Abstract
Cone snails are gastropod mollusks of the genus Conus that live in tropical marine habitats. They are predators that paralyze their prey by injection of venom containing a plethora of small, conformationally constrained peptides (conotoxins). We report the identification, characterization, and structure of a γ-carboxyglutamic acid-containing peptide, conotoxin ɛ-TxIX, isolated from the venom of the molluscivorous cone snail, Conus textile. The disulfide bonding pattern of the four cysteine residues, an unparalleled degree of posttranslational processing including bromination, hydroxylation, and glycosylation define a family of conotoxins that may target presynaptic Ca2+ channels or act on G protein-coupled presynaptic receptors via another mechanism. This conotoxin selectively reduces neurotransmitter release at an Aplysia cholinergic synapse by reducing the presynaptic influx of Ca2+ in a slow and reversible fashion. The three-dimensional structure, determined by two-dimensional 1H NMR spectroscopy, identifies an electronegative patch created by the side chains of two γ-carboxyglutamic acid residues that extend outward from a cavernous cleft. The glycosylated threonine and hydroxylated proline enclose a localized hydrophobic region centered on the brominated tryptophan residue within the constrained intercysteine region.
Marine snails of the genus Conus elaborate a series of conotoxins effecting paralysis of their prey (1). Conotoxins are invariably small (12–30 aa), conformationally constrained peptides with many of them possessing a stabilizing network of intramolecular disulfide bridges (1–3). These neurotoxins have a discriminatory ability to selectively target specific receptor subunits or ion channels with a very high affinity (for review see ref. 4 and refs. therein). This refined selectivity is associated with unique disulfide-bonding frameworks and specific amino acids positioned within hypervariable intercysteine regions. Conotoxins are generally classified on the basis of their neuropharmacological profile, including the α, μ, δ, κ, and ω classes, which specifically target the acetylcholine receptor, sodium channels (μ and δ), potassium channels, and voltage-sensitive calcium channels, respectively. In addition, conotoxins are classified structurally with respect to the arrangement of their cysteine residues, which define two-, three-, and four-loop intramolecular frameworks. Thus, multiple conotoxin families interacting at distinct pharmacological receptor targets are associated with a single structural class. The molecular diversity of these conotoxins is enhanced by the posttranslational modification of the hypermutable intercysteine regions. Some of the posttranslational modifications identified and characterized within the genus Conus include γ-carboxylation of glutamic acid residues, l-6-bromination of tryptophan residues, C-terminal amidation, and 4-trans-hydroxylation of proline residues (5–7).
The ω-conotoxins isolated from piscivorous and molluscivorous Conus possess determinants facilitating selective interactions with N, P, and Q Ca2+ channel subtypes that regulate neurotransmitter release from presynaptic neurons (3, 8). Typically, the ω-conotoxins possess a characteristic “four-loop Cys scaffold” that stabilizes a common tertiary structure facilitating this selective interaction with calcium ion channels (9, 10). Here we report the isolation, characterization, and structure determination of a conotoxin, conotoxin ɛ-TxIX. This peptide represents a structural class of conotoxin possessing a unique disulfide-bonding network, extensive posttranslational modification, and a unique ω-conotoxin-like neuropharmacological effect.
In an attempt to better understand the significance of this disulfide bonding arrangement in receptor recognition and the role played by each posttranslational modification, we have investigated the three-dimensional structure of the peptide. The unique, paired dyad arrangement of cysteine residues constrains the molecule in a conformation that is presumably responsible for the selective interaction demonstrated by this peptide. This constrained structure exposes two distinct recognition surfaces oriented in opposite directions, suggesting the need for each in receptor recognition and discrimination.
MATERIALS AND METHODS
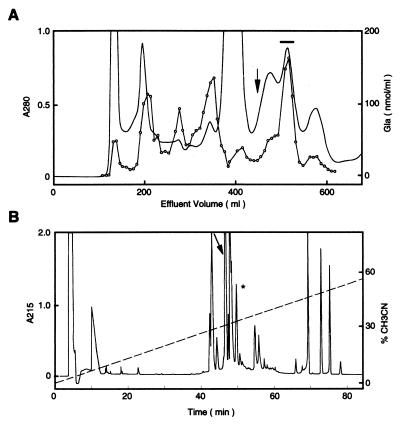
Conotoxin Purification and Characterization. Frozen cone snails (Conus textile) were obtained from Vietnam. Lyophilized venom extract (200 mg, from five snails) was dissolved in 0.2 M ammonium acetate (11) and chromatographed on a Sephadex G 50 superfine column (2.5 × 92 cm) equilibrated with 0.2 M ammonium acetate buffer (pH 7.5) and eluted with a flow rate of 9.2 ml/hour (Fig. 1A). The material in the major γ-carboxyglutamic acid (Gla)-containing peak was purified on a reverse-phase column (HyCrom C18, 5 μm; 10 × 250 mm) in 0.1% trifluoroacetic acid and eluted with an acetonitrile gradient (Fig. 1B). A total of 1.1 mg of peptide (560 nmol) was obtained. The amino acid composition was determined after acid hydrolysis, except for Gla, which was determined after alkaline hydrolysis. Monosaccharides were analyzed as described (12). N-terminal sequence determinations (Perkin–Elmer ABI Procise 494 sequencer) were performed before and after reduction and alkylation of the cysteine residues with vinyl pyridine (Sigma) by using a standard procedure (13). To identify Gla in a sequence, the carboxyl groups were first converted to methyl esters with methanolic HCl (14).
Figure 1.
Purification of ɛ-TxIX. (A) Elution profile of C. textile venom extract chromatographed on a Sephadex G 50 column. The major Gla-containing peak, ɛ-TxIX (denoted by a horizontal bar), elutes after one column volume (denoted by an arrow). (B) Isolation of ɛ-TxIX (peak denoted by arrow) by HPLC on a C18 column in 0.1% trifluoroacetic acid and eluted with an acetonitrile gradient. The peak denoted by ∗ contains a minor Gla-containing peptide (not analyzed). The material in the other peaks did not contain significant amounts of Gla.
For reduction and alkylation with iodoacetamide (Sigma), ɛ-TxIX (1 nmol) was dissolved in ammonium bicarbonate (100 mM), pH 7.8. After the addition of DTT (10 μl, 45 mM) (Sigma), the sample was incubated in the dark at 50°C for 2 hours in an argon atmosphere. The sample was cooled to room temperature, and iodoacetamide (10 μl, 100 mM) was added. This sample was incubated at room temperature for 2 hours in the presence of argon, and the peptide was purified by reverse-phase HPLC. S-carbamidomethylated ɛ-TxIX (500 pmol) was dissolved in 30 μl of 50 mM ammonium acetate (pH 6.0) and digested with carboxypeptidase Y at 37°C. Aliquots were removed after 6, 18, 24, and 48 hrs and analyzed directly by using matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS).
Mass Spectrometry.
MALDI-MS was performed on a Voyager-Elite MALDI–time-of-flight (TOF) mass spectrometer (PerSeptive Biosystems, Framingham, MA) equipped with delayed ion extraction. Spectra were acquired in positive-ion linear and reflector modes by using 2,5-dihydroxybenzoic acid (DHB), α-cyano-4-hydroxycinnamic acid (HCCA), and 2,4,6-trihydroxyacetophenone (THAP) mixed with nitrocellulose (Bio-Rad) as matrices. Sample preparation for MALDI-MS was performed by using the dried-droplet method for DHB, the sandwich method for HCCA, and the thin-layer method with THAP/nitrocellulose matrices as described (15).
Neurophysiology.
In preliminary experiments, 0.3–100 μg of ɛ-TxIX was injected intracerebrally into 32 mice less than 2 weeks old. These mice were observed for neurological signs and compared with 11 control mice that had been injected with saline. Neurophysiology experiments were performed on an identified cholinergic synapse of the buccal ganglion of Aplysia californica (16). Preparations were perfused with artificial seawater of the following composition (mM): NaCl, 460; KCl, 10; CaCl2, 11; MgCl2, 25; MgSO4, 28; Tris⋅HCl buffer, 10 (pH 7.8). The perfusion was stopped before the toxins were added to the bath.
Cells were impaled with two low-resistance (0.5–2.0 MΩ) microelectrodes filled with 3 M KCl. The generation of a presynaptic action potential produced an inhibitory postsynaptic current (IPSC) in the voltage-clamped postsynaptic neuron at −80 mV. The IPSC represents the summation of discrete miniature postsynaptic currents (MPSCs), which result from the release of acetylcholine (ACh) quanta by the presynaptic neuron. Postsynaptic responses were expressed as conductances (nS) by dividing their amplitude (nA) by the driving force (mV) (16). It was possible to study ACh release independent of spike conduction or ionic current modification (other than Ca2+), when pre- and postsynaptic neurons were simultaneously voltage-clamped to −50 and −80 mV, respectively. A 3-second square depolarization of the voltage-clamped presynaptic neuron induced a postsynaptic response, referred to as long duration-induced postsynaptic current (LDIPSC). The amplitude of evoked miniature postsynaptic currents and the number of quanta (Q) released by the presynaptic neuron were calculated from a statistical analysis of the fluctuations on the top of the LDIPSC (17).
Presynaptic Ca2+ currents were elicited by short depolarizing steps (20 ms) from a holding potential of −50 mV to a variety of test potentials. Tetrodotoxin at a final concentration of 100 μM was added to the bath to block the inward Na+ current, and outward K+ currents were blocked with extracellular application of tetraethylammonium (50 mM) and 4-aminopyridine (3 mM). Increasing the extracellular Ca2+ concentration to 55 mM enhanced the Ca2+ gradient. I/V curves were leakage-subtracted.
NMR Spectroscopy and Structure Calculations.
NMR spectra were recorded at 298.3 K and 285.3 K with a Bruker AMX-500 spectrometer. A sample containing 1.9 mg of conotoxin ɛ-TxIX was dissolved in 450 μl of H2O (2.7 mM), pH 5.60, 25 mM acetic-d3 acid-d, with 10% D2O as the deuterium lock signal. Samples prepared in D2O were initially lyophilized from 99.8% D2O and redissolved in 99.96% isotope-enriched D2O. All samples were pretreated with Chelex 100 at pH 7.5 to remove trace metal ions. Homonuclear Double Quantum Filtered Correlation Spectroscopy (DFQ-COSY) and Total Correlated Spectroscopy (TOCSY) experiments with MLEV-17 isotropic mixing times of 35 and 50 ms facilitated the spin system assignment (18–20). Nuclear Overhauser effect spectra collected at several mixing times facilitated identification of intermolecular nuclear Overhauser effects and the measurement of 3JHNα coupling constants (21). Structures were calculated by using DGII (InsightII, Molecular Simulations, Waltham, MA), using a combination of distance geometry and simulated annealing methodology. Structure determination employed a set of 159 nuclear Overhauser effect distance restraints (103 intraresidue, 36 sequential, 15 medium, and 5 long), 3 3JHNα coupling constants, and 4 χI torsion angles.
RESULTS
Conotoxin Purification and Characterization.
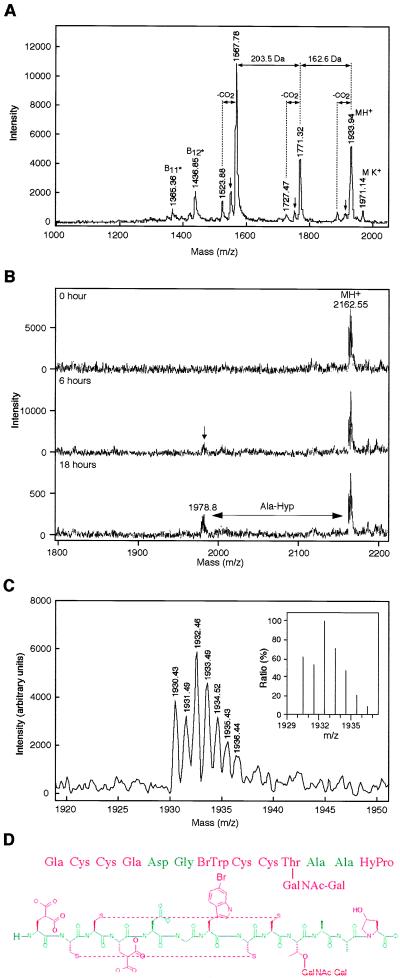
A major Gla-containing peptide, denoted ɛ-TxIX, has been purified from C. textile venom (Fig. 1). The peptide comprises 1–3% of the lyophilized crude venom. Peptide sequencing and MALDI mass spectrometry were used to deduce the sequence of this 13-residue peptide and ascertain the nature and position of each posttranslational modification (Fig. 2; Table 1). Reduction and alkylation of cysteine residues with iodoacetamide resulted in a mass increase of 232.2 Da (theoretical 232.24 Da) as compared with the native compound, consistent with four cysteine residues. The four cysteines are arranged pairwise in a framework not previously identified in conotoxins.
Figure 2.
Characterization of ɛ-TxIX. (A) Linear-mode MALDI mass spectrum of ɛ-TxIX obtained with 2,5-dihydroxybenzoic acid as matrix. The molecular ion at m/z 1,933.9 is accompanied by loss of CO2 (44 Da) that is consistent with the presence of Gla. The peaks at m/z 1,771.32 and 1,567.78 most likely arise by prompt fragmentation. The mass differences of 162.6 and 203.5 Da correspond to a loss of a galactose and an N-acetylgalactosamine residue, respectively, indicating the presence of an O-linked glycan. The N-terminal sequence ions B*11 and B*12 contain the intact N terminus of the peptide after loss of the glycan (indicated by ∗). They identify the sequential loss of hydroxyproline and alanine. The small arrows denote a loss of 18 Da, corresponding to loss of water via β-elimination. (B) Maldi-MS spectra of the carboxypeptidase Y digestion of the reduced S-carbimidomethylated ɛ-TxIX peptide, at the time points indicated. (C) Molecular ion region of the MALDI-MS spectrum of native ɛ-TxIX obtained in reflector mode by using 2,4,6-trihydroxyacetophenone as the matrix to reduce metastable decay. The isotope pattern matches exactly that calculated based on the assumed elementary composition (Inset). (D) Amino acid sequence, disulfide bond pairing (deduced by NMR spectroscopy) and posttranslational modifications of ɛ-TxIX.
Table 1.
Measured and calculated molecular masses for conotoxin ɛ-TxlX
| Species | Molecular mass, Da
|
|
|---|---|---|
| Monoisotopic | Average | |
| Native | ||
| Observed | 1,929.46 | 1,931.97 |
| Calculated | 1,929.42 | 1,931.78 |
| Carbamidomethylated | ||
| Observed | 2,161.54 | 2,164.25 |
| Calculated | 2,161.53 | 2,164.02 |
The calculated molecular mass was based on the amino acid sequence: Gla Cys Cys Gla Asp Gly BrTrp Cys Cys Thr(GalNAc-Gal) Ala Ala HyPro, where Gla = γ-carboxyglutamic acid, BrTrp = 6-bromotryptophan, Thr(GalNac-Gal) = N-acetylgalactosamine and galactosamine attached to threonine, and HyPro = 4-hydroxyproline. All cysteine residues are disulfide-bonded. The monoisotopic and average observed molecular masses were acquired in reflector and linear modes, respectively.
Peptide sequencing of underivatized and of reduced and alkylated (vinyl pyridine) material established the homogeneity of the peptide and allowed prompt identification of the amino acids in positions 2, 3, 5, 6, 8, 9, 11, and 12 and a tentative identification of the hydroxyproline in position 13 (Fig. 2D). Sequencing of methyl-esterified material was used to identify the Gla residues in postions 1 and 4. Amino acid analysis of alkaline hydrolysates showed that the peptide contains 2 mol of Gla per mol of peptide. Accordingly, the acid hydrolysates contained 2 mol of Glu per mol of peptide. Moreover, the acid hydrolysate also contained 1 mol of hydroxyproline per mol of peptide. The presence of Gla is also notable in the mass spectra, via decarboxylation of the peptide resulting in a loss of 44 Da for each Gla residue under the conditions used. The MS data also showed prompt fragmentation of the peptide with a concomitant loss of 162.6 and 203.5 Da, respectively, suggesting the presence of a disaccharide moiety consisting of an N-acetylhexosamine and a hexose moiety (Fig. 2A). Carbohydrate analysis identified 0.7 mol each of N-acetylgalactosamine and galactose per mol of peptide. After an 18-hour incubation with carboxypeptidase Y, the intensity of an ion signal (m/z 1978.8) that lacks the C-terminal hydroxyproline and alanine increased (Fig. 2B). With prolonged incubation times, the intensity of this peak increased with respect to the ion signal of the intact S-carbamidomethylated (m/z 2162.55) peptide. This is in agreement with a slowly removed C-terminal hydroxyproline residue, followed by a rapid release of the penultimate alanine residue and no further digestion, as we have observed previously for O-glycosylated proteins (unpublished data). Linkage of the N-acetylhexosamine and hexose moiety to the threonine residue is also consistent with the peptide sequencing, because no threonine was identified in postion 10, whereas the C-terminal Ala-Ala-HyPro sequence was readily identified. MS confirmed the linkage pattern of the disaccharide, as demonstrated in Fig. 2A, which shows loss of a hexose moiety (m/z 1771.32) followed by the sequential loss of N-acetylhexosamine (m/z 1567.78) from the molecular ion. Hence, the N-acetylhexosamine is the O-linked monosaccharide.
The UV and one-dimensional 1H NMR spectra of conotoxin ɛ-TxIX established the presence of a tryptophan ring (unpublished data). However, during sequence analysis, no phenylthiohydantoin (PTH) derivative of tryptophan was detected, but in position 7, a small peak from a compound with a slightly longer HPLC retention time than that of PTH-tryptophan was observed. These results combined with MALDI mass spectrometry data demonstrating that ɛ-TxIX possessed an isotopic pattern characteristic of bromine, indicated the presence of 6-bromotryptophan (bromine at the η2 position of the tryptophan indole ring), a modification recently identified in conopeptides (Fig. 2C) (6, 22, 23).
The molecular mass of ɛ-TxIX (Table 1) corresponds to that of the posttranslationally modified peptide shown in Fig. 2D, including four disulfide-linked cysteine residues, one brominated-tryptophan residue (bromotryptophan), two Gla residues, one threonine residue with a glycan consisting of one N-acetylhexosamine and one hexose moiety, and one hydroxyproline residue. Unlike most conopeptides, the C-terminal carboxyl group of ɛ-TxIX is not amidated. The sequence of the cDNA encoding ɛ-TxIX demonstrates that this is the result of proteolytic degradation, attributable to the loss of three C-terminal residues (K. Bush, unpublished result).
Neurophysiology.
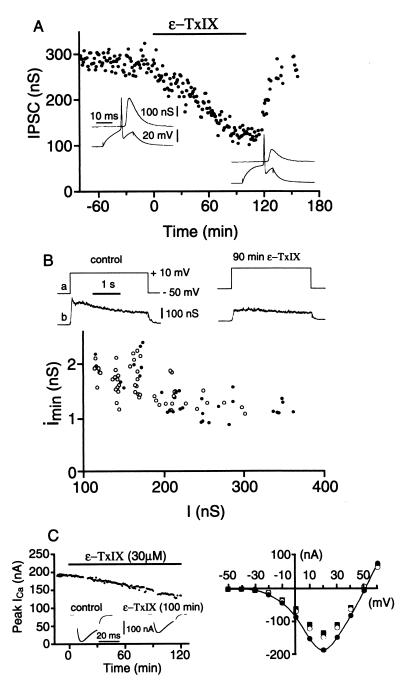
The biological activity of the peptide ɛ-TxIX was studied at an identified cholinergic synapse of the buccal ganglion of A. californica (16). The amplitude of the IPSC evoked by a presynaptic action potential decreased progressively to 40% of the initial value over ≈100 min, after the addition of 30 μM conotoxin ɛ-TxIX (Fig. 3A). Washing the preparation with artificial seawater reversed this effect. This reduction of a postsynaptic response may occur presynaptically via a decrease in the amount of neurotransmitter released or postsynaptically after a modification of the postsynaptic receptor-channel complex. At a neuronal synapse, the postsynaptic neuron receives several inputs, making it impossible to record a single MPSC. To circumvent this, we used the LDIPSC method, which uses a prolonged depolarization of the voltage-clamped presynaptic neuron, thus generating a complex postsynaptic response. Statistical analysis of this postsynaptic current identifies the number of ACh quanta released by the presynaptic neuron. Fig. 3B illustrates the decrease in the mean amplitude of the LDIPSC caused by a reduction in the number of released quanta after the application of ɛ-TxIX (60% reduction after 90 minutes). The specificity of this presynaptic effect was substantiated by the consistency observed in the mean amplitude of the MPSCs. The distribution of the mean amplitude of MPSCs with respect to the mean amplitude of the LDIPSC (17) was identical to control values after application of ɛ-TxIX (Fig. 3B). It has previously been established that the amplitude of the MPSCs depends on both the presynaptically released ACh quantum (number of ACh molecules released by exocytosis of a presynaptic vesicle) and the number of postsynaptic receptors activated by the quantum. The amplitude of the MPSCs was unaltered in the presence of ɛ-TxIX, suggesting that this peptide does not affect neurotransmitter release or postsynaptic receptors. Furthermore, the presynaptic Ca2+ current, which triggers ACh release, decreased to 70% with respect to control values, 120 minutes after the application of conotoxin ɛ-TxIX (Fig. 3C).
Figure 3.
Effects of ɛ-TxIX on synaptic transmission. (A) Bath-applied ɛ-TxIX (30 μM) reduced the amplitude of IPSC. Washing out the peptide led to a rapid recovery of synaptic transmission. Insets represent the postsynaptic responses (IPSCs, Upper traces) evoked by a presynaptic action potential (Lower traces) before (Left recordings) and after (Right recordings) 100 minutes ɛ-TxIX application. (B) LDIPSCs, evoked by a 3-second depolarization of the voltage-clamped presynaptic neuron, were decreased in the presence of ɛ-TxIX (30 μM). Control response: I = 250 nS; Imin = 1.2 nS = 12.25 ms; Q = 51 020 quanta; 90 minutes ɛ-TxIX: I = 142 nS, Imin= 1.7 nS = 11.8 ms; Q = 21 236 quanta. The graph shows the relationship between the mean amplitude of MPSCs (Imin, calculated from the above LDIPSCs) and the mean amplitude (I) of the LDIPSC before (●) and after 90 minutes of ɛ-TxIX application (○). (C, Left) Evolution of the peak presynaptic Ca2+ current after bath application of ɛ-TxIX (30 μM). Insets are examples of the Ca2+ current before (Left) and after (Right) ɛ-TxIX application. (Right) I/V curves representing the presynaptic Ca2+ current in the control situation (●), after 90 minutes (○) and 115 minutes (■) of ɛ-TxIX.
In addition, 32 mice were injected intracranially with ɛ-TxIX. Tremors and spastic gaits were observed. However, there were no generalized seizures or deaths, even at the highest concentration of peptide evaluated.
NMR Spectroscopy and Structure Calculations.
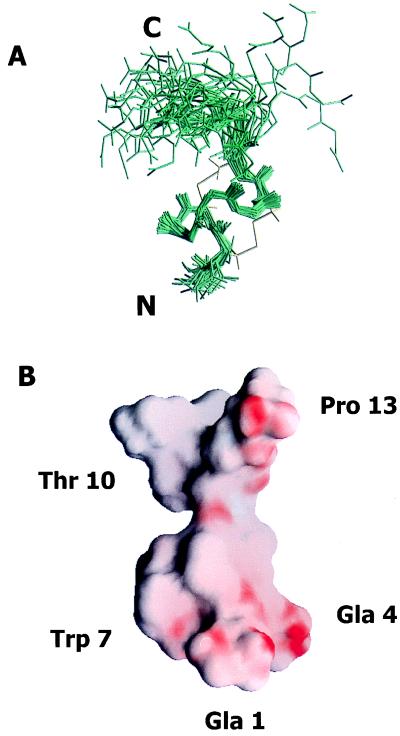
The three-dimensional structure of conotoxin ɛ-TxIX was calculated using distance geometry and simulated annealing methods. This data set included 159 distance restraints (103 intraresidue, 36 sequential, 15 medium, and 5 long range), 3 φ torsion angles, and 4 χ1 torsion angles. Of 40 structures generated, 38 converged (3 structures were discarded as mirror images) possessing no distance violation greater than 0.0015 Å and no nuclear Overhauser effect violation greater than 0.23 Å. The remaining 35 structures were superimposed over the well structured region from residue 2 and extending through to residue 10 (Fig. 4A). The pairwise rms deviation to the lowest energy structure is 0.46 ± 0.08 Å for the well defined backbone atoms between residues 2 and 10 of the 35 final structures.
Figure 4.
(A) Three-dimensional structure of ɛ-TxIX. Superimposition of the final 35 structures to the lowest energy conformer of ɛ-TxIX illustrating the (N, Cα, and carbonyl C) backbone atoms of all 13 residues. The peptide backbone pairwise rms deviation is 0.46 ± 0.08 Å over the well defined residues 2 through 10. The cysteine bond arrangement for the lowest energy conformer is identified in yellow. The N and C termini are labeled. (B) Surface view of the electrostatic potential of the lowest energy conformer of ɛ-TxIX illustrating the alignment of the negatively charged residues along one side of the molecule and protruding into the solvent. The surface of the molecule is colored using grasp (41) according to electrostatic potential, scaled from electronegative (red) to electropositive (blue). The Gla, hydroxyproline, bromotryptophan, and glycosylated threonine residues are also labeled.
The backbone of ɛ-TxIX is highly constrained by a novel intramolecular disulfide bonding arrangement (illustrated in Fig. 2D), producing a compact structure (Fig. 4A). The disulfide bond between Cys-2 and Cys-8 exists in two isomeric conformers, which increases the mobility of these residues without perturbing the overall conformation of the peptide. The rate of interconversion for this disulfide bond was investigated by using line shape analysis and was estimated to be in the intermediate exchange regime on the NMR time scale (1–10−4 s). The equivalent fractional population of each conformer further contributes to exchange broadening and coalescence of the proton resonances (24). The presence of bromine at the η2 position of the tryptophan indole ring and the hydroxyl group at the γ (4-trans) position of the C-terminal proline were confirmed by the notable absence of these protons in TOCSY spectra collected at low temperature. The N-acetylgalactosamine–galactose disaccharide linked to Thr-10 is very mobile, as demonstrated by the increased line width of this residue in the nuclear Overhauser effect (NOESY) spectra.
DISCUSSION
Many conotoxins demonstrate enhanced affinity and specificity for macromolecular receptors and ion channels, facilitating selective discrimination between receptor subtypes and different molecular isoforms of ligand-gated ion channels. The presence of a dense disulfide bonding network, a hypervariable intercysteine region, and posttranslational modifications illustrates the complexity of these diverse toxin molecules. We have purified and characterized a 13-residue conotoxin, conotoxin ɛ-TxIX, from the venom of the molluscivorous cone snail C. textile. This peptide, which has an unprecedented degree of posttranslational modifications, also possesses a novel disulfide bonding arrangement and a unique ability to regulate calcium influx.
Conotoxin ɛ-TxIX was extensively characterized by using peptide sequencing that allowed the identification of two Gla residues and a 4-trans-hydroxyproline residue. Since the discovery of Gla in 1974 and elucidation of its involvement in the Ca2+-mediated membrane interaction of the vitamin K-dependent blood clotting proteins, it has also been found in two mineralized tissue proteins (25–28). The discovery of Gla in toxins from invertebrate cone snails (conantokin-G, conantokin-T, conotoxin TxVIIA, and γ-conotoxin-PnVIIA) suggests a more general role for Gla in biology beyond processes involved in blood coagulation and tissue mineralization (5, 11, 29–31). This is also in accordance with the wide tissue distribution of the vitamin K-dependent carboxylase (32).
MALDI mass spectrometry and 1H NMR spectroscopy were used to identify the brominated tryptophan residue (residue 7), a posttranslationally modified amino acid identified in cone snail toxins, nonpeptidic marine natural products, and a peptide isolated from the morula cells of the marine organism ascidian (6, 22, 23, 33, 34). Brominated conotoxin, σ-conotoxin GVIIIA, was shown to selectively inactivate the 5-HT3 ligand-gated ion channel in a manner similar to the endogenous agonist 5-hydroxytryptamine (23).
ɛ-TxIX is a glycosylated peptide, isolated and characterized from cone snail venom, possessing an O-linked N-acetylhexosamine and a hexose moiety that regulates presynaptic Ca2+ influx. Recently, Craig et al. (35) identified an O-glycosylated conotoxin that inhibits voltage-gated K+ channels. O-linked carbohydrates are responsible for many functional roles within glycoproteins, including regulating protein expression, protein structure stabilization, and molecular recognition (36). The functional role of this disaccharide unit in the interaction of ɛ-TxIX with the Ca2+ channels or G-coupled receptors is now open to investigation.
The ω-conotoxins delineate a family of conotoxins known to selectively target voltage-sensitive Ca2+ channels, regulating Ca2+ entry into the presynaptic termini and thus controlling neurotransmitter release (3, 37). In the presence of conotoxin ɛ-TxIX, both the presynaptic Ca2+ influx and ACh release are reduced. It is widely accepted that the presynaptic Ca2+ influx is the main trigger of neurotransmitter release (38). The steep relationship between the presynaptic Ca2+ influx and evoked ACh release suggests that the observed 30% reduction in Ca2+ current after exposure to ɛ-TxIX for 120 minutes accounts for the 55% reduction of neurotransmitter release. We previously demonstrated (39, 40) that both histamine and ω-conotoxins reduce presynaptic Ca2+ current by 30–35%, which concomitantly reduces neurotransmitter release by 50–53%. Thus, transmitter release can be mainly attributed to this presynaptic Ca2+ influx as a result of inhibiting voltage-gated Ca2+ channels. However, we cannot discard the possibility that this peptide acts on another target such as G protein-coupled presynaptic receptors. The mode of presynaptic Ca2+ channel interaction and novel disulfide bonding arrangement, identifies ɛ-TxIX as a member of a new conotoxin family, ɛ-conotoxins.
As a first step toward structure–function studies, we have determined the solution structure of ɛ-TxIX by 1H NMR. The two disulfide bonds create a compact cysteine knot over a well defined region of the molecule. The electrostatic potential of the lowest energy structure of ɛ-TxIX illustrates the electronegativity clustered around Gla residues 1 and 4 and a hydrophobic area localized at Trp-7 (Fig. 4B). The two Gla residues at positions 1 and 4, the glycosylated threonine at position 10, and the hydroxyproline at position 13 protrude into the solvent and create a cavity within the surface of this peptide. It is conceivable that this negatively charged patch facilitates an electrostatic interaction with the receptor, thereby juxtaposing the brominated tryptophan to the active site of the receptor. This “dock and lock” modality of receptor-mediated interaction was postulated by Olivera and coworkers (4) to explain the refined specificity of the Conus peptides. The inherent flexibility of the disaccharide moiety may cause the weak, reversible receptor interaction exhibited by conotoxin ɛ-TxIX, perhaps by precluding the receptor-mediated interaction when the disaccharide moiety fills the cavity.
The breadth of posttranslational modifications and framework of disulfide bonds in ɛ-TxIX are unprecedented, raising questions regarding its mode of interaction with presynaptic Ca2+ channels or G protein-coupled receptors. It is plausible that the slow, reversible interaction with the Ca2+ channels is related to the abundance of this peptide and the fact that C. textile prey on relatively immobile mollusks rather than fish. Analysis of synthetic ɛ-TxIX analogues may delineate the role of specific posttranslational modifications in receptor-mediated ion channel interactions, helping to explain the refined receptor specificity and selectivity of conotoxins.
Acknowledgments
We thank Niclas G. Karlsson and Gunnar C. Hansson of the Department of Medical Biochemistry, University of Göteborg, Sweden for performing the carbohydrate analyses. This work was supported by grants from the National Institutes of Health to B.F. and B.C.F., from the Swedish Medical Research Council to J.S., from Association Française Contre les Myopathies to G.B., from Direction des Recherches Etudes et Techniques to P.F., and from the Danish Biotechnology Program to P.R.; J.S. was also supported by a Rand Fellowship from the Marine Biological Laboratory. A.C.R. was supported by a research fellowship from the Heart and Stroke Foundation of Canada. D.E.K. was supported by a grant from the Brazilian Postgraduate Federal Agency (CAPES).
ABBREVIATIONS
- Gla
carboxyglutamic acid
- ACh
acetylcholine
- IPSC
inhibitory postsynaptic currents
- MPSC
miniature PSC
- LDIPSC
long duration-induced PSC
- MALDI
matrix-assisted laser desorption/ionization
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the swissprot Protein Data Bank (accession no. P81755).
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY, 11973 (PDB ID code 1wct).
References
- 1.Olivera B M, Rivier J, Clark C, Ramilo C A, Corpuz G P, Abogadie F C, Mena E E, Woodward S R, Hillyard D R, Cruz L J. Science. 1990;249:257–263. doi: 10.1126/science.2165278. [DOI] [PubMed] [Google Scholar]
- 2.Gray W R, Olivera B M, Cruz L J. Annu Rev Biochem. 1988;57:665–700. doi: 10.1146/annurev.bi.57.070188.003313. [DOI] [PubMed] [Google Scholar]
- 3.Olivera B M, Miljanich G P, Ramachandran J, Adams M E. Annu Rev Biochem. 1994;63:823–867. doi: 10.1146/annurev.bi.63.070194.004135. [DOI] [PubMed] [Google Scholar]
- 4.Olivera B M. Mol Biol Cell. 1997;8:2101–2109. doi: 10.1091/mbc.8.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McIntosh J M, Olivera B M, Cruz L J, Gray W R. J Biol Chem. 1984;259:14343–14346. [PubMed] [Google Scholar]
- 6.Craig A G, Jimenez E C, Dykert J, Nielsen D B, Gulyas J, Abogadie F C, Porter J, Rivier J E, Cruz L J, Olivera B M, McIntosh J M. J Biol Chem. 1997;272:4689–4698. doi: 10.1074/jbc.272.8.4689. [DOI] [PubMed] [Google Scholar]
- 7.Sato S, Nakamura H, Ohizumi Y, Kobayashi J, Hirata Y. FEBS Lett. 1983;155:277–280. doi: 10.1016/0014-5793(82)80620-0. [DOI] [PubMed] [Google Scholar]
- 8.Adams M E, Myers R A, Imperial J S, Olivera B M. Biochemistry. 1993;32:12566–12570. doi: 10.1021/bi00210a003. [DOI] [PubMed] [Google Scholar]
- 9.Pallaghy P K, Nielsen K J, Craik D J, Norton R S. Protein Sci. 1994;3:1833–1839. doi: 10.1002/pro.5560031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narasimhan L, Singh J, Humblet C, Guruprasad K, Blundell T. Nat Struct Biol. 1994;1:850–852. doi: 10.1038/nsb1294-850. [DOI] [PubMed] [Google Scholar]
- 11.Fainzilber M, Gordon D, Hasson A, Spira M E, Zlotkin E. Eur J Biochem. 1991;202:589–595. doi: 10.1111/j.1432-1033.1991.tb16412.x. [DOI] [PubMed] [Google Scholar]
- 12.Karlsson N G, Hansson G C. Anal Biochem. 1995;224:538–541. doi: 10.1006/abio.1995.1084. [DOI] [PubMed] [Google Scholar]
- 13.Charbonneau H. In: A Practical Guide to Protein and Peptide Purification for Microsequencing. Matsudiara P T, editor. London: Academic; 1989. p. 22. [Google Scholar]
- 14.Cairns J R, Williamson M K, Price P A. Anal Biochem. 1991;199:93–97. doi: 10.1016/0003-2697(91)90274-w. [DOI] [PubMed] [Google Scholar]
- 15.Kussmann M, Lassing U, Sturmer C A, Przybylski M, Roepstorff P. J Mass Spectrom. 1997;32:483–493. doi: 10.1002/(SICI)1096-9888(199705)32:5<483::AID-JMS502>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 16.Mothet J P, Fossier P, Meunier F M, Stinnakre J, Tauc L, Baux G. J Physiol (London) 1998;507:405–414. doi: 10.1111/j.1469-7793.1998.405bt.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baux G, Tauc L. J Physiol (London) 1987;388:665–680. doi: 10.1113/jphysiol.1987.sp016637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rance M, Soerenson O W, Bodenhausen G, Wagner G, Ernst R R, Wuthrich K. Biochem Biophys Res Commun. 1983;117:479–486. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- 19.Braunschweiler L, Ernst R R. J Magn Reson. 1983;53:521–528. [Google Scholar]
- 20.Bax A D, Davis G. J Magn Reson. 1985;65:355–360. [Google Scholar]
- 21.Jeener J, Meirer B H, Bachman P, Ernst R R. J Chem Phys. 1979;71:4546–4553. [Google Scholar]
- 22.Jimenez E C, Craig A G, Watkins M, Hillyard D R, Gray W R, Gulyas J, Rivier J E, Cruz L J, Olivera B M. Biochemistry. 1997;36:989–994. doi: 10.1021/bi962840p. [DOI] [PubMed] [Google Scholar]
- 23.England L J, Imperial J, Jacobsen R, Craig A G, Gulyas J, Akhtar M, Rivier J, Julius D, Olivera B M. Science. 1998;281:575–578. doi: 10.1126/science.281.5376.575. [DOI] [PubMed] [Google Scholar]
- 24.Sudmeier J, Evelhoch J L, Jonsson N B H. J Magn Res. 1980;40:377–390. [Google Scholar]
- 25.Stenflo J, Suttie J W. Annu Rev Biochem. 1977;46:157–172. doi: 10.1146/annurev.bi.46.070177.001105. [DOI] [PubMed] [Google Scholar]
- 26.Furie B, Furie B C. Cell. 1988;53:505–518. doi: 10.1016/0092-8674(88)90567-3. [DOI] [PubMed] [Google Scholar]
- 27.Davie E W, Fujikawa K, Kisiel W. Biochemistry. 1991;30:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 28.Mann K G, Nesheim M E, Church W R, Haley P, Krishnaswamy S. Blood. 1990;76:1–16. [PubMed] [Google Scholar]
- 29.Hauschka P V, Mullun E A, Hintsch G, Jazwinski S. In: Current Advances in Vitamin K Research. Suttie J W, editor. Amsterdam: Elsevier; 1988. pp. 237–243. [Google Scholar]
- 30.Haack J A, Rivier J, Parks T N, Mena E E, Cruz L J, Olivera B M. J Biol Chem. 1990;265:6025–6029. [PubMed] [Google Scholar]
- 31.Nakamura T, Yu Z, Fainzilber M, Burlingame A L. Protein Sci. 1996;5:524–530. doi: 10.1002/pro.5560050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suttie J W. Annu Rev Biochem. 1985;54:459–477. doi: 10.1146/annurev.bi.54.070185.002331. [DOI] [PubMed] [Google Scholar]
- 33.Faulkner D J. Nat Prod Rep. 1998;15:113–158. doi: 10.1039/a815113y. [DOI] [PubMed] [Google Scholar]
- 34.Taylor S W, Kammerer B, Nicholson G J, Pusecker K, Walk T, Bayer E, Scippa S, de Vincentiis M. Arch Biochem Biophys. 1997;348:278–288. doi: 10.1006/abbi.1997.0371. [DOI] [PubMed] [Google Scholar]
- 35.Craig A G, Zafaralla G, Cruz L J, Santos A D, Hillyard D R, Dykert J, Rivier J E, Gray W R, Imerial J, DelaCruz R G, et al. Biochemistry. 1998;37:16019–16025. doi: 10.1021/bi981690a. [DOI] [PubMed] [Google Scholar]
- 36.Van den Steen P, Rudd P M, Dwek R A, Opdenakker G. Crit Rev Biochem Mol Biol. 1998;33:151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- 37.Olivera B M, Gray W R, Zeikus R, McIntosh J M, Varga J, Rivier J, de Santos V, Cruz L J. Science. 1985;230:1338–1343. doi: 10.1126/science.4071055. [DOI] [PubMed] [Google Scholar]
- 38.Fossier P, Tauc L, Baux G. Trends Neurosci. 1999;22:161–166. doi: 10.1016/s0166-2236(98)01307-1. [DOI] [PubMed] [Google Scholar]
- 39.Baux G, Fossier P, Tauc L. J Physiol (London) 1990;429:147–168. doi: 10.1113/jphysiol.1990.sp018249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fossier P, Baux G, Tauc L. Neuroscience. 1994;63:405–414. doi: 10.1016/0306-4522(94)90538-x. [DOI] [PubMed] [Google Scholar]
- 41.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]