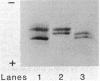
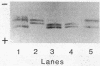
Abstract
The nitrogenase of the free-living, microaerobic, N2-fixing bacterium Azospirillum amazonense (strain Y1) was purified by chromatography on DEAE-52 cellulose, by heat treatment, and by preparative polyacrylamide gel electrophoresis. The specific nitrogenase activities were 2,400 nmol of C2H4 formed per min per mg of protein for dinitrogenase (MoFe protein) and 1,800 nmol of C2H4 formed per min per mg of protein for dinitrogenase reductase (Fe protein). The MoFe protein was composed of a minimum of 1,852 amino acid residues, had an isoelectric point of 5.2, and contained 2 atoms of Mo, 24 atoms of Fe, and 28 atoms of acid-labile sulfide per molecule. The Fe protein had 624 amino acid residues and an isoelectric point of 4.6 and contained four atoms of Fe and six atoms of acid-labile sulfide per molecule. The purified MoFe protein showed two subunits with molecular weights of 55,000 and 50,000. The purified Fe protein revealed two polypeptides on sodium dodecyl sulfate-polyacrylamide gel electrophoresis with apparent molecular weights of 35,000 and 31,000. The two Fe protein polypeptides were demonstrated with immunological techniques in the purified, highly active enzyme as well as in extracts. Also, Azotobacter vinelandii Fe protein showed two closely migrating polypeptides that migrated differently from the Fe protein polypeptides of Azospirillum brasilense or Rhodospirillum rubrum. The nitrogenase activity of Azospirillum amazonense Y1 was independent of Mn2+, and the addition of activating enzyme had no effect. No activating enzyme could be found in Azospirillum amazonense. Obviously, the nitrogenase system of Azospirillum amazonense Y1 is different from that of Azospirillum brasilense Sp7 and resembles the Azotobacter system.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BRUMBY P. E., MILLER R. W., MASSEY V. THE CONTENT AND POSSIBLE CATALYTIC SIGNIFICANCE OF LABILE SULFIDE IN SOME METALLOFLAVOPROTEINS. J Biol Chem. 1965 May;240:2222–2228. [PubMed] [Google Scholar]
- Bishop P. E., Jarlenski D. M., Hetherington D. R. Evidence for an alternative nitrogen fixation system in Azotobacter vinelandii. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7342–7346. doi: 10.1073/pnas.77.12.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braaksma A., Haaker H., Veeger C. Fully active Fe-protein of the nitrogenase from Azotobacter vinelandii contains at least eight iron atoms and eight sulphide atoms per molecule. Eur J Biochem. 1983 Jun 1;133(1):71–76. doi: 10.1111/j.1432-1033.1983.tb07430.x. [DOI] [PubMed] [Google Scholar]
- Burris R. H. Nitrogen fixation--assay methods and techniques. Methods Enzymol. 1972;24:415–431. doi: 10.1016/0076-6879(72)24088-5. [DOI] [PubMed] [Google Scholar]
- Emerich D. W., Burris R. H. Complementary functioning of the component proteins of nitrogenase from several bacteria. J Bacteriol. 1978 Jun;134(3):936–943. doi: 10.1128/jb.134.3.936-943.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOA J. A micro biuret method for protein determination; determination of total protein in cerebrospinal fluid. Scand J Clin Lab Invest. 1953;5(3):218–222. doi: 10.3109/00365515309094189. [DOI] [PubMed] [Google Scholar]
- Guth J. H., Burris R. H. Inhibition of nitrogenase-catalyzed NH3 formation by H2. Biochemistry. 1983 Oct 25;22(22):5111–5122. doi: 10.1021/bi00291a010. [DOI] [PubMed] [Google Scholar]
- Kanemoto R. H., Ludden P. W. Effect of ammonia, darkness, and phenazine methosulfate on whole-cell nitrogenase activity and Fe protein modification in Rhodospirillum rubrum. J Bacteriol. 1984 May;158(2):713–720. doi: 10.1128/jb.158.2.713-720.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klugkist J., Haaker H., Wassink H., Veeger C. The catalytic activity of nitrogenase in intact Azotobacter vinelandii cells. Eur J Biochem. 1985 Feb 1;146(3):509–515. doi: 10.1111/j.1432-1033.1985.tb08681.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Ludden P. W., Burris R. H. Activating factor for the iron protein of nitrogenase from Rhodospirillum rubrum. Science. 1976 Oct 22;194(4263):424–426. doi: 10.1126/science.824729. [DOI] [PubMed] [Google Scholar]
- Ludden P. W., Okon Y., Burris R. H. The nitrogenase system of Spirillum lipoferum. Biochem J. 1978 Sep 1;173(3):1001–1003. doi: 10.1042/bj1731001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger H., Shapiro M. B., Mosimann J. E., Vinton J. E. Assessment of compositional relatedness between proteins. Nature. 1968 Sep 14;219(5159):1166–1168. doi: 10.1038/2191166a0. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- OPIENSKA-BLAUTH J., CHAREZINSKI M., BERBEC H. A new, rapid method of determining tryptophan. Anal Biochem. 1963 Jul;6:69–76. doi: 10.1016/0003-2697(63)90009-5. [DOI] [PubMed] [Google Scholar]
- Pope M. R., Murrell S. A., Ludden P. W. Covalent modification of the iron protein of nitrogenase from Rhodospirillum rubrum by adenosine diphosphoribosylation of a specific arginine residue. Proc Natl Acad Sci U S A. 1985 May;82(10):3173–3177. doi: 10.1073/pnas.82.10.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice D., Mazur B. J., Haselkorn R. Isolation and physical mapping of nitrogen fixation genes from the cyanobacterium Anabaena 7120. J Biol Chem. 1982 Nov 10;257(21):13157–13163. [PubMed] [Google Scholar]
- Roberts G. P., MacNeil T., MacNeil D., Brill W. J. Regulation and characterization of protein products coded by the nif (nitrogen fixation) genes of Klebsiella pneumoniae. J Bacteriol. 1978 Oct;136(1):267–279. doi: 10.1128/jb.136.1.267-279.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saari L. L., Triplett E. W., Ludden P. W. Purification and properties of the activating enzyme for iron protein of nitrogenase from the photosynthetic bacterium Rhodospirillum rubrum. J Biol Chem. 1984 Dec 25;259(24):15502–15508. [PubMed] [Google Scholar]
- Scolnik P. A., Haselkorn R. Activation of extra copies of genes coding for nitrogenase in Rhodopseudomonas capsulata. Nature. 1984 Jan 19;307(5948):289–292. doi: 10.1038/307289a0. [DOI] [PubMed] [Google Scholar]
- Tarrand J. J., Krieg N. R., Döbereiner J. A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can J Microbiol. 1978 Aug;24(8):967–980. doi: 10.1139/m78-160. [DOI] [PubMed] [Google Scholar]
- Wrigley C. W. Analytical fractionation of plant and animal proteins by gel electrofocusing. J Chromatogr. 1968 Aug 27;36(3):362–365. doi: 10.1016/s0021-9673(01)92959-0. [DOI] [PubMed] [Google Scholar]
- van Berkum P., Bohlool B. B. Evaluation of nitrogen fixation by bacteria in association with roots of tropical grasses. Microbiol Rev. 1980 Sep;44(3):491–517. doi: 10.1128/mr.44.3.491-517.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]