each of which was amplified from more than one cell, and therefore represented an expanded CD8+ T cell clone. One of these clones (clone 1) was by far the most prominent. Its clonal rearrangement accounted for 28 of all 81 parenchymal CD8+ T cells (35%). The remaining CD8+ T cell clones were represented between one and five times among the 81 parenchymal CD8+ cells.
To identify potential differences in clonal composition and TCR repertoire between active and inactive lesions, two lesions were compared. Lesion 2 exhibited signs of active demyelination, and ORO+ macrophages throughout the affected area, whereas lesion 1, although heavily infiltrated by macrophages and T cells, was inactive (Fig. 1) and ORO− (described in Materials and Methods). The comparison was confined to the CD8+ subset because, in contrast to a large number of CD8+ cells, only very few CD4+ T cells could be identified in these two lesions. The frequency of clone 1 was about twofold higher in inactive lesion 1 compared with active lesion 2. The major CD8+ T cell clones (clones 1, 2, and 9) were found in lesion 1 as well as in lesion 2. In lesion 1, 15 additional rearrangements were obtained from CD3+ cells. Like those derived from CD8+ T cells, the majority could be assigned to clonal expansions (10/15 [67%]). Only members of CD8+ clones were encountered (clone 1, 6/15 [40%]; clone 2, 3/15; clone 19, 1/15).
The composition of CD8+ T cells infiltrating lesions 3 and 4, both of which showed active as well as inactive areas (Fig. 1), was similar to that of lesions 1 and 2. Clone 1 was the most prevalent clone in all four lesions. Likewise, members of most other clones were detected in at least two lesions.
Among 24 rearrangements obtained from single parenchymal CD8+ cells of case 2, 17 (71%) originated from 9 different clones, whereas only 7 sequences were unique (Table ).
The Population of CD4+ T Cells is more Heterogeneous than the CD8+ Population.
28 of 37 gene rearrangements amplified from parenchymal CD4+ T cells of case 1 were unique, whereas four rearrangements were obtained repeatedly and accounted for 9 of the 37 cells (24%; Table ). In case 2, 46 of the 52 parenchymal CD4+ T cells could not be assigned to any clonal expansion, whereas 6 (12%) belonged to three clones (Table ). Thus, parenchymal CD4+ T cells were more heterogeneous than parenchymal CD8+ T cells in both cases analyzed.
Most CD4+ and CD8+ Clones were Represented in Perivascular Infiltrates As Well As Among Parenchymal T Cells.
As shown in Table , 45% of the 86 potentially functional rearrangements amplified from samples of perivascular cells in case 1 could be assigned to clonal expansions. The major clones (more than three members detected) were all encountered in the parenchymal as well as in the perivascular location.
Of the 46 sequences derived from perivascular cells in case 2, 26% could be assigned to clonal expansions. Except for clone 5, the major clones (more than two members detected) were also represented in both locations in this case (Table ).
Region Sequences of Some CD8+ T Cells in Case 1 Showed Signs of Selection for Common Antigenic Specificities.
Potentially functional TCR-β gene rearrangements were analyzed for similarities of deduced CDR3 amino acid sequences and biased Vβ or Jβ gene segment usage, which might suggest selection for recognition of common epitopes. Analysis of CDR3 sequences of case 1 revealed that the V region amino acid sequence of CD8+ clone 7 was identical to an unique rearrangement amplified from perivascular sample A278 (Table ). This finding of clonally independent T cells (nucleotide sequences shown in Table ) with identical β chains within a limited number of cells strongly suggests that these cells were engaged in a response against the same epitope. Dominant clone 1 shared part of the NDN-encoded amino acid sequence motif (SGSG) of this β chain in the same position relative to the conserved cysteine of the Vβ segment, whereas unique sequence H518 shared all four amino acids. No recurrent CDR3 amino acid motifs were found in the sequences obtained from the remaining CD8+ or CD4+ T cells or from T cells from case 2.
Table 4.
Case 1: CDR3 Nucleotide and Deduced Amino Acid Sequences of TCR-β Gene Rearrangements Amplified from Micromanipulated T Cells
| Clone/sample | Vβ | VNDN | J | Jβ | CDR3 length | |
|---|---|---|---|---|---|---|
| A278 | 11S1 | CASSESGSG | EKLFFG | 1S4 | 10 | |
| TGTGCCAGCAGTGAATCCGGATCTGGC | GAAAAACTGTTTTTTGGC | |||||
| 7 | 11S1 | CASSESGSG | EKLFFG | 1S4 | 10 | |
| TGTGCCAGCAGTGAAAGTGGGTCGGGG | GAAAAACTGTTTTTTGGC | |||||
| 1 | 6S5 | CASSLSGQG | DYGYTF G | 1S2 | 11 | |
| H518 | 13S3 | CATGVS GSGG | EQFFG | 2S1 | 10 | |
Sequences A278 and H518 were both amplified from samples of perivascular cells (CD8+ clones 1 and 7 are introduced in Table ). CDR3 sequences are shown from the conserved cysteine of the Vβ to the conserved FG motif of the Jβ elements. Amino acids of the common SGSG motif are shown in bold. CDR3 lengths definition was as in Moss and Bell (reference 66).
16 of 37 CD4+ T cells (43%) amplified from case 1 harbored a potentially functional rearrangement incorporating Jβ1S1. Among unselected T cells from healthy individuals, only 9–15% have been reported to express rearrangements of this Jβ gene segment 34 35. Likewise, a control collection of 196 potentially functional gene rearrangements from peripheral blood of five healthy donors (20; this study, and data not shown) contained only 12% of rearrangements of Jβ1S1. Except for the overrepresentation of Jβ1S1 in rearrangements from CD4+ T cells of case 1, the distribution of Vβ and Jβ gene segments resembled that determined in the control collection in both cases analyzed, thus providing no further evidence that the micromanipulated cells were sampled from an antigen-selected population.
In Case 2, a CD8+ Clone Identified in Brain Tissue Was Also Detected in Blood Samples Obtained 12 and 31 mo after Brain Biopsy by CDR3 Spectratyping.
Two blood samples of case 2 obtained 12 and 31 mo after surgical removal of the lesion were available. To determine whether clonal expansions of T cells identified in the brain lesion were also encountered at a high frequency in peripheral blood T cells, and thus may in principle be amenable to isolation and cloning procedures, CDR3 spectratyping (Immunoscope) analyses were carried out. This PCR-based technique allows examination of length distributions in TCR-β transcripts from PBMCs, and thereby detection of expanded T cell clones. Clonal expansions stand out as prominent peaks from the overall CDR3 length distribution obtained for polyclonal T cell populations 36. This technique can detect clonal T cell populations among polyclonal T cells with a sensitivity of ∼1 in 5 × 103 depending on the clonal CDR3 length and the primer combination used 32 37.
FACS®-sorted CD4+ and CD8+ T cells were investigated. A prominent peak in the spectragram of the Vβ14-Jβ1S1 subpopulation of CD8+ T cells was shown (by cloning of the PCR product and sequencing of plasmid DNA from bacterial colonies) to reflect the presence of CD8+ clone 10 (Table ) in peripheral blood T cells at both time points analyzed (Fig. 2). No other clonal rearrangement identified in the brain sections was detected in blood samples by the spectratyping technique.
Figure 2.
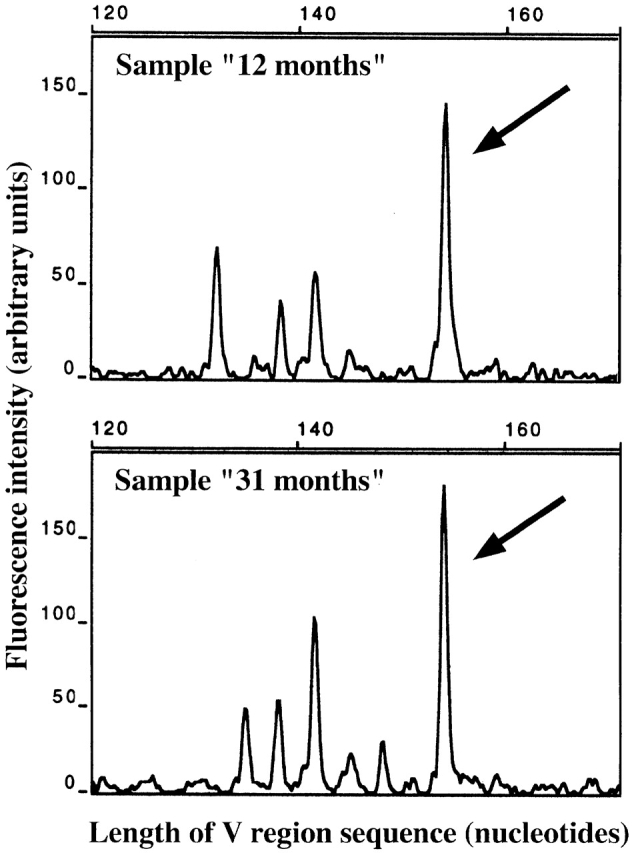
Detection of clone 10 (identified in brain tissue of case 2) in peripheral blood T cells by CDR3 spectratyping analysis. mRNA extracted from FACS®-sorted CD8+ T cells was reverse transcribed. TCR-β cDNA was amplified using 1 Cβ- and 26 Vβ-specific primers. PCR products were used as templates in runoff reactions with fluorescently labeled Jβ-specific primers, and runoff products were analyzed on an automated DNA sequencer. The products shown were amplified with the Vβ14-Cβ primer pair and labeled with an Jβ1S1-specific primer in the runoff reaction. The spectratypes correspond to the blood samples obtained 12 mo (top) and 31 mo (bottom) after brain surgery. The prominent peaks marked by the arrows potentially represented the rearrangement of clone 10 (same V-J combination and CDR3 length). Direct sequencing of the Vβ14-Jβ1S1 PCR products resulted in mixed sequences that potentially contained the CDR3 sequence of clone 10. The products were cloned into a plasmid vector. After transformation of Escherichia coli and sequence analysis of plasmid DNA from bacterial colonies, the V region sequence of clone 10 was identified in 7 of 26 colonies (“12 months”) and 7 of 12 colonies (“31 months”).
A Second CD8+ Clone Identified in Brain Tissue Was Also Detected in Blood Samples of Case 2 by Clone-specific PCR.
In a second approach, primers for specific nested single target amplification of the TCR-β gene rearrangements of three clones identified in brain tissue (CD8+ clones 2, 3, and 8; Table ) were designed. Clone-specific PCR was established using Jurkat cells stably transfected with a single copy of either clonal rearrangement. Each set of clone-specific primers was capable of detecting one single Jurkat control cell among 103 irrelevant T cells from a healthy donor (described in Materials and Methods), and thus with a similar sensitivity as the reverse transcriptase PCR–based spectratyping technique.
102 or 103 CD8+CD45RO+ cells (expanded CD8+ T cell clones were assumed to belong to the effector/memory compartment, and thus to be CD45RO+ 38) from both blood samples (12 mo and 31 mo) were sorted into each of several PCR tubes. 104 cells were analyzed in total for each clone and time point. The V region sequence of clone 8 was amplified from 6 of 10 and 8 of 10 samples of 103 cells (blood samples 12 mo and 31 mo, respectively) and from 2 of 20 samples containing 102 cells (sample 31 mo; Table ). PCR products were shown to represent the clonal V region sequences by sequencing. This indicates that the frequency of clone 8 was ∼1 in 103 peripheral blood CD8+CD45RO+ T cells at both time points analyzed. The clonal rearrangement of clone 2 was amplified only once from all samples analyzed, and clone 3 was not found at all (Table ).
Table 5.
Detection of Clone 8 (Identified in Brain Tissue of Case 2) in Peripheral Blood CD8+CD45RO+ T Cells by Clone-specific PCR
| Cell samples | |||
|---|---|---|---|
| Blood sample; no. of cells per PCR tube | PCR specific for V region of clone no. | Analyzed by clone-specific PCR | Positive for specific product |
| 12 mo; 103 | 2 | 10 | 0/10 |
| 31 mo; 103 | 2 | 10 | 1/10 |
| 12 mo; 103 | 3 | 9 | 0/9 |
| 12 mo; 5 × 102 | 3 | 1 | 0/1 |
| 12 mo; 102 | 3 | 5 | 0/5 |
| 31 mo; 103 | 3 | 10 | 0/10 |
| 12 mo; 103 | 8 | 10 | 6/10 |
| 31 mo; 103 | 8 | 10 | 8/10 |
| 31 mo; 102 | 8 | 20 | 2/20 |
Each set of primers consisted of the Vβ-specific primer and a nested pair of CDR3-specific primers. The clonal V region sequences were inserted into plasmid vectors, and the resulting constructs were stably transfected into Jurkat cells. To positively control single target amplification of the clonal V region sequence, single Jurkat control cells carrying a single copy of the respective V region DNA were sorted into tubes containing, as indicated, 103 or 102 irrelevant CD8+CD45RO+ T cells from a healthy donor. These samples were analyzed in parallel with the T cell samples from blood of case 2 under identical conditions (results of control amplifications are discussed in Materials and Methods).
Discussion
Clonal composition and TCR repertoire of CD4+ and CD8+ T cells infiltrating actively demyelinating brain lesions in two cases of MS were determined at the level of single cells. Suitable brain tissue was identified by screening autopsy and biopsy material of several cases for the presence of actively demyelinating lesions, using as an indicator the molecular composition of myelin degradation products in macrophages and an inflammatory activation marker on the macrophage surface 3. Active lesions, generally rare in MS brain material obtained postmortem 5 13, were detected in material of case 1. A large actively demyelinating lesion was also found in the biopsy of case 2, which likely represented an early stage of disease development, as the lesion was already resected 2 wk after the first clinical manifestation of MS. In this case, it was possible to relate findings in brain tissue to analyses of peripheral blood obtained after brain biopsy.
Frozen sections were stained for CD4 or CD8. In both cases, CD8+ T cells were far more numerous than CD4+ T cells in the parenchyma of all lesions studied. Some earlier studies described a preponderance of CD4+ over CD8+ T cells in MS lesions, whereas the opposite was reported by several other groups 13 14.
Individual CD4+ and CD8+ T cells were micromanipulated from the stained sections and subjected to single target amplification of TCR-β gene rearrangements using a complex mixture of Vβ- and Jβ-specific primers that was shown previously to reliably amplify the various V-J combinations from single cells 20. Amplified V region genes were directly sequenced. This strategy allows the characterization of T cell infiltrates in MS lesions with unprecedented resolution. In contrast to approaches based on the analysis of total RNA extracted from whole tissue specimens 14 15 16 17, in this study, sequences could be assigned to individual T cells in particular locations within the diseased tissue. Thus, it was possible to differentially investigate the composition of CD4+ and CD8+ T cell populations in perivascular aggregates or the parenchyma of active and inactive lesions. It should be noted that the combination of micromanipulation and single target amplification yields quantitative data, in contrast to semiquantitative reverse transcriptase PCR–based approaches used previously to study TCR repertoires in MS lesions 14 15 16 17 39, with the inherent problems of sequence representation. The frequency of detection of particular gene rearrangements in our analysis closely parallels the actual representation of the respective clone in the brain tissue.
Expansions of both CD4+ and CD8+ T cell clones were detected in the lesions. In both cases, the majority of the parenchymal CD8+ T cell population was composed of few clones. A background of polyclonal cells accounted for only 24–46% of all CD8+ cells in the five lesions analyzed. One CD8+ clone (clone 1) was particularly prominent accounting for 35% of the CD8+ cells in case 1. Among the parenchymal CD4+ cells of both cases, the fraction of cells accounted for by oligoclonal expansions was considerably lower. The majority of rearrangements (68–88%) could not be assigned to CD4+ T cell clones expanded in situ.
The finding of expanded T cell clones in MS lesions raises the question of their functional properties. CD4+ and CD8+ T cells may be involved in pathogenic immune responses directed against brain autoantigens and cause oligodendrocyte damage by direct cytolysis or cytokine secretion. The pathogenic potential of CD4+ T cells specific for myelin antigens is well established in animal models of demyelinating disease 7 40. Evidence for a pathogenic role of CD4+ T cells in MS was provided by the finding of increased frequencies of activated myelin-reactive T cells in the CSF of MS patients compared with blood of the patients or with other neurological diseases 41 42. Furthermore, the association of MS with particular MHC class II haplotypes suggests that presentation of antigen to CD4+ T cells may be an important pathogenic event 2 6. More recently, the role of CD8+ T cells in inflammatory myelin destruction was addressed in more detail (for a review, see reference 43). CD8+ T cells have been shown to contribute to tissue injury in animal models of inflammatory demyelination 44 45. CTLs specific for human myelin proteins were detected in blood of MS patients and healthy individuals 46 47. Human oligodendrocytes were shown to be capable of expressing MHC class I in vitro 48 49 and to be susceptible to lysis by myelin basic protein–specific CTLs 47. PLP-specific CD8+ T cells were shown to be capable of secreting proinflammatory chemokines 50 and of chemoattracting myelin-specific CD4+ T cells 51. In addition to pathogenic autoreactive T cells, MS lesions may contain T cell clones that do not promote but instead counteract the destructive process. In EAE, effects of both CD4+ and CD8+ regulatory T cells have been described 44 52 53 54 55.
In addition to autoreactive and regulatory T cell clones, clones not primarily connected to the autoimmune process may be present in the brain lesions. Activated T cells have the capacity to cross the blood–brain barrier and home to sites of inflammation irrespective of their antigenic specificity 56 57. Clonal expansions of CD8+ T cells were found in the blood of MS patients 12. However, the fraction of activated T cells, in particular the activated CD8+ subset, often contains large clonal expansions even in healthy individuals 36 58 59 60. Such oligoclonal expansions may be stable over years. They are preferentially found in elderly individuals and may represent chronic T cell responses to persistent infections 60. In rheumatoid arthritis but also in arthritis of nonimmunological pathogenesis (arthrosis), expansions of herpes virus–specific T cells have been detected in synovial fluid, suggesting that these cells were nonspecifically trapped at the site of inflammation 61. Therefore, the clonal expansions detected in MS lesions in this study may comprise clones not originally related to the pathogenic process.
The distribution of encephalitogenic T cells within central nervous system lesions at the peak of disease after induction of EAE by transfer of a myelin basic protein–specific Vβ8.2-expressing T cell line was investigated in Lewis rats 18 19. Although Vβ8.2+ cells constituted a minor fraction of α/β T cells in the perivascular infiltrates (reflecting the low frequency of Vβ8.2+ cells in the normal rat T cell repertoire), Vβ8.2+ cells were enriched among parenchymal T cells. One could therefore have speculated that in MS, parenchymal T cells may also differ fundamentally in repertoire and function from T cells in perivascular infiltrates, with a large fraction of the latter being nonspecifically attracted bystander cells, whereas most encephalitogenic T cells are located in the parenchyma. However, in this study, comparison of clonal composition and TCR repertoire of perivascular versus parenchymal T cells in MS lesions revealed no striking differences, as members of most larger clonal expansions were detected in both locations. The clonal rearrangement of the most prominent expansion in case 1, clone 1 (35% of CD8+ T cells), accounted for a similar fraction of parenchymal and perivascular CD8+ T cells (considering that roughly three quarters of perivascular T cells were CD8+).
Likewise, a comparison of the clonal composition of CD8+ T cells infiltrating an actively demyelinating versus an inactive lesion did not reveal striking differences except for a twofold-higher frequency of CD8+ clone 1 in the inactive lesion. Interpretations of this finding may be that clone 1 was a suppressive clone responsible for the downregulation of demyelinating activity in the inactive lesion 44 52 53. Alternatively, it may reflect a concept proposed by Steinman and colleagues 62 63 64 based on findings in EAE suggesting that pathogenetically relevant T cells may be encountered more frequently in inactive lesions where they may be less diluted by nonspecifically attracted cells.
In case 1, some of the gene rearrangements from CD8+ T cells showed sequence similarities that strongly suggested selection for common peptide specificities. It is tempting to speculate that these cells were actively involved in the disease process. However, nonspecific recruitment of T cell populations selected in responses unrelated to MS pathogenesis is an alternative explanation 61.
In summary, the T cell infiltrate in MS lesions of two cases was shown to be dominated by few CD8+ clones. CD4+ T cells were generally less numerous and more heterogeneous. Nevertheless, some clonal expansions were also detected in this population. In one case, comparison of TCR β-V region sequences provided evidence that some of the CD8+ T cells were selected for recognition of common epitopes. The spatial distribution of members of the CD4+ and CD8+ clones within the lesion and between lesions differing in demyelinating activity did not provide direct clues to the question of their pathogenetic relevance. Knowledge of the antigenic specificity of clones expanded in the brain lesions may help to elucidate their functional properties. Two of these clones were detected in peripheral blood of case 2 at two different time points after brain biopsy, one by CDR3 spectratyping, the other by a sensitive clone-specific PCR. Experiments aiming at the isolation of these clones by cultivation of CD8+ T cells from the blood of the patient are presently under way. After cloning by limiting dilution, detection of cells belonging to the clones in question should be possible by PCR. A complementary approach applicable to the analysis of clones from both cases aims at coamplification of both TCR-α and -β gene rearrangements from individual micromanipulated cells. Primer sets for the amplification of TCR-α gene rearrangements from single cells are currently being established. Cloning of amplified α and β V region sequences into TCR expression vectors should allow transfer of clonal receptor specificities to cell lines and subsequent in vitro analysis of antigenic specificities.
Acknowledgments
The authors thank Drs. E.D. Albert and E. Keller for the HLA typing, Dr. R. Küppers for his critical reading of the manuscript and valuable discussions, and D. Grimme for helpful comments. C. Göttlinger, J. Jesdinsky, A. Klöckner, and A. Fassbender are acknowledged for excellent technical assistance. We thank Drs. H.P. Dienes, P. Schirmacher, and M. Odenthal for their support and discussions.
This work was supported by Deutsche Forschungsgemeinschaft grants RA 131/5-1 and Go 514/4-1.
Footnotes
H. Babbe and A. Roers contributed equally to this work.
Abbreviations used in this paper: CSF, cerebrospinal fluid; EAE, experimental autoimmune encephalomyelitis; GFAP, glial fibrillary acidic protein; MS, multiple sclerosis; ORO, Oil Red O; PLP, proteolipid protein; WM, white matter.
References
- Martin R., McFarland H.F., McFarlin D.E. Immunological aspects of demyelinating diseases. Annu. Rev. Immunol. 1992;10:153–187. doi: 10.1146/annurev.iy.10.040192.001101. [DOI] [PubMed] [Google Scholar]
- Noseworthy J.H. Progress in determining the causes and treatment of multiple sclerosis Nature 3996738 Suppl.1999. A40 A47 [DOI] [PubMed] [Google Scholar]
- Lucchinetti C.F., Brück W., Rodriguez M., Lassmann H. Distinct patterns of multiple sclerosis pathology indicates heterogeneity on pathogenesis. Brain Pathol. 1996;6:259–274. doi: 10.1111/j.1750-3639.1996.tb00854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan C.F., Raine C.S. Mechanisms of immune injury in multiple sclerosis. Brain Pathol. 1996;6:243–257. doi: 10.1111/j.1750-3639.1996.tb00853.x. [DOI] [PubMed] [Google Scholar]
- Lassmann H., Raine C.S., Antel J., Prineas J.W. Immunopathology of multiple sclerosisreport on an international meeting held at the Institute of Neurology of the University of Vienna. J. Neuroimmunol. 1998;86:213–217. doi: 10.1016/s0165-5728(98)00031-9. [DOI] [PubMed] [Google Scholar]
- Steinman L. Multiple sclerosisa coordinated immunological attack against myelin in the central nervous system. Cell. 1996;85:299–302. doi: 10.1016/s0092-8674(00)81107-1. [DOI] [PubMed] [Google Scholar]
- Wekerle H., Kojima K., Lannes-Vieira J., Lassmann H., Linington C. Animal models Ann. Neurol. 36Suppl.1994. S47 S53 [DOI] [PubMed] [Google Scholar]
- Genain C.P., Cannella B., Hauser S.L., Raine C.S. Identification of autoantibodies associated with myelin damage in multiple sclerosis. Nat. Med. 1999;5:170–175. doi: 10.1038/5532. [DOI] [PubMed] [Google Scholar]
- Hafler D.A., Weiner H.L. Immunologic mechanisms and therapy in multiple sclerosis. Immunol. Rev. 1995;144:75–107. doi: 10.1111/j.1600-065x.1995.tb00066.x. [DOI] [PubMed] [Google Scholar]
- Hafler D.A., Duby A.D., Lee S.J., Benjamin D., Seidman J.G., Weiner H.L. Oligoclonal T lymphocytes in the cerebrospinal fluid of patients with multiple sclerosis. J. Exp. Med. 1988;167:1313–1322. doi: 10.1084/jem.167.4.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.J., Wucherpfennig K.W., Brod S.A., Benjamin D., Weiner H.L., Hafler D.A. Common T cell receptor V beta usage in oligoclonal T lymphocytes derived from cerebrospinal fluid and blood of patients with multiple sclerosis. Ann. Neurol. 1991;29:33–40. doi: 10.1002/ana.410290109. [DOI] [PubMed] [Google Scholar]
- Monteiro J., Hingorani R., Peroglizzi R., Apatoff B., Gregersen P.K. Oligoclonality of CD8+ T cells in multiple sclerosis. Autoimmunity. 1996;23:127–138. doi: 10.3109/08916939608995336. [DOI] [PubMed] [Google Scholar]
- Sobel R.A. T-lymphocyte subsets in the multiple sclerosis lesion. Res. Immunol. 1989;140:208–211. doi: 10.1016/0923-2494(89)90088-6. [DOI] [PubMed] [Google Scholar]
- Wucherpfennig K.W., Newcombe J., Li H., Keddy C., Cuzner M.L., Hafler D.A. T cell receptor V alpha-V beta repertoire and cytokine gene expression in active multiple sclerosis lesions. J. Exp. Med. 1992;175:993–1002. doi: 10.1084/jem.175.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenberg J.R., Stuart S., Begovich A.B., Bell R.B., Erlich H.A., Steinman L., Bernard C.C. Limited heterogeneity of rearranged T cell receptor V alpha transcripts in brains of multiple sclerosis patients Nature 345 1990. 344 346[published erratum at 353:94] [DOI] [PubMed] [Google Scholar]
- Birnbaum G., van Ness B. Quantitation of T cell receptor V beta chain expression on lymphocytes from blood, brain, and spinal fluid in patients with multiple sclerosis and other neurological diseases. Ann. Neurol. 1992;32:24–30. doi: 10.1002/ana.410320106. [DOI] [PubMed] [Google Scholar]
- Oksenberg J.R., Panzara M.A., Begovich A.B., Mitchell D., Erlich H.A., Murray R.S., Shimonkevitz R., Sherritt M., Rothbard J., Bernard C.C., Steinman L. Selection for T cell receptor V beta-D beta-J beta gene rearrangements with specificity for a myelin basic protein peptide in brain lesions of multiple sclerosis. Nature. 1993;362:68–70. doi: 10.1038/362068a0. [DOI] [PubMed] [Google Scholar]
- Tsuchida M., Matsumoto Y., Hirahara H., Hanawa H., Tomiyama K., Abo T. Preferential distribution of V beta 8.2-positive T cells in the central nervous system of rats with myelin basic protein-induced autoimmune encephalomyelitis. Eur. J. Immunol. 1993;23:2399–2406. doi: 10.1002/eji.1830231004. [DOI] [PubMed] [Google Scholar]
- Lannes-Vieira J., Gehrmann J., Kreutzberg G.W., Wekerle H. The inflammatory lesion of T cell line transferred experimental autoimmune encephalomyelitis of the Lewis ratdistinct nature of parenchymal and perivascular infiltrates. Acta Neuropathol. 1994;87:435–442. doi: 10.1007/BF00294169. [DOI] [PubMed] [Google Scholar]
- Roers A., Montesinos-Rongen M., Hansmann M.L., Rajewsky K., Küppers R. Amplification of TCRbeta gene rearrangements from micromanipulated single cellsT cells rosetting around Hodgkin and Reed-Sternberg cells in Hodgkin's disease are polyclonal. Eur. J. Immunol. 1998;28:2424–2431. doi: 10.1002/(SICI)1521-4141(199808)28:08<2424::AID-IMMU2424>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Blasczyk R., Wehling J., Weber M., Salama A. Sequence analysis of the 2nd intron revealed common sequence motifs providing the means for a unique sequencing based typing protocol of the HLA-A locus. Tissue Antigens. 1996;47:102–110. doi: 10.1111/j.1399-0039.1996.tb02521.x. [DOI] [PubMed] [Google Scholar]
- Pozzi S., Longo A., Ferrara G.B. HLA-B locus sequence-based typing. Tissue Antigens. 1999;53:275–281. doi: 10.1034/j.1399-0039.1999.530308.x. [DOI] [PubMed] [Google Scholar]
- Delfino L., Morabito A., Longo A., Ferrara G.B. HLA-C high resolution typinganalysis of exons 2 and 3 by sequence based typing and detection of polymorphisms in exons 1-5 by sequence specific primers. Tissue Antigens. 1998;52:251–259. doi: 10.1111/j.1399-0039.1998.tb03040.x. [DOI] [PubMed] [Google Scholar]
- Olerup O., Zetterquist H. HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hoursan alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens. 1992;39:225–235. doi: 10.1111/j.1399-0039.1992.tb01940.x. [DOI] [PubMed] [Google Scholar]
- Olerup O., Aldener A., Fogdell A. HLA-DQB1 and -DQA1 typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours. Tissue Antigens. 1993;41:119–134. doi: 10.1111/j.1399-0039.1993.tb01991.x. [DOI] [PubMed] [Google Scholar]
- Mierau R., Dick T., Bartz-Bazzanella P., Keller E., Albert E.D., Genth E. Strong association of dermatomyositis-specific Mi-2 autoantibodies with a tryptophan at position 9 of the HLA-DR beta chain. Arthritis Rheum. 1996;39:868–876. doi: 10.1002/art.1780390521. [DOI] [PubMed] [Google Scholar]
- Brück W., Porada P., Poser S., Rieckmann P., Hanefeld F., Kretzschmar H.A., Lassmann H. Monocyte/macrophage differentiation in early multiple sclerosis lesions. Ann. Neurol. 1995;38:788–796. doi: 10.1002/ana.410380514. [DOI] [PubMed] [Google Scholar]
- Brück W., Schmied M., Suchanek G., Bruck Y., Breitschopf H., Poser S., Piddlesden S., Lassmann H. Oligodendrocytes in the early course of multiple sclerosis. Ann. Neurol. 1994;35:65–73. doi: 10.1002/ana.410350111. [DOI] [PubMed] [Google Scholar]
- Ozawa K., Suchanek G., Breitschopf H., Brück W., Budka H., Jellinger K., Lassmann H. Patterns of oligodendroglia pathology in multiple sclerosis. Brain. 1994;117:1311–1322. doi: 10.1093/brain/117.6.1311. [DOI] [PubMed] [Google Scholar]
- Piddlesden S.J., Lassmann H., Zimprich F., Morgan B.P., Linington C. The demyelinating potential of antibodies to myelin oligodendrocyte glycoprotein is related to their ability to fix complement. Am. J. Pathol. 1993;143:555–564. [PMC free article] [PubMed] [Google Scholar]
- Küppers R., Zhao M., Hansmann M.L., Rajewsky K. Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:4955–4967. doi: 10.1002/j.1460-2075.1993.tb06189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebels N., Hofstetter H., Schmidt S., Brunner C., Wekerle H., Hohlfeld R. Repertoire dynamics of autoreactive T cells in multiple sclerosis patients and healthy subjectsepitope spreading versus clonal persistence. Brain. 2000;123:508–518. doi: 10.1093/brain/123.3.508. [DOI] [PubMed] [Google Scholar]
- Puisieux I., Even J., Pannetier C., Jotereau F., Favrot M., Kourilsky P. Oligoclonality of tumor-infiltrating lymphocytes from human melanomas. J. Immunol. 1994;153:2807–2818. [PubMed] [Google Scholar]
- Hall M.A., Lanchbury J.S. Healthy human T cell receptor beta-chain repertoire. Quantitative analysis and evidence for J beta-related effects on CDR3 structure and diversity. Hum. Immunol. 1995;43:207–218. doi: 10.1016/0198-8859(95)00013-t. [DOI] [PubMed] [Google Scholar]
- Rosenberg W.M., Moss P.A., Bell J.I. Variation in human T cell receptor V beta and J beta repertoireanalysis using anchor polymerase chain reaction. Eur. J. Immunol. 1992;22:541–549. doi: 10.1002/eji.1830220237. [DOI] [PubMed] [Google Scholar]
- Pannetier C., Even J., Kourilsky P. T cell repertoire diversity and clonal expansions in normal and clinical samples. Immunol. Today. 1995;16:176–181. doi: 10.1016/0167-5699(95)80117-0. [DOI] [PubMed] [Google Scholar]
- Cochet M., Pannetier C., Regnault A., Darche S., Leclerc C., Kourilsky P. Molecular detection and in vivo analysis of the specific T cell response to a protein antigen. Eur. J. Immunol. 1992;22:2639–2647. doi: 10.1002/eji.1830221025. [DOI] [PubMed] [Google Scholar]
- Dutton R.W., Bradley L.M., Swain S.L. T cell memory. Annu. Rev. Immunol. 1998;16:201–223. doi: 10.1146/annurev.immunol.16.1.201. [DOI] [PubMed] [Google Scholar]
- Shimonkevitz R., Murray R., Kotzin B. Characterization of T cell receptor V beta usage in the brain of a subject with multiple sclerosis. Ann. NY Acad. Sci. 1995;756:305–306. doi: 10.1111/j.1749-6632.1995.tb44527.x. [DOI] [PubMed] [Google Scholar]
- Lassmann H., Zimprich F., Rossler K., Vass K. Inflammation in the nervous system. Basic mechanisms and immunological concepts. Rev. Neurol. (Paris) 1991;147:763–781. [PubMed] [Google Scholar]
- Chou Y.K., Bourdette D.N., Offner H., Whitham R., Wang R.Y., Hashim G.A., Vandenbark A.A. Frequency of T cells specific for myelin basic protein and myelin proteolipid protein in blood and cerebrospinal fluid in multiple sclerosis. J. Neuroimmunol. 1992;38:105–113. doi: 10.1016/0165-5728(92)90095-3. [DOI] [PubMed] [Google Scholar]
- Zhang J., Markovic-Plese S., Lacet B., Raus J., Weiner H.L., Hafler D.A. Increased frequency of interleukin 2–responsive T cells specific for myelin basic protein and proteolipid protein in peripheral blood and cerebrospinal fluid of patients with multiple sclerosis. J. Exp. Med. 1994;179:973–984. doi: 10.1084/jem.179.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizler C., Bercovici N., Cornet A., Cambouris C., Liblau R.S. Role of autoreactive CD8+ T cells in organ-specific autoimmune diseasesinsight from transgenic mouse models. Immunol. Rev. 1999;169:81–92. doi: 10.1111/j.1600-065x.1999.tb01308.x. [DOI] [PubMed] [Google Scholar]
- Koh D.R., Fung-Leung W.P., Ho A., Gray D., Acha-Orbea H., Mak T.W. Less mortality but more relapses in experimental allergic encephalomyelitis in CD8−/− mice. Science. 1992;256:1210–1213. doi: 10.1126/science.256.5060.1210. [DOI] [PubMed] [Google Scholar]
- Murray P.D., Pavelko K.D., Leibowitz J., Lin X., Rodriguez M. CD4+ and CD8+ T cells make discrete contributions to demyelination and neurologic disease in a viral model of multiple sclerosis. J. Virol. 1998;72:7320–7329. doi: 10.1128/jvi.72.9.7320-7329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T., Parker K.C., Turner R.V., McFarland H.F., Coligan J.E., Biddison W.E. Autoreactive CD8+ T cell responses to human myelin protein-derived peptides. Proc. Natl. Acad. Sci. USA. 1994;91:10859–10863. doi: 10.1073/pnas.91.23.10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurewicz A., Biddison W.E., Antel J.P. MHC class I-restricted lysis of human oligodendrocytes by myelin basic protein peptide-specific CD8 T lymphocytes. J. Immunol. 1998;160:3056–3059. [PubMed] [Google Scholar]
- Grenier Y., Ruijs T.C., Robitaille Y., Olivier A., Antel J.P. Immunohistochemical studies of adult human glial cells. J. Neuroimmunol. 1989;21:103–115. doi: 10.1016/0165-5728(89)90166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa P.T., Ozato K., McFarlin D.E. Cell type-specific regulation of major histocompatibility complex (MHC) class I gene expression in astrocytes, oligodendrocytes, and neurons. Glia. 1993;8:201–207. doi: 10.1002/glia.440080307. [DOI] [PubMed] [Google Scholar]
- Biddison W.E., Taub D.D., Cruikshank W.W., Center D.M., Connor E.W., Honma K. Chemokine and matrix metalloproteinase secretion by myelin proteolipid protein-specific CD8+ T cellspotential roles in inflammation. J. Immunol. 1997;158:3046–3053. [PubMed] [Google Scholar]
- Biddison W.E., Cruikshank W.W., Center D.M., Pelfrey C.M., Taub D.D., Turner R.V. CD8+ myelin peptide-specific T cells can chemoattract CD4+ myelin peptide-specific T cellsimportance of IFN-inducible protein 10. J. Immunol. 1998;160:444–448. [PubMed] [Google Scholar]
- Sun D., Qin Y., Chluba J., Epplen J.T., Wekerle H. Suppression of experimentally induced autoimmune encephalomyelitis by cytolytic T-T cell interactions. Nature. 1988;332:843–845. doi: 10.1038/332843a0. [DOI] [PubMed] [Google Scholar]
- Jiang H., Zhang S.I., Pernis B. Role of CD8+ T cells in murine experimental allergic encephalomyelitis. Science. 1992;256:1213–1215. doi: 10.1126/science.256.5060.1213. [DOI] [PubMed] [Google Scholar]
- Olivares-Villagomez D., Wang Y., Lafaille J.J. Regulatory CD4+ T cells expressing endogenous T cell receptor chains protect myelin basic protein–specific transgenic mice from spontaneous autoimmune encephalomyelitis. J. Exp. Med. 1998;188:1883–1894. doi: 10.1084/jem.188.10.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Keere F., Tonegawa S. CD4+ T cells prevent spontaneous experimental autoimmune encephalomyelitis in anti–myelin basic protein T cell receptor transgenic mice. J. Exp. Med. 1998;188:1875–1882. doi: 10.1084/jem.188.10.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wekerle H., Linington C., Lassmann H., Meyermann R. Cellular immune reactivity within the CNS. Trends Neurosci. 1986;9:271–277. [Google Scholar]
- Hickey W.F., Hsu B.L., Kimura H. T-lymphocyte entry into the central nervous system. J. Neurosci. Res. 1991;28:254–260. doi: 10.1002/jnr.490280213. [DOI] [PubMed] [Google Scholar]
- Hingorani R., Choi I.H., Akolkar P., Gulwani-Akolkar B., Pergolizzi R., Silver J., Gregersen P.K. Clonal predominance of T cell receptors within the CD8+ CD45RO+ subset in normal human subjects. J. Immunol. 1993;151:5762–5769. [PubMed] [Google Scholar]
- Posnett D.N., Sinha R., Kabak S., Russo C. Clonal populations of T cells in normal elderly humansthe T cell equivalent to “benign monoclonal gammapathy.” J. Exp. Med 179 1994. 609 618[published erratum appears at 179:1077] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Szabo P., Manavalan J.S., Weksler M.E., Posnett D.N., Pannetier C., Kourilsky P., Even J. Expanded CD4+ and CD8+ T cell clones in elderly humans. J. Immunol. 1997;158:4493–4499. [PubMed] [Google Scholar]
- Scotet E., Peyrat M.A., Saulquin X., Retiere C., Couedel C., Davodeau F., Dulphy N., Toubert A., Bignon J.D., Lim A. Frequent enrichment for CD8 T cells reactive against common herpes viruses in chronic inflammatory lesionstowards a reassessment of the physiopathological significance of T cell clonal expansions found in autoimmune inflammatory processes. Eur. J. Immunol. 1999;29:973–985. doi: 10.1002/(SICI)1521-4141(199903)29:03<973::AID-IMMU973>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Karin N., Szafer F., Mitchell D., Gold D.P., Steinman L. Selective and nonselective stages in homing of T lymphocytes to the central nervous system during experimental allergic encephalomyelitis. J. Immunol. 1993;150:4116–4124. [PubMed] [Google Scholar]
- Brocke S., Gijbels K., Allegretta M., Ferber I., Piercy C., Blankenstein T., Martin R., Utz U., Karin N., Mitchell D. Treatment of experimental encephalomyelitis with a peptide analogue of myelin basic protein Nature 379 1996. 343 346[published erratum appears at 392:630] [DOI] [PubMed] [Google Scholar]
- Steinman L. A few autoreactive cells in an autoimmune infiltrate control a vast population of nonspecific cellsa tale of smart bombs and the infantry. Proc. Natl. Acad. Sci. USA. 1996;93:2253–2256. doi: 10.1073/pnas.93.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey E., Fowlkes B.J. The alpha beta versus gamma delta T cell lineage choice. Curr. Opin. Immunol. 1998;10:181–187. doi: 10.1016/s0952-7915(98)80247-1. [DOI] [PubMed] [Google Scholar]
- Moss P.A., Bell J.I. Comparative sequence analysis of the human T cell receptor TCRA and TCRB CDR3 regions. Hum. Immunol. 1996;48:32–38. doi: 10.1016/0198-8859(96)00084-5. [DOI] [PubMed] [Google Scholar]