Abstract
Many of the activating receptors on natural killer (NK) cells are multisubunit complexes composed of ligand-binding receptors that are noncovalently associated with membrane-bound signaling adaptor proteins, including CD3ζ, FcεRIγ, DAP12, and DAP10. Because the DAP10 and DAP12 genes are closely linked, expressed in NK cells, and have remarkably similar transmembrane segments, it was of interest to determine the specificity of their interactions with ligand-binding receptors and to examine their signaling properties. Despite their similarities, DAP10, DAP12, FcεRIγ, and CD3ζ form specific receptor complexes with their ligand-binding partners in NK cells and transfectants. The transmembrane regions of DAP10 and DAP12 are sufficient to confer specific association with their partners. Although cross-linking of either DAP10- or DAP12-associated receptors has been shown to be sufficient to trigger NK cell–mediated cytotoxicity against Fc receptor–bearing cells, substantial synergy was observed in the induction of cytokine production when both receptors were engaged. Activation of the Syk/ZAP70 tyrosine kinases by the immunoreceptor tyrosine-based activation motif–containing DAP12 adaptor and of the phosphatidylinositol 3-kinase pathway by the YxNM-containing DAP10 adaptor may play an important role in the stimulation of NK cells and T cells.
Keywords: DAP10, DAP12, immunoreceptor tyrosine-based activation motif, NKG2D, natural killer cell activation
Introduction
NK cells are a subset of lymphocytes that function as a critical component of innate immunity against intracellular and parasitic pathogens, and possibly tumors. Their effector functions are controlled by the opposing activity of activating and inhibitory cell surface receptors (for reviews, see references 1 and 2). Unlike T and B lymphocytes, in which the key activating receptors such as the T and B cell antigen receptors have been well characterized, the major stimulatory counterparts in NK cells are still being defined. Several genes in the killer cell Ig-like receptor (KIR), Ly49, and NKG2 families encode receptors that have short cytoplasmic domains lacking intrinsic signaling motifs (for reviews, see references 1 and 2). These receptors activate, rather than inhibit, NK cell cytotoxicity and cytokine production. However, similar to the situation with the TCR that signals via its association with the CD3 membrane adaptor molecules, these activating NK cell receptors noncovalently associate with DAP12, a CD3-like membrane adaptor containing an immunoreceptor tyrosine-based activation motif (ITAM) in its cytoplasmic domain 3 4 5 6 7. DAP12, a single gene on human chromosome 19q13.1 and mouse chromosome 7, encodes type I membrane proteins expressed as disulfide-bonded homodimers on NK cells, myeloid cells, and a subset of T cells 5. In NK cells, DAP12 associates with the several receptors for MHC class I, including KIR2DS2, Ly49D, Ly49H, and CD94/NKG2C 3 4 5 6 7.
Another membrane adaptor protein, DAP10, is encoded by a gene immediately adjacent to DAP12 in the genome, but in opposite transcriptional orientation 8. Although DAP10 has only 20% overall amino acid homology with DAP12, it is expressed as a disulfide-bonded homodimer on the surface of NK cells, myeloid cells, and a subset of T cells. Unlike DAP12, DAP10 does not have an ITAM in its cytoplasmic region. The cytoplasmic domain of human and mouse DAP10 has a YxxM motif, a potential src homology 2 (SH2) domain–binding site for the p85 regulatory subunit of the phosphatidylinositol (PI)3-kinase 9. DAP10 is noncovalently associated with NKG2D, a receptor on NK cells, TCR-γ/δ+ T cells, and CD8+ T cells that recognizes the nonclassical MHC class I molecules MICA and MICB 8 10.
Many of the activating immune receptors (e.g., TCR/CD3 on T cells, surface Ig on B cells, and FcR on NK cells and myeloid cells) are comprised of separate ligand-binding and signal-transducing subunits. An advantage of building receptors by the assembly of multiple subunits is the ability to mix and match those subunits to generate receptor diversity. Assuming that individual subunits have characteristic functional capabilities, such pairing allows a cell to construct different receptor complexes with different ligand specificity and/or signaling potential. In this report, we have examined the receptor specificity of the DAP10 and DAP12 adaptor proteins and the functional interplay between these receptor complexes.
Materials and Methods
cDNAs, Cells, and Transfectants.
cDNAs used were human KIR2DS2 11, NKG2C 12, Flag-DAP10 8, and Flag-DAP12 5, which were subcloned into either pMX-neo, pMX-puro, or pMX-pie (containing a puromycin resistance gene, an internal ribosomal entry site [IRES] element, and the enhanced green fluorescent protein [GFP] gene retroviral vectors 13). A cDNA containing the human CD8 leader segment, followed by the Flag epitope (DYKDDDDK) and joined to the extracellular domain of human FcεRIγ or CD3ζ was subcloned into the pMX-puro retroviral vector. A cDNA containing the human CD8 leader segment, followed by the Myc epitope (EQKLISEEDL) and joined to the extracellular domain of human DAP10, was subcloned into the pMX-puro-IRES-GFP retroviral vector. The transmembrane (TM) mutants of Flag-DAP10 (EC10-TM12-CY10) and Flag-DAP12 (EC12-TM10-CY12) were generated by standard PCR mutagenesis, in which the entire TM regions of DAP10 and DAP12 were swapped. The point mutations in the TM region of DAP10 were generated by using PCR. All mutations were confirmed by sequencing the cDNA. Retroviruses were generated by using the Phoenix packaging cell lines (gifts from G. Nolan, Stanford University, Palo Alto, CA 14). Ba/F3 cells (a gift from T. Kitamura, University of Tokyo, Tokyo, Japan) and NKL cells (provided by M. Robertson, University of Indianapolis, Indianapolis, IN 15) were infected, drug selected, and the resulting transfectants were isolated by flow cytometry using either GFP or specific Abs. Short-term polyclonal NK cell lines (uniformly CD3−CD56+) were established from the peripheral blood of healthy adult donors and cultured as described by Yssel et al. 16.
Abs and Flow Cytometry.
mAbs used were isotype-matched control mouse IgG mAb (Becton Dickinson), anti-KIR2D mAb DX27 17, anti-CD16 mAb Leu 11a (Becton Dickinson), anti-NKG2D mAb 1D11 10, anti-CD94 mAb DX22 17, anti-CD94/NKG2A/C mAb DX42, anti-CD3 mAb Leu 4 (Becton Dickinson), anti-Flag mAb M2 (Sigma-Aldrich), anti-myc mAb 9E10 (Sigma-Aldrich), and anti–human DAP12 mAb DX37. DX37 is an IgG1 mAb that was generated by immunizing BALB/c mice with a glutathione S-transferase fusion protein containing the entire cytoplasmic domain of human DAP12. DX42 is an IgG2b mAb that was generated by immunizing BALB/c mice with the mouse pro-B cell line Ba/F3 transfected with human CD94/NKG2C-DAP12. DX42 reacts with heterodimers of CD94 and NKG2A or CD94 and NKG2C, but does not bind to the isolated CD94, NKG2A, or NKG2C subunits. Affinity-purified rabbit anti-DAP10 and anti-DAP12 antisera were described previously 5 8. J. Bolen (DNAX Corp., Palo Alto, CA) provided rabbit anti-Syk antiserum. Antiphosphotyrosine mAb 4G10 was purchased from Upstate Biotechnology. FITC- and PE-conjugated goat anti–mouse Ig were purchased from Caltag. Immunofluorescence and flow cytometry were performed as described 18.
Surface Iodination, Biotinylation, and Immunoprecipitation.
Transfected Ba/F3 cells, polyclonal NK cell lines, and NKL cells were labeled with either 125I (Amersham Pharmacia Biotech) or sulfo-N-hydroxy-succinimide (NHS)-biotin (Pierce Chemical Co.) and solubilized in digitonin lysis buffer (pH 7.8, 1% digitonin, 0.12% Triton X-100, 150 mM NaCl, 20 mM triethanolamine, 0.01% NaN3, and protease inhibitors 19). Cell lysates were incubated on ice for 2 h with Pansorbin (Calbiochem) coated with rabbit anti–mouse Ig (Sigma-Aldrich) and the indicated mAbs. The resulting immune complexes were washed and analyzed by SDS-PAGE, and then either visualized by using a PhosphorImager (Molecular Dynamics) for 125I-labeled proteins or transferred to Immobilon membrane (Millipore), blocked, probed with horseradish peroxidase (HRP)-conjugated streptavidin (Amersham Pharmacia Biotech), and visualized by using a chemiluminescent substrate (Pierce Chemical Co.).
Cell Stimulation and Antiphosphotyrosine Blot.
Transfected Ba/F3 cells were incubated with either an isotype-matched control mAb, anti-KIR2DS2 mAb DX27, or anti-NKG2D mAb 1D11 on ice, washed, and then cross-linked with F(ab′)2 goat anti–mouse IgG (Jackson ImmunoResearch Laboratories) for 3 min at 37°C. Cells were lysed in buffer containing 1% NP-40 and protease inhibitors 5. Syk proteins were immunoprecipitated with rabbit anti-Sky antiserum and analyzed by SDS-PAGE and Western blot by using HRP-conjugated antiphosphotyrosine mAb (4G10; Upstate Biotechnology) and a chemiluminescent substrate (Pierce Chemical Co.).
Cytokine Secretion.
96-well flat-bottomed plates were coated (overnight at 10 μg/ml in PBS) with a goat anti–mouse Ig F(ab′)2, Fc portion–specific Ab (Jackson ImmunoResearch Laboratories). The wells were blocked with sterile PBS containing 0.5% BSA and incubated with various mAbs at the indicated concentrations. Wells were washed to remove excess Abs, and 105 cells were incubated for 20 h in 200 μl of IMDM (GIBCO BRL) supplemented with 200 U/ml recombinant human IL-2, 10% FCS, penicillin, streptomycin, and l-glutamine. Supernatants were harvested from triplicate wells, and the levels of secreted IFN-γ and GM-CSF were determined by ELISA, as described previously 20.
Cytotoxicity Assays.
NK cells were assayed in an Ab-redirected cytotoxicity against 51Cr-labeled FcR+ mouse P815 target cells in the presence or absence of anti–human CD56 mAb Leu 19 (used as a negative control) or anti–human NKG2D mAb 1D11, as described 10.
Results and Discussion
Specificity of the DAP10, DAP12, FcεRIγ, and CD3ζ Adaptor Proteins.
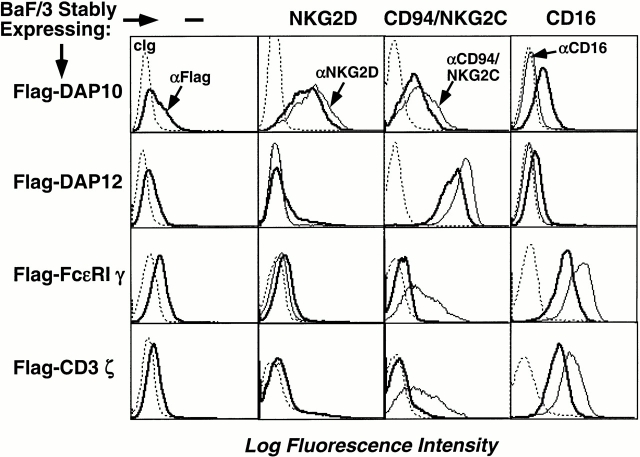
Many NK receptors, including KIR2DS, CD94/NKG2C, Ly49D, Ly49H, NKG2D, NKR-P1, NKp44, NKp46, and NKp30, are multisubunit receptor complexes that convey signals via the membrane adaptors DAP10, DAP12, FcεRIγ, or CD3ζ 5 6 7 8 21 22 23 24. All of these adaptors have an aspartic acid residue in their hydrophobic TM domains that is required for interaction with an oppositely charged, basic residue in their ligand-binding receptors. Because the TM regions of these adaptors are quite similar, studies were undertaken to determine whether these proteins are specific or promiscuous in their interactions with the ligand-binding receptors. A mouse pro-B cell line, Ba/F3, was infected with ecotropic retroviruses encoding human DAP10, DAP12, FcεRIγ, or CD3ζ (all contain Flag epitope tags on the NH2 terminus to permit detection on the cell surface). Consistent with prior results, transfection of these signaling adaptors alone into Ba/F3 cells did not permit efficient cell surface expression (Fig. 1), although Western blot analysis indicated abundant proteins in the cytoplasm (data not shown). When these cells were infected with retroviruses encoding human CD16, NKG2D, or CD94 and NKG2C, stable receptor complexes were present on the cell surface only when the physiologically relevant adaptor was coexpressed, as determined by detection of the Flag epitope of the adaptor protein on the plasma membrane (Fig. 1) and by coimmunoprecipitation of the adaptor and ligand-binding receptor (not shown). NKG2D paired with DAP10, but not with DAP12, FcεRIγ, or CD3ζ. CD16 paired with either FcεRIγ or CD3ζ, but not DAP10 or DAP12, and the CD94/NKG2C heterodimer associated only with DAP12 (Fig. 1). These data demonstrate the exquisite specificity in the adaptor–receptor interaction and indicate that the conserved aspartic acid residues in the TM domains of these adaptor proteins do not permit promiscuous pairing.
Figure 1.
Receptor-pairing specificity of DAP10, DAP12, FcεRIγ, and CD3ζ. BaF/3 cells were first transfected with either a control cDNA, human NKG2D, CD94 and NKG2C, or CD16, and stable transfectants were generated by maintaining the cells in appropriate drug selection. The Flag epitope–tagged DAP10, DAP12, FcεRIγ, or CD3ζ cDNA were then transfected into the indicated cell lines, and subsequent double or triple transfectants were selected by drug selection. Cells were analyzed by flow cytometry using the indicated Abs, and data are presented as histograms. Western blot analysis showed that the Flag-tagged adaptors were expressed comparably in all transfectants (data not shown). cIg, control Ig.
The oppositely charged residues in the TMs of the adaptor proteins (D) and receptors (R or K) may form salt bridges in the lipid bilayer, thus stabilizing these multisubunit receptor complexes in the plasma membrane. However, a salt bridge may not be requisite because both CD16 and its adaptor subunit FcεRIγ or CD3ζ possess an aspartic acid residue in their TM regions. Thus, interactions that are more complex dictate the nature and/or the orientation of the interaction sites within the TM segments of the receptors and adaptors. In addition, the relative position of the positively charged residues within the TM domains is different between DAP10 and NKG2D versus FcεRIγ or CD3ζ and its partner CD16. This different spacing may potentially contribute to the specific conformation for pairing with the corresponding subunit.
The TM Domains of DAP10 and DAP12 Confer Receptor Specificity.
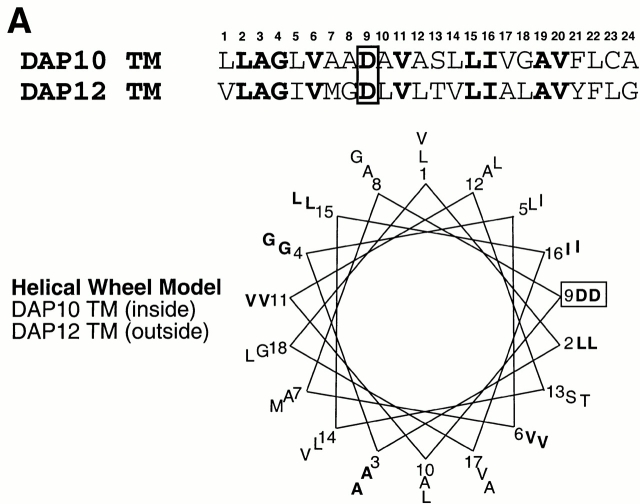
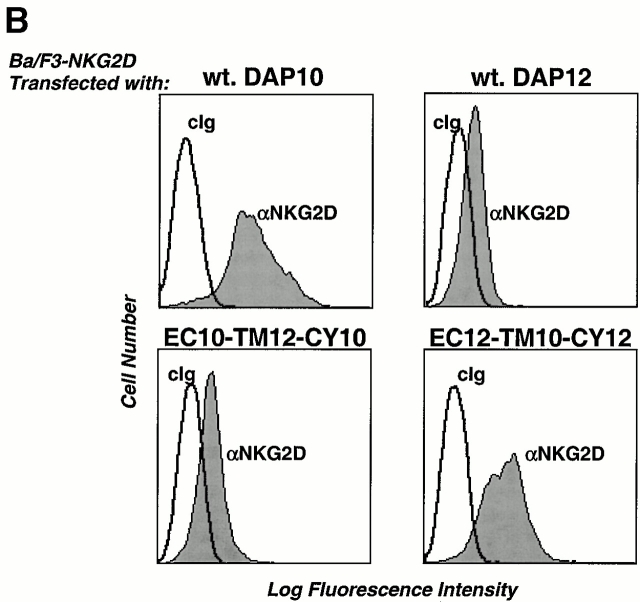
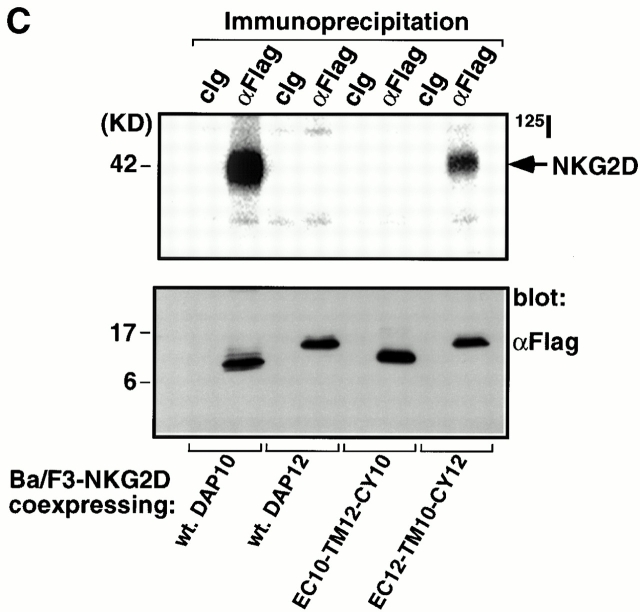
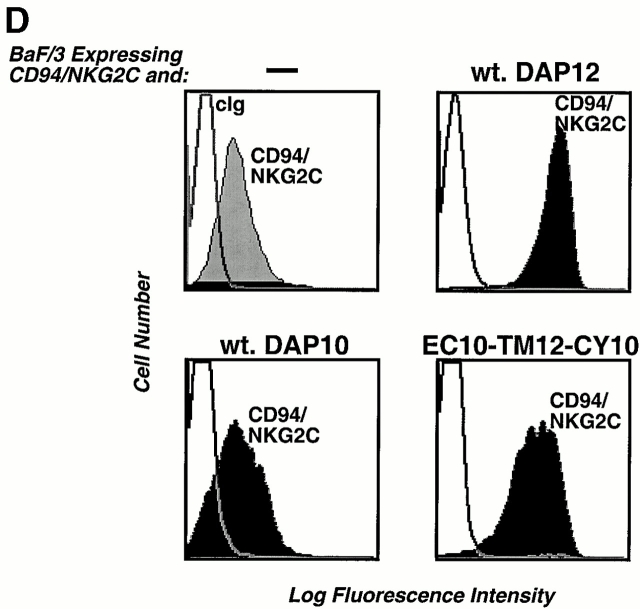
DAP10 and DAP12 both contain a pair of membrane-proximal extracellular cysteine residues that are likely required for the generation of disulfide-bonded dimers. Moreover, DAP10 and DAP12 have very homologous TM domains. The boundaries of these TM regions, the aspartic acid residues, and the relative positions of these charged residues within the TM regions are identical in DAP10 and DAP12 (Fig. 2 A). This raised the question of whether the TM domains of DAP10 and DAP12 confer specificity for their association with the ligand-binding receptor subunits. Preliminary studies with truncation mutants removing the entire cytoplasmic domains of DAP10 and DAP12 revealed that these regions were not required for the association with their ligand-binding partners (data not shown). Therefore, we generated chimeric molecules in which the entire TM domains were swapped between DAP10 and DAP12 (Fig. 2 A) and examined their ability to stabilize the surface expression of NKG2D and CD94/NKG2C. DAP10, DAP12, and the chimeric adaptors all contained a Flag epitope tag on the NH2 terminus for visualization by flow cytometry and by Western blot analysis. As shown in Fig. 2 B, only the wild-type (wt) DAP10 and the DAP12 chimera with the DAP10 TM region (EC12-TM10-CY12) were capable of stabilizing the expression of NKG2D on the cell surface. Cell surface iodination and immunoprecipitation with anti-Flag mAb further demonstrated that only coexpression of NKG2D with either the wt DAP10 or EC12-TM10-CY12 protein resulted in stable surface expression of the receptor complex (Fig. 2 C). Consistent with this conclusion, immunoprecipitation with anti-NKG2D mAb failed to reveal any 125I-labeled NKG2D glycoproteins when this molecule was cotransfected with DAP12 or the chimeric EC10-TM12-CY10 adaptor protein (data not shown). The DAP10 chimera with the DAP12 TM domain, although unable to form a receptor complex with NKG2D, was able to rescue the surface expression of CD94/NKG2C (Fig. 2 D). Similar results were also obtained when the chimeric EC10-TM12-CY10 adaptor protein was coexpressed with another DAP12-associated receptor, KIR2DS2 (data not shown). Taken together, these data demonstrate that, despite their striking similarity, the TM regions of DAP10 and DAP12, at least partially, confer specificity for their association with their ligand-binding partner subunits.
Figure 2.
The TM domains of DAP10 and DAP12 confer receptor-pairing specificity. (A) The alignment of the TMs of DAP10 and DAP12, and an illustration of the TM mutants used in the study. The consensus TM segments of DAP10 and DAP12 were predicted based on analysis of the protein structures using the SOSUI program, the DAS membrane predictor server, and the TMPred program. A predicted α-helical wheel diagram (http://www.bmm.icnet.uk/people/turcotte/Java/HelixWheel/) of the DAP10 and DAP12 TM regions is shown. Amino acid 19 in the TM region corresponds to the same relative location as amino acid 1 shown in the helix, amino acid 20 corresponds to amino acid 2, amino acid 21 corresponds to amino acid 3, amino acid 22 corresponds to amino acid 4, amino acid 23 corresponds to amino acid 5, and amino acid 24 corresponds to amino acid 6. The charged residues are boxed and the identities are in bold. (B) The TM of DAP10 confers specificity for its association with NKG2D. The Flag-tagged wt and TM mutants of DAP10 and DAP12 were transfected into a Ba/F3 transfectant expressing human NKG2D. Stable transfectants were generated by using drug selection. Cells were stained with anti-NKG2D mAb and analyzed by flow cytometry. Similar staining profiles were observed by using anti-Flag mAb M2 (data not shown). cIg, control Ig. (C) The double transfectants were surface labeled with 125I, lysed, and immunoprecipitated with either a control mAb or anti-Flag mAb M2. The top part of the gel was analyzed by autoradiography; the lower part was transferred to an Immobilon membrane and analyzed by Western blot using anti-Flag mAb M2. (D) The DAP12 TM confers specificity to pair with the DAP12 partner, CD94/NKG2C. A Ba/F3 transfectant expressing human CD94 and NKG2C was transfected with either a control vector, wt DAP12, wt DAP10, or EC10-TM12-CY10. Cells were stained with an mAb reactive with the heterodimers of CD94/NKG2A or CD94/NKG2C, and subsequently analyzed by flow cytometry.
So which TM amino acids determine the specificity? TM regions are most likely, on first principles, to exist as α-helices. Assuming that the interaction between the oppositely charged residues is crucial for a complex formation, one hypothesis is that residues on the face of the helix where the charged residues orient might be important in maintaining close contact between the adaptor and receptor, thus providing specific pairwise interactions in the lipid bilayer. As shown in Fig. 2 A, an α-helical model indicates that residues 1, 5, 10, 12, 13, 23, and 24 in these TM regions may lie on the same face as the aspartic acid residue and differ between DAP10 and DAP12. In particular, the amino acids at positions 1, 5, 10, 13, and 23 differed in DAP10 and DAP12, but are conserved in mice and humans. The amino acid at each of these five positions in DAP10 was changed by site-directed mutation to the corresponding amino acid present in DAP12, and these mutant molecules were tested for association with NKG2D or KIR2DS2. Remarkably, no single point mutation alone was able to confer partner chain specificity, suggesting that multiple contact points are required for interaction. It should be noted that stabilization of surface expression of NKG2D by the chimeric EC 12-TM10-CY12 adaptor was less efficient than that conferred by the wt DAP10 (Fig. 2b and Fig. c). Thus, although not required, the extracellular regions may cooperate with the TM domains to increase the specificity of the pairing.
DAP10 and DAP12 Form Distinct Receptor Complexes.
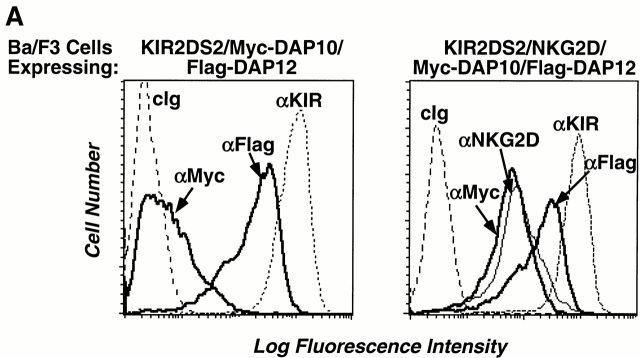
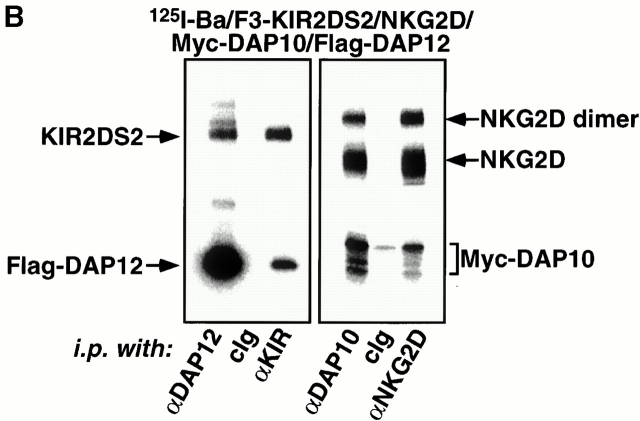
Reminiscent of the CD3 chains, DAP10 and DAP12 are also genetically linked and structurally homologous, thus raising the possibility that DAP10 and DAP12 may form a larger receptor complex in which they mediate different signaling functions. To address this possibility, Ba/F3 cells stably transduced with KIR2DS2 and a Flag epitope–tagged human DAP12 were infected with a retrovirus encoding Myc epitope–tagged human DAP10. As shown in Fig. 3 A, only minimal surface expression of Myc-DAP10 was detected in the triple transfectant, suggesting the absence of a trimolecular complex. Moreover, the expression of Myc-DAP10 did not alter the surface expression of Flag-DAP12 or KIR2DS2 (data not shown), again underscoring the specific interaction between DAP12 and KIR2DS2. Consistent with prior findings, infection of these cells with a retrovirus encoding human NKG2D led to a significant surface expression of both Myc-DAP10 and NKG2D (Fig. 3 A). These cells were labeled by surface iodination, lysed in digitonin detergent to preserve noncovalent interactions between the receptor subunits, and immunoprecipitated with Abs against the receptors or adaptor proteins. These studies revealed that distinct receptor complexes were formed by KIR2DS2/Flag-DAP12 and NKG2D/Myc-DAP10 (Fig. 3 B). There was no evidence for the existence of disulfide-bonded heterodimers between DAP10 and DAP12. Furthermore, KIR2DS2 was not coimmunoprecipitated with DAP10 or NKG2D. Similarly, NKG2D was not coimmunoprecipitated with DAP12 or KIR2DS2.
Figure 3.
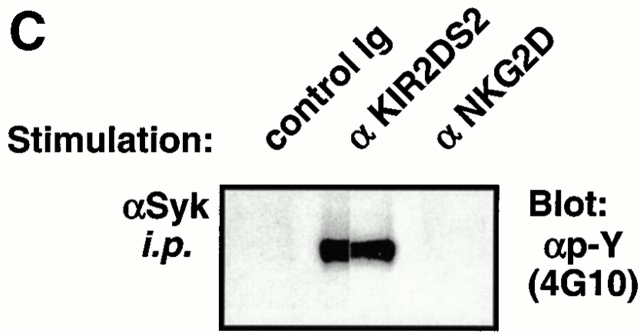
DAP10 and DAP12 form distinct receptor complexes in Ba/F3 transfectants. (A) A Ba/F3 transfectant expressing Flag-DAP12 and KIR2DS2 was transfected with Myc epitope–tagged DAP10 in an IRES-enhanced GFP–containing vector. Cells were sorted for GFP positive, maintained in appropriate drug selection, stained with the indicated mAbs, and analyzed by flow cytometry (left). cIg, control Ig. The triple transfectant was transfected with human NKG2D and sorted for NKG2D-positive cells. The KIR2DS2/NKG2D/Myc-DAP10/Flag-DAP12 transfectant was stained with the indicated mAb and analyzed by flow cytometry (right). (B) The KIR2DS2/NKG2D/Myc-DAP10/Flag-DAP12 Ba/F3 transfectant was surface labeled with 125I, lysed in digitonin buffer, and immunoprecipitated with either a control Ig, anti-DAP10 antiserum, anti-DAP12 antiserum, anti-KIR mAb DX27, or anti-NKG2D mAb 1D11. The resulting immune complexes were resolved by SDS-PAGE and analyzed by autoradiography. The heterogeneous migration pattern of Myc-DAP10 is likely due to O-link glycosylation of its extracellular domain. (C) The KIR2DS2/NKG2D/Myc-DAP10/Flag-DAP12 Ba/F3 transfectant was stimulated with a control mAb, anti-KIR mAb DX27, or anti-NKG2D mAb 1D11 for 3 min at 37°C. Anti-Syk Ab immune complexes were resolved by SDS-PAGE, transferred to Immobilon membrane, probed with HRP-conjugated antiphosphotyrosine (αp-Y) mAb 4G10, and developed by using a chemiluminescent substrate.
To further determine whether these complexes are physically distinct and functionally separate, the transfectants were stimulated with a control Ab, an anti-KIR mAb, or an anti-NKG2D mAb, and the tyrosine phosphorylation status of the protein tyrosine kinase Syk was evaluated. As shown in Fig. 3 C, triggering of KIR2DS2/Flag-DAP12, but not NKG2D/Myc-DAP10, resulted in tyrosine phosphorylation of Syk. These results using Ba/F3 transfectants were validated by using human polyclonal NK and T cell lines expressing NKG2D, in which cross-linking of NKG2D failed to phosphorylate Syk (data not shown). Collectively, these results suggest that KIR2DS2/DAP12 and NKG2D/DAP10 form separate receptors and activate distinct downstream signaling pathways.
DAP12 Complexes with Different Ligand-binding Receptors.
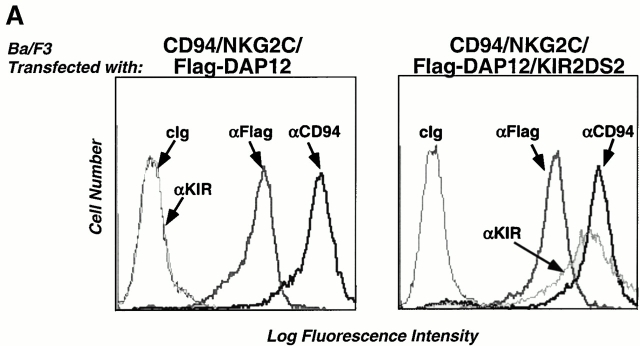
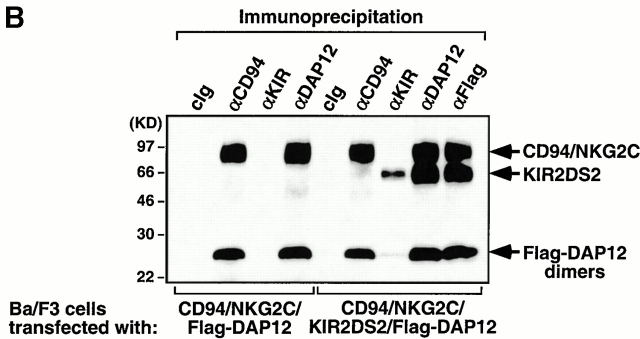
DAP12 associates with several different receptors in NK cells, monocytes, and dendritic cells. To examine whether these DAP12-containing receptors form higher order multisubunit complexes when present in the same cell, we established Ba/F3 cells stably expressing either CD94/NKG2C/Flag-DAP12 alone or both CD94/NKG2C/Flag-DAP12 and KIR2DS2 (Fig. 4 A). As previously shown 6, CD94, NKG2C, and Flag-DAP12 form a stable receptor complex on the cell surface (Fig. 4 A), evident by the coimmunoprecipitation of 125I-labeled CD94/NKG2C and Flag-DAP12 (Fig. 4 B). Transfection with KIR2DS2 resulted in efficient surface expression of KIR2DS2 and association between 125I-labeled KIR2DS2 and Flag-DAP12 (Fig. 4a and Fig. b). However, 125I-labeled CD94/NKG2C was not detected in the KIR2DS2 immunoprecipitates and vice versa, demonstrating that there is no detergent-stable higher order of complex assembled. Taken together, these results indicate that receptor complexes sharing the same signaling subunit, i.e., DAP12, are compartmentalized on the cell surface.
Figure 4.
DAP12 complexes with different ligand-binding receptors. (A) A Ba/F3 transfectant expressing human CD94/NKG2C was transfected with Flag-tagged DAP12 (left), and the resulting transfectant was further transfected with KIR2DS2. Cells were sorted for positive KIR expression, stained with the indicated mAbs, and analyzed by flow cytometry. cIg, control Ig. (B) The CD94/NKG2C/Flag-DAP12 and the CD94/NKG2C/KIR2DS2/Flag-DAP12 transfectants were surface biotinylated, lysed in digitonin lysis buffer, and the resulting lysates were immunoprecipitated with the indicated Abs. The immunoprecipitates were separated by SDS-PAGE, transferred to Immobilon membrane, probed with HRP-conjugated streptavidin, and visualized by using a chemiluminescent substrate. The KIR2DS2 mAb DX27 has been consistently shown to be a poor immunoprecipitating Ab (our unpublished observation).
DAP10 and DAP12 Form Distinct but Functionally Cooperative Receptor Complexes in NK Cells.
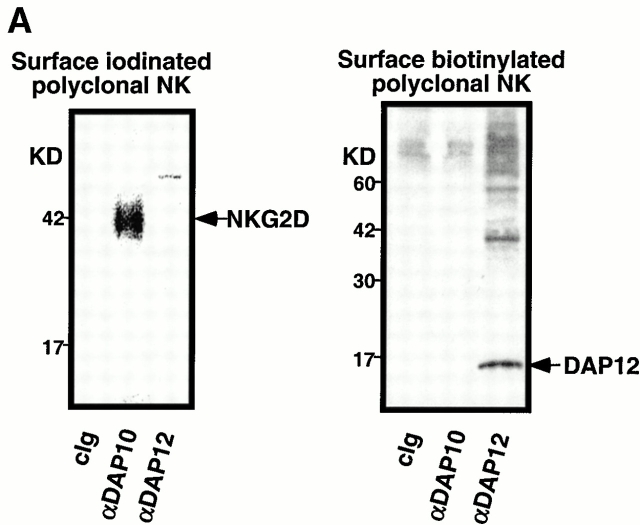
The NKG2D/DAP10- and DAP12-associated receptor complexes were further investigated by using polyclonal NK cell lines established from healthy adults. Results from a representative normal, polyclonal NK cell line are shown in Fig. 5 A. NK cells were surface labeled with either 125I or biotin, lysed in digitonin buffer, and immunoprecipitated with control Ig, anti-DAP10, or anti-DAP12. Anti-DAP10 exclusively coimmunoprecipitated a protein of ∼42 kD that labeled with 125I but did not label with biotin, consistent with the size and labeling properties of NKG2D (8; Fig. 5 A). In contrast, in this polyclonal NK cell line, anti-DAP12 did not coimmunoprecipitate any proteins efficiently labeled with 125I. However, biotinylated proteins were coimmunoprecipitated with anti-DAP12 mAb, along with biotin-labeled DAP12 proteins (Fig. 5 A). Because of the receptor heterogeneity in polyclonal NK cell lines and also due to the lack of specific serological reagents to discriminate between the activating and inhibitory isoforms of the KIR family, the nature of these biotin-labeled DAP12-associated proteins is undefined. Nonetheless, these experiments indicated that as with the Ba/F3 cell transfectants, DAP10 and DAP12 in normal, polyclonal NK cells associate with different receptors. There was no evidence for either 125I- or biotin-labeled receptors that coimmunoprecipitated with both DAP10 and DAP12.
Figure 5.
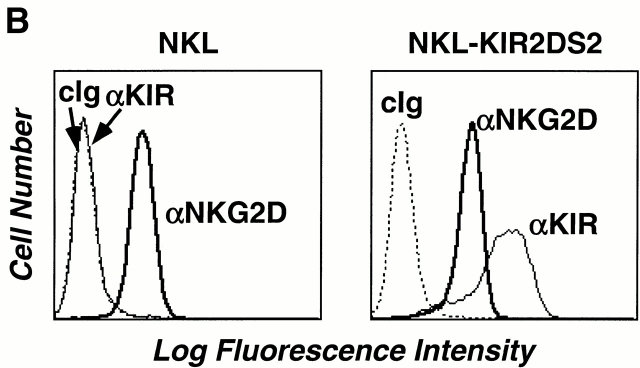
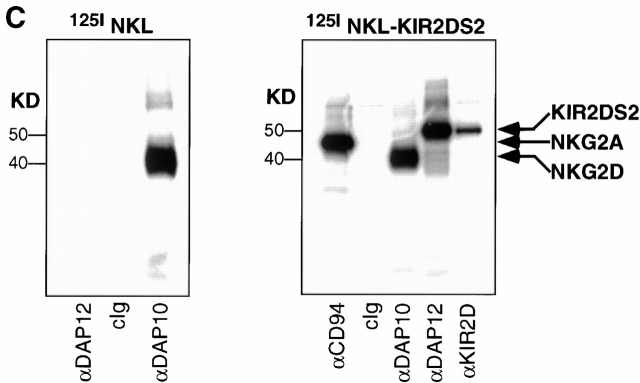
Separate DAP10 and DAP12 receptor complexes in polyclonal, normal NK cells and NKL cells. (A) A short-term polyclonal NK cell line established from a normal, healthy blood donor was labeled with either 125I (left) or biotin (right), lysed in 1% digitonin, and immunoprecipitated with the indicated Abs. Samples were analyzed by SDS-PAGE (reducing conditions) and visualized by autoradiography or chemiluminescence. Note that neither DAP10 nor DAP12 labels with 125I in NK cells, whereas DAP12, but not DAP10, labels with biotin. cIg, control Ig. (B) The parental NKL cell line was positive when stained with anti-NKG2D mAb 1D11, but was negative when stained with anti-KIR mAb DX27 (left). NKL cells were transfected with KIR2DS2, sorted for positive KIR expression, and maintained in neomycin (right). (C) Both NKL and the KIR2DS2+ NKL transfectant were 125I labeled, lysed in digitonin lysis buffer, immunoprecipitated with the indicated Abs, and the resulting immune complexes were analyzed by SDS-PAGE (reducing conditions) and autoradiography.
Because of the inherent heterogeneity of receptors expressed within a polyclonal NK cell population, further studies to explore the potential interactions between DAP10- and DAP12-associated receptors were performed using a homogeneous, clonal NK cell line, NKL. The NKG2D/DAP10 receptor complex on NKL cells has been shown to mediate both anti-NKG2D Ab–mediated redirected killing of FcR-bearing target cells and direct killing of MICA transfectants or MICA-expressing tumor cells 8 10. NKL also expresses an endogenous DAP12 adaptor protein, but an associated ligand-binding receptor has not been identified (our unpublished observation). Therefore, to examine potential interactions between the DAP10- and DAP12-associated receptor complexes, NKL cells were transduced with a retrovirus encoding KIR2DS2 (Fig. 5 B). The KIR2DS2/DAP12 receptor on NKL was capable of mediating potent anti-KIR mAb–redirected killing activity against FcR-bearing targets (data not shown). To determine whether DAP10 and DAP12 form distinct receptor complexes in NK cells, the KIR2DS2-transfected NKL cells were labeled with 125I, lysed, and immunoprecipitated with mAbs against DAP10, DAP12, KIR, or CD94. As shown previously 8 , 125I-labeled NKG2D was coimmunoprecipitated with an anti-DAP10 mAb (Fig. 5 C). In contrast, no 125I-labeled protein was found to associate with DAP12 in NKL. Although we cannot exclude the potential existence of DAP12-associated proteins that do not label efficiently with 125I, we nonetheless have consistently failed to detect any surface-labeled protein associating with both DAP10 and DAP12 in NKL cells or in other polyclonal NK lines (Fig. 5 A, and our unpublished observations). Expression of KIR2DS2 in NKL cells led to the surface expression of KIR2DS2 and the complex formation between DAP12 and KIR2DS2, as shown by surface labeling and coimmunoprecipitation experiments (Fig. 5 C).
Overall, these results using Ba/F3 cell transfectants and the NKL cell line have been validated by using polyclonal NK cells lines established from normal, healthy individuals (Fig. 5 A). In these experiments, there was no evidence for the presence on the cell surface of heterodimers composed of DAP10 and DAP12. In no case did we observe coimmunoprecipitation of NKG2D with DAP12 or any of the DAP12-associated receptors (e.g., KIR2DS or CD94/NKG2C) coimmunoprecipitating with DAP10. Furthermore, KIR2DS2, CD94/NKG2C, and NKG2D receptors have not been detected in immunoprecipitates from NK cells using either anti-CD3ζ or FcεRIγ (data not shown). Taken together, these data clearly demonstrate that DAP10 and DAP12 form distinct receptor complexes in NK cells.
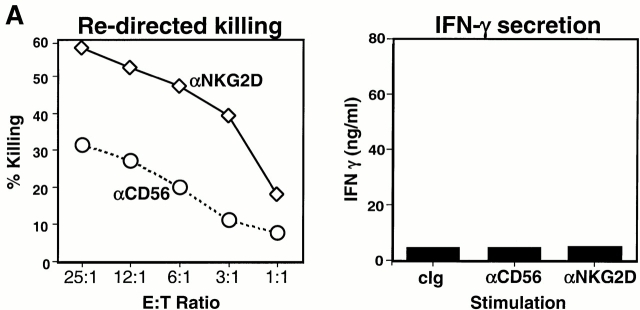
We have observed that anti-NKG2D mAb alone can trigger cytotoxicity mediated by normal, polyclonal NK cell lines (Fig. 6 A, left). By contrast, cross-linking of NKG2D alone, even at saturating concentrations of mAb, failed to induce substantial levels of cytokine production (Fig. 6 A, right). It is possible that these differential responses might be inherent to the different assays. In the cytotoxicity assays, other cell surface receptors on the effector and target cells (e.g., LFA-1 on the NK cells and intracellular adhesion molecules [ICAMs] on the target cells, or others) might be required to permit NKG2D function. Alternatively, because triggering NK cell–mediated cytotoxicity is a rapid event not requiring transcription or translation, the induction of cytokine production via NKG2D may be more stringent and require additional soluble or membrane-bound costimulators.
Figure 6.
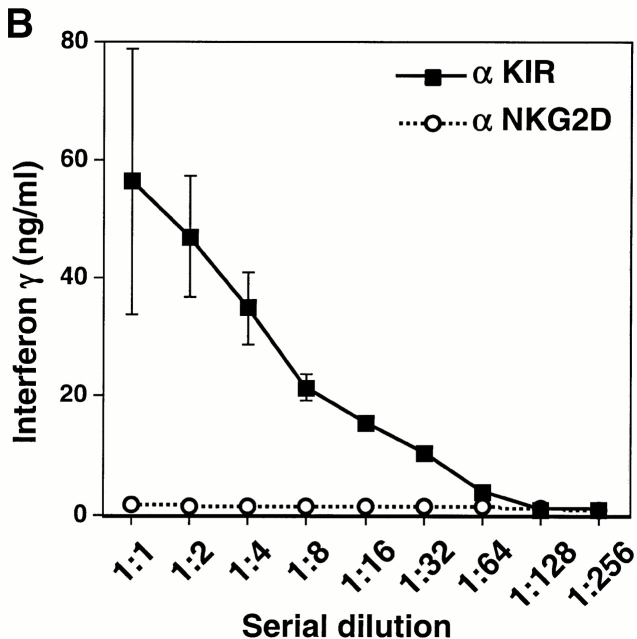
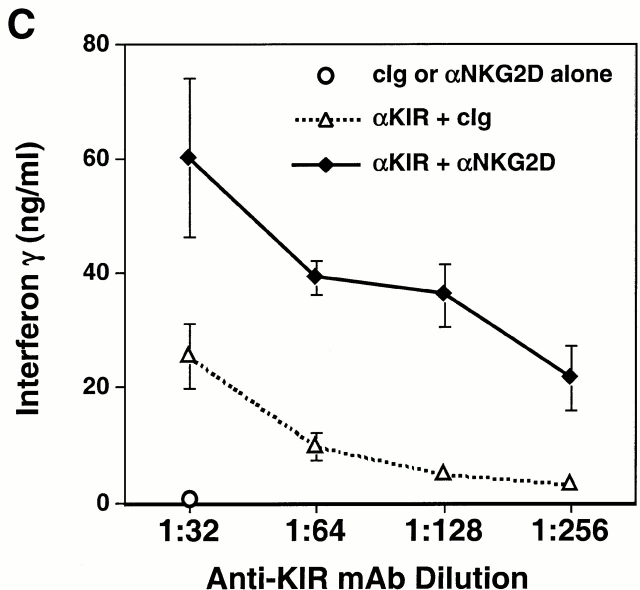
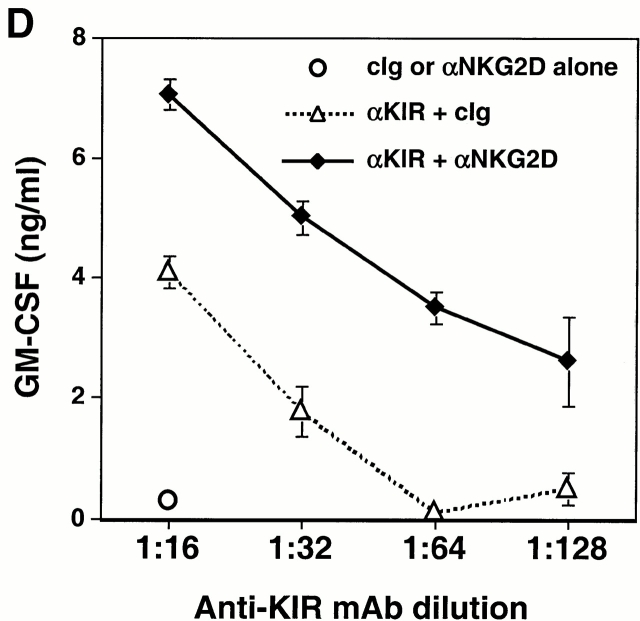
Functional cooperation between DAP10 and DAP12 receptor complexes. (A) A normal polyclonal NK cell line was assayed for Ab-redirected cytotoxicity against 51Cr-labeled FcR+ P815 target cells in the presence of anti-CD56 mAb Leu 19 (used as a negative control) or anti-NKG2D mAb 1D11 (at a 1:1,000 dilution of dialyzed ascites; left). In parallel, the normal polyclonal NK cell line was cultured for 20 h with immobilized control Ig (cIg), anti-CD56 mAb Leu 19, or anti-NKG2D mAb 1D11 (at 1:1,000 dilution of dialyzed ascites), and culture supernatants were analyzed by ELISA for IFN-γ (right). E:T, effector/target. Comparable results were obtained in two independent experiments using normal, polyclonal NK cell lines. (B) The KIR2DS2-expressing NKL cells were stimulated with immobilized anti-KIR mAb DX27 (starting at a concentration of 1 μg/ml) or anti-NKG2D mAb 1D11 (starting at a dilution of 1:1,000 dialyzed ascites). (C and D) KIR2DS2+ NKL cells were stimulated with a fixed dilution (1:5,000) of either control ascites or anti-NKG2D 1D11 ascites, together with either a control mAb or anti-KIR mAb DX27 at final concentrations of 31, 16, 8, and 4 ng/ml (C), or 62, 31, 16, and 8 ng/ml (D). Cells were stimulated in 96-well plates at 37°C for 20 h, and the supernatants were analyzed by ELISA for IFN-γ (C) or GM-CSF (D). Each stimulation condition was conducted in triplicate and the data were representative of three independent experiments.
There are 10 functional KIR genes with extensive allelic polymorphism that encode receptors that can be either activating or inhibitory 25. Within a polyclonal NK cell population, these are arrayed on overlapping subsets and some NK cell clones may simultaneously express four or more of these genes 26. Because the extracellular domains of the activating and inhibitory receptors are very similar, the existing mAbs against the molecules do not discriminate between them. Therefore, when polyclonal NK cells are incubated with an anti-KIR or anti-CD94 Ab, a combination of both activating and inhibitory receptors on different subsets are simultaneously engaged, often giving ambiguous results. For this reason, in order to address the issue of whether DAP12-associated receptors might cooperate with NKG2D/DAP10 in NK cell activation, experiments were performed with the well-characterized, homogeneous NK cell line NKL.
Cross-linking of the KIR2DS2/DAP12 receptor on NKL cells resulted in the dose-dependent secretion of IFN-γ and GM-CSF (Fig. 6 B), consistent with prior studies using anti-KIR mAbs to stimulate other NK cell lines and clones bearing activating KIR molecules 27. However, under suboptimal conditions in which few activating KIR molecules were engaged, marked synergy was observed between KIR2DS2/DAP12 and NKG2D/DAP10 in NKL cells in which coligation of both receptors showed a significant increase in the secretion of IFN-γ and GM-CSF (Fig. 6c and Fig. d). Taken together, the cooperative function between NKG2D and the ITAM-containing DAP12-associated receptors suggests that the NKG2D/DAP10 receptor may play an important costimulatory role in the activation of NK cells and T cells.
The NKG2D ligands, MICA and MICB, are expressed only in low levels on normal tissues, but are induced by stress or transformation, making them potentially important components in immune surveillance 28 29 30. The activation status of NK cells and T cells is believed to be constantly regulated by a delicate balance between activating and inhibitory receptors. Cells are kept in a resting state when this equilibrium is maintained in the normal microenvironment. However, induction or upregulation of MICA/B on unhealthy, abnormal cells due to transformation or infection may engage the NKG2D/DAP10 receptors on NK cells and cytotoxic T cells, disrupting this equilibrium.
Like CD28, which also activates the PI3-kinase signaling pathway 31 32 33, NKG2D/DAP10 may play a costimulatory role in the activation of NK cells and T cells. CD28 is an important costimulator for the activation of resting CD4+ and CD8+ T cells (for a review, see reference 34). However, the clearance of lymphocytic choriomeningitis virus by CTLs in CD28-deficient mice was normal 35, suggesting a mechanism of CD8+ T cell activation independent of CD28. Because DAP10 and CD28 both contain a YxxM signaling motif capable of mediating PI3-kinase activation, it is possible that NKG2D/DAP10 may serve as an alternative costimulator in the absence of CD28. In humans, a significant proportion of human CD8+ T cells lack CD28 36, but these cells do express NKG2D/DAP10. In addition, it is noteworthy that MICA/B are expressed on epithelial cells 28, which lack the ligands for CD28 (i.e., CD80 and CD86). Therefore, it is tempting to speculate that the NKG2D/DAP10 receptor may provide costimulation for NK cells and CTLs when they encounter virus-infected or transformed nonhematopoietic tissues.
Acknowledgments
We are grateful to M. Robertson for NKL cells, T. Kitamura and G. Nolan for retroviruses, J. Bolen for Abs, A. Bakker, A. Cerwenka, T. Hauser, and S. Tangye for helpful discussion and technical tips, and J. Katheiser, G. Burget, and M. Andonian for graphics.
This work is supported in part by the Sandler Family Supporting Foundation. T. Spies is supported by National Institutes of Health grant AI30581. Schering Plough Corporation supports DNAX Research Institute.
Footnotes
J. Wu's present address is Rigel, Inc., 240 East Grand Ave., South San Francisco, CA 94080.
Abbreviations used in this paper: GFP, green fluorescent protein; HRP, horseradish peroxidase; IRES, internal ribosomal entry site; ITAM, immunoreceptor tyrosine-based activation motif; KIR, killer cell inhibitory receptor; PI3-kinase, phosphatidylinositol 3-kinase; TM, transmembrane; wt, wild-type.
References
- Lanier L.L. Turning on natural killer cells. J. Exp. Med. 2000;191:1259–1262. doi: 10.1084/jem.191.8.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E.O. Regulation of immune responses through inhibitory receptors. Annu. Rev. Immunol. 1999;17:875–904. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- Mason L.H., Willette-Brown J., Anderson S.K., Gosselin P., Shores E.W., Love P.E., Ortaldo J.R., McVicar D.W. Characterization of an associated 16 kDa tyrosine phosphoprotein required for Ly-49D signal transduction. J. Immunol. 1998;160:4148–4152. [PubMed] [Google Scholar]
- Olcese L., Cambiaggi A., Semenzato G., Bottino C., Moretta A., Vivier E. Human killer cell activatory receptors for MHC class I molecules are included in a multimeric complex expressed by natural killer cells. J. Immunol. 1997;158:5083–5086. [PubMed] [Google Scholar]
- Lanier L.L., Corliss B.C., Wu J., Leong C., Phillips J.H. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- Lanier L.L., Corliss B., Wu J., Phillips J.H. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- Smith K.M., Wu J., Bakker A.B.H., Phillips J.H., Lanier L.L. Ly49D and Ly49H associate with mouse DAP12 and form activating receptors. J. Immunol. 1998;161:7–10. [PubMed] [Google Scholar]
- Wu J., Song Y., Bakker A.B.H., Bauer S., Groh V., Spies T., Lanier L.L., Phillips J.H. An activating receptor complex on natural killer and T cells formed by NKG2D and DAP10. Science. 1999;285:730–732. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- Songyang Z., Shoelson S.E., Chaudhuri M., Gish G., Pawson T., Haser W.G., King F., Roberts T., Ratnofsky S., Lechleider R.J. SH2 domains recognize specific phosphopeptide sequences. Cell. 1996;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- Bauer S., Groh V., Wu J., Steinle A., Phillips J.H., Lanier L.L., Spies T. Activation of natural killer cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–730. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- Colonna M., Samaridis J. Cloning of Ig-superfamily members associated with HLA-C and HLA-B recognition by human NK cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- Houchins J.P., Yabe T., McSherry C., Bach F.H. DNA sequence analysis of NKG2, a family of related cDNA clones encoding type II integral membrane proteins on human natural killer cells. J. Exp. Med. 1991;173:1017–1020. doi: 10.1084/jem.173.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onihsi M., Kinoshita S., Morikawa Y., Shibuya A., Phillips J., Lanier L.L., Gorman D.M., Nolan G.P., Miyajima A., Kitamura T. Applications of retrovirus-mediated expression cloning. Exp. Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- Kinsella T.M., Nolan G.P. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- Robertson M.J., Cochran K.J., Cameron C., Le J.-M., Tantravahi R., Ritz J. Characterization of a cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp. Hematol. 1996;24:406–415. [PubMed] [Google Scholar]
- Yssel H., de Vries J.E., Koken M., van Blitterswijk W., Spits H. Serum-free medium for the generation and the propagation of functional human cytotoxic and helper T cell clones. J. Immunol. Methods. 1984;72:219–227. doi: 10.1016/0022-1759(84)90450-2. [DOI] [PubMed] [Google Scholar]
- Phillips J.H., Chang C., Mattson J., Gumperz J.E., Parham P., Lanier L.L. CD94 and a novel associated protein (94AP) form a NK cell receptor involved in the recognition of HLA-A, HLA-B, and HLA-C allotypes. Immunity. 1996;5:163–172. doi: 10.1016/s1074-7613(00)80492-6. [DOI] [PubMed] [Google Scholar]
- Lanier L.L., Recktenwald D.J. Multicolor immunofluorescence and flow cytometry. Methods. 1991;2:192–199. [Google Scholar]
- Lanier L.L., Yu G., Phillips J.H. Co-association of CD3ζ with a receptor (CD16) for IgG Fc on human natural killer cells. Nature. 1989;342:803–805. doi: 10.1038/342803a0. [DOI] [PubMed] [Google Scholar]
- Tangye S.G., Phillips J.H., Lanier L.L., de Vries J.E., Aversa G. CD148a receptor-type protein tyrosine phosphatase involved in the regulation of human T cell activation. J. Immunol. 1998;161:3249–3255. [PubMed] [Google Scholar]
- Arase N., Arase H., Park S.Y., Ohno H., Ra C., Saito T. Association with FcRγ is essential for activation signal through NKR-P1 (CD161) in natural killer (NK) cells and NK1.1+ T cells. J. Exp. Med. 1997;186:1957–1963. doi: 10.1084/jem.186.12.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende D., Parolini S., Pessino A., Sivori S., Augugliaro R., Morelli L., Marcenaro E., Accame L., Malaspina A., Biassoni R. Identification and molecular characterization of NKP30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J. Exp. Med. 1999;190:1505–1516. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessino A., Sivori S., Bottino C., Malaspina A., Morelli L., Moretta L., Biassoni R., Moretta A. Molecular cloning of NKp46a novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J. Exp. Med. 1998;188:953–960. doi: 10.1084/jem.188.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale M., Bottino C., Sivori S., Sanseverino L., Castriconi R., Marcenaro E., Augugliaro R., Moretta L., Moretta A. NKp44, a novel triggering surface molecule specifically expressed by activated natural killer cells, is involved in non-major histocompatibility complex–restricted tumor cell lysis. J. Exp. Med. 1998;187:2065–2072. doi: 10.1084/jem.187.12.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.J., Torkar M., Haude A., Milne S., Jones T., Sheer D., Beck S., Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc. Natl. Acad. Sci. USA. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiante N.M., Uhrberg M., Shilling H.G., Lienert-Weidenbach K., Arnett K.L., D'Andrea A., Phillips J.H., Lanier L.L., Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- Bottino C., Sivori S., Vitale M., Cantoni C., Falco M., Pende D., Morelli L., Augugliaro R., Semenzato G., Biassoni R. A novel surface molecule homologous to the p58/p50 family of receptors is selectively expressed on a subset of human natural killer cells and induces both triggering of cell functions and proliferation. Eur. J. Immunol. 1996;26:1816–1824. doi: 10.1002/eji.1830260823. [DOI] [PubMed] [Google Scholar]
- Groh V., Bahram S., Bauer S., Herman A., Beauchamp M., Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc. Natl. Acad. Sci. USA. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh V., Steinle A., Bauer S., Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial γδ T cells. Science. 1998;279:1737–1740. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- Groh V., Rhinehart R., Secrist H., Bauer S., Grabstein K.H., Spies T. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc. Natl. Acad. Sci. USA. 1999;96:6879–6884. doi: 10.1073/pnas.96.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt K.E., Hicks C.M., Imboden J.B. Stimulation of CD28 triggers an association between CD28 and phosphatidylinositol 3-kinase in Jurkat T cells. J. Exp. Med. 1994;179:1071–1076. doi: 10.1084/jem.179.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages F., Ragueneau M., Rottapel R., Truneh A., Nunes J., Imbert J., Olive D. Binding of phosphatidyl-inositol-3-OH kinase to CD28 is required for T-cell signalling. Nature. 1994;369:327–329. doi: 10.1038/369327a0. [DOI] [PubMed] [Google Scholar]
- Rudd C.E., Janssen O., Cai Y.C., Da S.A., Raab M., Prasad K. Two-step TCRzeta/CD3-CD4 and CD28 signaling in T cellsSH2/SH3 domains, protein-tyrosine and lipid kinases. Immunol. Today. 1994;15:225–234. doi: 10.1016/0167-5699(94)90248-8. [DOI] [PubMed] [Google Scholar]
- Lenschow D.J., Walunas T.L., Bluestone J.A. CD28/B7 system of T cell costimulation. Annu. Rev. Immunol. 1996;14:233–258. doi: 10.1146/annurev.immunol.14.1.233. [DOI] [PubMed] [Google Scholar]
- Shahinian A., Pfeffer K., Lee K.P., Kundig T.M., Kishihara K., Wakeman A., Kawai K., Ohashi P.S., Thompson C.B., Mak T.W. Differential T cell costimulatory requirements in CD28-deficient mice. Science. 1993;261:609–612. doi: 10.1126/science.7688139. [DOI] [PubMed] [Google Scholar]
- Azuma M., Phillips J.H., Lanier L.L. CD28− T lymphocytesantigenic and functional properties. J. Immunol. 1993;150:1147–1159. [PubMed] [Google Scholar]