Abstract
Gene targeting studies have shown that T cell receptor (TCR)-β gene expression and recombination are inhibited after deletion of an enhancer (Eβ) located at the 3′ end of the ∼500-kb TCR-β locus. Using knockout mouse models, we have measured, at different regions throughout the TCR-β locus, the effects of Eβ deletion on molecular parameters believed to reflect epigenetic changes associated with the control of gene activation, including restriction endonuclease access to chromosomal DNA, germline transcription, DNA methylation, and histone H3 acetylation. Our results demonstrate that, in early developing thymocytes, Eβ contributes to major chromatin remodeling directed to an ∼25-kb upstream domain comprised of the Dβ-Jβ locus regions. Accordingly, treatment of Eβ-deleted thymocytes with the histone deacetylase inhibitor trichostatin A relieved the block in TCR-β gene expression and promoted recombination within the Dβ-Jβ loci. Unexpectedly, however, epigenetic processes at distal Vβ genes on the 5′ side of the locus and at the 3′ proximal Vβ14 gene appear to be less dependent on Eβ, suggesting that Eβ activity is confined to a discrete region of the TCR-β locus. These findings have implications with respect to the developmental control of TCR-β gene recombination, and the process of allelic exclusion at this locus.
Keywords: T cell receptor, thymus, lymphocyte differentiation, chromatin, T cell receptor rearrangement
Introduction
T cell development in the thymus involves sequential, receptor-based selection processes that drive cells through distinct cellular compartments: from CD4−CD8− double negative (DN) to CD4+CD8+ double positive (DP), and then CD4+CD8− or CD4−CD8+ single positive cells. During early α/β T cell differentiation, for example, the DN-to-DP cell stage transition represents a major developmental check-point (known as β selection), the passage through which depends on the expression of a productively rearranged TCR-β gene 1. In this study, we focus on the molecular events associated with TCR-β gene activation for expression and recombination in early developing T cells.
The TCR-β gene is assembled by V(D)J recombination from separate V, D, and J gene segments. Within the ∼500-kb (in the mouse) germline TCR-β locus, the Dβ and Jβ gene segments are located within a 15-kb region near the 3′ end of the locus, organized into two clusters, each comprised of one Dβ and seven Jβ segments (one of which is a pseudogene) and one constant (Cβ) region gene. In addition, ∼35 distinct Vβ genes occupy the 5′ side of the locus, spread over a region that extends from ∼200–450 kb upstream of the Dβ-Jβ-Cβ clusters, except for one (Vβ14) which lies 9.4 kb downstream, in the oppositely transcribed orientation 2.
V(D)J recombination at the TCR-β locus is strictly regulated with respect to lymphoid cell lineage and developmental stage 3. This process occurs in an ordered fashion in successive subpopulations of early DN thymocytes, defined as CD44+CD25+ and CD44−/loCD25+, commencing with Dβ-to-Jβ rearrangement followed by complete Vβ-to-DJβ assembly. Furthermore, whereas Dβ-to-Jβ joining is assumed to take place concurrently on both alleles, Vβ-to-DJβ joining apparently proceeds independently on each, in such a way that, after the first rearrangement, recombination on the opposite allele occurs only when the first joint is not productive (i.e., is out of frame and hence cannot yield a functional TCR-β protein). The arrest of Vβ recombination after productive Vβ-to-DJβ joining (which mediates TCR-β “allelic exclusion”) is tightly coupled to β selection and ensures that, in the vast majority of cases, individual developing T cells express a unique TCR-β chain 4.
Lineage- and developmental stage–specific gene expression within large genomic loci (including those encoding antigen receptors in lymphocytes) depends, to a large extent, on cis-acting transcriptional enhancers that act over long distances 5. A single enhancer (Eβ) has been described at the 3′ end of the TCR-β locus, flanked 5.9 kb upstream by the Dβ2-Jβ2-Cβ2 cluster and 2.9 kb downstream by the Vβ14 gene (for a review, see reference 2; see also Fig. 1 A). Transgenic mouse studies and deletional analysis by gene targeting have demonstrated that Eβ plays a critical role in activating expression and V(D)J recombination of linked TCR-β gene segments (for a review, see reference 3). Most strikingly, at the Eβ-deleted alleles, there is a severe defect in the formation of Dβ-to-Jβ recombination products which is yet more pronounced for the formation of Vβ-to-DJβ products: homozygous Eβ-deleted (Eβ−/−) mice show a block in T cell development resulting in the selective lack of α/β T cells 6 7.
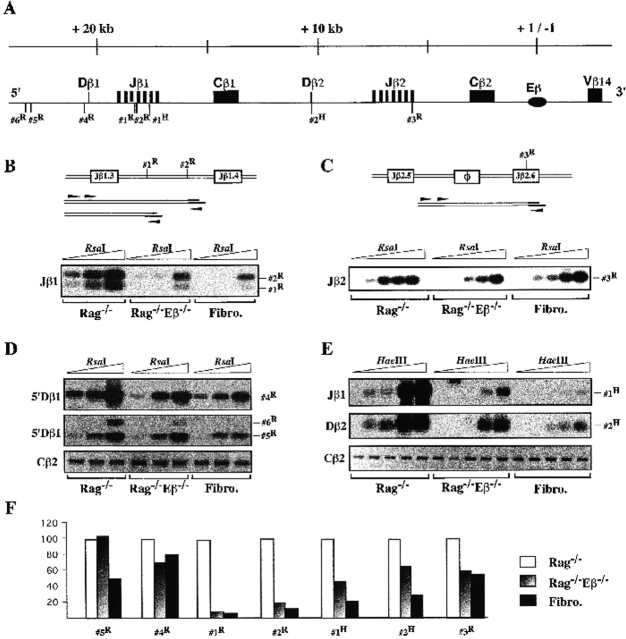
Figure 1.
Accessibility to restriction enzyme cleavage within the Dβ-Jβ loci. (A) The upper diagram shows the structural organization of the 3′ side of the TCR-β locus and relative location of the RsaI and HaeIII sites that have been analyzed. (B–E) A schematic representation of the restriction enzyme accessibility assays and the images generated are shown. Arrowheads in the schemes indicate the relative location of the forward and reverse primers used in the PCR assays. Thymocyte nuclei from Rag−/− mice (left lanes), Rag−/−Eβ−/− mice (middle lanes), and nuclei from NIH-3T3 fibroblasts (Fibro., right lanes) were treated with increasing amounts of the indicated enzyme (B and D: 1, 3, and 10 U of RsaI; C and E: 0.1, 0.3, 1, 3, and 10 U of RsaI or HaeIII). Enzyme cleavage at the indicated site(s) was analyzed by LM-PCR and Southern blotting. Cβ2 PCR amplifications using RsaI- or HaeIII-restricted, linker-ligated DNA are shown. (F) Accessibility level quantifications at the indicated restriction sites were obtained by PhosphorImager® scanning of the corresponding hybridizing bands and normalization for sample loading to the Cβ2 signals. Results in Rag−/− Eβ−/− thymocytes and normal fibroblasts are given as percentage of residual accessibility relative to that in the Rag−/− thymocytes to which the 100% value was attributed.
The above-mentioned studies are consistent with a model that postulates that V(D)J recombination activity at individual antigen receptor gene loci is controlled through the modulation of recombinase access to its target substrates, and that cis-acting regulatory elements, including enhancers, play a role in this process (the “accessibility model” of reference 8). Studies by Schlissel and colleagues have demonstrated that recombinational accessibility is indeed a property of lymphocyte chromatin by showing that the formation of double strand break (DSB) DNA recombination intermediates at chromosomal TCR and Ig loci by the recombination activating gene (RAG)-1 and RAG-2 components of the V(D)J recombinase is determined by the lineage and developmental stage of the original cells 9. The prevailing model assumes that, during lymphocyte development, repressive heterochromatin at a given antigen receptor gene segment and/or locus is disrupted in a cell lineage– and/or developmental stage–specific manner, hence allowing the V(D)J recombinase action.
Although it is generally agreed that lymphoid enhancers may participate in chromatin remodeling 10 11, the molecular mechanisms responsible are still poorly understood and other possibilities also exist to explain the effect of these elements in regulating V(D)J recombination 11. Moreover, the discrete domains that an individual enhancer can influence with respect to chromatin structure have not been precisely defined. Here we show that, in early DN T cells, the effect of Eβ deletion on chromatin structure exhibits an unanticipated degree of plasticity depending on the TCR-β region considered, with the influence of Eβ in chromatin remodeling being primarily restricted to the Dβ-Jβ gene clusters. We discuss these results with respect to Eβ function in the regulation of Dβ-to-Jβ versus Vβ-to-DJβ recombination.
Materials and Methods
Mice.
The mice used in this study, as well as mouse housing and analyzing conditions, were as described previously 11.
Thymocyte Preparation and Cell Cultures.
Thymocytes were separated from the stroma according to standard procedures. NIH-3T3 fibroblasts were maintained in DMEM (GIBCO BRL) supplemented with 10% FCS. Fibroblasts that were growing in log phase were used to prepare nuclei.
For treatment with the specific histone deacetylase inhibitor trichostatin A (TSA), thymic lobes were excised from fetal mice at day 14.5 postcoitum and placed on cellulose ester filters (Millipore) in 6-well plates containing DMEM supplemented with 10% FCS, penicillin, streptomycin, l-glutamine, and sodium pyruvate, without or with TSA (3 ng/ml; Sigma-Aldrich). The culture was carried out for 7 h at 37°C in a humidified, 10% CO2 atmosphere.
Restriction Endonuclease Accessibility Assays.
Cell nuclei were prepared as described previously 9 11. For restriction enzyme digests, 105 nuclei were treated with increasing concentrations of restriction endonuclease in digestion buffer consisting of 10 mM Tris, pH 7.9, 50 mM NaCl, 10 mM MgCl2, 4% glycerol. Digestion was performed on ice for 1 h. Nuclei were lysed in extraction buffer (10 mM Tris, pH 8, 100 mM NaCl, 1 mM EDTA, 0.5% SDS) and treated with proteinase K (final concentration of 0.4 mg/ml) at 56°C for 2 h. DNA was resuspended in 25 μl of H2O. Linker ligation, using 10 μl of the above preparation, and detection of the generated blunt ends by ligation-mediated (LM)-PCR were carried out as described previously 11. Quantification was performed by use of a PhosphorImager® (BAS 1000; Molecular Dynamics) and MacBAS software. The sequences of the TCR-β–specific oligonucleotide primers used in LM-PCR reactions are available upon request.
Reverse Transcription PCR.
Total RNA and cDNA preparation as well as analysis of T cell–specific transcripts by reverse transcription (RT)-PCR, using cDNA templates and locus-specific oligonucleotide primers, were performed as described (7; primer sequences available upon request).
Southern Blot Assay.
Preparation of genomic DNA, restriction enzyme digests, gel electrophoresis, Southern blotting, and hybridization procedures were performed as described previously 12. Hybridization signals were detected by autoradiography. The hybridizing probes used for hybridization were prepared by PCR amplification using genomic DNA from Rag−/− kidney as a template.
Bisulfite Sequencing.
Bisulfite treatment of genomic DNA was performed according to Zeschnigk et al. 13 with the following modifications. DNA was mixed with 2 vol of 2% LMP agarose (GIBCO BRL) dissolved in H2O, and the agarose/DNA mix was pipetted into chilled mineral oil to form agarose beads, which were then incubated overnight at 56°C in the presence of NaHSO3 (Sigma-Aldrich). The reaction was stopped by washing of the beads with 10 mM Tris, pH 8.0, 1 mM EDTA (TE; six times for 15 min) followed by the addition of 500 μl of 0.2 N NaOH (twice for 15 min), and then 1/5 vol of 1 M HCl. The beads were washed with 1 ml H2O (twice for 15 min) and used directly for PCR. Primers were designed to be complementary to the deaminated DNA strands within sequences that do not include CG dinucleotides. 5 μl of the PCR product was cloned into the vector PGEM-T (Promega). After bacterial transformation, DNA from individual clones was sequenced using the ABI PRISM™ 310 Genetic Analyzer (PerkinElmer).
Chromatin Immunoprecipitation PCR.
Chromatin fixation and immunoprecipitation were performed using the Acetyl-Histone H3 Immunoprecipitation (ChIP) Assay Kit (Upstate Biotechnology, Inc.) as recommended by the manufacturer. The input fraction corresponded to 10% of the chromatin solution before immunoprecipitation. After DNA purification, the input, unbound, and bound fractions were resuspended in 100 μl of TE, and analyzed by 25 cycles of PCR (40 s at 94°C, 40 s at 55°C, 40 s at 72°C) in 25-μl reactions. A twofold dilution series was typically used for PCR reactions with all TCR-β–specific primers (primer sequences available upon request). Oct-2– and TCR-δ enhancer (Eδ)-specific primers were designed according to McMurry et al. 14.
PCR Analysis of Dβ2-to-Jβ2 Rearrangements.
Oligonucleotide primers, PCR conditions, and analysis of the amplified products were as described previously 7.
Results
Differential endonuclease access to chromosomal DNA and the level of transcription and DNA methylation are molecular parameters widely used to monitor changes in chromatin architecture associated with the activation of gene expression 15 16. To study the effects of Eβ deletion on these parameters, we applied several molecular assays to comparatively analyze specific regions of the TCR-β locus (e.g., the Dβ-Jβ clusters and Vβ gene regions) in thymocytes from single knockout RAG-1 (Rag−/−) versus double knockout Rag−/−Eβ−/− mice. Due to a lack of V(D)J recombination, Rag−/− animals demonstrate no thymocyte development beyond the CD44−/loCD25+ DN cell stage 1. Thus, utilization of Rag−/− (and Rag−/− Eβ−/−) thymocytes offers two advantages: (a) the analyzed TCR-β locus is maintained in the germline configuration; and (b) the analyzed cells are enriched in thymocytes that are arrested at the developmental stage where TCR-β gene recombination normally takes place. Using CD4/CD8 and DN CD44/CD25 cell surface stainings and flow cytofluorometry, we have verified that the cell developmental block in Rag−/−Eβ−/− and Rag−/− thymi can be superimposed (data not shown).
Differential Effects of the Eβ Deletion on Accessibility of the Dβ-Jβ and Vβ Regions.
To examine the in vivo effects of Eβ deletion on the chromatin structure throughout TCR-β regions, we used a restriction enzyme accessibility assay. This involved incubating isolated cell nuclei with increasing concentrations of a restriction enzyme, then lysing them to prepare genomic DNA. Cleavage at the sites of interest was assayed by LM-PCR followed by Southern blotting (Fig. 1A–C). To assess their quality and the variability of sample loading, the linker-ligated DNAs were tested in parallel for amplification of a fragment within the Cβ2 constant region gene. By comparing amplified products from Rag−/− versus Rag−/−Eβ−/− thymic nuclei, the relative level of restriction enzyme cleavage at a given site can thus be evaluated. As a nonlymphoid control, we used nuclei from NIH-3T3 fibroblasts.
Fig. 1B–E, shows the results from the analysis of selected sites within the Dβ-Jβ regions. Whereas two RsaI sites located within the intronic sequences between the Jβ1.3 and Jβ1.4 gene segments (sites #1R and #2R; Fig. 1 B) and one RsaI site located within the Jβ2.6 gene segment (3R; Fig. 1 C) were readily cleaved at a low enzyme concentration in Rag−/− thymic nuclei, those in Rag−/−Eβ−/− nuclei were more resistant to cleavage, showing profiles largely comparable to those in fibroblast nuclei. After PhosphorImager® scanning of 32P emission from the hybridizing bands and correction for differences in sample loading, based on Cβ2 amplifications, we estimated that the levels of site #1R, #2R, and #3R cleavage in Rag−/−Eβ−/− thymocytes and normal fibroblasts were 3.2 and 2%, 20 and 12%, and 60 and 55%, respectively, of those in Rag−/− thymocytes (Fig. 1 F). Related percentages were obtained in two separate experiments. These results strongly suggest that the Dβ-Jβ loci demonstrate reduced levels of chromatin disruption in DN cells from the Eβ mutant mice (on average, <5% of those in Rag−/− thymocytes, once cleavage background in fibroblasts is taken into account) and, consequently, that Eβ critically impacts on the chromatin structure of these regions in early developing T cells. Reduced chromatin accessibility in the Rag−/−Eβ−/− thymocytes was consistently observed when testing two HaeIII sites (#1H and #2H) located 5′ of Jβ1.6 and within Dβ2, respectively (Fig. 1E and Fig. F). It is noteworthy that, in these instances, the cleavage assays also demonstrated larger differences between the Eβ mutant and fibroblast nuclei (percentages of 45 and 23%, and 64 and 38%, respectively), implying that Eβ may not be the only determinant of TCR-β gene accessibility in early T cells and/or that the action of Eβ may not be uniform across the entire length of the Dβ-Jβ clusters. Finally, a difference in chromosomal access depending on the presence or absence of Eβ was also observed when analyzing an RsaI site (#4R) located ∼540 bp 5′ of the Dβ1 gene segment (Fig. 1 D, top). However, the difference was attenuated when analyzing two such sites (#5R and #6R) located 2 and 2.15 kb, respectively, further upstream, with roughly equivalent cleavage levels observed in both Rag−/− and Rag−/−Eβ−/− thymic nuclei (Fig. 1 D, middle). These latter data suggest that, in DN thymocytes, the upper limit of Eβ control on chromosomal access may fall within the ∼2-kb sequences that separate these oppositely regulated sites.
Similar analyses of Vβ gene–containing regions demonstrated a lack of Eβ influence on chromosomal access. Thus, pairs of RsaI sites located in either the 3′ region of Vβ5.2 and downstream of this element (#7R and #8R; toward the 5′ end of the TCR-β locus) or within the leader exon and promoter region of Vβ14 (#9R and #10R; 3′ proximal to Eβ) exhibited equivalent levels of accessibility in Rag−/− and Rag−/−Eβ−/− thymic nuclei (Fig. 2). In these last two cases, enzyme cleavage in nuclei derived from thymocytes appeared to be significantly increased compared with those from fibroblasts, implying tissue-specific control of the underlying chromosomal structure. Similar results were obtained upon analysis of restriction sites within Vβ11, Vβ20, and Vβ18, three Vβ genes scattered further 3′ of Vβ5.2 (data not shown). Altogether, the above results demonstrate unexpected regional consequences of Eβ deletion on chromatin remodeling in early developing T cells, consisting of reduced accessibility of the upstream Dβ-Jβ clusters, whereas the Vβ gene regions still develop an accessible structure irrespective of their distance from this deletion.
Figure 2.
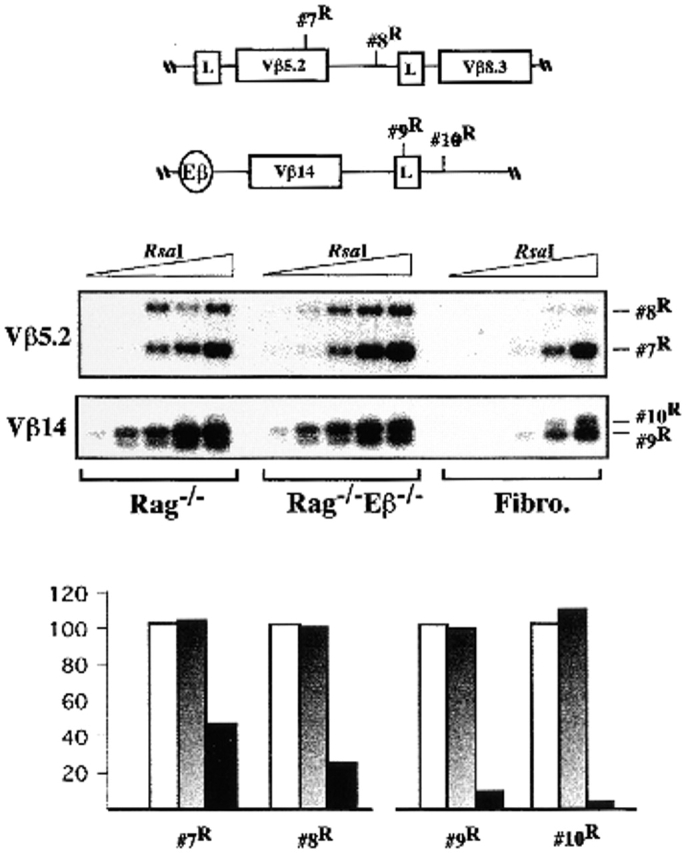
Accessibility to restriction enzyme cleavage within the Vβ5.2/Vβ8.3- and Vβ14-containing regions. Representative LM-POR experiments and accessibility level quantifications are shown, as outlined in the legend to Fig. 1B–F. The DNAs used in this experiment were the same as those used for analysis of the #3R site shown in Fig. 1 C.
TCR-β Gene Expression in Eβ−/− Thymocytes.
To analyze the consequences of Eβ deletion on TCR-β locus transcription, we used previously described RT-PCR assays 6 7. For these analyses, total RNA was prepared from Rag−/− and Rag−/−Eβ−/− thymocytes and, in addition, from thymocytes of heterozygous Eβ+/− and homozygous Eβ−/− mice as well as TCR-δ−/−Eβ−/− double knockout mice. Eβ+/− mice show a thymic cell profile similar to that of wild-type (wt) mice (e.g., comprised of ∼5% DN, ∼85% DP, and ∼10% single positive cells), whereas Eβ−/− mice have reduced thymi consisting of mainly DN and DP cells, varying from 55 to 65% for the former and 35 to 45% for the latter; thymocytes from the TCR-δ−/−Eβ−/− mice represent another model of Eβ-deleted, CD44−/loCD25+ DN-arrested T cells 7 11.
As expected, germline transcripts (GTs) initiating upstream of the Jβ1 and/or the Jβ2 gene clusters (the Jβ1 GT and Jβ2 GT) were found to be expressed in Rag−/− thymocytes (Fig. 3 A, top two panels, lane 1; note the heterogeneity in size of the Jβ1 GT, as reported previously [17, 18]). In striking contrast, neither transcript was detected using RNA from Rag−/−Eβ−/−, Eβ−/−, or TCR-δ−/−Eβ−/− thymocytes (Fig. 3 A, lanes 5–7). Using RAG-deficient blastocyst complementation and embryonic stem cells carrying both a wt and an Eβ-deleted TCR-β allele to generate chimeric mice, Alt and colleagues have reported a lack of detectable Jβ1 GT in the resulting thymocytes 6, a finding that is reproduced here using thymocytes from Eβ+/− mice carrying the germline-inherited Eβ deletion (Fig. 3 A, top, lane 4). In Eβ+/− thymocytes, the Dβ1-to-Jβ1 intervening sequences (to which the RT-PCR forward Jβ1 primer hybridizes) are present on the unrearranged mutant allele but are removed by several types of rearrangements on the wt allele (i.e., by Dβ1-to-Jβ1 joining and by any rearrangement involving an upstream Vβ and the downstream Dβ2-Jβ2 cluster). Altogether, these data strongly suggest that the Eβ deletion results in the cis-inhibition of germline transcription throughout the Jβ1 gene cluster. Presumably, the Jβ2 GT that can be found (at reduced levels) in Eβ+/− thymocytes (Fig. 3 A, middle, lane 4) are produced from wt alleles harboring a productive Vβ-to-DJβ rearrangement that involves the Dβ1 and Jβ1 gene segments. In agreement with this interpretation, Jβ2 GTs were not detected in Eβ+/− thymocytes derived from (Eβ−/− × NZW) mouse intercrosses (data not shown; NZW TCR-β alleles have a deletion covering the Dβ2 and Jβ2 elements 19). We conclude that, as a result of the Eβ deletion, germline transcription across the Dβ-Jβ loci is severely inhibited in early developing T cells.
Figure 3.
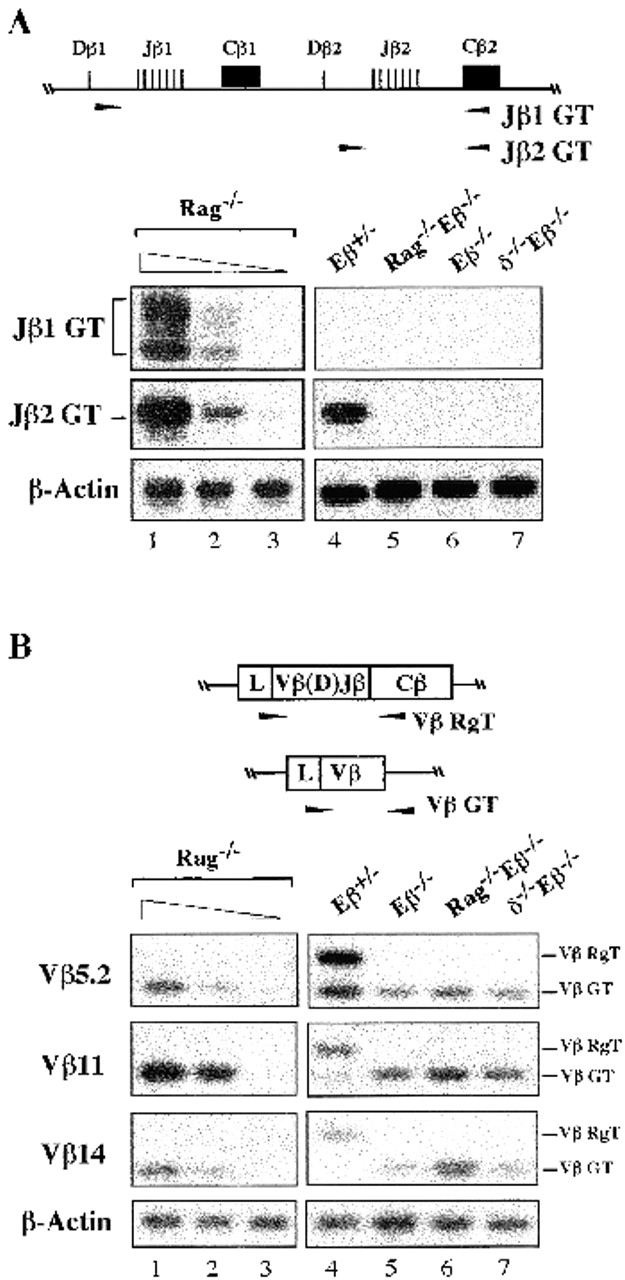
TCR-β locus expression in thymocytes from mutant mice. Total RNA from thymocytes of the indicated mouse lines was assayed by RT-PCR to analyze: (A), GTs initiated upstream of the unrearranged Jβ1 or Jβ2 clusters (Jβ1 GT and Jβ2 GT); or (B), Vβ5.2, Vβ11, and Vβ14 rearranged (Vβ RgT) and/or germline (Vβ GT) transcripts, as schematized above the corresponding panels. Arrowheads indicate the relative location of the primers used in the RT-PCR assays. Lanes 1–3 show a titration of Rag−/− thymocyte cDNA. Input cDNA was kept constant; lane 1: thymocyte cDNA undiluted; lanes 2 and 3: thymocyte cDNA diluted 1:3 and 1:30 with kidney cDNA. Amplified products from β-actin controls are shown.
The RT-PCR assays used to analyze Vβ gene transcription permit the simultaneous detection of germline Vβ transcripts (Vβ GTs), as seen for RNA from the Rag−/− thymocytes, as well as mature VβDJβCβ rearranged transcripts (Vβ RgTs) initiated from the rearranged loci, as seen for that from Eβ+/− thymocytes (Fig. 3 B, lanes 1 and 4). In line with previous findings 7, we found Vβ5.2, Vβ11, and Vβ14 GTs (but not Vβ RgTs) in thymocyte RNA from all animals carrying homozygously Eβ-deleted alleles, including the Eβ−/−, Rag−/−Eβ−/−, and TCR-δ−/−Eβ−/− mice, although Vβ GT was overall higher in the last two (for example, Fig. 3 B, lanes 5–7; consistent results were obtained in two separate RT-PCR experiments and in Northern blot analysis, data not shown). Strikingly, we estimated that the levels of Vβ GT in double knockout Rag−/−Eβ−/− and TCR-δ−/−Eβ−/− thymocytes were comparable to those in Rag−/− thymocytes (varying from ∼49 to ∼92%), whereas those in single knockout Eβ−/− thymocytes were significantly lower (varying from ∼12 to ∼25%) (Table ). We conclude that germline expression of the Vβ genes is, to a large extent, independent of Eβ. It is noteworthy that the differential levels of Vβ GT correlate with the presence of DP cells in the Eβ−/− but not the Rag−/−Eβ−/− and TCR-δ−/−Eβ−/− thymi (7; Mathieu, N., unpublished data), a situation that parallels previous reports of a downregulation of Vβ GT expression as thymocytes develop to the DP cell stage 20 21. Indeed, we observed such a downregulation of Vβ5.2 and Vβ11 GTs in thymocytes from both the Rag−/−Eβ−/− and TCR-δ−/−Eβ−/− mice after intraperitoneal injection of anti-CD3ε antibodies, a treatment that induces the DN-to-DP cell-transition in these animals (Mathieu, N., unpublished data; see also reference 22). Thus, developmental downmodulation of Vβ gene expression appears to be appropriately controlled in the absence of Eβ, underscoring the autonomous nature of the corresponding regions.
Table 1.
Levels of Vβ GT in Thymocytes from Eβ-deleted Animals Relative to Those in Rag−/− Mice
| Eβ2/− | Rag−/−Eβ2/− | TCR-δ2/−Eβ2/− | |
|---|---|---|---|
| Vβ5.2 | 25.2 ± 3.6 | 86.2 ± 5.1 | 66.2 ± 4.4 |
| Vβ11 | 12 ± 4.3 | 53.7 ± 6.3 | 49.3 ± 3.8 |
| Vβ14 | 16.7 ± 4.8 | 92 ± 9.4 | 52 ± 7.6 |
Quantification of the levels of the indicated Vβ GT in individual thymocyte populations was calculated after PhosphorImager®scanning of the Vβ-hybridizing images from two independent RT-PCR experiments, and correction for the amount of loaded sample as estimated by scanning of the β-actin images. Results are given as percentage ± SEM of residual transcription relative to that in the Rag−/− thymocytes to which the 100% value was attributed.
Effects of Eβ Deletion on CpG Methylation.
We performed Southern blot analyses using primarily the HpaII and MspI isoschizomers, which recognize and cleave the nucleotide sequence CCGG only when the central CG dinucleotide is unmethylated (HpaII) or irrespective of its methylation status (MspI). A total of nine HpaII/MspI sites have been investigated along the Dβ-Jβ clusters; a methyl-sensitive HhaI site, located 212 bp 5′ of Dβ1, was also analyzed. Fig. 4 A summarizes the results from these analyses, comparing genomic DNA from Rag−/− and Rag−/−Eβ−/− thymocytes (top diagram); Fig. 4 B shows representative data for both the Dβ1-Jβ1 and Dβ2-Jβ2 clusters (left and right panels, respectively). In all cases, kidney DNA from a Rag−/− mouse, used as a nonlymphoid control, was also analyzed (Fig. 4 B, and data not shown). We found that methyl-sensitive sites within the Dβ1-Jβ1 region, including the Dβ1 upstream-flanking HhaI site, while almost completely demethylated in Rag−/− thymocytes, remain methylated in those from Rag−/−Eβ−/− mice (for example, Fig. 4 B, left panels: see the opposite sensitivity to HpaII cleavage). In addition, within the Dβ2-Jβ2 region, DNA from Rag−/−Eβ−/− thymocytes was in general more highly methylated than that from Rag−/− thymocytes although, different from the Dβ1-Jβ1 analysis, significant demethylation was nevertheless observed at some sites with the former samples (for example, Fig. 4 B, right panels: see the partial cleavages resulting from HpaII digestion of Rag−/− Eβ−/− DNA). It should be noted that, within the Jβ2 region, kidney DNA was also found to be sensitive to HpaII cleavage (to a varying extent depending on the site; e.g., Fig. 4 B), raising concerns about the physiological significance of thymocyte DNA demethylation for the particular sites at this cluster. However, differential methylation levels throughout Jβ2-associated sequences at the Eβ-deleted versus wt alleles were further supported by results from separate experiments in which we used bisulfite genomic sequencing, a technique that permits the analysis of individual cytosines in genomic DNA (13; and Fig. 4 C). Ideally, analysis of individual cloned DNA permits the estimation of the percentage of methylated residues at a specific site. Thus, analysis of a Jβ2.6-containing fragment of 215 bp encompassing seven CG sites demonstrated consistent methylation for six, specifically in Eβ-deleted DNA, although methylation varied between the individual sites (Fig. 4 C, diagram). The fact that the methylation level at the HpaII/MspI site within Jβ2.6 noticeably differs upon comparison of the results of Southern blotting with the genomic sequencing experiments (it appears lower with the latter approach) most likely reflects a PCR bias in amplification of the bisulfite-converted sequences, as described previously 23.
Figure 4.
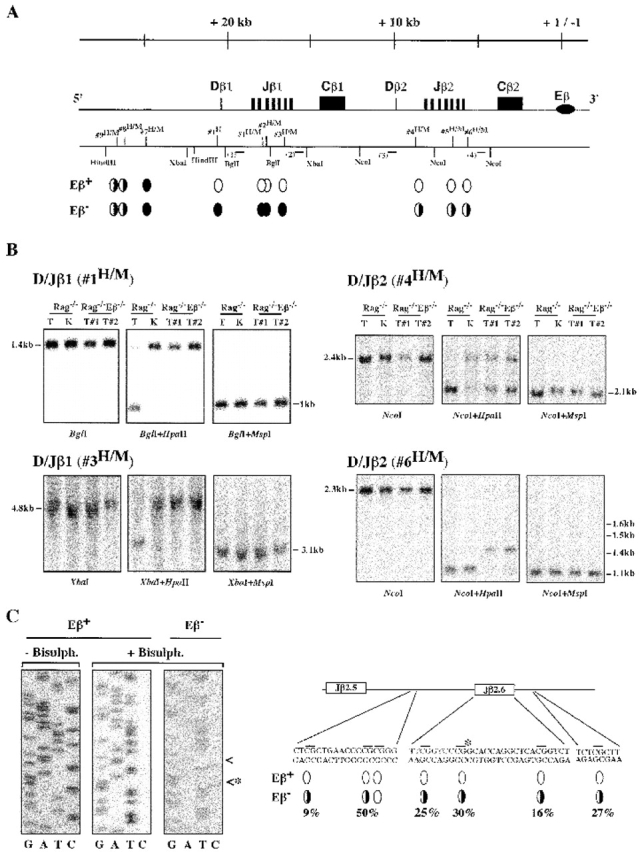
CpG methylation analyses within the Dβ-Jβ loci. (A) The relative location of the HpaII/MspI or HhaI restriction sites that have been analyzed is indicated. The relevant HindIII, XbaI, BglI, and NcoI restriction sites used in Southern blot analyses are also shown. The bottom part of the diagram summarizes the results of these assays. Eβ+ and Eβ−: results obtained when using Rag−/− and Rag−/−Eβ−/− thymocyte DNA, respectively. A filled or open oval underneath the corresponding site indicates hyper- or hypomethylation, respectively; partial methylation is indicated by the partially filled ovals. (B) Representative results of Southern blot analyses at specific HpaII/MspI sites, using genomic DNA from thymocytes of either one Rag−/− (T) or two Rag−/−Eβ−/− (T#1 and T#2) mice and appropriate single and double restriction enzyme digestion, as indicated. Kidney DNA (K) from a Rag−/− mouse was used as a nonlymphoid control. The size of the hybridizing fragments is indicated; the relative location of each probe used for hybridization of the individual blot is shown by a horizontal bar within the upper diagram. The 1.4-, 1.5-, and 1.6-kb hybridizing fragments seen in the D/Jβ2 (#6H/M) analyses correspond to additional HpaII/MspI restriction sites located in between sites #5H/M and #6H/M. (C) Genomic sequencing of a 215-bp DNA fragment overlapping with the Jβ2.6 gene segment. The sequences of untreated (−Bisulph.) and bisulfite-treated (+Bisulph.) DNAs are shown. The methylated cytosines that, within the Eβ− DNA, are resistant to thymidine conversion are indicated (arrowheads). The diagram on the right summarizes the results of these analyses (representing 4 [Eβ+] and 11 [Eβ−] sequencing reactions, respectively). Only the sequences that contain CG dinucleotides (horizontal bars) are shown; the asterisk indicates the central CG within the site #6H/M. The percentage of methylated residues which was found in bisulfite-converted DNA at six out of the seven analyzed CGs is indicated.
Southern blot analyses of TCR-β regions outside the Dβ/Jβ loci, however, demonstrated a general tendency towards hypermethylation, independent of the presence or absence of Eβ. Thus, HpaII/MspI sites within the far-upstream Dβ1 domain (i.e., located >2.5 kb 5′ of this segment) were found to be either completely (#7H/M) or partially (#8H/M and #9H/M) methylated using both Rag−/− and Rag−/−Eβ−/− thymocyte DNA (Fig. 4 A, and data not shown). Moreover, hypermethylation was also observed using DNA of both origins at HpaII/MspI sites located in the intergenic region between the Vβ5. 2 and Vβ8.3 genes and downstream of Vβ8.3, as well as at sites 3′ adjacent of the Vβ14 gene (Fig. 5; see the resistance to HpaII cleavage of sites 10H/M and 13H/M; also note the slight demethylation observed for kidney DNA at both sites). It might be argued that crucial demethylation events for Vβ gene activity in T cells rather involve CpGs located in the Vβ genes and/or associated promoter elements that have not been investigated in these assays (note that CpG average density within Vβ genes and surrounding sequences is low, and roughly similar to that in the vertebrate genome, ∼1 in every 100 bases 24, whereas, interestingly, it is higher within the Dβ-Jβ clusters, ∼1 in 50 bases). We do not favor the idea that CpGs internal to Vβ genes are regulated differently, however, based on the finding that methylation for both Rag−/− and Rag−/−Eβ−/− thymocyte DNA was also demonstrated by Southern blot analysis of two HpaII/MspI sites located within the coding exon of Vβ18 and internal intron of Vβ20, respectively (i.e., two genes that appear equally accessible to chromosomal enzyme cleavage) (data not shown).
Figure 5.
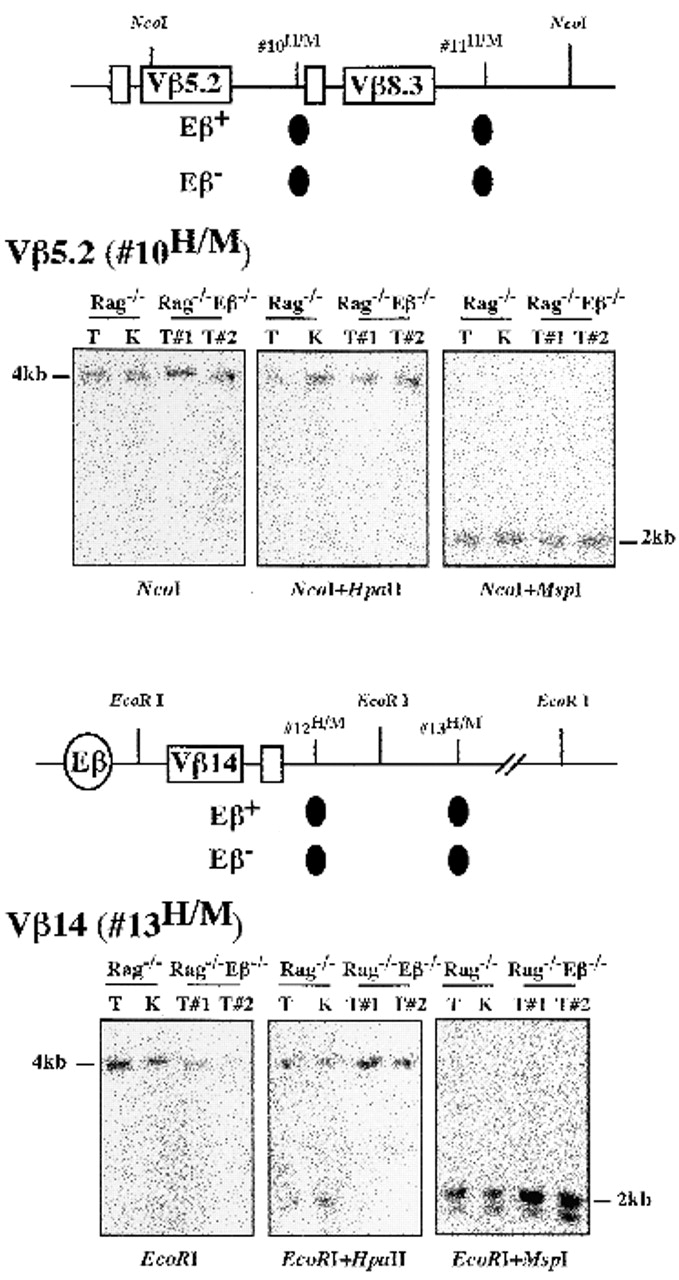
CpG methylation analyses within the Vβ5.2/Vβ8.3 and Vβ14 gene–containing regions. A summary of the results and representative Southern blot experiments at the indicated HpaII/MspI sites are shown, as outlined in the legend of Fig. 4A and Fig. B.
Effect of Eβ Deletion on Histone H3 Acetylation.
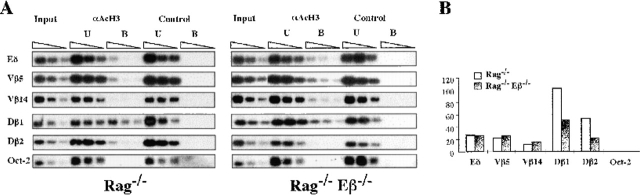
DNA methylation and histone deacetylase activities may operate along a common mechanistic pathway to repress gene expression 16. Moreover, a recent report has demonstrated a tight link between the histone H3 acetylation status of TCR-α/δ gene sequences and their V(D)J recombination potential 14. To test the effect of Eβ deletion on histone acetylation at discrete sites throughout the TCR-β locus, we have performed chromatin immunoprecipitation (CHIP)-PCR assays, using an antiacetylated H3 histone (αAcH3) antiserum and Rag−/− or Rag−/−Eβ−/− thymus chromatin. In addition to sites overlapping the Vβ5.2, Vβ14, Dβ1, and Dβ2 gene sequences, we also tested TCR-δ enhancer (Eδ) and Oct-2 gene sequences used here as hyper- and hypoacetylated controls, respectively (see reference 14). The relative enrichment of the particular sequences in acetylated chromatin was determined by measuring the PCR products in the αAcH3-bound fraction and normalizing to those in the input material (αAcH3-bound/input ratio).
Strikingly, whereas H3 acetylation was comparable for Vβ5.2 and Vβ14 (as well as for Eδ control) in both Rag−/− and Rag−/−Eβ−/− thymocytes, that for Dβ1 and Dβ2 sequences markedly differed depending on mouse origin, with lower levels specifically observed in Rag−/−Eβ−/− thymocytes (Fig. 6). Moreover, within a given sample, acetylation levels differed depending on the site analyzed, with significantly higher levels observed for the Dβ loci. Altogether, these results strongly suggest that, in early developing thymocytes, Eβ deletion predominantly affects histone acetylation within the Dβ-containing loci, with essentially no effect on acetylation at Vβ-containing loci. These differential effects are reminiscent of those observed in analyses for restriction enzyme access and germline transcription, as reported above. Moreover, Dβ versus Vβ histone H3 overacetylation at Eβ-containing alleles inversely correlates with our findings of opposite hypo- and hypermethylation profiles at these gene segments, in line with previous observations at various genomic loci. Finally, the residual amplification of transcriptionally inactive Dβ sequences in the αAcH3-bound fraction from Rag−/−Eβ−/− thymocytes (compared with the Oct-2 negative control; Fig. 6 B) indicates that Eβ activity is not the only determinant of histone acetylation at these sites. Indeed, tissue-specific genes may be acetylated in the cell type in which they are potentially active, independent of their transcriptional status 25.
Figure 6.
Histone H3 acetylation within TCR Vβ and Dβ locus regions. (A) H3 acetylation level was investigated in total thymocytes from the indicated mouse lines by chromatin immunoprecipitation (CHIP)-PCR assays. CHIP was performed with anti-AcH3 (αAcH3) antiserum or no antibody (control). Serial twofold dilutions of input, αAcH3-bound (B), and -unbound (U) fractions were analyzed by PCR. (B) H3 acetylation was quantified after PhosphorImager® scanning of Southern blots and plotting of B/input measurements.
TSA Treatment of Eβ-deleted Thymocytes Relieves the Block in Dβ/Jβ Gene Expression.
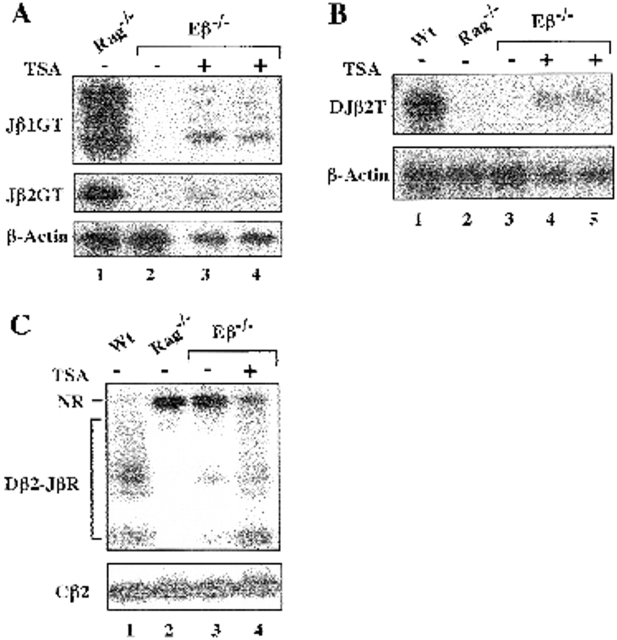
The above findings raise the question as to whether histone acetylation may be one factor that could compensate for the lack of Eβ in terms of chromatin structure and block in cis-recombination. To address this hypothesis, we tested Eβ−/− thymocytes for the production of Jβ1 and Jβ2 GTs after their culture in the presence of the histone deacetylase inhibitor TSA 26. Rag−/− and Eβ−/− thymocytes that were cultured in the absence of TSA were used as positive and negative controls, respectively. Because we have noticed that TSA treatment affects cell survival within single cell suspension of adult thymocytes, but has a less deleterious effect when using culture of fetal thymic lobes, the latter strategy was used as a source of thymocyte RNA. Strikingly, Jβ1 and Jβ2 GTs were specifically detected in TSA-treated thymocytes from two Eβ−/− animals whereas untreated thymocytes from an Eβ−/− littermate were found to be negative for Jβ GT production (Fig. 7 A). After quantification by PhosphorImager® scanning and correction for sample loading, we estimated the mean levels of TSA-induced Jβ1 and Jβ2 GTs in the Eβ−/− thymocytes at 38 ± 1.3 and 42.8 ± 2%, respectively, of those in the Rag−/− thymocytes. Moreover, DJβ2 transcripts encompassing the products of intermediate Dβ2-to-Jβ2 rearrangement were specifically detected in TSA-treated Eβ−/− fetal thymocytes (Fig. 7 B), accounting for >37% of those in untreated wt controls. The possibility that DJβ2 transcripts correspond, at least in part, to the expression of newly generated rearrangements in response to chromatin remodeling induced by TSA treatment (as opposed to the transcriptional activation of preexisting Dβ-to-Jβ joints that occur at low level in the Eβ−/− thymus [11]; see also Fig. 7 C, lane 3) was supported by further PCR analyses demonstrating increased level of Dβ2-to-Jβ2 rearrangement in genomic DNA from TSA-treated (compared with untreated) Eβ−/− thymocytes (Fig. 7 C, compare lanes 3 and 4). By PhosphorImager® scanning of 32P emission from the unrearranged Dβ2/Jβ2-containing fragments (the amount of which inversely reflects the rate of locus rearrangement involving the Dβ2-Jβ2 cluster) and correction for sample loading, we estimated the rearrangement level in TSA-treated thymocytes to be ∼7-fold over that in the untreated samples, still a level ∼4.5-fold below that in wt thymocytes. After thymic lobe cultures, cell yield and viability were similar for the TSA-treated and untreated thymocytes, making it unlikely that the observed effect is due to differences in cell survival and/or preferential expansion of a selected subpopulation (Mathieu, N., unpublished results). These findings, together with the above results, emphasize the Eβ function on the chromosomal structure of the Dβ-Jβ regions and bring support to the hypothesis that recruitment of a histone acetyltransferase (HAT)-containing complex and changes in histone acetylation may be significant features of Eβ-mediated activation of TCR-β locus expression and recombination.
Figure 7.
(A) Rescue of Jβ1/Jβ2 GTs upon TSA treatment of Eβ-deleted thymocytes. Thymocytes were prepared from either Rag−/− (Eβ+) or Eβ−/− (Eβ−) fetal thymic lobes (day 14.5 postcoitum) that have been cultured in the presence (+) or absence (−) of TSA. Jβ1 GTs and Jβ2 GTs were assayed by RT-PCR, as indicated in the legend to Fig. 3. Lanes 2–4 correspond to three individual Eβ−/− littermates. (B) Presence of DJβ2 transcripts (DJβ2T) in total RNA from TSA-treated Eβ−/− fetal thymocytes. Experimental procedures were as in A except that RNA from wt thymocytes was included as a positive control. (C) Dβ2-to-Jβ2 rearrangements (Dβ2-JβR) in genomic DNA isolated from the indicated fetal thymocytes were analyzed by PCR. NR, unrearranged Dβ2-Jβ2–containing fragments. Amplification of a Cβ2-containing fragment was carried out to control for sample loading.
Discussion
Compared with the wt situation, enhancer deletion at the TCR-β locus results in major epigenetic changes within the Dβ-Jβ region which thus exhibit opposite “open” (euchromatin) and “closed” (heterochromatin) structures, respectively. Unexpectedly, the Vβ gene–containing regions were relatively unaffected by the deletion, exhibiting a nuclease-sensitive, transcribed, acetylated, and yet hypermethylated profile whether Eβ was present or not. As discussed further below, the latter patterns may reflect a “specialized” chromosomal structure. Importantly, differential nuclease sensitivity and methylation profiles within the region 5′ of Dβ1 in the wt situation suggest that a border between open and closed—or specialized Vβ-like—chromatin lies proximal to this gene segment. Altogether, our data support a model in which Eβ activity during early T cell development results in regionalized chromatin remodeling directed to an upstream domain of ∼25 kb, with significant implications regarding Eβ function in regulating TCR-β gene recombination.
Dβ-Jβ Locus Recombination and Chromatin Opening by Eβ.
A function for Eβ in opening heterochromatinized Dβ-Jβ loci can explain, to a large extent, the defect in Dβ-to-Jβ rearrangement observed at Eβ-deleted alleles. Accordingly, using thymocyte DNA from Eβ-deleted, DN developmental knockout mouse models (such as the TCR-δ−/−Eβ−/− mouse), we have found that V(D)J recombinase–mediated DSB cleavages at the recombination signal sequences (RSS) flanking Dβ and Jβ gene segments are reduced up to ∼1/10 to 1/30 the level observed in thymi with an unaltered TCR-β locus 11. Significantly, our estimations on decrease in chromosomal accessibility at the Dβ-Jβ clusters in Rag−/−Eβ−/− thymocytes, based on quantification data from restriction enzyme cleavage assays, were mostly comprised within the same range. Based on a similar approach to study DSB cleavage at a transgenic minilocus, local chromatin remodeling by the TCR-δ gene enhancer has also been proposed to control Dδ-to-Jδ joining 10. Assuming that Eβ regulates Dβ-to-Jβ recombination in a similar fashion, it is, however, noteworthy that: (a) in vitro–generated DSBs can be produced at Dβ/Jβ RSS within chromosomal DNA from both Rag−/− and Rag−/− Eβ−/− thymi 11; and (b) in vivo–generated DSB intermediates at the Dβ and Jβ RSS are consistently found at a higher frequency than the DJβ rearranged products (11; Mathieu, N., unpublished data). Based on all of the available data, we favor the possibility that Eβ, in addition to a regulatory function via chromatin decondensation, may also play a role during the course of the Dβ-to-Jβ joining reaction (e.g., the stabilization and/or processing of recombination synaptic complexes; for discussion, see reference 11).
How does Eβ act to mediate this regional control of chromatin unfolding? In light of our estimation of the 5′ limit of Eβ action proximal to the Dβ1 gene segment, it is significant that specific defects in rearrangement and accessibility of this segment have recently been described after the genomic deletion of a 3-kb upstream DNA fragment 27. The deleted region, encompassing three DNase I hypersensitive sites (HS, usually associated with occupied regulatory sequences) present in DN cells 21, contains a minimal promoter adjacent to Dβ1 which is most active in T cell lines when linked with Eβ 17 18. Although our results cannot formally exclude a model in which Eβ and the Dβ1 upstream sites independently delimit and organize the intervening region by unidirectional activity, the prevailing view on the mechanisms of long-distance gene activation by enhancers 5 28, and the lower nuclease sensitivity that we observe for the region immediately 5′ of Dβ1 in the absence of Eβ (Fig. 1, site #4R), would argue rather that Eβ acts by providing a crucial activity to the Dβ1 promoter. In the simplest scheme, this may occur via interactions between transcription factors bound to each element (and to a putative promoter element associated with the Dβ2-Jβ2 cluster) which, as described in other systems, could themselves nucleate further contacts with chromatin modifying and/or remodeling activities (e.g., SWI/SNF-like complexes and/or histone acetyltransferases) and also bind to components of the basal transcription machinery. The resulting nucleoprotein structures, by favoring the propagation of chromatin changes across the intervening sequences, would ultimately permit their accessibility. The observed differential histone H3 acetylation profiles at Dβ loci of Eβ-containing versus Eβ-deleted alleles, and release of the regional block in Dβ/Jβ gene expression and recombination upon TSA treatment of Eβ−/− thymocytes (Fig. 6 and Fig. 7), are both compatible with this scenario. Importantly, the aforementioned model could accommodate an intrinsic boundary (or insulator) function for the Dβ promoter–associated sequences as well, analogous to the recently described property of certain promoters in Drosophila 29.
Our data do not indicate the exact function(s) of Eβ in promoting chromatin remodeling at the Dβ-Jβ loci. These may include an opening activity 30, a protective role against the spreading of repressive structures 31, and/or an influence on subnuclear localization 32. Also, the localized promotion of germline transcription, demethylation, and/or histone hyperacetylation may be involved since these activities are not separable in Rag−/− thymi and are downmodulated in Rag−/−Eβ−/− thymi (Fig. 3 and Fig. 4; methylation appears less strictly regulated at Jβ2 sequences, however). Recently, based on results from cell transfection experiments using an enhancerless construct that carries an inducible promoter, germline transcription has been proposed to be sufficient for heterochromatin disruption and rearrangement at Dβ and Jβ gene segments 33. However, a direct role for transcription remains to be demonstrated given all the examples in which, upon factor binding, chromatin remodeling occurs independently of RNA polymerase movement 34, a situation that could also be relevant to the control of V(D)J recombination 35.
A Putative Function for Eβ in Vβ Gene Recombination?
An unusual profile consisting of nuclease sensitivity, germline transcription, methylation, and intermediate level of histone acetylation was observed at the Vβ genes in both Rag−/− and Rag−/−Eβ−/− thymocytes. Thus, different from the Dβ-Jβ loci, establishment of a specific chromosomal structure at these regions depends on regulatory sequences distinct from Eβ, possibly involving the T cell–specific, Vβ promoter–associated elements 36. How does this structure impact on Vβ-to-DJβ rearrangement, and do these findings preclude any role for Eβ in regulating these recombination events? Contrary to Dβ-to-Jβ, Vβ-to-DJβ rearrangements are not found at Eβ-deleted alleles even when using sensitive PCR assays 6 7. Provocatively, DSBs at Vβ gene–flanking RSS are difficult to detect at Eβ-deleted and wt loci (Hempel, W.M., unpublished data), in striking contrast to endonuclease-mediated cleavage (e.g., Fig. 2). These findings may reflect a lower efficiency for Vβ-to-DJβ joining (relative to Dβ-to-Jβ joining) and/or, because V(D)J recombination involves coupled cleavage of pairs of gene segments (e.g., reference 10), the inaccessibility of partner RSS at the downstream Dβ-Jβ loci. Keeping in mind the differences between Dβ-to-Jβ and Vβ-to-DJβ assembly (see Introduction), it is therefore tempting to speculate on how these same factors may impact on the control of Vβ rearrangement in the wt situation where Eβ is active. First, the inherent inefficiency of V gene rearrangement, operating through epigenetic control, has been hypothesized to accommodate allelic exclusion at given antigen receptor genes 37. In this respect, the unusual profile (e.g., accessible/transcribed sequences are classically undermethylated) which we (this study) and others 20 have described at Vβ genes is intriguing. CpG methylation, in particular, could underlie a “unique” chromosomal structure to yield a system in which Vβ gene activation for recombination becomes stochastic and infrequent, and, ultimately, monoallelic. This would be consistent with data suggesting that, at individual Ig/TCR loci, V gene rearrangement in general, and allelic exclusion in particular, correlate with demethylation but not with transcription 4. Second, the interaction between Eβ- and Dβ-associated promoters may insulate the intervening, fully accessible, Dβ-to-Jβ recombining domains (see above). Thus, Vβ-to-DJβ joining would have to proceed through the casual relaxation of these boundary structures at either allele. This, for example, could involve competition for Eβ between the Vβ and Dβ promoters, similar to a model that has been proposed to explain developmental gene activation at multigene loci 28. The specialized structure at the Vβ genes would strongly disfavor the Vβ promoters in this competition, accounting for both the delay and allele specificity of Vβ gene recombination (i.e., relative to Dβ-to-Jβ assembly).
Methylation of Vβ-containing sequences may yet serve an additional function. It has been shown that methylated recombination substrates become highly resistant to V(D)J rearrangement after undergoing DNA replication, presumably by acquiring a fully repressed chromosomal structure 38. The intense cellular proliferation that is triggered by β selection 1 may similarly drive the unrearranged, methylated Vβ genes into a heterochromatin structure, thus terminating Vβ recombination and locking in TCR-β allelic exclusion as thymocytes develop towards the DP cell stage.
Apparently, in early T cells, the downstream Vβ14 gene presents the same profile of Eβ-independent regulation as that of the 5′ Vβ genes (Fig. 2, Fig. 3 B, 5, and 6). This is quite remarkable, given their differences in position relative to Eβ. Therefore, Vβ14 gene recombination may also be subject to gene competition as proposed above. Conceivably, at this location, the system may be further refined by the cis-acting, possibly blocking, activity of a regulatory element(s) located between Eβ and the Vβ14 promoter. Interestingly, T cell–specific hypersensitive (HS) sites have been described within the region that links Eβ to the Vβ14 gene 21 and, within a transgenic substrate, intron sequences in this region have been reported to function in regulating lineage-specific accessibility 3. However, it remains possible that, as thymocytes develop beyond the DN cell stage, Vβ14 may be regulated differently from the 5′ Vβ genes, as suggested by reports indicating that DP cell development results in augmented expression instead of an inhibition of this element 20 21. Additional studies will be necessary to further clarify this issue.
Acknowledgments
We thank Drs. M.S. Schlissel for critical reading of this manuscript, L. Rowen for providing the mouse TCR-β genomic sequence before publication, and C.J. Jolly for advice on bisulfite genomic sequencing.
This work was supported by institutional grants from Institut National de la Santé de la Recherche Médicale and the Centre National de la Recherche Scientifique, and by specific grants from the Association pour la Recherche sur le Cancer (ARC), the Commission of the European Communities, the Fondation Princesse Grace de Monaco, the Ligue Nationale Contre le Cancer (LNCC), and Rhone-Poulenc Pharmaceuticals.
Footnotes
W.M. Hempel's present address is Département de Biologie Cellulaire et Moléculaire, Institut de Recherche Jouveinal/Parke-Davis, 3-9 rue de la Loge, 94265 Fresnes Cedex, France.
Abbreviations used in this paper: CHIP, chromatin immunoprecipitation; DN, double negative; DP, double positive; DSB, double strand break; Eβ, TCR-β gene enhancer; GT, germline transcript; LM, ligation-mediated; RAG, recombination activating gene; RgT, rearranged transcript; RSS, recombination signal sequence(s); RT, reverse transcription; TSA, trichostatin A; wt, wild-type.
References
- Fehling H.J., von Boehmer H. Early αβ T cell development in the thymus of normal and genetically altered mice. Curr. Opin. Immunol. 1997;9:263–275. doi: 10.1016/s0952-7915(97)80146-x. [DOI] [PubMed] [Google Scholar]
- Hood L., Rowen L., Koop B.F. Human and mouse T-cell receptor locigenomics, evolution, diversity, and serendipity. Ann. NY Acad. Sci. 1995;758:390–412. doi: 10.1111/j.1749-6632.1995.tb24844.x. [DOI] [PubMed] [Google Scholar]
- Sleckman B.P., Bassing C.H., Bardon C.G., Okada A., Khor B., Bories J.C., Monroe R., Alt F.W. Accessibility control of variable region gene assembly during T-cell development. Immunol. Rev. 1998;165:121–130. doi: 10.1111/j.1600-065x.1998.tb01235.x. [DOI] [PubMed] [Google Scholar]
- Bergman Y. Allelic exclusion in B and T lymphopoiesis. Semin. Immunol. 1999;11:319–328. doi: 10.1006/smim.1999.0188. [DOI] [PubMed] [Google Scholar]
- Blackwood E.M., Kadonaga J.T. Going to distancea current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- Bories J.C., Demengeot J., Davidson L., Alt F.W. Gene-targeted deletion and replacement mutations of the T-cell receptor β-chain enhancerthe role of enhancer elements in controlling V(D)J recombinational accessibility. Proc. Natl. Acad. Sci. USA. 1996;93:7871–7876. doi: 10.1073/pnas.93.15.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier G., Watrin F., Naspetti M., Verthuy C., Naquet P., Ferrier P. Deletion of the mouse T-cell receptor β gene enhancer blocks αβ T-cell development. Proc. Natl. Acad. Sci. USA. 1996;93:7877–7881. doi: 10.1073/pnas.93.15.7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleckman B.P., Gorman J.R., Alt F.W. Accessibility control of antigen-receptor variable-region gene assemblyrole of cis-acting elements. Annu. Rev. Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- Stanhope-Baker P., Hudson K.M., Shaffer A.L., Constantinescu A., Schlissel M.S. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85:887–897. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- McMurry M.T., Hernandez-Munain C., Lauzurica P., Krangel M.S. Enhancer control of local accessibility to V(D)J recombinase. Mol. Cell. Biol. 1997;17:4553–4561. doi: 10.1128/mcb.17.8.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel W.M., Stanhope-Baker P., Mathieu N., Huang F., Schlissel M.S., Ferrier P. Enhancer control of V(D)J recombination at the TCRβ locusdifferential effects on DNA cleavage and joining. Genes Dev. 1998;12:2305–2317. doi: 10.1101/gad.12.15.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernex C., Capone M., Ferrier P. The V(D)J recombinational and transcriptional activities of the immunoglobulin heavy-chain intronic enhancer can be mediated through distinct protein-binding sites in a transgenic substrate. Mol. Cell. Biol. 1995;15:3217–3226. doi: 10.1128/mcb.15.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeschnigk M., Schmitz B., Dittrich B., Buiting K., Horsthemke B., Doerfler W. Imprinted segments in the human genomedifferent DNA methylation patterns in the Prader-Willi/Angelman syndrome region as determined by the genomic sequencing method. Hum. Mol. Genet. 1997;6:387–395. doi: 10.1093/hmg/6.3.387. [DOI] [PubMed] [Google Scholar]
- McMurry M.T., Krangel M.S. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- Razin A. CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikes M.L., Gomez R.J., Song J., Oltz E.M. A developmental stage-specific promoter directs germline transcription of DβJβ gene segments in precursor T lymphocytes. J. Immunol. 1998;161:1399–1405. [PubMed] [Google Scholar]
- Doty R.T., Xia D., Nguyen S.P., Hathaway T.R., Willerford D.M. Promoter element for transcription of unrearranged T-cell receptor β-chain gene in pro-T cells. Blood. 1999;93:3017–3025. [PubMed] [Google Scholar]
- Noonan D.J., Kofler R., Singer P.A., Cardenas G., Dixon F.J., Theofilopoulos A.N. Delineation of a defect in T cell receptor β genes of NZW mice predisposed to autoimmunity. J. Exp. Med. 1986;163:644–653. doi: 10.1084/jem.163.3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo M., Shinkai Y. Regulation of Vβ germline transcription in RAG-deficient mice by the CD3ε-mediated signalsimplications of Vβ transcriptional regulation in TCR β allelic exclusion. Int. Immunol. 1998;10:553–560. doi: 10.1093/intimm/10.5.553. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S., Whitehurst C.E., Schwenk F., Chen J. Biochemical and functional analyses of chromatin changes at the TCR-β gene locus during CD4−CD8− to CD4+CD8+ thymocyte differentiation. J. Immunol. 1998;160:1256–1267. [PubMed] [Google Scholar]
- Levelt C.N., Mombaerts P., Iglesias A., Tonegawa S., Eichmann K. Restoration of early thymocyte differentiation in T-cell receptor β-chain-deficient mutant mice by transmembrane signaling through CD3ε. Proc. Natl. Acad. Sci. USA. 1993;90:11401–11405. doi: 10.1073/pnas.90.23.11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirzaker C., Millar D.S., Paul C.L., Warnecke P.M., Harrison J., Vincent P.C., Frommer M., Clark S.J. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res. 1997;57:2229–2237. [PubMed] [Google Scholar]
- Bird A.P. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- O'Neil L.P., Turner B.M. Histone H4 acetylation distinguishes coding regions of the human genome for heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Kijima M., Akita M., Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- Whitehurst C.E., Chattopadhyay S., Chen J. Control of V(D)J recombinational accessibility of the Dβ1 gene segment at the TCRβ locus by a germline promoter. Immunity. 1999;10:313–322. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
- Bulger M., Groudine M. Looping versus linkingtoward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Dorsett D. Distant liaisonslong-range enhancer-promoter interactions in Drosophila . Curr. Opin. Genet. Dev. 1999;9:505–514. doi: 10.1016/s0959-437x(99)00002-7. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Forrester W.C., Fernández-Herrero L.A., Laible G., Dull M., Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- Festenstein R., Tolaini M., Corbella P., Mamalaki C., Parrington J., Fox M., Miliou A., Jones M., Kioussis D. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- Francastel C., Walters M.C., Groudine M., Martin D.I.K. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centromeric heterochromatin. Cell. 1999;99:259–269. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- Sikes M.L., Suarez C.C., Oltz E.M. Regulation of V(D)J recombination by transcriptional promoters. Mol. Cell. Biol. 1999;19:2773–2781. doi: 10.1128/mcb.19.4.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S.A., Kingston R.E. Disruption of downstream chromatin directed by a transcriptional activator. Genes Dev. 1997;11:3116–3121. doi: 10.1101/gad.11.23.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S.R., Baltimore D. Chromatin remodeling directly activates V(D)J recombination. Proc. Natl. Acad. Sci. USA. 1999;96:10788–10793. doi: 10.1073/pnas.96.19.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle J.P., Haus-Seuffert P., Woltering C., Stelzer G., Meisterernst M. A conserved tissue-specific structure at a human T-cell receptor β-chain core promoter. Mol. Cell. Biol. 1997;17:4220–4229. doi: 10.1128/mcb.17.8.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner S.L., Seder R.A. Dealing from the evolutionary pawnshophow lymphocytes make decisions. Immunity. 1999;11:1–10. doi: 10.1016/s1074-7613(00)80076-x. [DOI] [PubMed] [Google Scholar]
- Hsieh C.-L., Lieber M.R. CpG methylated minichromosomes become inaccessible for V(D)J recombination after undergoing replication. EMBO (Eur. Mol. Biol. Organ.) J. 1992;11:315–325. doi: 10.1002/j.1460-2075.1992.tb05054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]