Abstract
Proinflammatory stimuli induce the rapid and transient translocation of nuclear factor (NF)-κB to the nucleus, where it activates transcription from several genes, including those encoding inflammatory cytokines and chemokines, adhesion molecules, and cytoprotective proteins. Using chromatin immunoprecipitation, we show that after an acute stimulation two distinct waves of NF-κB recruitment to target promoters occur: a fast recruitment to constitutively and immediately accessible (CIA) promoters and a late recruitment to promoters requiring stimulus-dependent modifications in chromatin structure to make NF-κB sites accessible (promoters with regulated and late accessibility [RLA]). Our results suggest that a mechanism of specificity in NF-κB–dependent transcriptional responses relies on the ability of individual stimuli to make RLA promoters accessible to NF-κB before its rapid extrusion from the nucleus.
Keywords: NF-κB, Rel, histone acetylation, chromatin immunoprecipitation, lipopolysaccharide
Introduction
Nuclear factor (NF)-κB is a family of transcription factors which exist in most vertebrates cell types as homo- and heterodimers of five structurally related Rel/NF-κB proteins 1 2. In nonstimulated cells, NF-κB is sequestered in the cytoplasm in an inactive form via interaction with inhibitory proteins, the inhibitors of NF-κB (IκBs; reference 3). The three most important IκBs, IκBα, IκBβ, and IκBε 4 5 6 7, bind the Rel homology domain (RHD) of NF-κB dimers and mask their nuclear localization signal (NLS; 8 9 10 11). Upon stimulation with various agonists such as bacterial products, proinflammatory cytokines, viral proteins, and radiations, IκBs are rapidly phosphorylated at two serines located in their NH2-terminal regulatory region, ubiquitinated, and degraded 12 13. The exposure of the NLS determines a rapid nuclear translocation of NF-κB, which then binds target decanucleotide sequences and activates transcription of several genes, including those encoding proinflammatory chemokines and cytokines, adhesion molecules, antiapoptotic or cytoprotective genes, and several others 2 14. After NF-κB–dependent resynthesis, IκBα enters the nucleus, enhances NF-κB removal from DNA 15, and takes it back to the cytoplasm, thus restoring the inducible cytoplasmic pool of NF-κB 16. In the last years, several important breakthroughs have clarified the essential cytoplasmic steps of NF-κB regulation: a multisubunit IκB–kinase complex receiving signals from several upstream pathways and capable of directly phosphorylating the IκBs at the regulatory NH2-terminal serines has been identified 17, and its components have been cloned (18 19 20 21 22; for reviews see references 23 and 24). In addition, the ubiquitin–ligase complex that binds and ubiquitinates the IκBs has been identified and shown to be a member of the SKp1-Cullin-F-box (SCF)-type E3 ubiquitin ligases 23 25 26.
However, the advanced knowledge of the molecular mechanisms regulating NF-κB activity at the cytoplasmic level (i.e., phosphorylation and degradation of IκBs) contrasts sharply with the paucity of information on the mechanisms governing NF-κB activity in the nucleus. We studied how NF-κB is recruited to target promoters and enhancers in intact cells using a very powerful and increasingly popular technique, chromatin immunoprecipitation (ChIP; reference 27). In this study, we show that in lipopolysaccharide-stimulated macrophages, recruitment of NF-κB to target genes occurs in two temporally distinct phases. A subset of target genes whose promoter is already heavily acetylated before stimulation is constitutively and immediately accessible to NF-κB and is transcribed immediately after NF-κB recruitment (constitutively and immediately accessible genes [CIA genes]). In contrast, other target genes (that we indicate as genes with regulated and late accessibility [RLA genes]) are not immediately accessible to NF-κB. Recruitment of NF-κB to their promoters occurs between 90 and 120 min after nuclear entry and is preceded by the formation of an initial transcription factors complex that directs the hyperacetylation of the promoter and makes it accessible to NF-κB. The dependency of NF-κB recruitment to RLA promoters on stimulus-induced modifications in chromatin structure is a mechanism allowing a highly selective activation of these genes in response to a subset of NF-κB activators.
Materials and Methods
Cell Culture.
The murine macrophage cell line Raw 264.7 was obtained from American Type Culture Collection. For all the experiments described here, cells were used between the 4th and 10th passage.
Antibodies.
The anti p65 (c-20) affinity-purified rabbit polyclonal antibody and the anti-cJun (H79) antibody were obtained from Santa Cruz Biotechnology, Inc. The anti-IκBα mouse monoclonal antibody is from Imgenex. The antiacetylated histone H4 antiserum was obtained from Upstate Biotechnology.
ChIP Assay.
After stimulation, cells were fixed by adding directly to the medium formaldehyde (HCHO; from a 37% HCHO/10% methanol stock; Calbiochem) to a final concentration of 1%. After 10 min, ice-cold PBS was added and plates were transferred on ice, washed extensively with PBS, and scraped. After centrifugation, cells were lysed for 5 min in L1 buffer (50 mM Tris, pH 8.0, 2 mM EDTA, 0.1% NP-40, 10% glycerol) supplemented with proteases inhibitors. Nuclei were pelleted at 3,000 rpm in microfuge and resuspended in L2 buffer (50 mM Tris, pH 8.0, 1% SDS, 5 mM EDTA). Chromatin was sheared by sonication (4 × 12 s at one-fifth of the maximum potency), centrifuged to pellet debris, and diluted 10 times in dilution buffer (50 mM Tris, pH 8.0, 0.5% NP-40, 0.2 M NaCl, 0.5 mM EDTA). Extracts were precleared for 3 h with 80 μl of a 50% suspension of salmon sperm–saturated protein A (ss protein A). Immunoprecipitations were carried out at 4°C overnight. Immune complexes were collected with ss protein A and washed three times (5 min each) with high salt buffer (washing buffer: 20 mM Tris, pH 8.0, 0.1% SDS, 1% NP-40, 2 mM EDTA, 500 mM NaCl) and three times with low salt buffer (1× Tris/EDTA [TE]). Immune complexes were extracted in 1× TE containing 1% SDS, and protein–DNA cross-links were reverted by heating at 65°C overnight. After proteinase K digestion (100 μg, 1 h at 50°C), DNA was extracted by phenol–chloroform and ethanol precipitated. About 1/20 of the immunoprecipitated DNA was used in each PCR. Sequences of promoter-specific primers and more detailed technical information are available upon request.
Results
Kinetic Analysis of NF-κB Activation and Recruitment to the IκBα Promoter.
Stimulation of Raw 264.7 murine macrophages with lipopolysaccharide determines a fast degradation of IκBα, which is immediately followed by nuclear translocation of NF-κB and appearance of NF-κB DNA binding activity (Fig. 1 a), which lasts for about 4 h. We used the ChIP assay 27 to measure NF-κB recruitment to specific promoters during the time in which this transcription factor is present in the nucleus. After stimulation, cells were fixed in formaldehyde, which promotes the formation of stable protein–DNA cross-links and therefore preserves relevant protein–DNA interactions. After lysis, extracts were sonicated to shear chromatin into fragments of an average size between 500 and 1,500 bp and aliquots of the extracts were immunoprecipitated with an affinity-purified antibody directed against p65/RelA (which in the form of heterodimers with p50 is the most abundant NF-κB species in these cells). After extensive and stringent washing, protein–DNA complexes were extracted, cross-links were reverted by heating, and released DNA was analyzed by semiquantitative PCR to detect an enrichment of specific promoters in the immunoprecipitates. We first analyzed p65/RelA recruitment to the IκBα promoter, a canonical NF-κB–dependent gene that is rapidly switched on after NF-κB activation (as indicated by the appearance of resynthesized IκBα at about 30 min after stimulation; see Fig. 1 a). Recruitment of p65/RelA to the IκBα promoter is extremely fast, being observed immediately after NF-κB entry in the nucleus (Fig. 1 b). Interestingly, removal of p65/RelA from this promoter is also very rapid, being most of the NF-κB binding to the IκBα gene lost in <30 min from the stimulation. At this time point, accumulation of newly synthesized IκBα has just started and the overall NF-κB DNA binding activity (as detected by electrophoretic mobility shift assay [EMSA]) is at its peak. Therefore, it is extremely unlikely that removal of p65/RelA from this specific gene depends on newly synthesized IκBα. Interestingly, this promoter undergoes a very fast and transient deacetylation after stimulation (data not shown), but whether it is directly responsible for or facilitates NF-κB removal is to be determined. The histone H2A gene promoter (which has constitutive activity and no NF-κB sites) and the IL-2 promoter (which has an NF-κB site but is active only in T cells) were not found in the p65/RelA immunoprecipitates, thus suggesting the specificity of IκBα promoter enrichment. In addition, when a matched control antibody (anti-IκB kinase α [IKKα]) was used in the ChIP assay, no IκBα promoter was immunoprecipitated (Fig. 1 b).
Figure 1.
Early and transient recruitment of NF-κB to the IκBα promoter. (a) Kin-etic of NF-κB activation by LPS in Raw 264.7 macrophages. Cells were stimulated with 100 ng/ml LPS (Escherichia coli 055:B5; Sigma-Aldrich) for the indicated times. Detergent lysates were prepared and analyzed for NF-κB activation. The anti-IκBα immunoblot shows IκBα degradation and resynthesis, which is completed between 50 and 60 min from the stimulation. Anti-IκBa monoclonal antibody is from Imgenex. The blot was reprobed with anti-p65 (Santa Cruz Biotechnology, Inc.) to verify equal loading of the samples (second panel from the top). NF-κB DNA-binding activity in LPS-stimulated cells was analyzed by EMSA (n.s., non specific). Cytoplasmic vs. nuclear p65/RelA NF-κB levels in stimulated Raw 264.7 cells were analyzed in fractionated extracts (cy, cytoplasmic fraction; n, nuclear fraction). W.B., Western blot (b) Recruitment of p65/RelA to the IκBα promoter in LPS-stimulated Raw 264.7 cells. After stimulation, cells were fixed with 1% formaldehyde and lysed. The nuclear fraction was sonicated to shear chromatin in 0.5- to 1.5-kb fragments and immunoprecipitated with an anti-p65 affinity-purified rabbit polyclonal antibody (C-20; Santa Cruz Biotechnology, Inc.). p65-coprecipitating DNA was analyzed by semiquantitative PCR with promoter-specific primers amplifying the IκBα promoter, the H2A histone promoter, or the IL-2 promoter. In a control ChIP, an anti-IKKα affinity-purified rabbit polyclonal antibody (Santa Cruz Biotechnology, Inc.) was used.
Two Temporally Distinct Waves of NF-κB Recruitment to Target Genes.
We next analyzed p65/RelA recruitment to other LPS-inducible NF-κB–dependent genes (Table ). The promoter of the gene for the macrophage inflammatory protein 2 (MIP-2), a proinflammatory macrophage-derived chemokine with chemotactic activity towards neutrophils, contains a canonical NF-κB site and has been shown to be dependent on p65/RelA for induction 28. Similarly to the IκBα gene, p65/RelA recruitment to the MIP-2 promoter occurs rapidly, and no significant time gap could be observed between NF-κB entry in the nucleus and recruitment to MIP-2 (Fig. 2 a). Transcriptional induction of this gene also occurs very rapidly. However, in this case p65/RelA persists on the promoter for a much longer time (>3 h) and it is still detectable after completion of IκBα resynthesis. Manganese superoxide dismutase (MnSOD) is a prototype antiapoptotic NF-κB–inducible protein which protects mitochondria from superoxide radical–mediated damage. The MnSOD intronic enhancer contains a canonical NF-κB site (Table ) that is responsible for the NF-κB dependency of MnSOD expression 29; also in this case p65/RelA is recruited quickly and persistently, and transcriptional induction occurs shortly after initial NF-κB binding (Fig. 2 a). Thus, NF-κB persists on both MnSOD enhancer and MIP-2 promoter long after IκBα resynthesis has been completed and the overall NF-κB DNA binding activity has significantly declined. The apparent dissociation between IκBα resynthesis and NF-κB persistence on target promoters suggests that under physiological conditions, newly synthesized IκBα is not sufficient to promote NF-κB removal from target genes. A potential mechanism explaining the persistence of NF-κB on target sequences after completion of IκBα resynthesis is represented by the protection of NF-κB–DNA interaction by newly synthesized, unphosphorylated IκBβ: as proposed 6 30, this molecule may interact with DNA-bound NF-κB without displacing it and conversely affording protection from newly synthesized IκBα at the promoter level.
Table 1.
NF-κB Sites in the Promoters/Enhancers Under Investigation
| Gene | NF-κB site | Position |
|---|---|---|
| IκBα | GGGGAAGTCC | −260 |
| GGGAATTTCC | −70 | |
| MnSOD | GGGAATAGCC | |
| MIP-2 | GGGAATTTCC | |
| MCP-1 | GGGAACTTCC | −2,348 |
| GGGAATTTCC | −2,352 | |
| IL-6 | GGGATTTTCC | −73 |
| RANTES | GGGAGTTTCC | −78 |
| GGGCACTTCC | −58 |
Figure 2.
Two waves of NF-κB recruitment to target promoters. (a) ChIP assay with an anti-p65 antibody shows that NF-κB recruitment to MnSOD intronic enhancer and MIP-2 promoter occurs rapidly; in both cases binding persists for >3 h. Conversely, recruitment to the RANTES, MCP-1, and IL-6 promoters (b) occurs relatively late during the stimulation, when the overall NF-κB DNA binding activity is already markedly reduced (see EMSA in Fig. 1 a). The kinetic of induction of each mRNA (evaluated by reverse transcription [RT]-PCR) is also shown.
In sharp contrast with these three genes, which appear to be immediately accessible to NF-κB, other promoters displayed a significantly different behavior. NF-κB recruitment to the promoters of the regulated upon activation, normal T cell expressed and secreted (RANTES) chemokine, the macrophage chemoattractant protein 1 (MCP-1), and the IL-6 genes occurred with a markedly slower kinetic, i.e., between 90 and 120 min from stimulation (Fig. 2 b). A good correlation between NF-κB recruitment and accumulation of the corresponding mRNA was found, thus suggesting that high level transcription of these genes does not occur before NF-κB recruitment. This is in agreement with previous reports indicating that transcriptional induction of these genes is indeed NF-κB dependent (see among the others references 31 32 33). It is interesting to note that when NF-κB is recruited to these promoters, the overall NF-κB DNA binding activity is significantly reduced with respect to the early phases of stimulation (see Fig. 1 a). Thus, an extremely important implication of these results is that total NF-κB binding activity detected by EMSA does not reflect the specific NF-κB activity on individual promoters.
Late NF-κB Recruitment to Target Promoters Is Preceded by Stimulus-induced Histone H4 Hyperacetylation.
One possibility to consider is that the affinity of NF-κB for the recognition sites contained in the MCP-1, IL-6, and RANTES promoters is low, and that efficient binding may require cooperative interactions with other transcription factors, thus resulting in a slower recruitment of NF-κB to these genes. However, sequence analysis shows that the number and predicted affinity of the NF-κB sites contained in the MCP-1, IL-6, and RANTES promoters is not significantly different from those in MIP-2, MnSOD, and IκBα (Table ). In particular, the highly conserved guanines at positions 1, 2, and 3, and the cytosines at positions 9 and 10, are present in all sites 34. Position 8 is a thymine in all sites except in MnSOD. The central nucleotides, which minimally influences the overall binding affinity, do not show particular trends in the two groups of genes. Finally, the sequence of the NF-κB site in MIP-2 (a gene with immediate NF-κB recruitment) is identical to the sequence of one of the two NF-κB sites in MCP-1 (a gene with delayed NF-κB recruitment). Overall, this suggests that the observed differences in the kinetic of NF-κB recruitment to the genes under investigation do not arise from differences in the number or affinity of the NF-κB sites.
To explain the delay between NF-κB nuclear entry and recruitment to MCP-1, RANTES, and IL-6 promoters, we hypothesized that signal-induced changes in chromatin structure may be required to make these promoters accessible to NF-κB. Modification of chromatin structure is accomplished by the action of two distinct groups of enzymatic activities, namely chromatin remodeling and covalent modifications of N-terminal histone tails. ATP-dependent chromatin remodeling complexes act by modifying the spatial relationship between nucleosomes and DNA, either through changing the position of specific nucleosomes (sliding) or through altering their three-dimensional structure (for recent reviews, see references 34 and 35). Covalent modifications of the N-terminal tails of histones H3 and H4 consist of acetylation, phosphorylation, methylation, and other less studied modifications (for reviews, see references 36 37 38). As the N-terminal tail domains of histones protrude out of the nucleosome, they provide an extended surface for interactions with other nucleosomes, thus modulating chromatin folding. At the same time, histone tails may act as receptors for enzymes implicated in signal transduction to chromatin and/or transcriptional regulation (discussed in reference 39). Therefore, covalent modifications of the N-terminal tails of core histones can affect both folding of the chromatin (and consequently accessibility of the underlying DNA) and recruitment to nucleosomes of enzymes or proteins implicated in transcriptional regulation and/or in modulation of chromatin structure.
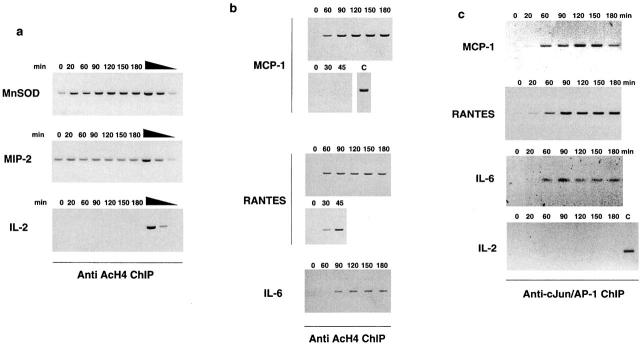
The levels of core histones acetylation, which reflects the balance of histone acetyltransferase (HAT) and histone deacetylase (HDAC) activities on a specific gene, have long been correlated with the transcription status of many genes, being hyperacetylated histones usually associated with transcriptionally active genes 38 39. We therefore used a ChIP assay with an antiacetylated histone H4 antibody to measure the levels of acetylation of the promoters under investigation. The early bound and constitutively accessible promoters (IκBα, MIP-2, and the MnSOD enhancer) were all characterized by high basal levels of histone H4 acetylation (Fig. 3 a, and data not shown). Recruitment of NF-κB, which has previously been shown to contribute to stimulus-dependent acetylation of the human IFN-β promoter 40, is associated with a further increase in acetylation of the MnSOD enhancer, whereas acetylation of the IκBα and MIP-2 promoter was not augmented (Fig. 3 b, and data not shown).
Figure 3.
Stimulus-induced histone H4 hyperacetylation of a subset of NF-κB target promoters precedes NF-κB recruitment. Basal and stimulated levels of histone H4 acetylation were evaluated by ChIP analysis using an anti-AcH4 antiserum (Upstate Biotechnology). Both MnSOD intronic enhancer and MIP-2 promoter were found to display constitutively high levels of histone H4 acetylation (a) which in the case of MnSOD (but not of MIP-2) were further increased upon stimulation. The PCR was performed in the linear range of the amplification as verified using serial dilutions (1, 1:10, 1:100) of the DNA extracted from immunoprecipitation supernatant. The IL-2 gene promoter, which is not active in macrophages, has a constitutively very low level of H4 acetylation, which is not affected by stimulation. Differently from these two NF-κB–dependent promoters, prestimulation histone H4 acetylation levels in the MCP-1, RANTES, and IL-6 promoters were extremely low (b) but increased upon LPS treatment. Recruitment of NF-κB to these promo-ters and accumulation of the corresponding transcripts can be detected only after extensive acetylation has occurred (compare the kinetics shown in b with those in Fig. 2 b). The 0–45-min kinetic comes from a separate experiment. As a positive control (C) for MCP-1 hyperacetylation, the 90-min time point of this experiment is shown. (c) cJun/AP1 recruitment to MCP-1, RANTES, and IL-6 promoters precedes NF-κB recruitment and probably contributes to recruitment of histone acetyltransferases. Anti-cJun ChIP was carried out with a rabbit polyclonal antibody (H79; Santa Cruz Biotechnology, Inc.).
Conversely, prestimulation levels of histone H4 acetylation in the RANTES, MCP-1, and IL-6 promoters were extremely low. However, a clear increase in the acetylation levels of these promoters was found to occur between 20 and 60 min after stimulation, being faster in the case of RANTES in comparison to MCP-1 and IL-6 (Fig. 3 b). From a quantitative point of view, the MCP-1 and RANTES promoters underwent a stronger hyperacetylation than the IL-6 promoter. Comparison of the kinetic of p65/RelA recruitment to these promoters (Fig. 2 b) with the kinetic of stimulus-induced acetylation (Fig. 3 b) indicates that NF-κB binding to these sequences occurs only after extensive hyperacetylation has occurred. We therefore considered the possibility that a transcription factors complex not containing NF-κB initially forms on these promoters and directs hyperacetylation and eventually other conformational changes that make the chromatin subsequently accessible to NF-κB. All of the promoters under study contain one or more c-Jun/AP1 DNA binding site. ChIP analysis of c-Jun recruitment to DNA (Fig. 3 c) shows that recruitment to MCP-1, RANTES, and IL-6 promoters starts at around 20 min, peaks at 60 min, and then slowly declines. Thus, an initial phase in the activation of these late promoters can be identified, in which c-Jun and probably other transcription factors are recruited and direct local histones acetylation and chromatin modifications but not transcriptional initiation. A similar multistep activation of an inducible promoter, the HO endonuclease promoter in yeast, has been described 41. In the HO promoter, transcription factor Swi5p recruits the Swi/Snf chromatin remodeling complex, which in turn makes the promoter accessible to the SAGA acetyltransferase multisubunit complex; only after these sequence of events is completed, transcription factor SBF is recruited and transcription is initiated. A sequential recruitment of chromatin modifying and general transcription factors to the IFN-β promoter in response to viral infection has also been described recently 42.
Constitutive or Regulated Accessibility of NF-κB–dependent Promoters.
To test if signal-induced modifications in chromatin structure are required for MCP-1, RANTES, and IL-6 promoters to become accessible to NF-κB, we transfected Raw 264.7 with a p65/RelA expression vector. Under these conditions, p65 exceeds endogenous IκBs levels and freely enters the nucleus. We found that although transfected p65 could effectively bind the MIP-2 promoter and the MnSOD enhancer in unstimulated cells, it was not recruited to either the MCP-1, RANTES, or IL-6 promoters (Fig. 4 a, and data not shown), thus indicating that these promoters have to undergo modifications to become accessible to NF-κB. Additional evidence that signal-induced modifications of these promoters facilitates NF-κB binding comes from the analysis of IFN-γ effects. IFN-γ enhances the response of macrophages to LPS, in part by activating transcription factors such as IFN regulatory factor (IRF)1 and signal transducer and activator of transcription (STAT)1, which cooperate with c-Jun, NF-κB, and other transcription factors on target promoters. We found that IFN-γ pretreatment allows NF-κB to be recruited to the MCP-1 promoter with a considerably faster kinetic than in nonprestimulated cells (20 min vs. 90 min; Fig. 4 b). Analysis of histone H4 acetylation shows that IFN-γ induces a significant increase in MCP-1 promoter acetylation levels (Fig. 4 b), possibly through stimulation of IRF-1 binding and activity. Similar results were obtained with the RANTES promoter (data not shown).
Figure 4.
A subset of NF-κB–dependent promoters is not constitutively accessible to NF-κB, but accessibility can be increased by adequate stimulation. (a) Raw 264.7 macrophages were transfected with a p65 expression vector or empty vector. Under these experimental conditions, transfected p65 is constitutively nuclear (and can activate transcription from naked cotransfected DNA). ChIP assay with an anti-p65 antibody shows that p65 is recruited only to MnSOD and MIP-2 promoters but not on the MCP-1 and RANTES promoters. Stimulation with LPS for 2 h is followed by NF-κB recruitment to the latter promoters. (b) IFN-γ pretreatment induces hyperacetylation of the MCP-1 promoter and makes it immediately accessible to NF-κB. Raw 264.7 cells were prestimulated with IFN-γ for 2 h and then stimulated with LPS for 20 or 40 min. ChIP assay was performed with anti-p65 antibody or anti-AcH4 serum. Control dilutions of IP supernatant in the anti-AcH4 ChIP are shown. Similar results were obtained with the RANTES promoter.
Discussion
In this study, we have shown that a chromatin-dependent regulatory mechanism generates two distinct classes of NF-κB–activatable genes: those containing CIA NF-κB sites (CIA genes) and those that have to be conformationally modified to become accessible to NF-κB before it is taken back to the cytoplasm (RLA genes). CIA promoters recruit NF-κB immediately after its nuclear entry and rapidly direct transcriptional activation. As they are constitutively accessible, they will recruit NF-κB no matter what is the stimulus that induced its nuclear translocation (and as a matter of fact they are also able to recruit transfected p65/RelA in the absence of any stimulation). Conversely, RLA genes are not accessible to NF-κB unless properly modified in response to stimulation. As transcription of these genes is NF-κB dependent, they will be transcribed only if changes in accessibility take place before the termination of the response, i.e., before NF-κB extrusion from the nucleus. Keeping a subset of NF-κB–dependent promoters in a NF-κB–inaccessible state that can be quickly reversed upon proper stimulation is a strategy with a tremendous regulatory impact, as it contributes to determine the panel of genes that will be transcribed in response to individual stimuli with a similar NF-κB–activating capacity. A considerable number of NF-κB inducers (ranging from bacterial molecules to radiations) with very different biological activities is known: mechanisms like the one described here participate in the configuration of a specific program of gene expression in response to each activator. In support of this model, in Raw 264.7, TNF-α has a similar potency to LPS in terms of ability to activate NF-κB; however, whereas TNF-α is a strong inducer of the CIA genes (IκBα, MIP-2, and MnSOD), it does not induce hyperacetylation and transcription of either of the three RLA genes tested (namely MCP-1, RANTES, and IL-6) (unpublished data).
The mechanism(s) responsible for the fast and stimulus-specific modification in RLA promoters accessibility to NF-κB requires further investigation. We have shown (Fig. 3) that RLA promoters are hyperacetylated before NF-κB recruitment. Although acetylation of N-terminal histone tails may represent by itself the direct cause of the increased accessibility of target genes to NF-κB, it is possible and likely that chromatin remodeling events are implicated in the generation of an accessible state as well. Therefore, we favor a cautious and parsimonious interpretation of our results: we consider stimulus-induced hyperacetylation of histone H4 just as a marker of ongoing changes in RLA promoters' structure. In this light, we can reasonably assume that RLA promoters are modified (as indicated by an increase in H4 acetylation) in response to stimulation before NF-κB is recruited. It could be speculated that the kinetic differences between p65 recruitment and histone H4 hyperacetylation may only reflect differences in the sensitivity of the ChIP assay, as the anti-acetyl H4 ChIP is more sensitive than the anti-p65 ChIP; indeed, many more histone H4 molecules than p65 molecules are likely to be present on a single promoter at a given time. Thus, the faster kinetic of histone H4 hyperacetylation compared with p65 recruitment may reflect a technical limitation of the anti-p65 ChIP assay rather than a true kinetic difference. However, the sensitivity of the ChIP assay can be increased just by increasing the number of PCR cycles in the last step of the assay. Even when the anti-p65 ChIP was carried out with eight PCR cycles more than the anti-acetyl H4 ChIP (40 vs. 32), we could not find any detectable p65 recruitment to either the MCP-1, RANTES, or IL-6 promoters before 90 min, thus substantially ruling out this possibility (data not shown). Moreover, when the ChIP assay was performed with antibodies against cJun/AP1 (which is supposed to be present on target promoters at a frequency similar to that of p65/RelA, and therefore lower than that of histone H4), its recruitment to RLA promoters was found to be faster than the increase in histone H4 acetylation.
However, a detailed characterization of nucleosomal organization before and after stimulation will have to be performed at the level of each individual promoter to define the molecular mechanisms responsible for the increase in NF-κB accessibility. Recent detailed studies have reported that in murine macrophages LPS induces a selective remodeling of a single, strategically positioned nucleosome encompassing the promoter elements required for the induction of the p40 subunit of IL-12 43. This event, which is associated with a mild increase in histone H4 acetylation, occurs independently of the cRel subunit of NF-κB, which is required for transcriptional induction of the IL-12 p40 gene 44. Indeed, IL-12 p40 promoter remodeling can be observed also in cRel−/− macrophages 45; as the remodelled nucleosome covers the cRel binding site, it is likely that remodeling precedes cRel recruitment and is required to make the cRel binding site accessible (although this has not been directly tested by the authors). Thus, according to our classification, we could include the NF-κB(cRel)–dependent IL-12 p40 gene among the RLA genes.
It must be also considered that other factors in addition to nucleosome positioning are likely to affect NF-κB access to promoters. For instance, accessibility of a nucleosome-free NF-κB binding site may be reduced/abrogated by histone H1 (linker histone) molecules lying upon it (discussed in references 46 and 47). Moreover, a nucleosome-free segment of DNA may be poorly accessible due to its intrinsic bending properties. On the other hand, DNA-histone contacts in nucleosomes can be altered by chromatin-remodeling complexes in a way that makes nucleosomal DNA accessible 35. Therefore, several factors in addition to nucleosome positioning have to be considered that may positively or negatively affect access of NF-κB to its binding sites and thus generate the biphasic recruitment we describe here.
It is implied in our model that distinct NF-κB activators differ in their ability to make RLA genes accessible to NF-κB. For instance, in the cellular system described in this study, TNF-α was found to be unable to modify accessibility to NF-κB of the RLA genes tested, whereas all of them were found to undergo changes in NF-κB accessibility in response to LPS. A molecular mechanism responsible for these differences may be represented by the exclusive induction by a subset of NF-κB activators of specific signal transduction pathways and transcription factors required to generate changes in promoters' accessibility to NF-κB. However, a plausible alternative is that kinetic and quantitative differences in the induction of the same signal transduction pathways and transcription factors by different stimuli may generate dramatically different effect at the chromatin level (unpublished data).
Another important issue that will have to be investigated is how chromatin access to different Rel/NF-κB dimers (for instance cRel- or RelB-containing NF-κB complexes) is regulated. The main limitations to this studies is definitely the availability of antibodies capable of immunoprecipitating the different Rel proteins from formalin-fixed lysates.
Given the requirement for the different NF-κB subunits in the transcriptional induction of genes implicated in immune defense, cytoprotection, and cell growth, determining how chromatin accessibility to different NF-κB dimers is modulated in response to different activators and at the level of different genes may provide in the future the molecular basis for therapeutical interventions aiming at selectively blocking the induction of specific NF-κB–dependent genes, without affecting the induction of the others.
Acknowledgments
Antonio Lanzavecchia, Marcus Thelen, Michael Karin, and Roberto Gherzi are gratefully acknowledged for useful suggestions on the manuscript. We would also like to thank Eitan Shaulian for the ChIP protocol.
Footnotes
S. Saccani and S. Pantano contributed equally to this work.
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; CIA, constitutively and immediately accessible; EMSA, electrophoretic mobility shift assay; IκB, inhibitor of NF-κB; MCP-1, macrophage chemoattractant protein 1; MIP-2, macrophage inflammatory protein 2; MnSOD, manganese superoxide dismutase; NF-κB, nuclear factor κB; RANTES, regulated upon activation, normal T cell expressed and secreted; RLA, regulated and late accessibility.
References
- Baldwin A.S., Jr. The NF-kappa B and I kappa B proteinsnew discoveries and insights. Annu. Rev. Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Ghosh S., May M.J., Kopp E.B. NF-kappa B and Rel proteinsevolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Baeuerle P.A., Baltimore D. I kappa Ba specific inhibitor of the NF-kappa B transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- Davis N., Ghosh S., Simmons D.L., Tempst P., Liou H.C., Baltimore D., Bose H.R., Jr. Rel-associated pp40an inhibitor of the rel family of transcription factors. Science. 1991;253:1268–1271. doi: 10.1126/science.1891714. [DOI] [PubMed] [Google Scholar]
- Haskill S., Beg A.A., Tompkins S.M., Morris J.S., Yurochko A.D., Sampson-Johannes A., Mondal K., Ralph P., Baldwin A.S., Jr. Characterization of an immediate-early gene induced in adherent monocytes that encodes I kappa B-like activity. Cell. 1991;65:1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- Thompson J.E., Phillips R.J., Erdjument-Bromage H., Tempst P., Ghosh S. I kappa B-beta regulates the persistent response in a biphasic activation of NF-kappa B. Cell. 1995;80:573–582. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]
- Whiteside S.T., Epinat J.C., Rice N.R., Israel A. I kappa B epsilon, a novel member of the I kappa B family, controls RelA and cRel NF-kappa B activity. EMBO J. 1997;16:1413–1426. doi: 10.1093/emboj/16.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg A.A., Ruben S.M., Scheinman R.I., Haskill S., Rosen C.A., Baldwin A.S., Jr. I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa Ba mechanism for cytoplasmic retention Genes Dev. 6 1992. 1899 1913[published erratum at 6:2664–2665] [DOI] [PubMed] [Google Scholar]
- Ganchi P.A., Sun S.C., Greene W.C., Ballard D.W. I kappa B/MAD-3 masks the nuclear localization signal of NF-kappa B p65 and requires the transactivation domain to inhibit NF-kappa B p65 DNA binding. Mol. Biol. Cell. 1992;3:1339–1352. doi: 10.1091/mbc.3.12.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatada E.N., Nieters A., Wulczyn F.G., Naumann M., Meyer R., Nucifora G., McKeithan T.W., Scheidereit C. The ankyrin repeat domains of the NF-kappa B precursor p105 and the protooncogene bcl-3 act as specific inhibitors of NF-kappa B DNA binding. Proc. Natl. Acad. Sci. USA. 1992;89:2489–2493. doi: 10.1073/pnas.89.6.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel U., Henkel T., Silva M.S., Baeuerle P.A. Nuclear uptake control of NF-kappa B by MAD-3, an I kappa B protein present in the nucleus. EMBO J. 1993;12:201–211. doi: 10.1002/j.1460-2075.1993.tb05646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg A.A., Finco T.S., Nantermet P.V., Baldwin A.S., Jr. Tumor necrosis factor and interleukin-1 lead to phosphorylation and loss of I kappa B alphaa mechanism for NF-kappa B activation. Mol. Cell. Biol. 1993;13:3301–3310. doi: 10.1128/mcb.13.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel T., Machleidt T., Alkalay I., Kronke M., Ben-Neriah Y., Baeuerle P.A. Rapid proteolysis of I kappa B-alpha is necessary for activation of transcription factor NF-kappa B. Nature. 1993;365:182–185. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- Mayo M.W., Baldwin A.S. The transcription factor NF-kappaBcontrol of oncogenesis and cancer therapy resistance. Biochim. Biophys. Acta. 2000;1470:M55–M62. doi: 10.1016/s0304-419x(00)00002-0. [DOI] [PubMed] [Google Scholar]
- Zabel U., Baeuerle P.A. Purified human I kappa B can rapidly dissociate the complex of the NF- kappa B transcription factor with its cognate DNA. Cell. 1990;61:255–265. doi: 10.1016/0092-8674(90)90806-p. [DOI] [PubMed] [Google Scholar]
- Arenzana-Seisdedos F., Thompson J., Rodriguez M.S., Bachelerie F., Thomas D., Hay R.T. Inducible nuclear expression of newly synthesized I kappa B alpha negatively regulates DNA-binding and transcriptional activities of NF- kappa B. Mol. Cell. Biol. 1995;15:2689–2696. doi: 10.1128/mcb.15.5.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., Parent L., Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination- dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- DiDonato J.A., Hayakawa M., Rothwarf D.M., Zandi E., Karin M. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- Regnier C.H., Song H.Y., Gao X., Goeddel D.V., Cao Z., Rothe M. Identification and characterization of an IkappaB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- Zandi E., Rothwarf D.M., Delhase M., Hayakawa M., Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- Woronicz J.D., Gao X., Cao Z., Rothe M., Goeddel D.V. IkappaB kinase-betaNF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- Mercurio F., Zhu H., Murray B.W., Shevchenko A., Bennett B.L., Li J., Young D.B., Barbosa M., Mann M., Manning A., Rao A. IKK-1 and IKK-2cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- Karin M., Ben-Neriah Y. Phosphorylation meets ubiquitinationthe control of NF-[kappa]B activity. Annu. Rev. Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- Israel A. The IKK complexan integrator of all signals that activate NF-kappaB? Trends Cell Biol. 2000;10:129–133. doi: 10.1016/s0962-8924(00)01729-3. [DOI] [PubMed] [Google Scholar]
- Yaron A., Hatzubai A., Davis M., Lavon I., Amit S., Manning A.M., Andersen J.S., Mann M., Mercurio F., Ben-Neriah Y. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- Maniatis T. A ubiquitin ligase complex essential for the NF-kappaB, Wnt/Wingless, and Hedgehog signaling pathways. Genes Dev. 1999;13:505–510. doi: 10.1101/gad.13.5.505. [DOI] [PubMed] [Google Scholar]
- Hecht A., Grunstein M. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. [DOI] [PubMed] [Google Scholar]
- Ouaaz F., Li M., Beg A.A. A critical role for the RelA subunit of nuclear factor kappaB in regulation of multiple immune-response genes and in Fas-induced cell death. J. Exp. Med. 1999;189:999–1004. doi: 10.1084/jem.189.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.L., Ping D., Boss J.M. Tumor necrosis factor alpha and interleukin-1beta regulate the murine manganese superoxide dismutase gene through a complex intronic enhancer involving C/EBP-beta and NF-kappaB. Mol. Cell. Biol. 1997;17:6970–6981. doi: 10.1128/mcb.17.12.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyang H., Phillips R., Douglas I., Ghosh S. Role of unphosphorylated, newly synthesized I kappa B beta in persistent activation of NF-kappa B. Mol. Cell. Biol. 1996;16:5444–5449. doi: 10.1128/mcb.16.10.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura T.S., Kempiak S.J., Nel A.E. Activation of the human RANTES gene promoter in a macrophage cell line by lipopolysaccharide is dependent on stress-activated protein kinases and the IkappaB kinase cascadeimplications for exacerbation of allergic inflammation by environmental pollutants. Clin. Immunol. 1999;90:287–301. doi: 10.1006/clim.1998.4659. [DOI] [PubMed] [Google Scholar]
- Georganas C., Liu H., Perlman H., Hoffmann A., Thimmapaya B., Pope R.M. Regulation of IL-6 and IL-8 expression in rheumatoid arthritis synovial fibroblaststhe dominant role for NF-kappaB but not C/EBPss or c-Jun. J. Immunol. 2000;165:7199–7206. doi: 10.4049/jimmunol.165.12.7199. [DOI] [PubMed] [Google Scholar]
- Ping D., Jones P.L., Boss J.M. TNF regulates the in vivo occupancy of both distal and proximal regulatory regions of the MCP-1/JE gene. Immunity. 1996;4:455–469. doi: 10.1016/s1074-7613(00)80412-4. [DOI] [PubMed] [Google Scholar]
- Grilli M., Chiu J.J., Lenardo M.J. NF-kappa B and Relparticipants in a multiform transcriptional regulatory system. Int. Rev. Cytol. 1993;143:1–62. doi: 10.1016/s0074-7696(08)61873-2. [DOI] [PubMed] [Google Scholar]
- Aalfs J.D., Kingston R.E. What does “chromatin remodeling” mean? Trends Biochem. Sci. 2000;25:548–555. doi: 10.1016/s0968-0004(00)01689-3. [DOI] [PubMed] [Google Scholar]
- Kornberg R.D., Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Cheung W.L., Briggs S.D., Allis C.D. Acetylation and chromosomal functions. Curr. Opin. Cell Biol. 2000;12:326–333. doi: 10.1016/s0955-0674(00)00096-x. [DOI] [PubMed] [Google Scholar]
- Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Merika M., Williams A.J., Chen G., Collins T., Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- Cosma M.P., Tanaka T., Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Agalioti T., Lomvardas S., Parekh B., Yie J., Maniatis T., Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Weinmann A.S., Plevy S.E., Smale S.T. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- Sanjabi S., Hoffmann A., Liou H.C., Baltimore D., Smale S.T. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc. Natl. Acad. Sci. USA. 2000;97:12705–12710. doi: 10.1073/pnas.230436397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann A.S., Mitchell D.M., Sanjabi S., Bradley M.N., Hoffmann A., Liou H.-C., Smale S.T. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat. Immunol. 2001;2:51–57. doi: 10.1038/83168. [DOI] [PubMed] [Google Scholar]
- Roth S.Y., Allis C.D. Chromatin condensationdoes histone H1 dephosphorylation play a role? Trends Biochem. Sci. 1992;17:93–98. doi: 10.1016/0968-0004(92)90243-3. [DOI] [PubMed] [Google Scholar]
- Lee H.L., Archer T.K. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J. 1998;17:1454–1466. doi: 10.1093/emboj/17.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]